Abstract
The occurrence of pleiotropic mutants that are defective in both antibiotic production and aerial mycelium formation is peculiar to streptomycetes. Pleiotropic mutant KSB was isolated from wild-type Streptomyces kasugaensis A1R6, which produces kasugamycin, an antifungal aminoglycoside antibiotic. A 9.3-kb DNA fragment was cloned from the chromosomal DNA of strain A1R6 by complementary restoration of kasugamycin production and aerial hypha formation to mutant KSB. Complementation experiments with deletion plasmids and subsequent DNA analysis indicated that orf5, encoding 90 amino acids, was responsible for the restoration. A protein homology search revealed that orf5 was a homolog of rpoZ, the gene that is known to encode RNA polymerase subunit omega (ω), thus leading to the conclusion that orf5 was rpoZ in S. kasugaensis. The pleiotropy of mutant KSB was attributed to a 2-bp frameshift deletion in the rpoZ region of mutant KSB, which probably resulted in a truncated, incomplete ω of 47 amino acids. Furthermore, rpoZ-disrupted mutant R6D4 obtained from strain A1R6 by insertion of Tn5 aphII into the middle of the rpoZ-coding region produced neither kasugamycin nor aerial mycelia, similar to mutant KSB. When rpoZ of S. kasugaensis and Streptomyces coelicolor, whose deduced products differed in the sixth amino acid residue, were introduced into mutant R6D4 via a plasmid, both transformants produced kasugamycin and aerial hyphae without significant differences. This study established that rpoZ is required for kasugamycin production and aerial mycelium formation in S. kasugaensis and responsible for pleiotropy.
Members of the genus Streptomyces produce an enormous variety of biologically active antibiotics and morphologically differentiate in a similar manner to eukaryotic fungi. Mature colonies of these gram-positive prokaryotic bacteria show an upper layer of aerial mycelia bearing spores that covers a lower layer of vegetative mycelia growing on the surface of agar medium. The production of antibiotics and formation of aerial hyphae, which appear to be independent events, have been considered genetically correlated based on the occurrence of pleiotropic mutants that neither produce antibiotics nor form aerial mycelia (8, 17). Hence, the pleiotropic regulation has provided a subject of special interests in Streptomyces genetics.
A variety of pleiotropic mutants such as bldA, bldB, and bldD mutants have been obtained from Streptomyces coelicolor together with many other types of bld mutants that fail to form aerial hyphae. Molecular cloning of the responsible genes has demonstrated transcriptional and translational regulation of pleiotropy. The bldA gene encodes leucyl tRNA for a rarely used UUA codon in streptomycetes (24, 26). Since the rare TTA codon is present in the regulatory genes for actinorhodin and undecylprodigiosin syntheses in S. coelicolor and for streptomycin and aerial mycelium productions in Streptomyces griseus, bldA mutations result in the pleiotropic phenotype in the streptomycetes (12, 23). Furthermore, the findings that bldB and bldD encode 98 and 167 amino acids, respectively, with putative motifs of helix-turn-helix, support transcriptional regulation of pleiotropy (10, 11, 36). Meanwhile, studies on A-factor, the pleiotropic autoregulator that controls streptomycin biosynthesis and resistance together with the morphogenesis in S. griseus, have revealed the regulatory cascade triggered by A-factor (34). The binding of A-factor to its receptor protein ArpA relieves its repression to adpA that regulates the streptomycin-specific transcriptional activator strR, and the derepression leads to the activation of the genes for streptomycin biosynthesis and resistance. In addition, adsA that is involved in aerial mycelium formation is under the regulation of the cascade (42).
We present here evidence that rpoZ, the gene that is known to encode the RNA polymerase (RNAP) subunit omega (ω), is closely related to pleiotropy in Streptomyces kasugaensis, i.e., the production of kasugamycin, an aminoglycoside antibiotic that protects rice plants from infection with the fungus Piricularia oryzae, and the formation of aerial mycelium. S. kasugaensis MB273, isolated from soil, carries three pock-forming plasmids (1, 2), and the host-vector system for gene cloning and analysis has been established (31, 35). We isolated pleiotropic mutant KSB that produces neither kasugamycin nor aerial mycelium after mutagenesis of S. kasugaensis A1R6, a wild-type strain, and subsequently cloned a 9.3-kb DNA fragment by phenotypic complementation. Our results establish that rpoZ and consequently its product ω play a physiologically pivotal role on antibiotic production and morphogenesis in S. kasugaensis.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
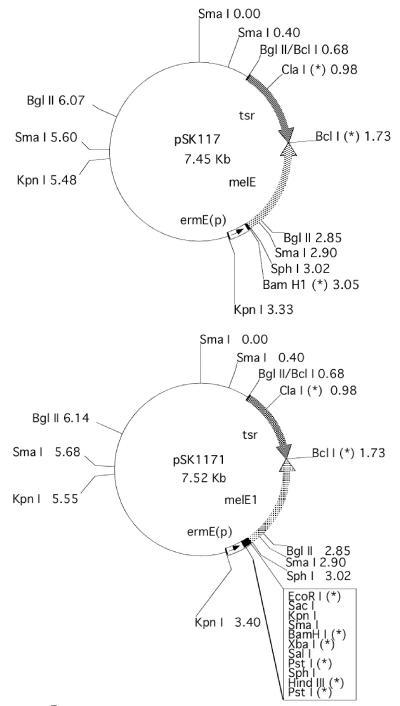
S. kasugaensis A1R6, a wild-type, plasmid-free derivative of strain MB273 isolated from soil, and GMY medium were described previously (1, 35). S. coelicolor M145 and Streptomyces lividans TK21 were kindly provided by D. A. Hopwood. Multicopy Streptomyces plasmids pSK117, pSK1171 (as illustrated in Fig. 1), and pSK1172 and the E. coli plasmid pUC18 were used for construction of plasmids using S. lividans TK21 and Escherichia coli JM109 as host strains. Plasmid pSK117 was constructed by insertion of a hybrid tyrosinase gene (melE) and the thiostrepton resistance gene (tsr) into pSK11-ΔSK10 (31), a derivative of S. kasugaensis multicopy plasmid pSK1 (1). The melE gene consists of the promoter of the erythromycin resistance gene (ermE) (4) and the tyrosinase gene (melC1 and melC2) (3, 25). The melE1 gene of pSK1171 was constructed by insertion of the 72-bp multiple-cloning site in pIJ486 (39) into melE between the ermE promoter and the tyrosinase gene. Plasmid pSK1172 is identical to pSK1171 except that the multiple-cloning site in the hybrid tyrosinase gene is opposite in direction. DNA manipulations in Streptomyces were carried out as described in the manuals of Hopwood et al. (16) and Kieser et al. (21).
FIG. 1.
Restriction map of pSK117 and pSK1171. Asterisks represent unique restriction sites in the plasmid, and ermE(p) indicates the promoter region of ermE harboring ermEp1 and ermEp2 (4). Both plasmids carry the hybrid tyrosinase genes melE and melE1 as described in Materials and Methods.
Aerial mycelium formation and kasugamycin production.
Aerial mycelium formation was observed after S. kasugaensis strains were incubated on GMY medium at 30°C for 7 to 10 days. Kasugamycin production was assayed by the agar diffusion method with Pseudomonas fluorescens IFO15334, which is particularly susceptible to kasugamycin. Shaken cultures of S. kasugaensis strains grown in GPY medium (31) were inoculated onto agar plugs consisting of maltose, 25 g/liter; Bacto Yeast extract (Difco), 2 g/liter; and Phytone peptone (BBL), 10 g/liter (pH 7.0). The plugs were incubated at 30°C for 5 days and transferred onto P. fluorescens-seeded kasugamycin bioassay medium consisting of glucose, 1 g/liter; Bacto Yeast extract (Difco), 2 g/liter; Polypepton (Wako Chemical Co., Ltd.), 5 g/liter; and agar, 8 g/liter (pH 7.0). Subsequently, the medium was incubated at 30°C for about 40 h for detection of kasugamycin production.
Construction of deletion plasmids: pAK3521, pAK3451, pAK3371, pAK3621, and pAK3631.
The respective desired fragment was isolated from the cloned 9.3-kb KpnI-BclI fragment by digestion with appropriate restriction enzymes and inserted into pUC18. As no HindIII site was present in the cloned region, the resulting pUC18-derivative plasmid was digested by HindIII and ligated into a unique HindIII site of pSK1171 as described above. Of two possible plasmids regarding the insertional direction of the pUC18-derivative, the plasmid in which the desired fragment was located in the downstream region of the ermE promoter plus the whole pUC18 DNA was selected. DNA manipulations were conducted using E. coli JM109 as a host.
DNA sequencing and protein homology search.
Nine deletion mutants of the 3.9-kb BamHI-SmaI region subcloned in pAK3541 were obtained using a deletion kit (Deletion Kit for Kilo-Sequence) from Takara Shuzo Co., Ltd., for DNA sequencing. Specifically, an ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Perkin-Elmer Corp.) was used, and the samples were analyzed on an Applied Biosystems model 377 sequencer. DNA sequence was analyzed for open reading frames with FramePlot 2.3.2 (www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) (19), and database searches were performed with BLAST (www.ncbi.nlm.nih.gov/BLAST/) for identification of homologs to deduced amino acid sequences. In addition, BLAST searches of the S. coelicolor database were conducted using The Sanger Institute website at www.sanger.ac.uk/Projects/S_coelicolor/blast_server.shtml.
Construction of pAK522 and pAK531.
For amplification of the 1.57-kb fragment covering the regions of orf3 to orf5, a PCR was performed with the upstream and downstream primers containing BamHI adaptor sequences at 5′ ends (underlined), i.e., 5′-GCGGATCCTGTTCTCGCTGACTCCGGCGCG-3′ and 5′-GCGGATCCCCGAGCCTCGGTCCGTACGTAT-3′, and the chromosomal DNA of S. kasugaensis as a template. Using a reaction kit (KOD-plus-DNA polymerase) from Toyobo Co., Ltd., the reaction mixture was prepared according to the manufacturer's recommendations. The reaction program consisted of 3 min of initial heating at 98°C and 40 subsequent cycles of heating for 15 s at 98°C, 30 s at 65°C, and 60 s at 72°C. For construction of pAK522, the PCR product was digested with EcoRI and BamHI, and the resulting 0.79-kb fragment containing rpoZ with 5′-truncated orf4 was inserted into pSK1171.
For amplification of the 0.82-kb rpoZ-encoding region of S. coelicolor, which corresponded to the 0.79-kb EcoRI-BamHI subcloned in pAK522, PCR amplification was carried out with the upstream and downstream primers containing HindIII and EcoRI adaptor sequences at 5′ ends (underlined), i.e., 5′-ATAAGCTTACAAGCTGATCGCCAACGGC-3′ and 5′-ACGAATTCTTCGGACTCGCTCTACGACC-3′, and the chromosomal DNA of S. coelicolor M145 as a template. The primers were designed based on information on S. coelicolor cosmid 9C5 (AL357523), whose sequence data were produced by the S. coelicolor Sequencing Group at The Sanger Institute. PCR amplification was performed under the same conditions as described above, and the PCR product was digested with HindIII and EcoRI and inserted into pSK1172 for construction of pAK531. DNA manipulations were carried out using S. lividans TK21 as a host.
Construction of E. coli-Streptomyces vector pAK3741 for disruption of rpoZ.
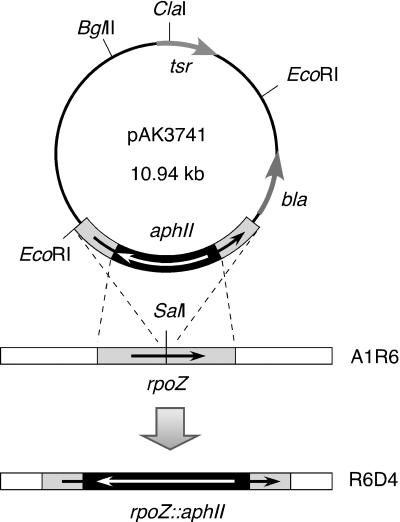
Plasmid pAK364, a pUC18 derivative carrying the 1.2-kb rpoZ-encoding EcoRI-SmaI region where a SalI site was uniquely present in the middle, was digested with SalI. A 1.5-kb SalI-digested fragment carrying Tn5 aphII was inserted into the SalI site of pAK364 opposite in direction for generation of pAK374. Subsequently, EcoRI-digested pAK374 was ligated into a unique EcoRI site of pSK21-K2 (31) for construction of pAK3741 (see Fig. 6). DNA manipulations were performed using E. coli JM109 as a host.
FIG. 6.
Restriction map of pAK3741 used for generation of disruptant mutant R6D4. A shuttle vector pAK3741 composed of 5.57- and 5.37-kb EcoRI digests of S. kasugaensis plasmid pSK21-K2 carrying tsr (31) and pAK374, whose construction is detailed in Materials and Methods. In the plasmid, bla indicates β-lactamase originating from pUC18. As illustrated, a homologous recombination was expected to occur between aphII-disrupted rpoZ and chromosomal rpoZ of wild-type strain A1R6.
Isolation of rpoZ-disrupted mutants.
Transformants of strain A1R6 with pAK3741 were incubated with constant shaking in GPY medium supplemented with kanamycin (50 μg/ml) at 30°C for 4 days. An aliquot of the culture was transferred to fresh GPY medium and cultivated for another 4 days. The same cultivation procedure was repeated three times. Kanamycin-resistant and thiostrepton-susceptible colonies were isolated on GMY medium from the resulting culture and subjected to colony PCR (20) for detection of disruptant mutants of rpoZ. PCR was performed with a reaction kit (TaKaRa La Taq with GC buffer) from Takara Shuzo Co., Ltd. The reaction mixture containing the same primers used for construction of pAK522 was prepared according to the manufacturer's recommendations. The reaction program consisted of 3 min of initial heating at 98°C and 40 subsequent cycles of heating for 20 s at 98°C, 30 s at 55°C, and 2 min at 72°C.
Nucleotide sequence accession no.
The nucleotide sequence data for the 3,911-bp region containing orf1 through orf6 from strain A1R6 and 271-bp rpoZ-encoding region from mutant KSB have been deposited in DDBJ, EMBL, and GenBank under the accession numbers AB081073 and AB081074, respectively.
RESULTS
Isolation of pleiotropic mutant KSB.
Pleiotropic mutant KSB that neither produces kasugamycin nor forms aerial mycelium was isolated after mutagenesis of wild-type strain A1R6 with N-methyl-N′-nitro-N-nitrosoguanidine (NTG). The mutant KSB retained its aerial mycelium-minus phenotype when grown on modified GMY medium containing mannitol instead of glycerol or in close proximity to wild-type strain A1R6 on the same agar medium. Hence, the mutant turned out to be genetically different from a group of S. coelicolor bld mutants (e.g., bldA, bldC, bldD, bldG, and bldH mutants) as explained in detail in Discussion.
Cloning of DNA fragment that restores aerial mycelium and kasugamycin production to mutant KSB.
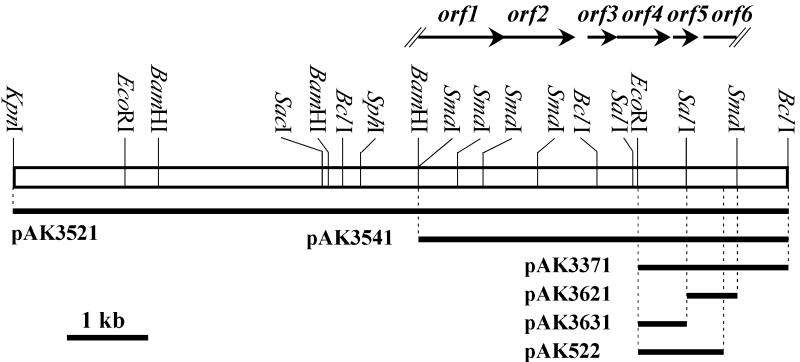
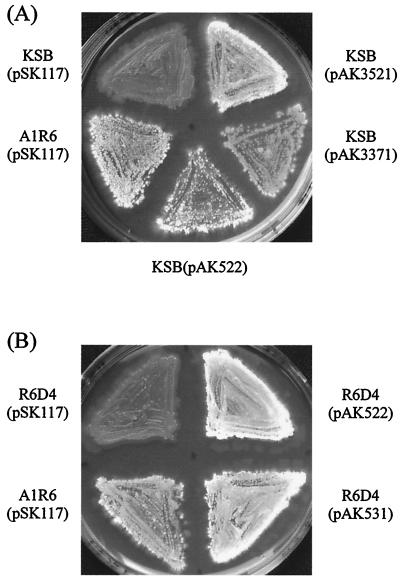
From the chromosomal DNA of strain A1R6, we cloned an approximately 9.3-kb KpnI-BclI DNA fragment that complemented the antibiotic- and aerial mycelium-minus phenotype of the mutant and subsequently examined the phenotype of KSB transformants with five deletion plasmids, pAK3521, pAK3541, pAK3371, pAK3621, and pAK3631 (Fig. 2). The transformants with pAK3521, pAK3541, and pAK3371 restored both kasugamycin production (Fig. 3A) and aerial mycelium formation (Fig. 4A), whereas those with pAK3621 and pAK3631 showed no complementation. Specifically, however, the restored kasugamycin production varied with the plasmids, probably because of the mutations in mutant KSB as detailed in Discussion.
FIG. 2.
Restriction map of 9.3-kb cloned DNA fragment. Solid arrows above the map indicate open reading frames predicted by FramePlot analysis (19) on the 3.9-kb BamHI-SmaI sequenced region. Solid lines below the map denote the DNA regions subcloned into the respective deletion plasmids.
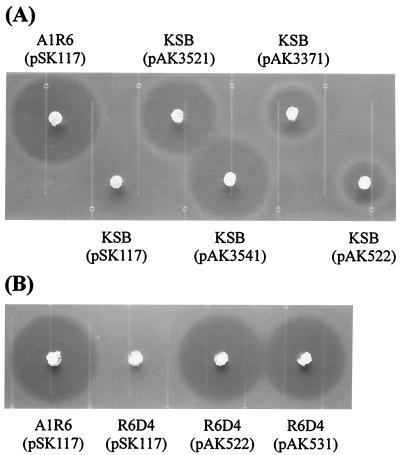
FIG. 3.
Kasugamycin production by transformants of mutant KSB (A) and disruptant mutant R6D4 (B). Bioassays using agar plugs were performed for detection of kasugamycin production by the transformants as detailed in Materials and Methods. The halos formed around the plugs indicate the accumulation of kasugamycin by the transformants. The transformants of wild-type strain A1R6 with pSK117 were used as a positive control. (A) From left to right, strain A1R6 carrying pSK117; and mutant KSBs harboring pSK117, pAK3521, pAK3541, pAK3371, and pAK522. (B) From left to right, strain A1R6 carrying pSK117 and mutant R6D4s harboring pSK117, pAK522, and pAK531.
FIG. 4.
Aerial mycelium formation by transformants of mutant KSB (A) and disruptant mutant R6D4 (B). Aerial hypha formation was examined qualitatively by inoculation of the transformants on GMY media as described in Materials and Methods. The transformants of wild-type strain A1R6 with pSK117 were used as a positive control. (A) Clockwise, from top left, mutant KSBs carrying pSK117, pAK3521, pAK3371, and pAK522 and strain A1R6 harboring pSK117. (B) Clockwise, from top left, disruptant mutant R6D4s carrying pSK117, pAK522, and pAK531 and strain A1R6 harboring pSK117.
Determination of base sequence of cloned DNA and homology search of orf5.
We determined the nucleotide sequences of the BamHI-SmaI fragment subcloned in pAK3541, and FramePlot analysis of the sequences predicted six open reading frames, i.e., 5′-truncated orf1, orf2 (the deduced sequence of 283 amino acids), orf3 (107), orf4 (205), and orf5 (90) and 3′-truncated orf6 as illustrated in Fig. 2.
BLAST searches revealed that the predicted 90-amino-acid product of orf5 showed a similarity to bacterial RNAP subunit ω. We concluded therefore that orf5 was rpoZ in S. kasugaensis. S. kasugaensis ω exhibited 32% identity with the amino acid sequence (91 amino acids) of E. coli ω, and as presented in Fig. 5, the protein was highly homologous with ω from actinomycetes, i.e., the 90-amino-acid S. coelicolor ω (99% identity), the 110- and 133-amino-acid Mycobacterium tuberculosis ω for strains H37Rv and CDC1551 (72% identities), and the 110-amino-acid Mycobacterium leprae ω (73% identity). In particular, S. coelicolor ω differed from S. kasugaensis ω in only the sixth amino acid, i.e., serine for S. coelicolor and threonine for S. kasugaensis.
FIG. 5.
Multiple alignment of the amino acid sequences of subunit of ω in S. kasugaensis. With the ClustalX program (available at inn-prot.weizmann.ac.il/software/ClustalX.html), sequences of ω subunits from the following genome-sequenced actinomycetes were analyzed: S. coelicolor (CAB93358), M. tuberculosis H37Rv (CAB02173), M. tuberculosis CDC1551 (AAK45700), and M. leprae (CAC30050). Asterisks indicate fully conserved amino acids, colons represent that one of the “strong” groups of amino acids (such as STA, MILV, NDEQ, HY, and NHQK) is fully conserved, and dots indicate that one of the “weak” groups of amino acids (such as STPA, ATV, and SGND) is fully conserved.
Complementation of pleiotropic phenotype of mutant KSB by rpoZ.
We amplified the 1.57-kb segment carrying orf3 through rpoZ by PCR, digested the product with EcoRI and BamHI, and inserted the resulting 0.79-kb segment coding for 5′-truncated orf4 and rpoZ into pSK1171 to construct pAK522 (Fig. 2) as detailed in Materials and Methods. The transformants of KSB with pAK522 produced both of kasugamycin (Fig. 3A) and aerial hypha (Fig. 4A), which indicated that rpoZ was defective in mutant KSB. For construction of pAK522, the rpoZ-coding fragment was inserted in the opposite direction downstream of the ermE promoter containing ermEp1 and ermEp2 in order that possible influences of the promoters should be avoided. Despite the complementary expression of rpoZ in the transformants, we have failed to identify any typical streptomycete promoter sequences in the upstream region of the gene. However, there still remained the possibility of transcriptional readthrough from the tsr promoters.
Isolation of rpoZ-disrupted mutant R6D4 and phenotypic complementation by rpoZ.
We constructed pAK3741 by inserting Tn5 aphII conferring resistance to kanamycin into a unique SalI site located in the middle of rpoZ from S. kasugaensis (Fig. 6). As described in Materials and Methods, we isolated disruptant mutant R6D4, in which the kanamycin resistance gene was inserted into the SalI site of the chromosomal rpoZ of wild-type strain A1R6 in the opposite direction.
We found that disruptant mutant R6D4 neither produced kasugamycin nor formed aerial mycelium, in a very similar manner to strain KSB. In addition, as similarly observed with mutant KSB, mutant R6D4 retained its aerial mycelium-minus phenotype when grown on modified GMY medium containing mannitol as a sole carbon source or in close proximity to wild-type strain A1R6. The transformants of mutant R6D4 with pAK522 carrying rpoZ exhibited kasugamycin synthesis (Fig. 3B) and morphological differentiation (Fig. 4B). It should be noted that the antibiotic accumulation by the transformant harboring pAK522 was similar to that by wild-type strain A1R6 carrying pSK117, in contrast to that by the KSB transformant with the same plasmid.
Sequencing analysis of rpoZ region in mutant KSB.
We determined the nucleotide sequence of the rpoZ region in mutant KSB and found a 2-bp (GC) deletion in the middle of the gene, which probably resulted in a truncated, incomplete ω protein of 47 amino acids (Fig. 7). Taken together with the observation on rpoZ-disrupted mutant R6D4, we conclude that a defective rpoZ results in the pleiotropic phenotype of the S. kasugaensis mutants. In other words, rpoZ is crucially related to antibiotic production and morphogenesis in S. kasugaensis.
FIG. 7.
Comparison of nucleotide sequences of rpoZ from strain A1R6 and mutant KSB. Hyphens in the rpoZ-encoding region from mutant KSB indicate gaps in the nucleotide sequence.
Complementation of S. kasugaensis rpoZ-disrupted mutant R6D4 by S. coelicolor rpoZ.
We amplified the 0.82-kb rpoZ-encoding region of S. coelicolor M145 by PCR and inserted the PCR product into pSK1172 to produce pAK531 as detailed in Materials and Methods. The cloned region corresponded to the 0.79-kb rpoZ-encoding region of S. kasugaensis subcloned in pAK522. The transformants of disruptant mutant R6D4 with pAK531 harboring S. coelicolor rpoZ formed aerial mycelium (Fig. 4B) and produced kasugamycin, similar to transformants of parental strain A1R6 with pSK117 and mutant R6D4 with pAK522 carrying S. kasugaensis rpoZ (Fig. 3B). These results indicate that rpoZ of S. coelicolor is functionally expressed and substitutes for the indigenous rpoZ in S. kasugaensis.
DISCUSSION
We have discovered in this study that the rpoZ gene that encodes the RNAP ω subunit is required for antibiotic production and morphogenesis in S. kasugaensis. The rpoZ genes have been found in all the sequenced genomes of free-living bacteria. In E. coli, ω binds to the β′ subunit to form the RNAP core enzyme, consisting of four subunits (2α, β, β′, and ω) (13). Moreover, bacterial ω subunits (including S. coelicolor ω) which are structural and functional homologs of archaeal RNAP subunit RpoK and eukaryotic RNAP I, II, and III subunit RPB6, are suggested to facilitate association with the α2β assembly through binding to β′ (29). Apart from the physical findings, however, little has been known about a physiological effect of bacterial ω except that an ω-null mutant of E. coli exhibits the slow-growth phenotype and induces the molecular chaperonin GroEL to structurally maintain RNAP devoid of ω (30).
By cloning and subsequent complementation experiments using pleiotropic S. kasugaensis mutant KSB whose phenotype is aerial-mycelium- and kasugamycin-minus, we found that the responsible gene is orf5, which encodes 90 amino acids. On the basis of the results with BLAST searches, we conclude that orf5 is the rpoZ encoding RNAP subunit ω in S. kasugaensis. Disruption of rpoZ of wild-type strain A1R6 by insertion of aphII produced mutant R6D4, which neither produced kasugamycin nor formed aerial mycelium in a phenotypically similar manner to mutant KSB. Furthermore, we showed that the pleiotropic phenotype of mutant KSB is due to a 2-bp frameshift deletion in the rpoZ-coding region, which probably results in a truncated, incomplete ω subunit of 47 amino acids. These results verify the involvement of rpoZ in the antibiotic and aerial hypha production in S. kasugaensis. Subunits ω of S. kasugaensis and S. coelicolor are conserved in constitutive amino acids except for the sixth amino acid residue, and rpoZ from S. coelicolor rescued defective rpoZ of S. kasugaensis mutant, which may indicate the possible involvement of rpoZ in the pleiotropic events of S. coelicolor as well. Previously, a gene designated rpoZ that encodes the sporulation-controlling σ factor of 278 amino acids, a homolog of S. coelicolor whiG, was cloned from Streptomyces aureofaciens (22). Neither ω of S. kasugaensis nor that of S. coelicolor show any similarity to the rpoZ-encoding product of S. aureofaciens. Southern hybridization analysis indicated no highly homologous DNA region with rpoZ in S. kasugaensis genome (data not shown) and a homology search of S. coelicolor database revealed no homologs of S. kasugaensis ω except that from the indigenous gene, which might indicate that rpoZ is solely present in the genomes of S. kasugaensis and S. coelicolor.
The complementary restoration of kasugamycin production as observed in Fig. 3 clarified a genetic difference between NTG-derived mutant KSB and rpoZ-disrupted mutant R6D4. Introduction of rpoZ alone via pAK522 rescued the disruptant mutant to accumulate the antibiotic to a level of kasugamycin accumulation similar to that of wild-type strain A1R6. Meanwhile, pAK3541 or pAK3521 returned mutant KSB to produce kasugamycin to a similar level of kasugamycin accumulation to strain A1R6, but pAK522 or pAK3371 to accumulate the antibiotic to a far lower level. Plasmids pAK3451 and pAK3521 bore three complete orf2, orf3, and orf4 in common together with rpoZ, and rpoZ resided 5.8 kb and 10.5 kb downstream from the ermE promoter, respectively. Furthermore, a plausible terminator located in the 193-bp region between orf2 and orf3 (data not shown) was positioned upstream of rpoZ in both plasmids. The rpoZ expression was therefore thought to remain unaffected by transcriptional readthrough from the ermE promoter. Additionally, in pAK522 and pAK3371, rpoZ resided downstream of the tsr promoter in the same orientation. Taken together, it is likely that other gene(s) was impaired in mutant KSB together with rpoZ and that a combination of orf2, orf3, or orf4 with rpoZ turned the mutant into a wild-type producer of kasugamycin.
Many genetic studies have been conducted on the pleiotropy in streptomycetes, but none has reported the importance of rpoZ and its product ω. Moreover, on the possible involvement of ω in bacterial physiology, the slow-growth phenotype of an E. coli ω deletion mutant (30) is the only indication presented to date. We believe that the results presented here provide a new insight into studies on bacterial rpoZ and RNAP ω.
Initially, we speculated that mutant KSB might be included in a group of bld mutants isolated from S. coelicolor, such as bldA, bldB, bldC, bldD, bldG, and bldH mutants, that lack glucose catabolite repression of galactose utilization (37). Of the bld mutants, bldA, bldC, bldD, bldG, and bldH mutants display the aerial-mycelium-plus phenotype when mannitol is replaced by glucose in minimum agar medium. Extracellular molecules such as SapB relate to the aerial mycelium formation in S. coelicolor (32, 40, 41), and according to the extracellular complementation, the bld mutants except for bldB mutants can be sorted into a hierarchical grouping (33). The bldB mutants that are catabolite derepressed for genes for glycerol utilization and agar decomposition fail to fit into the grouping (33, 40) and neither sporulate nor produce antibiotics, regardless of carbon sources (7, 28). The phenotypic observations on bldB mutants are very similar to mutants KSB and R6D4. However, we found no homology between the 90-amino-acid ω of S. kasugaensis and 98-amino-acid BldB of S. coelicolor. Although the presence of bld genes has not been examined in S. kasugaensis, these results indicate that rpoZ is involved in the pleiotropy of S. kasugaensis in a manner different from that of the bld genes of S. coelicolor.
Intracellular accumulation of ppGpp above certain limits correlates with the onset of antibiotic biosynthesis in various Streptomyces species (6, 18, 27). The disruption of relA coding for ppGpp synthetase turns the wild-type phenotype of S. coelicolor into the pleiotropic phenotype on some agar media (5). Physically, ppGpp binds to the N-terminal portion of β′ (38) together with the C-terminal of β in E. coli RNAP (9). Although ω binds to β′, we are skeptical about a possible physiological connection of ppGpp with ω in the pleiotropy of S. kasugaensis, because an ω-null mutant of E. coli is known to exhibit a normal wild-type stringent response (14).
Although an ω-less mutant of E. coli grows to a lower cell density than the normal strain (30), mutant R6D4 reached the same levels of cell growth as wild-type strain A1R6 in GPY and other liquid media (data not shown), which may indicate that ω is not required for growth of S. kasugaensis. However, as reported with E. coli RNAP, the N-terminal domain of ω with 52 of 91 amino acids is capable of assembling a functional RNAP core enzyme (15). To further study the relation of rpoZ and its product ω with the growth of S. kasugaensis, we need to construct ω-null mutants.
As the rpoZ genes are present in all bacteria, it seems unlikely that rpoZ of S. kasugaensis controls directly and simultaneously both kasugamycin production and aerial mycelium formation. It is of great importance therefore to examine whether a functional role of S. kasugaensis ω is fundamentally equivalent to E. coli ω. We will study the detailed mechanism how rpoZ is involved in the streptomycete-specific events in S. kasugaensis and investigate possible involvement of the gene in the pleiotropy in S. coelicolor that produces polyketide antibiotics, a class structurally different from the aminoglycoside antibiotics.
Acknowledgments
We thank Yasuo Fukagawa for his critical reading of the manuscript and helpful comments and discussions. We are also very grateful to Arnold L. Demain for his encouragement to this study and his invaluable comments on the manuscript.
REFERENCES
- 1.Akagawa, H., K. Kawaguchi, and M. Ichihara. 1984. Plasmids of Streptomyces kasugaensis MB273: their pock formation, their dispensable endonuclease cleavage sites for pock formation, and transformation of S. kasugaensis MB273 by them. J. Antibiot. 37:1016-1025. [DOI] [PubMed] [Google Scholar]
- 2.Akagawa, H., Y. Takano, and K. Kawaguchi. 1987. Characterization of a natural cointegrate of the pock-forming plasmids pSK1∗ and pSK2∗ of Streptomyces kasugaensis MB273. J. Gen. Microbiol. 133:1941-1949. [DOI] [PubMed] [Google Scholar]
- 3.Bernan, V., D. Filpula, W. Herver, M. Bibb, and E. Katz. 1985. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene 37:101-110. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., G. R. Janssen, and J. M. Ward. 1986. Cloning and analysis of the promoter region of the erythromycin-resistance gene (ermE) of Streptomyces erythraeus. Gene 41:E357-E368. [DOI] [PubMed] [Google Scholar]
- 5.Chakraburtty, R., and M. J. Bibb. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraburtty, R., J. White, E. Takano, and M. Bibb. 1996. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2). Mol. Microbiol. 19:357-368. [DOI] [PubMed] [Google Scholar]
- 7.Champness, W. C. 1988. New loci required for Streptomyces coelicolor morphological and physiological differentiation. J. Bacteriol. 170:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chater, K. F., and M. J. Bibb. 1997. Regulation of bacterial antibiotic production, p. 59-105. In H. Kleinkauf and H. von Döhren (ed.), Biotechnology, vol. 7. VCH, Weinheim, Germany. [Google Scholar]
- 9.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the β-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279-287. [DOI] [PubMed] [Google Scholar]
- 10.Elliot, M., F. Damji, R. Passantino, K. Chater, and B. Leskiw. 1998. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 180:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliot, M., and B. Leskiw. 1999. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J. Bacteriol. 181:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 13.Gentry, D., and R. R. Burgess. 1993. Cross-linking of Escherichia coli RNA polymerase subunits: identification of β′ as the binding site of ω. Biochemistry 32:11224-11227. [DOI] [PubMed] [Google Scholar]
- 14.Gentry, D., H. Xiao, R. Burgess, and M. Cashel. 1991. The omega subunit of Escherichia coli K-12 RNA polymerase is not required for stringent RNA control in vivo. J. Bacteriol. 173:3901-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, P., A. Ishihama, and D. Chatterji. 2001. Escherichia coli RNA polymerase subunit ω and its N-terminal domain bind full-length β′ to facilitate incorporation into the α2β subassembly. Eur. J. Biochem. 268:4621-4627. [DOI] [PubMed] [Google Scholar]
- 16.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces, a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 17.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377-398. [DOI] [PubMed] [Google Scholar]
- 18.Hoyt, S., and H. G. Jones. 1999. relA is required for actinomycin production in Streptomyces antibioticus. J. Bacteriol. 181:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the Frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, J., N. Tsuchizaki, M. Yoshida, D. Ishiyama, and K. Hotta. 2000. Colony PCR for detection of specific DNA sequences in actinomycetes. Actinomycetologica 14:1-5. [Google Scholar]
- 21.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 22.Kormanec, J., L. Potúčková, and B. Řežuchová. 1994. The Streptomyces aureofaciens homologue of the whiG encoding a putative sigma factor essential for sporulation. Gene 143:101-103. [DOI] [PubMed] [Google Scholar]
- 23.Kwak, J., L. A. McCue, and K. E. Kendrick. 1996. Identification of bldA mutants of Streptomyces griseus. Gene 171:75-78. [DOI] [PubMed] [Google Scholar]
- 24.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y.-H. W., B.-F. Chen, S.-Y. Wu, W.-M. Leu, J.-J. Lin, C. W. Chen, and S. J. Lo. 1988. A trans-acting gene is required for the phenotypic expression of a tyrosinase gene in Streptomyces. Gene 65:71-81. [DOI] [PubMed] [Google Scholar]
- 26.Leskiw, B. K., E. J. Lawlor, J. M. Fernandez-Abalos, and K. F. Chater. 1991. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA 88:2461-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Costa, O. H., M. A. Fernández-Moreno, and F. Malpartida. 1998. The relA/spoT-homologous gene in Streptomyces coelicolor encodes both ribosome-dependent (p)ppGpp-synthesizing and -degrading activities. J. Bacteriol. 180:4123-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 29.Minakhin, L., S. Bhagat, A. Brunning, E. A. Campbell, S. A. Darst, R. H. Ebright, and K. Severinov. 2001. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 98:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee, K., H. Nagai, N. Shimamoto, and D. Chatterji. 1999. GroEL is involved in activation of Escherichia coli RNA polymerase devoid of the ω subunit in vivo. Eur. J. Biochem. 266:228-235. [DOI] [PubMed] [Google Scholar]
- 31.Nabeshima, S., Y. Hotta, and M. Okanishi. 1984. Construction of plasmid vectors from Streptomyces kasugaensis plasmids, pSK1 and pSK2. J. Antibiot. 37:1026-1037. [DOI] [PubMed] [Google Scholar]
- 32.Nodwell, J. R., and R. Losick. 1998. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. J. Bacteriol. 180:1334-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nodwell, J. R., K. McGovern, and R. Losick. 1996. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Mol. Microbiol. 22:881-893. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi, Y., S. Kamayema, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 35.Okanishi, M., K. Katagiri, T. Furumai, K. Takeda, K. Kawaguchi, M. Saitoh, and S. Nabeshima. 1983. Basic techniques for DNA cloning and conditions required for streptomycetes as a host. J. Antibiot. 36:99-108. [DOI] [PubMed] [Google Scholar]
- 36.Pope, M. K., B. Green, and J. Westpheling. 1998. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J. Bacteriol. 180:1556-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pope, M. K., B. D. Green, and J. Westpheling. 1996. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell-cell signalling. Mol. Microbiol. 19:747-756. [DOI] [PubMed] [Google Scholar]
- 38.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the β′-subunit. J. Biol. Chem. 276:1220-1225. [DOI] [PubMed] [Google Scholar]
- 39.Ward, J. M., G. R. Janssen, T. Kieser, and M. J. Bibb. 1986. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 40.Willey, J., R. Santamaria, J. Guijarro, M. Geistlich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641-650. [DOI] [PubMed] [Google Scholar]
- 41.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 42.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]