Abstract
Expression of the Bacillus subtilis pyrG gene, which encodes CTP synthetase, is repressed by cytidine nucleotides. Regulation involves a termination-antitermination mechanism acting at a transcription terminator located within the 5′ untranslated pyrG leader sequence. Deletion and substitution mutagenesis of a series of pyrG′-lacZ transcriptional fusions integrated into the B. subtilis chromosome demonstrated that only the terminator stem-loop and two specific 4- to 6-nucleotide RNA sequences were required for derepression of pyrG by starvation for cytidine nucleotides. The first sequence, GGGC/U, comprises the first four nucleotides at the 5′ end of the pyrG transcript, and the second, GCUCCC, forms the first six nucleotides of the 5′ strand of the terminator stem. All of the nucleotides lying between the two required RNA sequences can be deleted without loss of regulation. We propose that an as-yet-unidentified regulatory protein binds to these two RNA segments and prevents termination of transcription in the pyrG leader region when intracellular CTP levels are low.
The Bacillus subtilis pyrG gene, which encodes CTP synthetase, lies in the chromosome far from the pyr operon, which encodes all of the other genes of de novo pyrimidine nucleotide biosynthesis. Furthermore, pyrG is regulated by a mechanism that is entirely distinct from the regulation of the pyr operon (8). Expression of the pyr operon is governed by the PyrR attenuation protein, which promotes transcription termination at three sites located in the 5′ end of the operon when uridine nucleotide levels are elevated (for a review, see reference 11). Regulation of pyrG is independent of PyrR. In a previous study (8) we demonstrated that B. subtilis pyrG is specifically repressed by elevated levels of a cytidine nucleotide, presumably CTP, and that this repression involves a transcription terminator (attenuator) that lies within the 179-nucleotide (nt) pyrG 5′ leader sequence. We proposed that a trans-acting antitermination protein prevents termination in the pyrG leader, except when elevated levels of a cytidine nucleotide are present. The site or sites at which this putative regulatory protein binds to pyrG RNA were not identified. However, we pointed out that the 5′ leader sequences of the pyrG genes from several gram-positive bacteria contained three highly conserved 6-nt sequences (8). The first of these, GGGCU/AC, comprise the first six nucleotides of the B. subtilis pyrG transcript. The second and third conserved sequences, GCUCCC and GGGAGC, are complementary and are predicted to form the base of the stem of the terminator stem-loop.
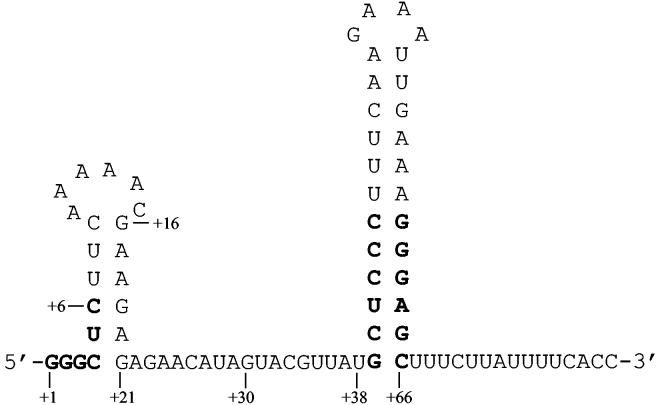
In the present study, substitution and deletion mutagenesis of the pyrG 5′ leader within pyrG′-lacZ transcriptional fusions (8) was used to identify the elements of primary RNA sequence and secondary structure that are required for normal repression of pyrG expression by cytidine. The parent strain, QM401, contained the native pyrG sequence from nt −49 to +81 fused to lacZ. This pyrG sequence specifies the pyrG promoter, the transcription start site, and all of the nucleotides of the pyrG transcript through the transcription termination sequence previously shown to be required for repression of pyrG (8) (Fig. 1). (Note that the previous numbering of the pyrG transcript [8] has been shifted by 1 nt in the present report. Mapping of the start of transcription by primer extension indicated that the 5′ sequence of the pyrG transcript was 5′-GGCUCUUCAAAA, but the results presented here, particularly experiments with strain QM416 [Table 1], strongly indicate that the transcript actually begins with the sequence 5′-GGGCUCUUAAAA.) The plasmid used to generate strain QM401 was constructed by PCR amplification of part of plasmid pJH4133 (12). The plasmids used to generate all of the mutant pyrG′-lacZ fusions were constructed by the same method, in which two long oligonucleotides that contained the desired mutations were chemically synthesized. Each oligonucleotide was designed to contain the desired mutation and the desired restriction site at its 5′ end. The two oligonucleotides, which were complementary to each other by about 20 nt at their 3′ ends, were annealed and filled in with the Klenow fragment of DNA polymerase I. Full-length extended products were purified and digested with EcoRI and BamHI. The parent pyrG′-lacZ fusion construct and all mutant forms of it were inserted into the integration plasmid pDH32 (3) at its EcoRI and BamHI sites and integrated as single copies into B. subtilis strain HC-11 (pyrB::Sptr [4]) at the amyE locus as previously described (8). Regulation of pyrG′-lacZ expression in these fusion-integrant strains, which require pyrimidines for growth, was studied by growth and measurement of β-galactosidase activity in cells grown on cytidine (repressing conditions) and on orotate, which is a poor pyrimidine source (derepressing conditions), as in our previous studies (8). Strain QM401 exhibited 15-fold regulation of pyrG′-lacZ expression by cytidine (Table 1), which was similar to that observed previously with other pyrG′-lacZ fusions (8).
FIG. 1.
Nucleotide sequence at the 5′ end of the B. subtilis pyrG 5′ untranslated leader RNA. DNA specifying the pyrG promoter and this leader RNA was fused to lacZ for the studies of pyrG expression in this work. Three RNA sequences found to be conserved in the pyrG leader sequences of several gram-positive bacteria (8) are shown in boldface. The computer program MFOLD (7, 15) predicts the secondary structure shown. The transcription terminator stem-loop following nucleotide +38 was previously shown to be essential for repression of pyrG expression by cytidine (8). Numbering of the sequence is based on the G nucleotide at +1 as the probable start site of transcription (see the text).
TABLE 1.
Mutagenesis of the conserved sequence at the 5′ end of the pyrG leader
| Strain | Mutation | Mutation position | β-Galactosidase activity (Miller units)
|
||
|---|---|---|---|---|---|
| +Cytidine | +Orotate | Ratio | |||
| QM401 | None | 40 ± 4 | 600 ± 34 | 15 | |
| QM403 | GGG to AAA | +1 to +3 | <1 | <1 | |
| QM416 | GGG to AGG | +1 | <1 | 1.4 ± 0.6 | |
| QM404 | GGG to GAG | +2 | 5 ± 0.4 | 1.3 ± 0.2 | 0.3 |
| QM405 | GGG to GGA | +3 | <1 | <1 | |
| QM412 | C to A | +4 | 100 ± 36 | 200 ± 51 | 2 |
| QM413 | C to U | +4 | 14 ± 2 | 550 ± 92 | 39 |
| QM417 | U to G | +5 | 34 ± 2 | 1,000 ± 300 | 29 |
| QM417C | U to C | +5 | 30 ± 2 | 700 ± 140 | 23 |
| QM414 | AAA +1 insertion | 12 ± 3 | 90 ± 21 | 8 | |
| QM408 | QM405 with terminator deletion | 150 ± 30 | 300 ± 50 | 2 | |
The sequence GGGC at the 5′ end of the pyrG transcript is essential for normal regulation.
The sequence of the first 6 nt in the putative pyrG transcripts from several gram-positive bacteria is strongly conserved (8), so we determined the effects of substitution mutagenesis of these nucleotides on pyrG′-lacZ expression (Table 1). Replacement of nucleotides +1 through +3 (GGG) with A residues, either all three together or each individually, reduced expression to extremely low levels and prevented derepression by pyrimidine starvation. The most obvious interpretation of these results is that replacement of the G nucleotides prevents antitermination, so that the downstream termination functions under all growth conditions. Alternative possibilities were that these substitutions cause greatly reduced transcription or greatly reduced stability of the pyrG′-lacZ transcripts. These possibilities were eliminated by two experiments. First, three A nucleotides were inserted immediately 5′ to the normal GGG transcription start site (strain QM414, Table 1). Reasonable levels of expression were obtained with this mutant fusion, and it retained partial regulation by cytidine. Second, deletion of the attenuation terminator (nucleotides from +38 to +79) from strain QM405 restored expression in strain QM408, although, as expected, expression was not regulated by cytidine (Table 1). (The twofold residual apparent regulation of strain QM408 was seen with other mutants in which the terminator was deleted and has been attributed to a nonspecific artifact of slow growth on orotate [8]). The results obtained with QM408 indicated that the extremely low levels of expression’s by mutants in which one or all of the 5′ GGG’s were replaced with A’s was a consequence of failure of an antitermination mechanism.
The nucleotide at position +4 in the pyrG transcript is C in all of the pyrG sequences we compared (8). Replacement of this C by U (strain QM413) permitted normal regulation of a pyrG′-lacZ fusion-integrant, but replacement by A (strain QM412) abolished normal regulation. In the latter case, expression was relatively high under both repressing and derepressing conditions; that is, this mutation permitted antitermination even when the medium contained high levels of cytidine. Replacement of the U at nucleotide +5 with either G or C (QM417 and QM417C) yielded fusions that displayed normal regulation. We did not construct mutations of the C nucleotide at +6, but it was previously shown that replacement of this nucleotide by U had no effect on regulation of a different pyrG′-lacZ fusion-integrant (8). These results, together with those presented in the next section, which indicate that the nucleotides +5 and +6 can be deleted without loss of normal regulation, lead to the conclusion that normal regulation requires only the sequence GGGC/U at the 5′ end of the pyrG transcript.
Nucleotides between the 5′-GGGC sequence and the pyrG attenuation terminator are not required for normal regulation.
We next sought to determine whether there was a requirement for specific nucleotide sequences or RNA secondary structures lying between the conserved GGGC sequence at the 5′ end of pyrG transcripts and the terminator stem-loop (nt +39 to +66), which contains six conserved base pairs at the base of its stem. The RNA secondary structure prediction computer program MFOLD (7, 15) predicted the possible presence of a small stem-loop between nt +4 and +21 (Fig. 1), although this secondary structure was not consistently predicted for the pyrG leader RNAs from other species. Substitution of nt +16 through +21 with nucleotides that would prevent base pairing with nt +4 through +9 (Table 2, QM406) had no significant effect on regulation. This result ruled out participation of this putative stem-loop or the specific sequence GAAGAG at positions +16 to +21 in antitermination control of pyrG. We previously excluded another potential antiterminator stem-loop (formed by base pairing of nt +23 to +30 with nt +35 to +42) from participation in pyrG regulation by substitution of the nucleotides from +22 to +30 with nucleotides that cannot base pair with the +35 to +42 sequence (8). Since the specific nucleotide sequence from +16 to +30 is not important for pyrG regulation, we deleted these nucleotides and observed that the resultant pyrG′-lacZ fusion was still regulated normally (repression ratio of 12). Further stepwise deletions of the nucleotides between the conserved GGGC sequence at the 5′ end of the pyrG transcript and the terminator at +39 to +67 were conducted, and characterization of the resultant pyrG′-lacZ fusions demonstrated that, surprisingly, all of the nucleotides between the U at +7 and the G at +39, a nucleotide that participates in the first of six conserved base pairs of the terminator stem, could be deleted without significant loss in normal regulation (Table 2, QM411). Additional deletion of C at +6 and U at +5 yielded fusions with somewhat reduced overall expression but normal or even higher-than-normal repression ratios (Table 2, QM415 and QM420). Further deletion of one additional nucleotide, the C residue at +4, yielded a fusion that was expressed at very low activity but appeared to retain some regulation by cytidine (strain QM421). We conclude that there are no specific RNA sequences or secondary structures between +4 and +39 that participate in the antitermination control of pyrG expression.
TABLE 2.
Mutagenesis of the linker region between the 5′ conserved sequence and the terminator in the pyrG leader
| Strain | Mutation | β-Galactosidase activity (Miller units)
|
||
|---|---|---|---|---|
| +Cytidine | +Orotate | Ratio | ||
| QM401 | None | 40 ± 4 | 600 ± 34 | 15 |
| QM406 | +16 to +21 substitution with ACCACA | 25 ± 4 | 300 ± 8 | 12 |
| QM411 | +7 to +38 deletion | 40 ± 5 | 600 ± 140 | 15 |
| QM415 | +6 to +38 deletion | 4 ± 1.3 | 80 ± 25 | 20 |
| QM420 | +5 to +38 deletion | 10 ± 1 | 240 ± 14 | 24 |
| QM421 | +4 to +38 deletion | 0.6 ± 0.5 | 6 ± 0.5 | 10 |
| QM422 | +16 to +21 substitution with GCUCCC | 0.2 ± 0.1 | 65 ± 5 | 325 |
| QM423 | +16 to +21 substitution with GGGAGC | 70 ± 10 | 1200 ± 160 | 17 |
The sequence, but not the position, of six conserved base pairs of the pyrG leader terminator is important for normal regulation.
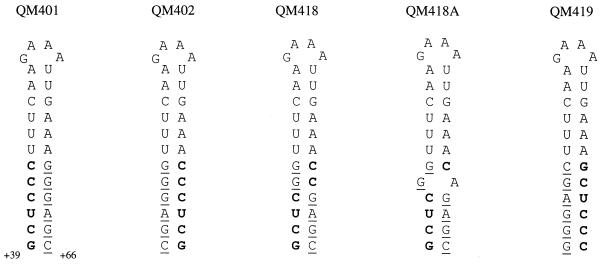
Three mutant versions of the pyrG leader terminator were studied to test whether the specific sequence and positions of the six conserved base pairs at the base of its stem were important for normal regulation (Fig. 2 and Table 3). In each of the mutants the numbers of GC and AU base pairs were unchanged, and the calculated stability (7, 15) of the mutant terminator stem-loops was not significantly altered. Reversing each of the base pairs, i.e., replacing each GC with a CG and vice versa and exchanging UA for AU, resulted in low-level, unregulated expression in strain QM402. A more conservative change, reversal of just two GC base pairs in the stem in strain QM418, gave the same phenotype, although with even weaker expression. It appeared that the mutant pyrG′-lacZ fusions in strains QM402 and QM418 were incapable of antitermination during cytidine starvation. A direct exchange from the 5′ strand to the 3′ strand of the terminator stem and vice versa of the two conserved 6-nt sequences in the terminator stem that preserved their 5′-to-3′ sequence identity (strain QM419) yielded a fusion that retained nearly normal regulation. Together, these observations indicate that the entire GCUCCC sequence in the terminator stem and presumably its complementary sequence are needed for normal antitermination of pyrG, but their positions can be reversed.
FIG. 2.
Mutagenesis of the conserved sequences in the terminator stem of the pyrG leader. Nucleotides shown in boldface are those that are found at +39 to +44 in the 5′ stem of the wild-type pyrG terminator; underlined nucleotides are those that are found at +61 to +66 in the 3′ stem of the wild-type terminator. (Also see Table 3.)
TABLE 3.
Mutagenesis of the conserved sequences in the terminator stem of the pyrG leader
| Strain | β-Galactosidase activity (Miller units)
|
Ratio | |
|---|---|---|---|
| +Cytidine | +Orotate | ||
| QM401 | 40 ± 4 | 600 ± 34 | 15 |
| QM402 | 30 ± 13 | 40 ± 19 | 1 |
| QM418 | 4 ± 0.7 | 3 ± 0.6 | 1 |
| QM418A | 250 ± 40 | 1300 ± 160 | 5 |
| QM419 | 40 ± 12 | 400 ± 94 | 10 |
During preparation of the plasmid for construction of strain QM418, we recovered another plasmid in which one of the desired GC mutations had been replaced with a GA base pair. This pyrG′-lacZ fusion was integrated into strain HC-11, and the regulation profile of the resultant strain, QM418A, was analyzed (Table 3). Expression of lacZ in strain QM418A was markedly elevated, and repression by cytidine was reduced. These findings can be readily rationalized from the reduced stability of the terminator stem-loop in this mutant, which we calculated (7, 15) to be about 30% less stable than the wild-type stem-loop.
The conserved GCUCCC sequence in the pyrG leader terminator is specifically involved in regulation of repression.
From the preceding studies, we formulated the hypothesis that a putative pyrG antitermination regulatory protein might act by binding to the conserved GGGC sequence at the 5′ end of the pyrG transcript and to the conserved GCUCCC sequence that forms the 5′ strand of the terminator stem-loop when cytidine nucleotides are at low concentrations. Such binding would prevent the GCUCCC sequence from base pairing with its complementary sequence in the 3′ strand of the terminator, resulting in antitermination. We tested this hypothesis by construction of a mutant pyrG′-lacZ fusion in which the 6-nt sequence from +16 to +21, which we had already shown could be replaced by other nucleotides in strain QM406, was replaced with a second copy of the GCUCCC sequence. We reasoned that such a transcript might bind to the antiterminator protein under conditions of cytidine starvation and prevent or reduce normal antitermination, since the site that binds the GCUCCC sequence in the terminator would already be occupied. That is, we expected expression of the corresponding pyrG′-lacZ fusion (strain QM422) to be low under derepressing conditions. This expectation was met (Table 2). Expression of β-galactosidase during growth on orotate was only 65 Miller units, a value much lower than seen in strain QM401 or most other strains that displayed normal regulation. This level was higher than that generally observed with most repressed strains, however, which suggests that correct binding of the pyrG leader to the regulatory protein could occur some of the time, even when a competing copy of the GCUCCC sequence was present. Expression of pyrG in QM422 under repressing conditions was extremely low; we suggest the following explanation for this. If high levels of cytidine nucleotides act simply by causing the hypothetical regulatory protein to fail to bind the pyrG leader RNA, expression of strain QM422 grown in the presence of cytidine might be expected to be similar to that of strain QM401 grown under the same conditions. However, we believe that the levels of cytidine nucleotides inside cells grown with cytidine are not high enough to repress pyrG expression in QM401 to the maximum possible extent. Rather, some of the putative regulatory protein probably remains bound, causing some residual antitermination. In strain QM422, this remaining antitermination is largely prevented by the extra GCUCCC sequence in the pyrG leader.
To test whether the putative antitermination protein might also bind to the third conserved sequence in the pyrG leader, we constructed fusion strain QM423, in which this sequence, GGGAGC, replaced the normal sequence at nt +16 to +21, i.e., was substituted in the same position as that of the GCUCCC sequence in QM422. Expression of the pyrG′-lacZ fusion and repression by cytidine in QM423 were essentially the same as in the wild-type strain, QM401 (Table 2).
Our experiments demonstrate that two short cis-acting elements of the pyrG leader mRNA are crucial for normal regulation of the gene by cytidine. These are the GGGC/U sequence at the 5′ end of the mRNA and the GCUCCC sequence that forms the first 6 bp on the 5′ side of the stem of the pyrG leader transcription terminator. It is likely that the GGGAGC sequence in the 3′ strand of the terminator stem is conserved in various species simply because it forms the complement to the essential GCUCCC sequence. No nucleotides or secondary structures between these two segments are required for normal regulation. These conclusions do not fully define the mechanism of antitermination control of pyrG expression, but they allow many potential mechanisms to be excluded, particularly those mechanisms that do not involve the participation of trans-acting regulatory proteins. The excluded mechanisms are as follows. (i) There is no reasonable upstream RNA antiterminator stem-loop in the pyrG leader as seen for the B. subtilis pyr (13) and trp (1) operons. One can imagine base pairing of the GGG segment (nt +1 to +3) with the CCC segment in the second conserved region acting as an antiterminator, but the stability of such a structure would be so much lower than that of the terminator stem-loop that it could not form an effective antiterminator. (ii) There is no possibility for a coupled transcription-translation regulatory mechanism as found with the Escherichia coli pyrBI or pyrE-rph operon (5), because there is no open reading frame or ribosome binding site in the essential elements of the pyrG leader. (iii) The sequence of the 5′ end of the pyrG leader is also incompatible with a regulatory mechanism involving UTP-sensitive reiterative RNA synthesis, as described for E. coli pyrBI (6) and codBA (9), or a variant of it that would involve CTP-dependent reiterative RNA synthesis. (iv) Finally, a mechanism involving nucleotide pool-sensitive transcriptional start site switching, as described for the pyrC and pyrD genes in E. coli and Salmonella enterica serovar Typhimurium (10, 14), seems implausible. One could imagine that the site of pyrG transcription initiation might shift from the G at +1 to the C at +4 under conditions of low-GTP and high-CTP pools. This would prevent any possible base pairing of the 5′ end of the mRNA with the GCUCCC sequence in the terminator stem and allow for greater termination. However, as noted above, this potential antiterminator structure is too weak to be effective. Furthermore, such a mechanism is not compatible with the properties of several of our mutant pyrG-lacZ fusions.
Our experiments on the cis-acting elements required for antitermination control of the pyrG gene are most readily explained by a mechanism in which these elements interact specifically with a trans-acting regulatory protein that governs antitermination in response to the intracellular cytidine nucleotide pool size. The simplest mode of action for such a protein would be that it binds to the pyrG leader mRNA at both the 5′-GGGC sequence and the GCUCCC sequence of the terminator stem-loop when the CTP concentration is low. This binding would prevent the GCUCCC sequence from base pairing with its complementary sequence in the 3′ stem of the terminator, leading to antitermination and read-through of the pyrG coding region. High concentrations of CTP are predicted to cause the antitermination protein to fail to bind to the pyrG mRNA and allow formation of the terminator stem-loop. This simple model explains most of our observations in this report, but a few are not accounted for. Specifically, the model does not explain why replacement of the first three G residues by A residues (Table 1, strains QM403, QM404, QM405, and QM416) caused almost total loss of pyrG expression, whereas replacement of the C nucleotide at +4 with an A nucleotide (strain QM412) gave relatively high, unregulated expression. If the GGGC sequence were needed only for binding of a regulatory protein, all five mutations would have been expected to give similar phenotypes. The GGGC sequence at the 5′ end of the pyrG transcript appears to be involved in antitermination in a more complex fashion than simply as a determinant of binding of a regulatory protein. Accordingly, we propose that the putative antitermination protein recognizes and binds to the two pyrG leader sequences but that it may remain bound during transcription and could interact with RNA polymerase and possibly other transcription factors, such as NusA and the template DNA, as well. The mode of action of such a protein might resemble some aspects of the action of the phage lambda N or Q antitermination protein (2). The action of cytidine nucleotides would be to control the extent to which the GCUCCC sequence is prevented from binding to its complementary sequence in the 3′ stem of the terminator, leading to antitermination. We suggest that this interaction is modulated not only by CTP levels but also by interactions with the GGGC sequence of the pyrG mRNA.
There is at present no direct evidence for the existence of this proposed trans-acting pyrG regulatory protein. A search for the gene encoding such a protein, using transposon mutagenesis, is under way in our laboratory. If such a gene can be identified and the protein it encodes can be isolated, direct testing of our proposed antitermination mechanism will be possible.
Acknowledgments
We acknowledge John Cronan for his critical review of the manuscript.
This research was supported by Public Health Service grant GM47112 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 183:5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the λ paradigm updated. Mol. Microbiol. 18:191-200. [DOI] [PubMed] [Google Scholar]
- 3.Grandoni, J. A., S. B. Fulmer, V. Brizzio, S. A. Zahler, and J. M. Calvo. 1993. Regions of the Bacillus subtilis ilv-leu operon involved in regulation by leucine. J. Bacteriol. 175:7581-7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu, P., and R. L. Switzer. 1995. Evidence for substrate stabilization in regulation of the degradation of Bacillus subtilis aspartate transcarbamylase in vivo. Arch. Biochem. Biophys. 316:260-266. [DOI] [PubMed] [Google Scholar]
- 5.Landick, R., C. L. Turnbough, Jr., and C. Yanofsky. 1996. Transcription attenuation, p. 1263-1286. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 6.Liu, C., L. S. Heath, and C. L. Turnbough. 1994. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8:2904-2912. [DOI] [PubMed] [Google Scholar]
- 7.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 8.Meng, Q., and R. L. Switzer. 2001. Regulation of transcription of the Bacillus subtilis pyrG gene encoding cytidine triphosphate synthetase. J. Bacteriol. 183:5513-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi, F., and C. L. Turnbough, Jr. 1995. Regulation of codAB operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching. J. Mol. Biol. 254:552-565. [DOI] [PubMed] [Google Scholar]
- 10.Sørensen, K. I., K. E. Baker, R. A. Kelln, and J. Neuhard. 1993. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J. Bacteriol. 175:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Switzer, R. L., R. J. Turner, and Y. Lu. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acids Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 12.Trach, K., J. W. Chapman, P. Piggot, D. LeCoq, and J. A. Hoch. 1988. Complete sequence and transcriptional analysis of the spo0F region of the Bacillus subtilis chromosome. J. Bacteriol. 170:4194-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson, H. R., C. D. Archer, J. Liu, and C. L. Turnbough, Jr. 1992. Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J. Bacteriol. 174:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.