Abstract
A set of 30 mutants exhibiting reduced production of the phenazine poison pyocyanin were isolated following transposon mutagenesis of Pseudomonas aeruginosa PAO1. The mutants could be subdivided into those with defects in the primary phenazine biosynthetic pathway and those with more pleiotropic defects. The largest set of pleiotropic mutations blocked the production of the extracellular Pseudomonas quinolone signal (PQS), a molecule required for the synthesis of secondary metabolites and extracellular enzymes. Most of these pqs mutations affected genes which appear to encode PQS biosynthetic functions, although a transcriptional regulator and an apparent response effector were also represented. Two of the genes required for PQS synthesis (phnA and phnB) had previously been assumed to encode phenazine biosynthetic functions. The transcription of one of the genes required for PQS synthesis (PA2587/pqsH) was regulated by the LasI/R quorum-sensing system, thereby linking quorum sensing and PQS regulation. Others of the pleiotropic phenazine-minus mutations appear to inactivate novel components of the quorum-sensing regulatory network, including one regulator (np20) previously shown to be required for virulence in neutropenic mice.
A complex network of regulatory factors governs the production of secondary metabolites and other virulence factors in the opportunistic pathogen Pseudomonas aeruginosa. This network regulates gene expression in response to stimuli such as growth phase, culture density, and oxygen and iron availability (12, 26, 28, 37). Central components of the network are the las and rhl quorum-sensing systems, which activate gene expression in response to culture density (13). Each system is made up of two genes, one encoding an enzyme which produces a specific acylated homoserine lactone autoinducer (lasI/rhlI), and a second encoding a transcriptional activator that binds the corresponding autoinducer (lasR/rhlR). The las system directs expression of virulence factors such as elastases A and B and alkaline protease (16, 25). The rhl system directs expression of rhamnolipid biosynthesis enzymes, pyocyanin biosynthesis enzymes, and hydrogen cyanide synthase (3, 24, 28). In addition, LasI/R regulates expression of both itself and rhlI (1, 26). The las and rhl systems together have been shown to influence the expression of over two hundred genes (36).
Recently, a third signaling system based on 2-heptyl-3-hydroxy-4-quinolone, designated the Pseudomonas quinolone signal (PQS), has been shown to be a part of the quorum-sensing regulatory network in P. aeruginosa (27). The production of PQS depends on lasR (27), and exogenous PQS strongly induces expression of elastase B and rhlI in a lasR mutant background (22). These results place PQS between the las and rhl quorum-sensing systems in the quorum-sensing regulatory network (22).
We have described a process (“paralytic killing”) in which P. aeruginosa PAO1 rapidly kills the nematode Caenorhabditis elegans by cyanide poisoning (8, 14). Previous studies of a different P. aeruginosa strain (PA14) had implicated a different poison, the phenazine pyocyanin, in nematode killing (20). Pyocyanin is a redox cycling agent synthesized as a secondary metabolite from chorismate by the phz gene products (21). Phenazines have been used as electron acceptors for cyanide production in vitro (2, 6), and it appeared possible that they could be required for cyanide production and nematode killing by strain PAO1. To help address this issue, we isolated and characterized PAO1 mutants defective in phenazine production. Unexpectedly, the majority of the mutants we found appear to be defective in the regulation of phenazine synthesis rather than in the biosynthetic pathway itself. This study presents an analysis of the regulatory mutants and shows that the most common class is defective in PQS signaling.
MATERIALS AND METHODS
Strains, plasmids, growth media, and culture conditions.
The P. aeruginosa strains used were PAO1 (17) from L. Passador and B. Iglewski (MPAO1) and S. Lory (PAO1seq) and those listed in Tables 1 and 2. Chromosomal transposon insertion mutations were transferred between strains by transformation using a technique to be described in detail elsewhere (N. Benkers and C. Manoil, unpublished studies). Briefly, recipient strains carrying a plasmid encoding phage lambda red recombination functions were electroporated with chromosomal DNA isolated from donor strains, with selection for transposon antibiotic-resistant transformants. Transformants were cured of the plasmid carrying lambda red prior to further analysis. The lacZ transcriptional gene fusion alleles used to construct the strains for Tables 2 and 3 were isolated by Whiteley et al. (36) as transposon insertions in genes qsc101, qsc118, qsc128, qsc105, qsc131, and qsc135. The Escherichia coli strains used were DH5α (30) for plasmid construction and SM10λpir (32) for conjugal suicide plasmid delivery. The growth media used were brain heart infusion (BHI) agar (Difco), low-phosphate succinate minimal medium (LPSM) (7), L agar (30), and L broth. For visual analysis of β-galactosidase activity, L agar was supplemented with 50-μg/ml 5-bromo-4-chloro-3-indolyl galactoside (X-Gal). Plasmids were maintained in P. aeruginosa in medium supplemented with 200-μg/ml carbenicillin and in E. coli in medium supplemented with 100 μg/ml ampicillin. To construct plasmid pLG10, cosmid 122 (containing the phnAB genomic region) from the laboratory of S. Lory was first digested with HindIII and NheI. The 10,301-bp, phnAB-containing fragment from this digestion was gel purified (QIAgen kit) and DNA ligase joined to the gel-purified 4,532-bp fragment from pUCP18 (31) cleaved with HindIII and XbaI. To construct plasmid pLG12, the 6,084-bp PstI fragment from pLG10 was cloned into the PstI site of pUCP18. To construct plasmid pLG14, pLG12 was digested with AscI and HindIII and treated with Klenow enzyme (New England Biolabs). The larger fragment was gel purified and self-ligated. To construct plasmid pLG16, pLG10 was digested with SacI and AscI and treated with mung bean nuclease (New England Biolabs). The larger fragment was then gel purified and self-ligated. All constructs were confirmed by restriction analysis. Standard molecular biology protocols were used (30).
TABLE 1.
Mutants defective in pyocyanin productiona
| Strain | Parent | Insert site | Gene | Description | Source or reference |
|---|---|---|---|---|---|
| phz1 region | |||||
| MP601 | MPAO1 | 4,712,812 | phzM | Pyocyanin biosynthesis | This study |
| MP651 | PAO1seq | 4,714,312 | phzA1 | Core phenazine biosynthetic operon | This study |
| *MP705 | MPAO1 | 4,714,312 | phzA1 | Core phenazine biosynthetic operon | This studyb |
| MP602 | MPAO1 | 4,715,531 | phzC1 | Core phenazine biosynthetic operon | This study |
| MP652 | PAO1seq | 4,718,219 | phzE1 | Core phenazine biosynthetic operon | This study |
| *MP706 | MPAO1 | 4,718,219 | phzE1 | Core phenazine biosynthetic operon | This studyb |
| MP653 | PAO1seq | 4,720,699 | phzS | Pyocyanin biosynthesis | This study |
| *MP702 | MPAO1 | 4,720,699 | phzS | Pyocyanin biosynthesis | This studyb |
| phnAB region | |||||
| MP654 | PA01seq | 1,081,164 | PA0998 (pqsC) | β-Ketoacyl-acyl protein synthase | This study |
| *MP703 | MPAO1 | 1,081,164 | PA0998 (pqsC) | β-Ketoacyl-acyl protein synthase | This studyb |
| MP603 | MPAO1 | 1,081,418 | PA0998 (pqsC) | β-Ketoacyl-acyl protein synthase | This study |
| MP604 | MPAO1 | 1,081,470 | PA0998 (pqsC) | β-Ketoacyl-acyl protein synthase | This study |
| MP655 | PAO1seq | 1,082,732 | PA0999 (pqsD) | β-Ketoacyl-acyl protein synthase | This study |
| *MP704 | MPAO1 | 1,082,732 | PA0999 (pqsD) | β-Ketoacyl-acyl protein synthase | This studyb |
| MP605 | MPAO1 | 1,082,972 | PA1000 (pqsE) | Hypothetical | This study |
| MP710 | MPAO1 | NDc | phnA | Anthranilate synthase. | This study |
| MP551 | MPAO1 | 1,086,674 | PA1003 (pqsR) | lysR-like transcriptional regulator | 14 |
| PA2587 region | |||||
| MP562 | MPAO1 | 2,927,500 | PA2587 (pqsH) | FAD-dependent monooxygenase | 14 |
| MP606 | MPAO1 | 2,928,247 | PA2587/PA2588 | Between genes | This study |
| Known regulators | |||||
| MP701 | MPAO1 | ND | lasR | lasR quorum-sensing regulator | 15; this study |
| MP607 | MPAO1 | 3,889,853 | rhll/R | rhl quorum-sensing regulatory locus | This study |
| MP608 | MPAO1 | 1,013,341 | gacS | Global regulator | This study |
| MP502 | MPAO1 | 1,015,249 | gacS | Global regulator | 14 |
| Putative regulators | |||||
| MP501 | MPAO1 | 4,423,808 | PA3946 | Similar to B. pertussis virulence gene regulator bvgS | 14 |
| MP609 | MPAO1 | 5,302,623 | dksA | Repressor of rhll | This study |
| MP553 | MPAO1 | 5,304,386 | PA4725 | Putative two-component sensor kinase | 14 |
| MP552 | MPAO1 | 5,304,811 | PA4725 | Putative two-component sensor kinase | 14 |
| MP656 | PAO1seq | 5,306,302 | PA4725 | Putative two-component sensor kinase | This study |
| MP610 | MPAO1 | 5,482,018 | PA4886 | Putative two-component sensor kinase. | This study |
| MP611 | MPAO1 | 6,192,554 | np20 | Similar to E. coli zinc uptake regulator Zur | This study |
| Miscellaneous | |||||
| MP612 | MPAO1 | 447,569 | PA0406 | Hypothetical protein | This study |
| MP613 | MPAO1 | 448,033 | PA0406 | Hypothetical protein | This study |
| MP614 | MPAO1 | 812,055 | PA0744 | Probable enoyl-CoA hydratase/isomerase | This study |
| MP504 | MPAO1 | 812,969 | PA0745 | Probable enoyl-CoA hydratase/isomerase | 14 |
| MP615 | MPAO1 | 3,395,202 | PA3031/PA3032 | Between genes | This study |
All mutants correspond to insertions of ISphoA/hah-Tc except the phnA and lasR mutants, which were constructed by in vitro methods.
Strain was derived by transferring into the chromosome of MPAO1 the insertion allele from the strain listed immediately above in the list.
ND, not determined
TABLE 2.
β-Galactosidase activities in strains carrying chromosomal lacZ reporter fusions to quorum-sensing-controlled genesa
| Mutant gene |
phzC1-lacZ
|
hcnB-lacZ
|
pqsH-lacZ
|
phzD2-lacZ
|
rhlI-lacZ
|
cytC-lacZ
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Activity | (SEM) | nc | % Activity | (SEM) | n | % Activity | (SEM) | n | % Activity | (SEM) | n | % Activity | (SEM) | n | % Activity | (SEM) | n | |
| w-t | (100) | (8) | 13b | (100) | (8) | 12b | (100) | (3) | 10b | (100) | (6) | 12b | (100) | (2) | 9b | (100) | (8) | 10b |
| pqsC | 16 | (3) | 3 | 43 | (4) | 3 | 85 | (4) | 2 | 117 | (16) | 2 | 91 | (5) | 2 | 71 | (13) | 2 |
| pqsD | 4 | (0) | 3 | 30 | (5) | 2 | 82 | (1) | 2 | 140 | (9) | 2 | 84 | (7) | 2 | 65 | (10) | 2 |
| pqsE | 3 | (0) | 3 | 30 | (3) | 2 | 84 | (2) | 2 | 121 | (11) | 2 | 88 | (4) | 4b | 74 | (12) | 2 |
| phnA | 14 | (2) | 3 | 38 | (3) | 2 | 83 | (4) | 2 | 108 | (20) | 2 | 89 | (6) | 2 | 75 | (17) | 2 |
| pqsR | 5 | (1) | 8b | 27 | (0) | 5b | 80 | (2) | 5b | 148 | (14) | 4b | 88 | (3) | 5b | 76 | (9) | 3 |
| pqsH | 8 | (2) | 5b | 32 | (1) | 3 | NDd | 128 | (14) | 5b | 84 | (4) | 4b | 72 | (6) | 5b | ||
| lasR | 1 | (0) | 3 | 7 | (2) | 8b | 9 | (2) | 3 | 5 | (1) | 4b | 5 | (0) | 4b | 49 | (4) | 4b |
| np20 | ND | 6 | (1) | 6b | 6 | (0) | 6b | 4 | (0) | 7b | 5 | (0) | 6b | 61 | (2) | 7b | ||
| PA3946 | 162 | (17) | 8b | 20 | (1) | 5b | 47 | (1) | 4b | 85 | (4) | 7b | 58 | (1) | 4b | 64 | (5) | 3 |
The chromosomal qsc-gene-lacZ reporter alleles tested are listed at the top of each data column. The gene mutated in each strain is indicated in the far left column. β-Galactosidase activity is shown for each reporter allele as a percentage of the average activity in the wild-type background. 100% β-galactosidase activity represents, for phzC1-lacZ, 813 Miller units; for hcnB-lacZ, 534 units; for pqsH-lacZ, 268 units; for phzD2-lacZ, 77 units; for rlhI-lacZ, 2,000 units; and for cytC-lacZ, 95 units.
Value determined from analysis of duplicate constructed strains.
n, number of independent assays performed.
ND, not done.
TABLE 3.
Induction of phzC1-lacZ expression by extracellular complementation
| Adjacent straina |
phzC1-lacZ fusion straina,b
|
||||||
|---|---|---|---|---|---|---|---|
| MP882.1 (pqsC) | MP883.1 (pqsD) | MP884.1 (pqsE) | MP894.1 (phnA) | MP885.2 (pqsR) | MP886.1 (pqsH) | MP892.1 (lasR) | |
| MPAO1 (wild-type) | + | ++ | − | + | +/− | +++ | ++++ |
| MP703 (pqsC) | − | − | − | − | − | − | − |
| MP704 (pqsD) | − | − | − | − | − | + | − |
| MP605 (pqsE) | + | ++ | − | + | +/− | +++ | ++++ |
| MP710 (phnA) | − | − | − | − | − | − | − |
| MP551 (pqsR) | − | − | − | − | − | − | +/− |
| MP562 (pqsH) | + | + | − | +/− | +/− | − | +++ |
| MP701 (lasR) | +/− | + | − | − | − | − | − |
| MP607 (rhlR/I) | +/− | ++ | − | − | − | +++ | ++++ |
| MP611 (np20) | − | − | − | − | − | − | − |
The gene mutated in each strain is indicated paranthetically
Increased β-galactosidase activity over background levels as measured by X-Gal hydrolysis was assessed visually and ranged from slight (+/−) to extensive (++++).
Transposon mutagenesis and mutant screening.
Transposon ISphoA/hah-Tc mutagenesis was carried out as described previously (14). Lawns of mutants derived from strain MPAO1 were screened visually for pigmentation defects after 24 h of growth at 37°C on 3.5-cm-diameter BHI agar plates. Small cultures (200 μl) of mutants of PAO1seq were screened visually for pigmentation defects after 2 days of growth at room temperature in a 96-well format in LPSM.
DNA sequencing.
The chromosomal DNA flanking the transposon insertions was sequenced as described previously (14), except that primer TnphoA-II (5′-GTGCAGTAATATCGCCCTGAGCA-3′) replaced primer MTN5I.1, primer HAH-1 (5′-ATCCCCCTGGATGGAAAACGG-3′) replaced primer MTN5O.1, and primer HAH-2 (5′-AAACGGGAAAGGTTCCGTCCA-3′) replaced primer MTN5S.1. Chromosomal locations were determined by BLAST analysis of the transposon-adjacent chromosomal DNA sequences compared with the complete strain PAO1 genome sequence (obtained at www.pseudomonas.com).
Nematode killing, β-galactosidase, pyocyanin and PQS assays.
Nematode killing assays were carried out as described previously (14). β-Galactosidase assays for Table 2 were carried out as described previously (23) after growth with aeration at 37°C of 5 ml cultures in L broth. All cultures were inoculated from fresh colonies and adjusted to an optical density at 600 nm (OD600) of 0.02 before incubation. To assess the optimal growth point for comparing gene expression levels, β-galactosidase levels were monitored over 15 h of growth. For all six reporter alleles in the wild-type background, the β-galactosidase levels reached a linear rate of increase by early stationary phase (∼10 h of incubation), and the slopes remained constant, even after 15 h of incubation (data not shown). The results were reproducible in multiple trials, and all subsequent β-galactosidase assays were carried out at the 13-h time point. For extracellular complementation analysis, strains were patched directly next to one another on L agar containing 50-μg/ml X-Gal. Increased β-galactosidase activity from the complementation was scored visually at adjacent regions of growth after 2 days of incubation at 37°C. For Table 4, pyocyanin extracted from agar medium was assayed as previously described (14). For Table 5, samples of culture supernatants (4 ml) from 5-ml cultures grown with aeration for 16 h at 37°C from an initial inoculum at OD600 of 0.02 were extracted with 3 ml of chloroform; 2.5 ml of the chloroform phase was then further extracted with 0.5 ml of 0.2 N HCl, and the OD520 of the aqueous phase was measured (14). For PQS assays, bacteria from plates grown overnight were inoculated into 5 ml of Luria-Bertani (LB) broth and cultures were incubated at 37°C with shaking (260 rpm) for approximately 6 h. Cells were then subcultured into 1 ml of LB broth at an A660 of 0.02 and grown for 24 h at 37°C with shaking (260 rpm). After growth, 300 μl of culture was extracted twice with 900 μl of acidified ethyl acetate (27) by vigorously vortexing for 30 s, followed by centrifugation at 16,000 × g for 5 min. An 800-μl aliquot of the upper organic layer from each extraction was transferred to a single microcentrifuge tube and allowed to dry overnight at room temperature. The following day, dried extracts were either analyzed by thin-layer chromatography (TLC) or stored at −20°C until analysis. For TLC analysis, dried extracts were resuspended in 50 μl of a 1:1 acidified ethyl acetate-acetonitrile mixture by vortexing and pulse spinning multiple times. Ten microliters of each extract was loaded onto a Silica Gel 60 F254 (10 by 20 cm; EM Science), along with synthetic PQS. Chromatography was performed with a solvent mixture containing a 17:2:1 ratio of methlyene chloride-acetonitrile-1,4-dioxane. When the solvent front neared the top of the plates, the plates were photographed under long-wave UV light (365 nm) using a Polaroid camera with ISO 3000 black-and-white film.
TABLE 4.
Pyocyanin production, nematode killing, and PQS production by P. aeruginosa mutants
| Strain | Mutant gene | Pyocyanina | % Nematode killinga | PQS |
|---|---|---|---|---|
| MPAO1 | (1.00) | 98 (2) | +++ | |
| phz1 cluster | ||||
| MP601 | phzM | 0.01 (0.00) | 100 (0) | +++ |
| MP705 | phzA1 | 0.04 (0.01) | 99 (1) | +++ |
| MP706 | phzE1 | 0.17 (0.02) | 99 (1) | +++ |
| MP702 | phzS | 0.02 (0.00) | 78 (11) | +++ |
| phnAB region | ||||
| MP703 | PA0998 (pqsC) | 0.04 (0.00) | 36 (8) | − |
| MP704 | PA0999 (pqsD) | 0.02 (0.01) | 14 (2) | − |
| MP605 | PA1000 (pqsE) | 0.01 (0.01) | 25 (8) | +++ |
| MP710 | phnA | 0.04 (0.02) | 37 (8) | − |
| MP551 | PA1003 (pqsR) | 0.01 (0.00) | 12 (10) | − |
| PA2587 region (MP562) | PA2587 (pqsH) | 0.02 (0.01) | 9 (4) | − |
| Known regulators | ||||
| MP701 | lasR | 0.01 (0.00) | 2 (1) | +/− |
| MP607 | rhll/R | 0.02 (0.00) | 11 (8) | +++ |
| MP502 | gacS | 0.10 (0.01) | 50 (5) | +++ |
| Putative regulators | ||||
| MP501 | PA3946 | 0.31 (0.02) | 22 (7) | +++ |
| MP552 | PA4725 | 0.19 (0.04) | 19 (8) | +++ |
| MP610 | PA4886 | 0.29 (0.03) | 40 (6) | +++ |
| MP611 | np20 | 0.00 (0.00) | 4 (3) | +/− |
| Miscellaneous | ||||
| MP613 | PA0406 | 0.07 (0.00) | 48 (17) | + |
| MP504 | PA0745 | 0.03 (0.01) | 50 (13) | + |
| MP615 | PA3031/PA3032 | 0.03 (0.01) | 1 (1) | + |
Values listed are averages from three separate assays. Pyocyanin recoveries relative to the parent strain (MPAO1) are presented. Numbers in parentheses indicate standard errors of the mean.
TABLE 5.
Pyocyanin production by phnAB-region mutants carrying plasmids
| Plasmid | pqs genes on plasmid | Straina
|
|||||
|---|---|---|---|---|---|---|---|
| MPAO1 | MP703 (pqsC) | MP704 (pqsD) | MP605 (pqsE) | MP710 (phnA) | MP551 (pqsR/mvfR) | ||
| pUCP18 | 0.29 (0.05) | 0.02 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.01 (0.00) | 0.01 (0.00) | |
| pLG10 | pqsABCDE | 0.93 (0.10) | 0.78 (0.04) | 0.82 (0.06) | 0.80 (0.09) | 0.77 (0.05) | 0.00 (0.00) |
| pLG12 | pqsABCD | 0.13 (0.02) | 0.01 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.01 (0.00) | 0.00 (0.00) |
| pLG14 | pqsABC | 0.17 (0.04) | 0.01 (0.00) | 0.01 (0.01) | 0.02 (0.01) | 0.02 | 0.01 (0.00) |
| pLG16 | pqsABC | 0.35 | 0.02 | 0.01 | 0.01 | 0.02 | 0.01 |
The gene mutated in each strain is indicated. Pyocyanin was assayed as described previously (14), and its recovery is expressed in absorbance units. Standard errors of the mean are shown where multiple independent assays were performed.
RESULTS
Isolation of P. aeruginosa mutants deficient in pyocyanin production.
We identified transposon insertion mutants of P. aeruginosa strain PAO1 defective in production of the blue phenazine pigment pyocyanin by visual screening (Materials and Methods). In the process of carrying out the screen, we observed that different isolates of wild-type PAO1 behaved differently with respect to pyocyanin production. During liquid growth in LPSM, a medium that induces strong pyocyanin production (7), the PAO1 strain whose genome had been sequenced (designated PAO1seq) (33) produced significantly more pyocyanin than did our laboratory strain of PAO1 (designated MPAO1, obtained from L. Passador and B. Iglewski) (data not shown). When grown on BHI agar, MPAO1 produced much more pyocyanin than did PAO1seq. We initially screened for mutants in the PAO1seq strain (2,800 ISphoA/hah-Tc transposon insertion mutants screened) and switched to screening in our laboratory strain once the conditions promoting its pyocyanin production were identified (2,100 ISphoA/hah-Tc transposon insertion mutants screened) (Materials and Methods).
Twenty-one pyocyanin-deficient mutants which formed normal-size colonies on L agar were isolated (Table 1). Seven additional pyocyanin-deficient mutants obtained in a previous screen (14) and two inactivating mutations constructed in vitro (in lasR and phnA) were also represented in the set of mutants analyzed (Table 1). We determined the transposon insertion sites for the new mutants by PCR amplification and sequencing of the genomic DNA flanking each transposon insertion, followed by BLAST analysis against the completed PAO1 genome sequence.
A variety of genes were affected in the mutant set, including clusters in the phz1 and phnAB regions (Table 1). The phz1 region (genes phzM, phzA1 through phzG1, and phzS) encodes pyocyanin biosynthetic enzymes (21). The phnA and phnB genes encode an anthranilate synthase homologue originally proposed to encode a phenazine biosynthetic function (10), a role which has been questioned in a recent report (21). Numerous regulatory genes were also identified in the mutant set, including three (lasR, rhlI, and gacS) known to be a part of the quorum-sensing regulatory network. Four more novel putative regulatory genes were also identified, including a homologue of the E. coli zinc uptake regulator Zur (np20) and three putative two-component regulatory genes (PA3946, PA4725, and PA4886). (PA gene numbers refer to ORF designations assigned to unnamed genes in the PAO1 genome sequence [www.pseudomonas.com].) Additional potential regulators were also identified (Table 1).
Since the two supposedly identical parent strains (MPAO1 and PAO1seq) displayed different pyocyanin production characteristics, it was essential for further analysis that the new mutations be characterized in an isogenic strain background. The transposon insertion alleles generated in PAO1seq were therefore transferred into the chromosome of strain MPAO1 by transformation (Materials and Methods). The resulting MPAO1 derivatives are marked with asterisks (Table 1). The inactivating alleles of lasR (from strain PAO-R1 [15]) and phnA (S. L. McKnight and E. C. Pesci, unpublished data) were also transferred into the MPAO1 strain background (Table 1).
Pyocyanin is not required for nematode killing.
We measured pyocyanin production and nematode killing for a subset of the pyocyanin-deficient mutants, representing insertions in most of the genes identified in the mutant set (Table 4). Previous studies had shown that hydrogen cyanide is necessary and sufficient for the killing (14). Strains carrying mutations in genes of the core phenazine biosynthetic locus (phzM, phzA1, phzE1, and phzS) (21) killed worms efficiently in spite of severe defects in pyocyanin production (Table 4), implying that pyocyanin (and probably other phenazines) is not essential for cyanide production in P. aeruginosa. Similar results were obtained in studies of several phenazine-deficient mutants of other Pseudomonas species (C. Cosma, D. Mavrodi, C. Manoil, and L. Thomashow, unpublished results). Remarkably, all of the pyocyanin-deficient mutants affecting genes outside of the phz1 locus failed to kill worms efficiently and are thus apparently pleiotropically defective in both cyanide and pyocyanin production. Direct assays of HCN production (14) and expression of HCN biosynthetic gene-lacZ fusions (see below) provide additional evidence of this pleiotropy.
Identification of genes required for PQS production.
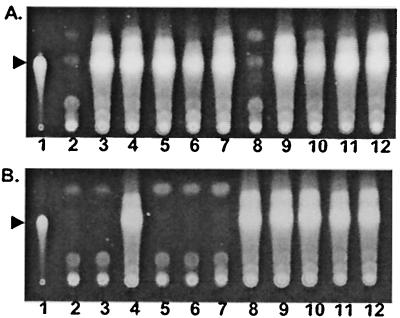
The recently identified PQS is known to be required for phenazine production in P. aeruginosa (21, 22). Since one of the genes we identified, PA2587, had been found to be required for PQS production (E. C. Pesci, unpublished), we examined production of PQS by the other pyocyanin-deficient mutants. PQS production was assayed using TLC under conditions which distinguish PQS from the acylated homoserine lactone autoinducers (Fig. 1 and Table 4). Although most of the strains produced PQS at levels comparable to the wild-type strain MPAO1, seven were negative, including most of the strains with mutations in the phnAB region (in genes PA0998, PA0999, phnA, and PA1003/mvfR), PA2587, lasR, and np20. Detectable spots with an Rf similar to PQS were observed for the lasR and np20 mutants, but these spots were purple as opposed to the fluorescent blue of authentic PQS (data not shown).
FIG. 1.
PQS production by P. aeruginosa strains. PQS samples extracted from 24-h cultures were analyzed by TLC. (A) Lanes: 1, 50 ng of PQS; 2, strain MP701 (lasR); 3, strain MP607 (rhlR/I); 4, strain MP502 (gacS); 5, strain MP501 (PA3946); 6, strain MP552 (PA4725); 7, strain MP610 (PA4886); 8, strain MP611 (np20); 9, strain MP613 (PA0406); 10, strain MP504 (PA0745); 11, strain MP615 (PA3031/3032); 12, strain MPAO1. (B) Lanes: 1, 50 ng of PQS; 2, strain MP703 (pqsC); 3, strain MP704 (pqsD); 4, strain MP605 (pqsE); 5, strain MP710 (phnA); 6, strain MP551 (pqsR/mvfR); 7, strain MP562 (pqsH); 8, strain MP601 (phzM); 9, strain MP705 (phzA1); 10, strain MP706 (phzE1); 11, strain MP702 (phzS); 12, strain MPAO1. The arrowhead in each panel indicates the position of PQS.
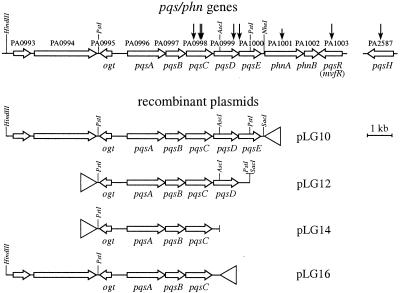
The finding that mutations in several genes in the phnAB region prevented PQS production suggested that this region may encode PQS biosynthetic functions. The genomic organization of the region suggests a possible five-gene operon made up of genes PA0996 to PA1000 (Fig. 2). Based on the results presented here and in an independent study (9) confirming that these genes play essential roles in PQS biosynthesis and signaling, we propose naming the genes pqsABCDE. We propose the designations pqsH for the unlinked gene PA2587 and pqsR for PA1003, which encodes a member of the LysR family of transcriptional regulators corresponding to strain PA14 MvfR (5).
FIG. 2.
Map of the phnAB genomic region and plasmids used for complementation analysis. The vertical arrows in the map of the pqs and phn genes indicate the locations of insertions in strains MP703, MP603, MP604, MP704, MP605, MP710, MP551, and MP562, respectively (reading left to right). The phnA mutation in MP710 is a constructed deletion carrying a Tcr cassette. The chromosomal regions present in recombinant plasmids pLG10, pLG12, pLG14, and pLG16 are shown, with the open triangle indicating the orientation of the Plac promoter in the pUCP18 vector.
Although most of the mutations in the phnAB region prevented PQS production, the insertion in pqsE produced a substance comigrating with PQS (Fig. 1). This result suggests that pqsE is not required for PQS synthesis, even though it is required for phenazine and cyanide production. The PQS-deficient and pyocyanin-deficient phenotypes cotransformed with the pqsE insertion mutation (data not shown), implying that they were not due to an undetected secondary mutation in the original mutant. While pqsE may play a role in pyocyanin production that is unrelated to PQS signaling, it appears more likely, given its genetic context, that pqsE is either responsible for a modification in the PQS biosynthetic pathway that is not distinguishable by our assay or that pqsE is required for the cellular response to PQS. Studies presented below support the second of these alternatives.
Complementation analysis.
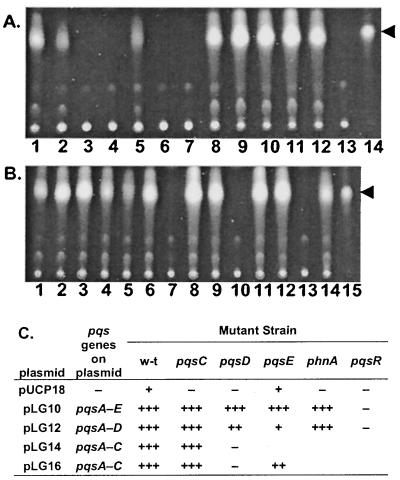
We carried out complementation studies to investigate whether polar effects were responsible for the phnAB-region mutant phenotypes. Four plasmids were constructed (Fig. 2): pLG10 (which carries pqsABCDE), pLG12 (pqsABCD), pLG14 (pqsABC) and pLG16 (pqsABC). Selected phnAB region mutants carrying these plasmids were examined for PQS and pyocyanin production. Plasmids pLG10 (pqsABCDE) and pLG12 (pqsABCD) fully restored PQS production to all the mutants tested except the pqsR (mvfR) mutant (Fig. 3). These results demonstrate that polar effects on phnA and phnB were not responsible for the PQS defects observed in the pqsC and pqsD mutants and confirm that the pqsABCDE operon is itself required for PQS production. Plasmids pLG14 (pqsABC) and pLG16 (pqsABC) restored PQS production to the pqsC mutant but not to the pqsD mutant (Fig. 3), indicating that pqsC and pqsD are both needed for PQS biosynthesis.
FIG. 3.
Complementation of Pqs− mutants. PQS samples extracted from 24-h cultures of strains carrying different pqs genes in plasmids were analyzed by TLC. (A) Lane 1, strain MPAO1 grown without a plasmid; lanes 2 to 7, strains MPAO1, MP703 (pqsC), MP704 (pqsD), MP605 (pqsE), MP710 (phnA), and MP551 (mvfR), respectively, carrying plasmid pUCP18; lanes 8 to 13, the same strains as the previous six lanes, but carrying plasmid pLG10; lane 14, 50 ng of PQS. (B) Lane 1, strain MPAO1 grown without a plasmid; lanes 2 to 7, strains MPAO1, MP703 (pqsC), MP704 (pqsD), MP605 (pqsE), MP710 (phnA), and MP551 (pqsR), respectively, carrying plasmid pLG12; lanes 8 to 10, strains MPA01, MP703 (pqsC), and MP704 (pqsD), respectively, carrying plasmid pLG14; lanes 11 to 14, strains MPAO1, MP703 (pqsC), MP704 (pqsD), and MP605 (pqsE), respectively, carrying plasmid pLG16; lane 15, 50 ng of PQS. The arrowhead indicates the position of PQS. (C) Summary of complementation results shown in panels A and B.
The pattern of complementation of the pyocyanin production defects differed from that of the PQS defects (Table 5). Although, as expected, plasmid pLG10 restored pyocyanin production to all the mutants tested except the pqsR (mvfR) mutant, none of the other plasmids restored pyocyanin production to any of the mutants. Since none of the other plasmids carry the pqsE gene (Fig. 2A), the results imply that pqsE is required for pyocyanin (but not PQS) production and that the mutations in pqsC and pqsD are polar on pqsE.
Surprisingly, although none of the complementing plasmids carried the phnA gene, they all complemented the PQS defect (and pLG10 complemented the pyocyanin defect) of a phnA mutant (Fig. 3 and Table 5). phnA and phnB encode an anthranilate synthase (10), and anthranilate is a precursor of PQS (4). Since there are five anthranilate synthase homologues in addition to PhnAB encoded in the P. aeruginosa genome (10, 11; Pesci, unpublished), it is possible that multicopy expression of the plasmid-borne pqsA-D leads to PQS production using anthranilate synthesized by one of the other enzymes. Recruitment of anthranilate produced by another anthranilate synthetase may also account for why the presence of pLG10 (which lacks phnAB) leads to significantly increased production of PQS and pyocyanin over wild-type in the strains examined (including the phnA mutant) (Table 5 and Fig. 3). Anthranilate synthase redundancy could also help account for the finding that the effect of phnA inactivation on PQS production depends critically on strain background (Pesci, unpublished).
Transcriptional analysis of mutants.
We examined the effects of pqs and other pleiotropic mutations on the expression of six quorum sensing-controlled (qsc) lacZ transcriptional gene fusions isolated by Whiteley et al. (36). The reporter gene fusions were transferred into the chromosomes of the wild-type (MPAO1) and pyocyanin-deficient mutant strains by transformation (Materials and Methods). The six fusions used for the analysis (Table 2, top row) represent three of the four major classes of quorum sensing-regulated genes identified in the earlier study (36) and include phzC1 (phenazine biosynthesis), hcnB (cyanide biosynthesis), and pqsH (PQS synthesis).
We assessed the optimal growth phase at which to compare the qsc gene expression levels in the various mutants by monitoring expression of each of the six fusion alleles in the wild-type (MPAO1) strain background during growth in liquid culture. By mid-stationary phase, all six strains showed significant and reproducible β-galactosidase activity (data not shown). We therefore chose a time point in mid-stationary phase for analyzing expression levels in the various mutant backgrounds (Materials and Methods). For most combinations, two independent constructions were assayed. The β-galactosidase levels measured in the different reporter strains are shown in Table 2, with the values of the mutant strains expressed relative to the corresponding wild-type values.
The mutations in genes implicated in PQS signaling (pqsCDE, phnA, pqsR, and pqsH) all reduced transcription of both phzC1 (more than sixfold) and hcnB (about threefold), results compatible with their decreased pyocyanin production and nematode killing (Table 4). The pqs mutations did not significantly reduce expression of the other four lacZ fusions examined. Thus, for all six fusions, the effects on expression were similar for mutations in the different pqs genes, a finding compatible with the hypothesis that all participate in a common regulatory process.
We were surprised that none of the mutations in the genes required for PQS synthesis significantly reduced rhlI transcription (Table 2). This finding appears to contradict the previous finding that PQS supplied exogenously to a lasR mutant greatly enhanced rhlI transcription from a multicopy plasmid (22). Since the latter result was obtained at a later growth phase than that examined here, the two findings could reflect growth-stage-dependent differences in rhlI transcription. To test this possibility, we examined rhlI transcription in some of our mutant strains at the same late growth point that was used for the plasmid-based assay (18 h of growth). However, we observed that even at the later growth point, rhlI transcription in the pqsC, pqsD, pqsE, phnA, pqsR, and pqsH mutant backgrounds was at 85% (2%), 94% (0%), 88%, 92%, 91% (0%), and 89% (8%) of wild-type levels, respectively (standard errors given in parentheses). Our results thus indicate that loss of PQS does not prevent rhlI transcription.
The lasR and np20 mutations were highly pleiotropic, causing greater than 10-fold reductions in the activities of the majority of the reporter fusions analyzed, including the pqsH-lacZ fusion. The lasR and np20 mutant defects in producing PQS can thus be at least partially explained by reduced pqsH expression. Of the three novel two-component regulators (Table 1), only the mutation in PA3946 showed significant reductions in qsc fusion expression, reducing transcription of hcnB fivefold and that of pqsH twofold (Table 2 and data not shown).
Extracellular complementation.
To further investigate the roles that the various pqs genes play in PQS signaling, we tested whether diffusible substances (e.g., PQS) produced by the mutants could induce transcription of phenazine biosynthetic genes in other mutant strains. Wild-type MPAO1 or PQS-defective mutants were grown adjacent to mutants carrying the chromosomal phzC1-lacZ transcriptional reporter on LB agar containing X-Gal indicator, and expression of the gene fusion was scored by visual inspection (Table 3).
Most of the findings were in agreement with the results of the PQS assays presented above. Strains shown to produce PQS (wild-type, pqsE, and rhlI/R) increased β-galactosidase expression in nearly all of the phzC1-lacZ fusion strains except that carrying the pqsE mutation. The responses of the pqsH and lasR mutant strains were particularly strong. The failure of the pqsE mutant strain to be induced is compatible with a role of PqsE in the response to PQS, as suggested above. (The findings also indicate that the polarity of the pqsC and pqsD mutations on pqsE [see above] may be incomplete.) The failure of most of the PQS-nonproducing mutants (pqsC, pqsD, phnA, and pqsR) to significantly complement any of the phzC1-lacZ fusion strains is also in accord with expectations. On the other hand, the relatively strong extracellular complementation by the pqsH mutant of several of the PQS-deficient fusion strains is not simply explained and suggests the action of a diffusible PQS intermediate (i.e., analogous to auxotroph cross-feeding).
The behavior of several regulatory mutations was also in accord with the PQS assays. Both lasR and np20 mutations greatly reduced extracellular complementation of the phzC1-lacZ fusion strains, whereas the rhlI/R mutation showed little reduction relative to wild type.
DISCUSSION
This report presents a genetic analysis of pyocyanin production in P. aeruginosa. Pyocyanin is a redox cycling phenazine poison produced as a secondary metabolite from chorismate by a multistep pathway (21). Pyocyanin production is highly regulated, being subject to control by two quorum-sensing systems (Las and Rhl), a novel extracellular quinolone signal (PQS), and other components of the quorum-sensing regulatory network (3, 19, 22, 29, 36).
We initially set out to determine whether pyocyanin or other phenazine compounds were required for cyanide-mediated nematode killing by P. aeruginosa PAO1. Although our previous experiments had shown that hydrogen cyanide is sufficient for the killing (14), studies with a different strain of P. aeruginosa (PA14) had revealed a partial correlation between killing of nematodes and production of pyocyanin (20). Biochemical studies had also shown that under some conditions phenazines participate in the synthesis of cyanide (2, 6). We therefore identified PAO1 mutants showing reduced production of pyocyanin and tested them for killing. Several of the mutants with severe defects in pyocyanin production due to mutations inactivating the biosynthetic pathway showed normal nematode killing, indicating that pyocyanin is not required for nematode killing by P. aeruginosa PAO1.
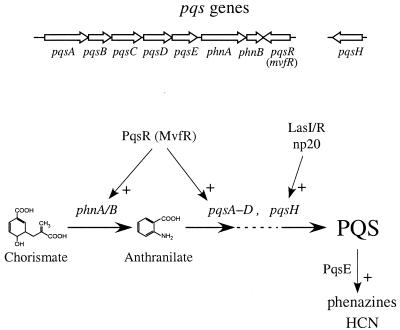
Remarkably, the majority of the mutations leading to pyocyanin defects did not alter biosynthetic pathway genes and were pleiotropically defective in both phenazine production and nematode killing (cyanide production). Three mutations (in lasR, rhlI/R, and gacS) inactivated components of the quorum-sensing regulatory network already known to be required for production of the two substances. However, the largest class of pleiotropic mutations was found to cause dramatic reductions in the production of the quinolone signal (PQS). These genes included a cluster in the phnAB region (PA0998, PA0999, and PA1003) and an unlinked gene (PA2587). A phnA deletion mutant constructed in vitro was also defective in PQS synthesis. A different study has demonstrated that two other phnAB region genes (PA0996 and PA0997) are also required for PQS synthesis (9). Another gene in the phnAB cluster (PA1000) was not required for PQS production, but may participate in the cellular response to PQS (see below). The new genes required for the production and action of PQS were named pqsA-R (Fig. 2 and 4).
FIG. 4.
PQS synthesis and regulation. The genomic organization of the pqs genes and a model for PQS biosynthesis and regulation are shown. We propose that PQS is synthesized from chorismate via anthranilate by the phnAB, pqsABCD, and pqsH gene products as indicated. Points of regulation by PqsR, LasI/R, np20, and PqsE based on the results presented in this and a previous report (5) are shown. The PqsR (MvfR) regulation of pqsABCDE is suggested by the finding that a plasmid carrying pqsABCDE complements a phnA mutant but not a pqsR mutant for PQS production (Fig. 3C).
PQS biosynthetic functions of several of the pqs products are suggested by their sequence homologies (Fig. 4). PhnA and PhnB presumably synthesize the anthranilate precursor of PQS from chorismate (4, 10). Gene pqsA encodes a product homologous to benzoate coenzyme A ligase, which may be involved in activating anthranilate for PQS synthesis. Genes pqsB, pqsC, and pqsD encode proteins homologous to β-keto-acyl-acyl carrier protein synthases and are presumably involved in the production of a long chain hydrocarbon which reacts with anthranilate in the PQS biosynthetic pathway (4). Gene pqsE is not homologous to any defined proteins and our results indicate that it is not required for PQS synthesis. Gene pqsH encodes a putative FAD-dependent monooxygenase that may be responsible for the addition of the hydroxyl group to PQS.
Although phnA and phnB were originally assumed to encode an anthranilate synthetase comprising part of the phenazine biosynthetic pathway (10), recent studies by Mavrodi et al. (21) imply that they are unlikely to participate directly in the formation of the phenazine nucleus. The requirement of phnAB for PQS biosynthesis indicates that rather than playing a direct role in phenazine biosynthesis, the defect of phnAB mutants in pyocyanin production is most likely due to the highly pleiotropic phenotype caused by reduced PQS signaling. The observation that the lysR regulator encoded by mvfR (pqsR) in strain PA14 is required for the expression of phnAB also accounts directly for why the gene is required for the production of multiple virulence factors (5). An earlier study showed that PA14 mvfR was required for production of the Las system autoinducer and/or PQS (the assay used did not distinguish between the two substances), although the nature of the requirement was not clear (5).
To further characterize the Pqs proteins and other functions needed for pyocyanin production, we examined the effects of the corresponding mutations on the expression of several quorum sensing-regulated lacZ transcriptional gene fusions described by Whitely et al. (36). Several conclusions emerged from this analysis. First, mutations in the genes required for PQS production (pqsC, pqsD, phnA, pqsR, and pqsH) and response (pqsE) reduced expression of pyocyanin and hydrogen cyanide biosynthetic functions (phzC1 and hcnB), indicating that reduced transcription accounts for the defects in the production of both substances. These findings are compatible with models proposing that PQS signaling controls expression of genes regulated by the rhl quorum-sensing system (22). Second, transcription of pqsH was severely reduced in the lasR mutant background. Since pqsH is required for PQS synthesis, this finding accounts for a previous observation that PQS production requires LasR function (27) and provides a specific link between the Las and PQS regulatory systems. Third, mutations in the putative regulators np20 and PA3946 reduced transcription of a number of quorum sensing-controlled genes, showing that these functions participate in the quorum-sensing regulatory network. The np20 mutant exhibited a particularly strong phenotype, suggesting that it functions above PQS signaling in the network. np20 is a homologue of the E. coli zinc uptake regulator Zur, whose expression is induced by respiratory mucus from cystic fibrosis patients and which is required for virulence in neutropenic mice (34, 35). PA3946 is homologous to Bordetella pertussis bvgS, a primary regulator of virulence genes (18).
The role of pqsE differs from that of the other pqs genes. Loss of pqsE function causes defects in pyocyanin production, worm killing, and expression of quorum sensing-regulated lacZ gene fusions, defects which parallel those observed for the other pqs mutants. However, pqsE is not required for PQS biosynthesis as assessed by chromatography and extracellular complementation. The findings suggest a role for PqsE in the cellular response to PQS. Such a role is supported by the finding that pqsE mutants are not complemented extracellularly by wild-type and other strains for phzC1-lacZ expression.
In conclusion, this study has helped specify functions required for the action of the novel extracellular quinolone signaling system in P. aeruginosa. In addition, this work has identified several new regulators which appear to belong to the quorum-sensing regulatory network. Although the biological meaning of the formidable complexity of this network remains elusive, specifying the components which make it up constitutes an important step in approaching this goal.
Acknowledgments
We thank Marvin Whiteley and E. Peter Greenberg for providing reporter strains, Matt Wolfgang and Steve Lory for cosmids, Linda Thomashow and Dmitri Mavrodi for helpful discussions and strains, Chris Cosma for sharing unpublished data, and David D'Argenio for helpful discussions.
This work was supported in part by a Cystic Fibrosis Research Development Program postdoctoral fellowship (to L.A.G.), by National Institutes of Health grant R01-AI46682 (to E.C.P.), and by a grant from the Cystic Fibrosis Foundation (to C.M).
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumer, C., and D. Haas. 2000. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173:170-177. [DOI] [PubMed] [Google Scholar]
- 3.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calfee, M. W., J. P. Coleman, and E. C. Pesci. 2001. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:11633-11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 98:14613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castric, C. A. 1981. The metabolism of hydrogen cyanide by bacteria, p. 233-261. In B. Vennesland, E. E. Conn, C. J. Knowles, J. Westley, and F. Wissing (ed.), Cyanide in biology. Academic Press, New York, N.Y.
- 7.Cox, C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Argenio, D., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Essar, D. W., L. Eberly, A. Hadero, and I. P. Crawford. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essar, D. W., L. Eberly, C. Y. Han, and I. P. Crawford. 1990. DNA sequences and characterization of four early genes of the tryptophan pathway in Pseudomonas aeruginosa. J. Bacteriol. 172:853-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 13.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 14.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinnear, S. M., R. R. Marques, and N. H. Carbonetti. 2001. Differential regulation of Bvg-activated virulence factors plays a role in Bordetella pertussis pathogenicity. Infect. Immun. 69:1983-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 20.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 21.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. F. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 26.Pesci, E. C., and B. H. Iglewski. 1997. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 5:132-135. [DOI] [PubMed] [Google Scholar]
- 27.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 32.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 34.Wang, J., S. Lory, R. Ramphal, and S. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 35.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]