Abstract
Two distinctive colony morphologies were noted in a collection of Pseudomonas aeruginosa transposon insertion mutants. One set of mutants formed wrinkled colonies of autoaggregating cells. Suppressor analysis of a subset of these mutants showed that this was due to the action of the regulator WspR and linked this regulator (and the chemosensory pathway to which it belongs) to genes that encode a putative fimbrial adhesin required for biofilm formation. WspR homologs, related in part by a shared GGDEF domain, regulate cell surface factors, including aggregative fimbriae and exopolysaccharides, in diverse bacteria. The second set of distinctive insertion mutants formed colonies that lysed at their center. Strains with the most pronounced lysis overproduced the Pseudomonas quinolone signal (PQS), an extracellular signal that interacts with quorum sensing. Autolysis was suppressed by mutation of genes required for PQS biosynthesis, and in one suppressed mutant, autolysis was restored by addition of synthetic PQS. The mechanism of autolysis may involve activation of the endogenous prophage and phage-related pyocins in the genome of strain PAO1. The fact that PQS levels correlated with autolysis suggests a fine balance in natural populations of P. aeruginosa between survival of the many and persistence of the few.
Pseudomonas aeruginosa, although readily defined taxonomically (69), shows no evidence of being constrained to a particular bacterial lifestyle (72). Indeed, this opportunistic pathogen proliferates within hosts as varied as plants, insects, nematodes, and mammals and interacts with amoebae and yeasts as well (13, 16, 31, 44, 58). The broad host range of P. aeruginosa can be used both to uncover the role of specific virulence factors and to characterize new regulatory systems controlling virulence (10, 12, 13, 16, 21, 44, 58). One study identified transposon insertion mutants of P. aeruginosa strain PAO1 that were less virulent in the fruit fly model host (16). Most such strains had an insertion in a chemotaxis-like regulatory locus (PA0408 to PA0417) required for surface spreading by twitching motility and controlling unknown factors required for efficient fly killing. Based only on their distinctive compact colony morphology, a large collection of these mutants was assembled for testing in the fly model.
In this same study (16), two even more striking colony morphologies were noted, and strains of each type were saved. One set of mutants formed wrinkled colonies and had additional properties described for autoaggregating cells of Pseudomonas fluorescens (59, 68) and Salmonella enterica serovar Typhimurium (61, 62, 83). The second set of mutants formed colonies with visible lysis, a characteristic noted in some of the earliest descriptions of P. aeruginosa isolates (5, 6, 26, 32), but for which the molecular basis has been elusive. Since autoaggregation and autolysis are likely to reflect fundamental aspects of the biology of P. aeruginosa, the genetic basis of the two mutant colony morphologies was explored. Autoaggregation, in the subset of strains analyzed, was found to require the GGDEF-type (28) response regulator WspR. Autolysis, in strains with the most pronounced phenotype, correlated with the level of the Pseudomonas quinolone signal (PQS), an extracellular signal (9, 49, 56) that interacts with the quorum sensing regulatory hierarchy.
MATERIALS AND METHODS
Strains and culture conditions.
P. aeruginosa strain PAO1 (from the laboratory of B. Iglewski) was grown with Luria-Bertani (LB) medium (with NaCl at 8 g/liter) at 37°C on 1.5% agar or in 5-ml cultures unless otherwise noted (all percentages indicate wt/vol). Medium ingredients (Difco) included brain heart infusion, nutrient broth, Bacto Agar, tryptone peptone, tryptic soy broth, and yeast extract. The wrinkled-colony phenotype of mutant cells was most evident after growth with the richest media. To observe swarming motility, cells were grown on 0.5% agar with 0.8% nutrient broth and 0.5% glucose (60), while for swimming motility, cells were grown in 0.3% agar with LB medium with NaCl at 8 g/liter and without the yeast extract. Freshly poured agar for motility tests was left at room temperature for 24 to 36 h before use. For PQS analysis, cells were grown with peptone tryptic soy broth (PTSB) (49).
Derivatives of plasmid pVSP61 carrying wspR19 or wspR9 (P. J. Goymer, S. G. Kahn, and P. B. Rainey, unpublished data) were transferred into P. aeruginosa by conjugation (as described below for transposon mutagenesis of the wspF10 mutant). These plasmids were maintained using kanamycin at 500 μg/ml, and expression of the cloned genes from the plasmid lac promoter was constitutive in P. aeruginosa because of the absence of LacI. Plasmid pUCP18 (64) and derivatives were transferred into P. aeruginosa by transformation and maintained using carbenicillin at 200 μg/ml.
Growth curves were determined from experiments performed in triplicate: 35 ml of LB medium in a 300-ml Erlenmeyer flask with baffles was inoculated with 1 ml of a fresh 12-h culture and then incubated with shaking at 37°C for 3 days. The CFU in the cultures over time were determined by plating dilutions of 100-μl samples on LB agar.
Transposon mutagenesis.
To provide a genetic background in the colony morphology mutants for a second round of mutagenesis with ISphoA/hah-Tc (16, 21), the transposable element was excised from the chromosome, as described previously (45), by introduction of a plasmid expressing the Cre recombinase. Subsequent recombination between loxP sites flanking the transposable element left a chromosomal insertion of 189 bp and restored sensitivity to tetracycline (45). Insertional mutagenesis in the resulting strains was performed as in a previous study (16), but with the following modifications. Cultures of mating recipient cells were started with fresh single colonies of the wspF mutant grown at 23°C or of the pqsL mutant grown at 37°C. These cultures were incubated at 42°C without shaking for approximately 20 h. Colonies of cells in which a chromosomal insertion suppressed the mutant phenotype were identified after growth for 21 h at 37 and 30°C, respectively, for wspF mutants and pqsL mutants. The DNA sequence of the region flanking the insertion site was determined as before (16), and the identity of the mutated gene was determined by using data and annotations from the Pseudomonas Genome Project (http://www.pseudomonas.com).
PQS detection and quantitation.
PQS analysis has been described previously (9, 56). Freshly grown colonies were used to inoculate 1-ml cultures. After growth with PTSB for 20 h at 37°C, the entire culture was extracted twice with three volumes of acidified ethyl acetate and with centrifugation at 10,000 × g (extraction of spent culture supernatants gives equivalent results). The ethyl acetate fractions from each extraction were pooled and analyzed by thin-layer chromatography (TLC). TLC plates (20 by 20 cm; Silica Gel 60 F254; EM Science) were soaked in 5% KH2PO4 and activated at 100°C for 1 h. Sample extracts were separated on TLC plates using 17:2:1 methylene chloride-acetonitrile-dioxane as the solvent, conditions previously determined to be optimal for separation of PQS (56). TLC plates were then dried, illuminated with long-wave UV light, and photographed with a Polaroid 3000ISO camera and Polaroid 667 black-and-white film. PQS was quantitated by densitometry using ImageQuant software (Molecular Dynamics).
For autolysis restoration by extracellular complementation, spent culture supernatants were sterilized using Millipore filters (0.22-μm pore size), and sterility was confirmed by testing aliquots for growth. Synthetic PQS, synthesized previously by Jarad Milbank (56), was dissolved in 1:1 ethyl acetate-acetonitrile for addition to 6-mm-diameter paper disks (Becton Dickinson).
RESULTS
Autoaggregation in mutants of P. aeruginosa strain PAO1.
Out of approximately 6,000 insertion mutants, previously generated using ISphoA/hah-Tc (16), six formed wrinkled colonies as shown in Fig. 1. DNA sequencing of the region flanking the insertion was successful for five of the six mutants, revealing three insertions in gene PA3703, one in gene PA0171, and one in gene PA2933 (gene numbers are from the Pseudomonas Genome Project). Different insertions in some of the same genes were also identified in wrinkled mutants obtained from transposon mutagenesis in separate studies: two insertions in PA3703, one in PA0171, and one in PA1121. DNA sequence analysis suggests that PA3703 encodes a CheB-like methylesterase and that PA2933 encodes an efflux protein of the major facilitator superfamily. The potential function of the remaining two genes is unknown. Gene PA3703, with five insertions, lies within a gene cluster predicted to encode a chemotaxis-like signal transduction cascade (72). This cluster is homologous to the wsp cluster from P. fluorescens that is also involved in autoaggregation and wrinkled-colony morphology (68; E. Bantinaki, A. J. Spiers, and P. B. Rainey, unpublished data). The P. aeruginosa homologs were consequently given the wsp designation, with PA3703 corresponding to wspF (Fig. 2), and the phenotype associated with one insertion, wspF10, was characterized in more detail.
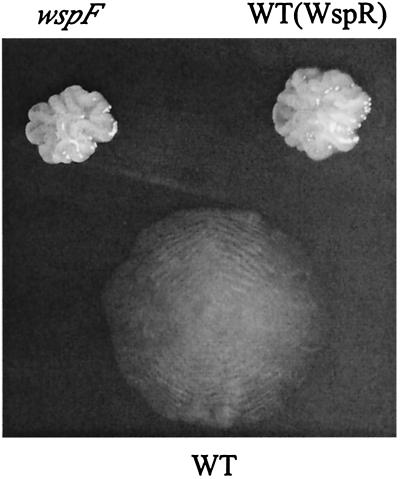
FIG. 1.
Colony morphology of the wild-type P. aeruginosa strain PAO1 (WT), the wspF10 insertion mutant, and strain PAO1 carrying a plasmid expressing the P. fluorescens constitutively active WspR19 [WT(WspR)]. Each strain was incubated separately on brain heart infusion agar, and after 3 days at 23°C, a colony of each of the two wrinkled mutants was transferred using forceps to the agar surface adjacent to a colony of strain PAO1. The WT colony had a diameter of approximately 0.3 cm.
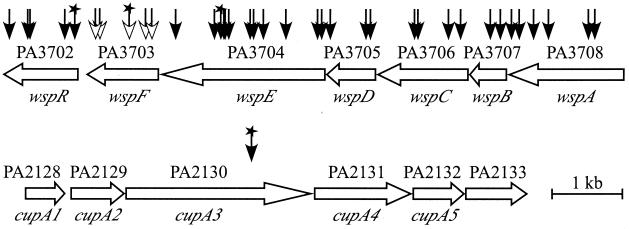
FIG. 2.
P. aeruginosa genes in two chromosomal loci (distinguished by gene numbering) involved in autoaggregation and wrinkled-colony morphology. Vertical arrows indicate the sites of insertion of the ISphoA/hah-Tc transposable element. Unfilled arrowheads denote insertions, all in wspF, that confer autoaggregation. Stars indicate insertions in strains that were characterized in more detail. For suppressor analysis of the strain with the wspF10 insertion, the transposon was excised from wspF (leaving a 189-bp insertion) and transposon mutagenesis of this strain was repeated: filled arrowheads denote insertions that suppress autoaggregation, either completely (insertions in the wsp genes) or partially (the insertion in cupA3). The one exception was the strain with the wspE65 insertion, in which autoaggregation was suppressed at 37°C but not at 23°C.
The wspF10 mutant did not spread by twitching motility and formed wrinkled colonies that grew mostly vertically (Fig. 1). Cells of this mutant adhered so tightly to each other that the entire wrinkled colony was easily transferred with forceps from one agar surface to another, as was done to generate Fig. 1. In contrast, nonaggregative wild-type cells spread using type IV pili for twitching motility (65) and formed flatter colonies (Fig. 1). The wspF10 mutant also failed to spread by either of two forms of flagellum-based motility (Fig. 3): swarming motility on the agar surface using self-generated biosurfactant or swimming within the agar (40, 60). Indeed, under conditions favoring swimming, the wspF10 mutant grew predominately as a wrinkled mass protruding above the agar (Fig. 4), in stark contrast to the diffuse distribution of wild-type cells (Fig. 3).
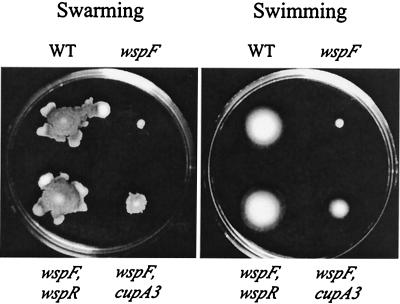
FIG. 3.
Swarming motility (on the surface of 0.5% agar after 24 h at 37°C) and swimming motility (within 0.3% agar after 12 h at 37°C) of strain PAO1 (WT), the wspF10 mutant, and strains in which autoaggregation caused by an insertion in wspF was suppressed by an insertion in either wspR or cupA3 (Fig. 2).
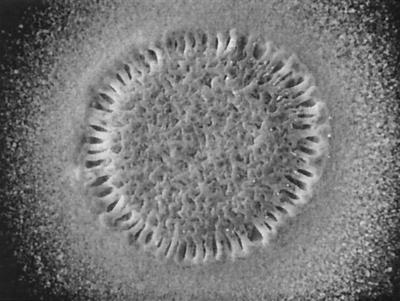
FIG. 4.
Autoaggregation of the wspF10 mutant growing in and above swimming-motility agar after 48 h at 23°C. The agar was inoculated using a toothpick with wspF10 cells from a diffusely turbid culture grown at 42°C, a temperature that suppresses clumping (inoculation with cells grown at 23°C on an agar surface yielded equivalent results). Cells have dispersed to a maximum diameter of 0.5 cm; cells of strain PAO1 incubated in the same way disperse evenly over a zone approximately 4 cm in diameter.
Additional phenotypes of the wspF10 mutant (not shown) suggested even closer parallels with properties of autoaggregating cells of P. fluorescens (59, 68) and S. enterica serovar Typhimurium (61, 62, 83). These phenotypes included increased bacterial lawn hydrophobicity, evident because wspF10 mutant cells formed lawns with a dry appearance upon which water droplets persisted as tight beads, while wild-type cells formed lawns with a wet appearance upon which water droplets collapsed. In addition, the wspF10 mutant grew in standing LB cultures primarily as a pellicle at 23°C and with diffuse turbidity at 42°C, indicating that autoaggregation was suppressed by higher growth temperatures. After prolonged incubation, autoaggregating cultures or colonies of the wspF10 mutant were commonly overgrown by phenotypic revertants. A similar autoaggregation phenotype was described for spontaneous mutants of P. aeruginosa strain 57RP (17) and strain PAK (3), but the genes responsible were not identified.
WspR controls autoaggregation.
Analysis of the wrinkled-colony phenotype of P. fluorescens implicated WspR (68), a protein related to the Caulobacter crescentus PleD global regulator controlling the transition between motile swarmer cell and nonmotile stalked cell (28). To test whether a similar regulatory system operated in P. aeruginosa, a constitutively active WspR from P. fluorescens (WspR19) (Goymer et al., unpublished) was expressed in strain PAO1. The resulting colonies were wrinkled and identical in appearance to those of the wspF10 mutant (Fig. 1). Furthermore, expression of a dominant negative mutant WspR from P. fluorescens (WspR9) (Goymer et al., unpublished) had no apparent effect on colonies of strain PAO1 but completely suppressed the wrinkled phenotype of wspF10 mutant colonies (data not shown). The latter colonies, however, were slightly smaller than those of the wild-type strain, suggesting that some autoaggregation remained.
WspF is mutated in five strains forming wrinkled colonies, including the wspF10 mutant (Fig. 2). WspF is 30% identical (over 335 amino acids) to CheB, a methylesterase from Escherichia coli. In E. coli, CheB removes a methyl group from methyl-accepting chemotaxis proteins, reducing activity in the signal transduction cascade during chemotaxis (71). Assuming a similar function in P. aeruginosa, inactivation of WspF would result in hypermethylation of the methyl-accepting chemotaxis protein (WspA). This would cause constitutive activation of the Wsp chemosensory pathway and ultimately of the response regulator WspR, the predicted target of the Wsp phosphorelay (68). Since WspR controls the structural components necessary for the wrinkled-colony morphology (68; Goymer et al., unpublished), this would mimic the effect of expressing the heterologous constitutively active WspR.
To test this hypothesis, the transposon in the wspF10 mutant was excised from the chromosome, leaving an insertion of 189 bp (see Materials and Methods) which maintained the wrinkled phenotype. A second round of transposon mutagenesis was then performed in this strain to identify genes required for the wrinkled-colony morphology. Approximately 60,000 insertion mutants were screened, yielding 95 suppressed double mutants. The insertion site was sequenced in those 39 suppressed mutants that were independently obtained, 38 of which had a wild-type colony morphology at 37°C. The one remaining mutant was only partly suppressed for autoaggregation. Of the 38 strains, 34 carried an insertion in the wsp genes (Fig. 2), and all with the exception of three identical insertions in wspR were unique.
Mutation of wspR completely suppressed colony wrinkledness and restored swarming and swimming motility (Fig. 3), consistent with WspR being the ultimate target of the Wsp phosphorelay and the proximate cause of the wrinkled phenotype. The same conclusion was reached recently based on the fact that a mutation in either wspR or the wspABCDEF operon eliminated wrinkledness in a P. fluorescens strain with a spontaneous mutation in wspF (A. J. Spiers, Z. Robinson, and P. B. Rainey, unpublished data). The strain with the wspE65 insertion (Fig. 2) provided further evidence that autoaggregation was favored by lower growth temperatures. Of the double mutants forming colonies with a wild-type appearance at 37°C, only this strain formed wrinkled colonies at 23°C, suggesting that the wspE65 insertion partly inactivated wspE and that this leaky mutation was sufficient to suppress the wrinkled phenotype only at higher growth temperatures.
Additional genes involved in autoaggregation.
The one remaining mutant, in which autoaggregation at 37°C was only partly suppressed, carried an insertion in PA2130 (Fig. 2). Although growth curves indicated that this mutant grew at a wild-type rate in cultures shaken at 37°C, the mutant formed unwrinkled colonies on LB agar slightly smaller than those formed by the wild-type strain (not shown) and was partly impaired in swarming and swimming motility (Fig. 3). Excision of the transposon in this strain (again leaving a 189-bp insertion) restored the wrinkled-colony phenotype, suggesting that the transposon in PA2130 had a polar effect on expression of downstream genes. PA2130, recently designated cupA3 in P. aeruginosa strain PAK, lies within a gene cluster (Fig. 2) that is predicted to encode a novel fimbrial adhesin (14, 75), and insertion mutations within this cluster prevent biofilm formation (75). In addition, at least two genes in this cluster, PA2128 and PA2129, are differentially regulated during growth as a biofilm (81), and PA2128 is related to the gene encoding the subunit of aggregative fimbriae which mediate biofilm formation and the wrinkled-colony phenotype in Salmonella (61, 62, 75, 83).
Autolysis in mutants of P. aeruginosa strain PAO1.
Among the approximately 6,000 transposon insertion mutants previously generated (16), not only were the six that formed wrinkled colonies notable, but also three that formed colonies that lysed at their centers. In the areas of highest cell density in lawns of these three strains, plaque-like clearings developed, expanding and merging to form zones of confluent lysis (Fig. 5). The plaque-like clearings and the metallic, iridescent sheen associated with the lysed areas are both properties noted in early descriptions of P. aeruginosa isolates (5, 6, 26, 32) but absent in strain PAO1 (Fig. 5). In a recent collection of P. aeruginosa isolates from cystic fibrosis patients, 31% (59 of 191) had visible lysis (S. Miller, personal communication).
FIG. 5.
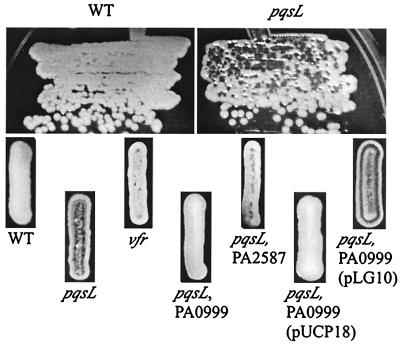
Autolysis visible as plaque-like clearings (darker zones) in areas of dense growth on LB agar after 18 h at 37°C (top row) or in toothpick-generated streaks after 18 h at 37°C followed by 6 days at 23°C (bottom row). Visible autolysis is absent in P. aeruginosa strain PAO1 (WT), in contrast to the mutant with the pqsL64 insertion or an insertion in vfr. In double-mutant strains, autolysis caused by an insertion in pqsL is suppressed by an insertion in PA0999 or PA2587. The transposon was excised from the chromosome in the former mutant, leaving an insertion of 189 bp and resulting in an insertion of 63 amino acids in the protein encoded by PA0999. Autolysis in this strain was restored by complementation with plasmid pLG10 (22) carrying PA0996 to PA1000, but the vector pUCP18 had no effect.
In the two insertion mutants with the most pronounced lysis, DNA sequencing revealed two different insertions in PA4190, here designated pqsL, and the phenotype associated with the pqsL64 insertion was characterized in more detail (Fig. 5 and 6). The pqsL gene is predicted to encode a member of a family of monooxygenases in P. aeruginosa (72) that include products of pobA and ubiH, genes which encode hydroxylases involved in p-hydroxybenzoate catabolism and ubiquinone biosynthesis, respectively. PA4190 is also related to three quorum-sensing-controlled genes (80): PA2587 (qsc105), PA3328 (qsc125), and PA4217 (qsc132), of which the third gene encodes the hydroxylase PhzS involved in phenazine biosynthesis (48). The one remaining mutant had less pronounced lysis and carried an insertion in the gene encoding Vfr (Fig. 5). Vfr is a homolog of the E. coli cyclic AMP receptor protein CRP (4, 78) and acts at the top of the quorum sensing regulatory hierarchy (1, 4). An insertion in PA4190 was also identified in a mutant with visible lysis obtained from transposon mutagenesis in a separate study, but no more strains with this phenotype were found by screening an additional 3,000 transposon insertion mutants.
FIG. 6.
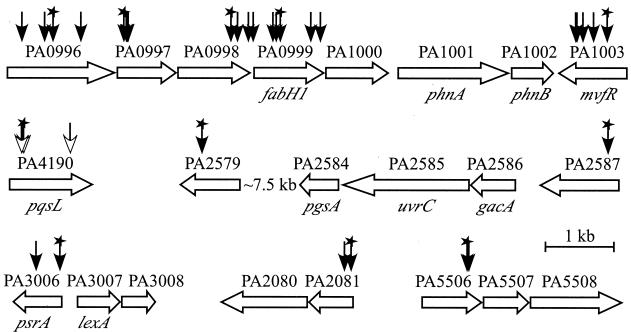
P. aeruginosa genes in six chromosomal loci (distinguished by gene numbering) involved in autolysis. Vertical arrows indicate the sites of insertion of the ISphoA/hah-Tc transposable element. Unfilled arrowheads denote insertions, all in pqsL, that confer autolysis. Stars indicate insertions in strains that were characterized in more detail. For suppressor analysis of the strain with the pqsL64 insertion, the transposon was excised from pqsL (leaving a 189-bp insertion) and transposon mutagenesis was repeated: filled arrowheads denote insertions that suppress autolysis, either completely (insertions in PA0996 to PA1003) or partially.
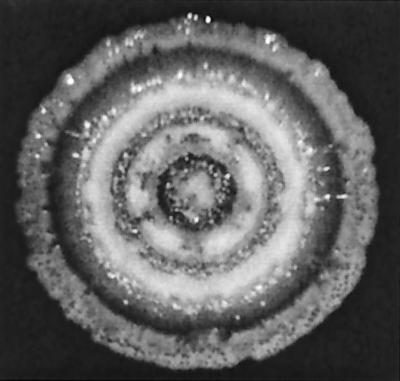
Colonies of the pqsL64 mutant had a particularly distinctive morphology after prolonged incubation. As the band of peripheral cells spread outward from the central lysed area, the center of the band itself developed plaque-like holes which coalesced, forming concentric zones of lysis as this process repeated (Fig. 7). The same pattern of lysis was observed for pqsL64 mutant colonies incubated in the dark, indicating a different mechanism of cell death than that by which visible light kills E. coli ubiH (visB) mutant cells (51). In liquid culture, the pqsL64 mutant grew like the wild-type strain for the first 24 h, reaching a density of approximately 1010 CFU/ml, and then both strains lost viability over time. However, by 72 h, the cultures of the pqsL64 mutant had lost an average of four times more CFU than those of the wild-type strain, with CFU dropping by a factor of 48 versus 12, respectively. No phenotypic revertants were noted. The relative subtlety of this mutant phenotype suggested that high cell density, or another condition required for the pronounced lysis during surface growth, was not completely reproduced by growth in liquid.
FIG. 7.
Concentric zones of lysis (darker bands) formed during growth of the pqsL64 mutant on LB agar. The agar was inoculated with 20 μl of 10 mM MgSO4 containing cells of the pqsL64 mutant from six small colonies from LB agar. After 36 h at 30°C, the plate was sealed with Parafilm, inverted, and incubated for 25 days at 23°C. The colony has grown to a diameter of 3.5 cm, and the plaque-like holes in the outermost band of cells have not yet coalesced. No lysis was visible in colonies of wild-type cells generated in the same way (as also seen in Fig. 5).
Autolysis is suppressed by mutations that reduce the level of PQS.
The colony phenotype of the pqsL64 mutant suggested that autolysis would be as amenable to genetic analysis as autoaggregation. The transposon in the pqsL64 mutant was excised, with the remaining 189-bp chromosomal insertion preserving the phenotype, and transposon mutagenesis of this strain was repeated. Out of approximately 20,000 insertion mutants, 37 strains formed colonies in which visible lysis was either reduced or absent, and these strains had a wild-type growth rate based on colony size (400 strains that had delayed lysis because of slower growth were not analyzed further). The insertion site was sequenced in those 30 suppressed mutants that were independently obtained: 22 strains with no apparent lysis (typified by one with an insertion in PA0999 in Fig. 5) had a unique insertion in a single chromosomal region (Fig. 6). The remaining eight strains with reduced lysis (typified by one with an insertion in PA2587 in Fig. 5) had unique insertions more dispersed around the chromosome (Fig. 6).
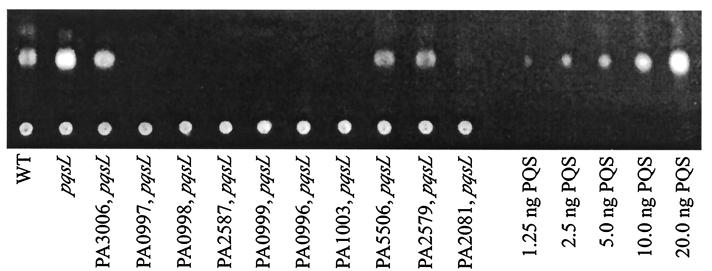
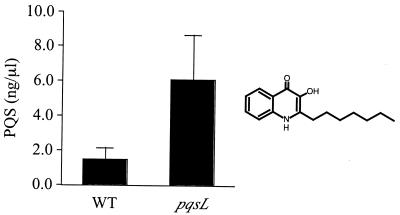
The 22 strains with autolysis completely suppressed carried insertions in PA0996, PA0997, PA0998, PA0999, and PA1003 (Fig. 6). PA1003, designated mvfR in P. aeruginosa strain PA14 (10), encodes a transcriptional regulator required for production of multiple virulence factors, including pyocyanin, elastase, and cyanide. PA1003 is consequently required for full virulence in diverse model hosts (10, 21). Recently, both mvfR (10) and the operon including PA0996 to PA0999 (22) were found to be involved in biosynthesis of PQS, an extracellular signal in the quorum sensing hierarchy (9, 49, 56). Measurement of the amount of PQS made by the strains in this study revealed that the pqsL64 mutant produced approximately four times as much PQS as the wild-type strain, and all the suppressor mutations reduced this amount (Fig. 8 and 9). Those strains in which autolysis was completely suppressed made no detectable PQS (Fig. 8). In a mutant with a nonpolar suppressor mutation in PA0999, autolysis was restored by complementation with a plasmid containing PA0996 to PA1000 (22), while the vector alone had no effect (Fig. 5).
FIG. 8.
TLC analysis of PQS produced by P. aeruginosa strain PAO1 (WT), the pqsL64 mutant, and derivatives of the latter strain in which autolysis is suppressed (Fig. 6). Extracts of 1-ml overnight cultures were analyzed by TLC using conditions previously optimized for separation and visualization of PQS (see Materials and Methods). Various quantities of synthetic PQS (56) were analyzed for comparison. Doubling the amount of sample extract in the TLC analysis yielded equivalent results.
FIG. 9.
PQS in cultures of P. aeruginosa strain PAO1 (WT) and the pqsL64 mutant. PQS was separated by TLC as described in the legend for Fig. 8 and quantitated by densitometry. Relative PQS concentration was determined by comparing spots associated with sample extracts with those generated by known amounts of PQS. Data are the averages (error bars, standard deviations) of three independent experiments. PQS (2-heptyl-3-hydroxy-4-quinolone) is shown on the right.
Autolysis in the PQS-overproducing pqsL64 mutant occurred in the areas of highest cell density during surface growth (Fig. 5), consistent with a causal role for PQS, an extracellular compound whose production peaks in late stationary phase (49). Supporting this connection, lysis of the partially suppressed PA2587 mutant was triggered by addition of synthetic PQS (Fig. 10). Further implicating PQS, DNA sequence analysis of one mutant with partially suppressed autolysis suggested that the affected gene, PA2579 (Fig. 6), encodes tryptophan 2,3-dioxygenase. This is the first enzyme in the kynurenine pathway from tryptophan to anthranilate (53), which is a precursor of PQS (9). Partial suppression of autolysis by mutation of psrA (41) (Fig. 6), encoding an activator for expression of the stationary-phase sigma factor RpoS, also implicates stationary-phase processes.
FIG. 10.
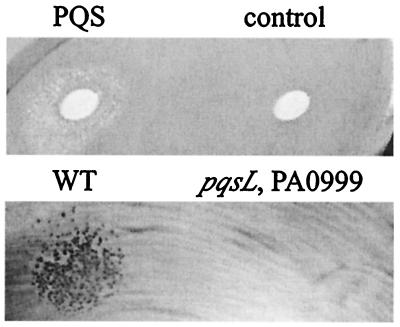
Extracellular complementation of autolysis in a lawn of cells of a suppressed mutant. Fifteen microliters of solvent, either containing 15 μg of synthetic PQS (PQS) or alone (control), was added to a filter disk on the lawn (top row); 10 μl of sterile spent culture supernatant from 24-h cultures, either of P. aeruginosa strain PAO1 (WT) or of a double mutant in which autolysis is fully suppressed by an insertion in PA0999 (pqsL, PA0999), was added directly to the lawn (bottom row). The lysed area appears lighter in the top panel due to reflected light. The lawn was formed using a 12-h culture of a double mutant (in which autolysis caused by an insertion in pqsL is partially suppressed by an insertion in PA2587) that was diluted 10-fold to delay the density-dependent lysis during subsequent surface growth. One hundred microliters of the dilution was spread on LB agar, and after the various additions, the lawn was incubated for 24 h at 37°C. Three independent experiments yielded equivalent results.
Although PQS may be required for the full autolysis observed for the pqsL64 mutant, it appears not to be the sole factor involved. In the partially suppressed mutants (with lysis still evident), PQS levels are equivalent to that in the wild-type strain or are not detectable, as in the PA2587 mutant (Fig. 8). P. aeruginosa is known to make multiple quinolones in addition to PQS (8, 42), and some that also require PA0996 to PA0999 for biosynthesis may account for the residual lysis. Indeed, lysis of the partially suppressed PA2587 mutant could be triggered with spent culture supernatant from the wild-type strain, but not with supernatant from a mutant carrying an insertion in PA0999 (Fig. 10).
DISCUSSION
Intracellular signaling and autoaggregation.
The wrinkled-colony phenotype has been analyzed for cells of P. fluorescens (59, 68), S. enterica serovar Typhimurium (61, 62, 83), and now P. aeruginosa. These studies have uncovered the role in autoaggregation played by regulatory proteins that are related to C. crescentus PleD (28), in part because of a shared GGDEF domain (23, 28). The genetic basis of autoaggregation, which was revealed using wrinkled colonies of mutant cells, is likely to apply to multicellular behavior of wild-type bacteria as well. Autoaggregation of the plague bacterium Yersinia pestis, for instance, requires the GGDEF-containing protein HmsT (35). Clumping cells of this organism block food intake of both nematode worms (15) and fleas (30). In the latter case, this results in increased transmission by the flea vector (30). For P. aeruginosa, aggregation caused by the GGDEF-containing WspR is linked to regulation of cup genes (Fig. 2) that encode a putative fimbrial adhesin required in wild-type cells for biofilm formation (75). The role of WspR in biofilm formation could include controlling not only adhesion to foreign surfaces but also an alignment of bacterial cells that could act as a regulatory signal itself, as shown to occur during Myxococcus xanthus differentiation (38).
Proteins with a GGDEF domain control expression of a conserved set of products in cells forming wrinkled colonies, including a cellulose-like polymer (68, 83) and aggregative fimbriae (61, 62, 83). P. aeruginosa strain PAO1, however, appears not to have the known cellulose biosynthesis genes: the P. fluorescens wss operon (68) or the P. putida and S. enterica serovar Typhimurium bcs operon (83). Instead, WspR could participate in regulation of glycogen expression in P. aeruginosa. Glycogen is an intracellular polymer that is involved in E. coli and Salmonella biofilm formation (7, 34). As in these organisms, glycogen could function as a store of glucose (7, 34) for use as a precursor for the extracellular polysaccharides that comprise the protective matrix of P. aeruginosa biofilms (25, 29, 81). Glycogen metabolism and expression of aggregative fimbriae are coordinately regulated in E. coli (34). In P. aeruginosa, genes differentially regulated during biofilm growth (81) include PA2128 and PA2129 in the cupA fimbrial gene cluster (Fig. 2) and PA2160. DNA sequence analysis suggests that PA2160 encodes the glycogen catabolic enzyme GlgX (82), and several putative glycogen biosynthesis genes are closely linked to PA2160 on the chromosome (72).
The mechanism by which regulatory proteins with a GGDEF domain exert their effect is not yet clear, but there is growing evidence that the domain encodes a nucleotide cyclase (2, 55, 63, 73). The first such evidence was the requirement of this domain for biosynthesis of cyclic diguanylate, the intracellular signal regulating production of extracellular cellulose in Gluconacetobacter xylinum (63). The prediction that this novel signal might have more widespread functions in bacteria (63) was subsequently supported by bacterial genome sequences. GGDEF-containing proteins are not only widespread but also abundant in individual genomes, with 33 such proteins encoded in P. aeruginosa strain PAO1 alone (14, 23).
In this study, mutants with the wrinkled-colony phenotype carried insertions in three different loci (wspF, PA0171, and PA1121) that are each closely linked to a gene for a protein with a GGDEF domain (wspR, PA0169, and PA1120, respectively). Furthermore, PA2133 in the cupA operon (Fig. 2) is predicted to encode a protein in which the presence of an EAL domain (23, 50) suggests a phosphodiesterase activity for degrading the cyclic diguanylate signal (23, 73). An EAL domain is also present in the recently described regulator PvrR, which is involved in autoaggregation and the formation of a small, rough colony variant of P. aeruginosa strain PA14 (18). These observations support an intracellular signaling system in P. aeruginosa that is mediated by GGDEF-containing proteins and that plays a key role in controlling the adhesiveness of the bacterial cell surface.
Extracellular signaling and autolysis.
The pqsL64 mutant, identified because of its pronounced autolysis, overproduces PQS. The link between PQS and the monooxygenase that pqsL is predicted to encode is not yet clear. The fact that autolysis is partially suppressed by mutation of the homologous monooxygenase predicted to be encoded by PA2587 is suggestive. PA2587 could encode the enzyme for the final step in PQS biosynthesis, addition of the hydroxyl group (Fig. 9). Therefore, the homologous pqsL could encode an enzyme that also acts on PQS, and the pqsL64 mutation could increase PQS levels by interrupting a pathway for PQS degradation (49) or modification.
Although there is a correlation between suppression of autolysis and decreased PQS production by the pqsL64 mutant, the mechanism of autolysis also remains unknown. P. aeruginosa quinolones have antibacterial activity, a property discovered because of the ability of this bacterium to prevent anthrax in a mixed infection, a line of investigation begun by Pasteur (8, 27). The efficacy of quinolone antibiotics may be due in part to their induction of endogenous prophage via the bacterial SOS response (20, 57), perhaps the same phenomenon triggered by overproduction of PQS in the pqsL64 mutant. The genome of P. aeruginosa strain PAO1 contains a filamentous bacteriophage (72) whose genes are highly upregulated during biofilm growth (81). In addition, inserted between the two anthranilate synthase subunit genes, trpE and trpG, is a gene cluster encoding bacteriocins believed to be evolved phage tails (52) and whose induction shares elements with the SOS response (47). Closely linked to this gene cluster (PA0616 to PA0648) is vfr (PA0652). The vfr gene is the other locus in this study whose mutation is associated with visible autolysis (Fig. 5). This is perhaps analogous to the situation in E. coli where mutation of crp shifts the balance from lysogeny to lysis (33), given that vfr and crp are homologs (4, 78), and suggests an alternate pathway for autolysis in P. aeruginosa.
The autolysis associated with PQS overproduction notwithstanding, PQS is likely to play a fundamental role in tuning cellular physiology. PQS strongly induces the rhl quorum-sensing system (49), and a balance between expression of the las and rhl systems weighted towards rhl is associated with growth of P. aeruginosa cells as a resistant biofilm outside of and within a human host (66). Furthermore, PQS was originally discovered because it induced expression of the quorum-sensing-regulated lasB gene that encodes the secreted virulence factor elastase (56). The LasB protease, however, is also required within the bacterial cell to activate the nucleotide diphosphate kinase Ndk (36). In the activated form, Ndk generates GTP, used as the substrate for RelA to generate the signal for the stringent response to starvation (11, 36, 37). Further supporting such a metabolic link, the quorum-sensing system can be activated by starvation either physiologically (77) or by mutation (76). This situation may underlie the selection pressure for compensatory mutations in certain quorum-sensing mutants (3, 4).
GTP is in particularly high demand during biosynthesis of alginate (11, 36, 37), which is secreted as part of a stress response, and which contributes to persistence of mucoid P. aeruginosa cells in a biofilm in chronic infections (25, 29). Consistent with a delicate equilibrium (26) between this protective response and autolysis, each controlled by PQS levels, P. aeruginosa strain PAO1 cells forming colonies with visible lysis (74) were repeatedly recovered during chemostat growth with media that stimulated alginate production. Such an equilibrium could be maintained in part at the level of transport. PQS appears to be the substrate of at least one multidrug efflux pump (39, 54), including a transport system whose expression varies between isolates of strain PAO1 depending on the allele of mexT (39, 46).
Even autolysis, which might seem unambiguously detrimental to a unicellular organism, may be an adaptive behavior mediated by PQS. There is increasing evidence for programmed death pathways in bacteria, some based on an extracellular signal and influenced by the stringent response (19, 43, 70). Whether or not cell death is their primary function (24) remains uncertain, but an outcome in common between these pathways is persistence of a fraction of the bacterial population (19, 43, 70). Such persistence may be reflected in the cycles of lysis seen for the pqsL mutant in this study (Fig. 7). Persisting cells could benefit from the DNA released by lysed cells, since extracellular DNA was recently shown to be required by P. aeruginosa for formation of a protective biofilm (79).
P. aeruginosa diversity.
The myriad capabilities of P. aeruginosa have been revealed in part by examining single strains under a variety of conditions, including models of infection in diverse hosts. In a complementary approach applied in this and other Pseudomonas studies (17, 59, 68), mutants with a gain of function were characterized on the basis of their unique colony morphology. The readily visible phenotype associated with P. aeruginosa mutants with enhanced autolysis and enhanced autoaggregation will facilitate analysis of extracellular signaling by PQS and intracellular signaling mediated by WspR and perhaps other GGDEF domain-containing regulators. It remains a continual challenge to reveal the breadth and sources of Pseudomonas diversity (67), but the knowledge gained is likely to fundamentally contribute to our understanding of the evolutionary arms race between bacteria and their hosts.
Acknowledgments
This work was supported by a Cystic Fibrosis Research Development Program postdoctoral fellowship (to D.A.D.), by a Cystic Fibrosis Foundation Research Grant (grant PESCI99I0) and National Institutes of Health grant RO1-AI46682 (to E.C.P.), and by the University of Oxford and the Biotechnology and Biological Sciences Research Council (to P.B.R.).
We acknowledge Colin Manoil, who instigated transposon mutagenesis of P. aeruginosa strain PAO1, which led to the discovery of the colony morphology mutants. We thank Mitchell Brittnacher for invaluable computer assistance and Leo Pallanck for the use of the microscope and digital camera.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 3.Beatson, S. A., C. B. Whitchurch, A. B. T. Semmler, and J. S. Mattick. 2002. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 184:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berk, R. S. 1963. Nutritional studies on the “auto-plaque” phenomenon in Pseudomonas aeruginosa. J. Bacteriol. 86:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berk, R. S. 1965. Effect of antibacterial agents on the autoplaque phenomenon of Pseudomonas aeruginosa. Can. J. Microbiol. 11:213-219. [DOI] [PubMed] [Google Scholar]
- 7.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. García-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191:31-36. [DOI] [PubMed] [Google Scholar]
- 8.Budzikiewicz, H. 1993. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol. Rev. 104:209-228. [DOI] [PubMed] [Google Scholar]
- 9.Calfee, M. W., J. P. Coleman, and E. C. Pesci. 2001. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:11633-11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 98:14613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarty, A. M. 1998. Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell signalling and polysaccharide synthesis. Mol. Microbiol. 28:875-882. [DOI] [PubMed] [Google Scholar]
- 12.Chugani, S. A., M. Whitely, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie, M. Benghezal, C. van Delden, L. Kocjancic-Curty, and T. Köhler. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184:3027-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croft, L., S. A. Beatson, C. B. Whitchurch, B. Huang, R. L. Blakeley, and J. S. Mattick. 2000. An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways, and structures. Microbiology 146:2351-2364. [DOI] [PubMed] [Google Scholar]
- 15.Darby, C., J. W. Hsu, N. Ghori, and S. Falkow. 2002. Plague bacteria biofilm blocks food intake. Nature 417:243-244. [DOI] [PubMed] [Google Scholar]
- 16.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 19.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 20.Froshauer, S., A. M. Silvia, M. Chidambaram, B. Sharma, and G. M. Weinstock. 1996. Sensitization of bacteria to danofloxacin by temperate prophages. Antimicrob. Agents Chemother. 40:1561-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher, L. A., S. L. McKnight, M. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed]
- 23.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadley, P. 1924. Transmissible lysis of Bacillus pyocyaneus. J. Infect. Dis. 34:260-304. [Google Scholar]
- 27.Hays, E. E., I. C. Wells, P. A. Katzman, C. K. Cain, F. A. Jacobs, S. A. Thayer, E. A. Doisy, W. L. Gaby, E. C. Roberts, R. D. Muir, C. J. Carroll, L. R. Jones, and N. J. Wade. 1945. Antibiotic substances produced by Pseudomonas aeruginosa. J. Biol. Chem. 159:725-750. [Google Scholar]
- 28.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentzer, M., G. M. Teitzel, G. L. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 31.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 32.Holloway, B. W. 1969. Genetics of Pseudomonas. Bacteriol. Rev. 33:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong, J.-S., G. R. Smith, and B. N. Ames. 1971. Adenosine 3′:5′-cyclic monophosphate concentration in the bacterial host regulates the viral decision between lysogeny and lysis. Proc. Natl. Acad. Sci. USA 68:2258-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, H. A., J. W. Lillard, Jr., and R. D. Perry. 1999. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145:2117-2128. [DOI] [PubMed] [Google Scholar]
- 36.Kamath, S., V. Kapatral, and A. M. Chakrabarty. 1998. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol. Microbiol. 30:933-941. [DOI] [PubMed] [Google Scholar]
- 37.Kim, H.-Y., D. Schlictman, S. Shankar, Z. Xie, A. M. Chakrabarty, and A. Kornberg. 1998. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol. Microbiol. 27:717-725. [DOI] [PubMed] [Google Scholar]
- 38.Kim, S. K., and D. Kaiser. 1990. Cell alignment required in differentiation of Myxococcus xanthus. Science 249:926-928. [DOI] [PubMed] [Google Scholar]
- 39.Köhler, T., C. van Delden, L. K. Curty, M. M. Hamzehpour, and J.-C. Pechère. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhler, T., L. K. Curty, F. Barja, C. van Delden, and J.-C. Pechère. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leisinger, T., and R. Margraff. 1979. Secondary metabolites of the fluorescent pseudomonads. Microbiol. Rev. 43:422-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahajan-Miklos, S., M.-W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 45.Manoil, C. 2000. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 326:35-47. [DOI] [PubMed] [Google Scholar]
- 46.Maseda, H., K. Saito, A. Nakajima, and T. Nakae. 2000. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192:107-112. [DOI] [PubMed] [Google Scholar]
- 47.Matsui, H., Y. Sano, H. Ishihara, and T. Shinomiya. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J. Bacteriol. 175:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merkel, T. J., C. Barros, and S. Stibitz. 1998. Characterization of the bvgR locus of Bordetella pertussis. J. Bacteriol. 180:1682-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakahigashi, K., K. Miyamoto, K. Nishimura, and H. Inokuchi. 1992. Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J. Bacteriol. 174:7352-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakayama, K., K. Takashima, H. Ishihara, T. Shinomiya, M. Kageyama, S. Kanaya, M. Ohnishi, T. Murata, H. Mori, and T. Hayashi. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol. Microbiol. 38:213-231. [DOI] [PubMed] [Google Scholar]
- 53.Palleroni, N. J., and R. Y. Stanier. 1964. Regulatory mechanisms governing synthesis of the enzymes for tryptophan oxidation by Pseudomonas fluorescens. J. Gen. Microbiol. 35:319-334. [DOI] [PubMed] [Google Scholar]
- 54.Pearson, J. P. 2002. Early activation of quorum sensing. J. Bacteriol. 184:2569-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei, J., and N. V. Grishin. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210-216. [DOI] [PubMed] [Google Scholar]
- 56.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips, I., E. Culebras, F. Moreno, and F. Baquero. 1987. Induction of the SOS response by new 4-quinolones. J. Antimicrob. Chemother. 20:631-638. [DOI] [PubMed] [Google Scholar]
- 58.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rainey, P. B., and M. Travisano. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69-72. [DOI] [PubMed] [Google Scholar]
- 60.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Psudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Römling, U., M. Rohde, A. Olsén, S. Normark, and J. Reinköster. 2000. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 62.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behavior of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 63.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 64.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 65.Semmler, A. B. T., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 66.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. A. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 67.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 68.Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano, and P. B. Rainey. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161:33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanier, R. Y., N. J. Palleroni, and M. Douderoff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 70.Steinmoen, H., E. Knutsen, and L. S. Håvarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stock, J. B., and M. G. Surette. 1996. Chemotaxis, p. 1103-1129. In F. C. Neidhart, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaecter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 72.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 73.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Terry, J. M., S. E. Piña, and S. J. Mattingly. 1991. Environmental conditions which influence mucoid conversion in Pseudomonas aeruginosa PAO1. Infect. Immun. 59:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Delden, C., E. C. Pesci, J. P. Pearson, and B. H. Iglewski. 1998. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect. Immun. 66:4499-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Delden, C., R. Comte, and M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.West, S. E. H., and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487.. [DOI] [PubMed] [Google Scholar]
- 80.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 82.Yang, H., M. Y. Liu, and T. Romeo. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J. Bacteriol. 178:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]