Abstract
Bacterial surface motility works by retraction of surface-attached type IV pili. This retraction requires the PilT protein, a member of a large family of putative NTPases from type II and IV secretion systems. In this study, the PilT homologue from the thermophilic eubacterium Aquifex aeolicus was cloned, overexpressed, and purified. A. aeolicus PilT was shown to be a thermostable ATPase with a specific activity of 15.7 nmol of ATP hydrolyzed/min/mg of protein. This activity was abolished when a conserved lysine in the nucleotide-binding motif was altered. The substrate specificity was low; UTP, CTP, ATP, GTP, dATP, and dGTP served as substrates, UTP having the highest activity of these in vitro. Based on sedimentation equilibrium and size exclusion chromatography, PilT was identified as a ≈5- to 6-subunit oligomer. Potential implications of the NTPase activity of PilT in pilus retraction are discussed.
The ability to move across surfaces is critical to the life style of many bacteria. Social gliding motility in Myxococcus xanthus, gliding motility in Synechocystis spp., and twitching motility in gram-negative mammalian pathogens such as Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa, and Dichelobacter nodosus and the plant pathogens Xylella fastidiosa and Pseudomonas syringae are mechanistically indistinguishable forms of surface motility all driven by the retraction of filamentous type IV pili (3, 16, 20, 21, 30, 34). Pili attached at their distal tips to a solid surface such as a latex bead, another bacterial cell, or a eukaryotic host are pulled in as grappling hooks (21) at a speed of approximately 1 μm/s in N. gonorrhoeae and P. aeruginosa (21, 30). This retraction requires the PilT protein; although pilT mutants are hyperfimbriated, they are nonmotile and do not retract their pili (5, 21, 40, 41).
The roles of PilT extend beyond pilus-dependent surface motility. Loss of functional PilT leads to defects in colonization and invasion of host cells by N. gonorrhoeae (22) and P. aeruginosa (8). Naturally competent Neisseria and Pseudomonas stutzeri species lose the ability to take up DNA in pilT strains (14, 41). P. aeruginosa strains with motility protein defects form abnormal biofilms (G. A. O'Toole and K. Gibbs, personal communication). Additionally, phototaxis in Synechocystis sp. strain PCC6803 requires a PilT protein (4). Thus, PilT plays an important role in pathogenic and symbiotic motility and signaling.
The protein sequence of PilT includes canonical nucleotide-binding motifs (Fig. 1A) (37). The Walker A box, or phosphate-binding loop (GxxxxGKT/S), is the standard signature motif used to predict nucleotide-binding proteins. These residues adopt a β-strand-turn-α-helix motif in which the glycines allow the turn while the invariant lysine and hydroxyl side chains form salt bridges or hydrogen bonds with the β- and γ-phosphates of the bound nucleotide and requisite divalent cation. The Walker B motif is less well defined but requires a carboxylate side chain to maintain proper geometry and activate a solvent molecule for the hydrolysis reaction.
FIG. 1.
PilT sequence and purification. (A) Conservation of PilT and putative type II secretion ATPase family members. A. aeolicus, P. aeruginosa, and N. gonorrhoeae PilT, A. aeolicus and P. aeruginosa PilU, P. aeruginosa PilB (putative NTPase required for pilus assembly), and P. aeruginosa XcpR (type II secretion putative nucleotide-binding protein) were aligned with the GCG package program PILEUP. The sequence of amino acids 83 to 348 of A. aeolicus PilT is shown. Identities or conservative changes among at least six of these seven are in bold uppercase letters, while identities or conservative changes among all three PilT proteins are in uppercase. The Walker A motif, ASP box, Walker B motif, and HIS boxes (shaded, in that order) identify the putative type II and type IV secretion ATPase families (26). The K149Q mutation is noted with an asterisk (see text). An N-terminal extension of approximately 200 amino acids is not shown for PilB and XcpR. Pair wise amino acid identity for PilT is 68% for the P. aeruginosa and N. gonorrhoeae proteins and 51% for the P. aeruginosa and A. aeolicus proteins. (B) Purification of A. aeolicus PilT. TL, total cell lysate from E. coli BL21(DE3)/pet23a(+)AaPilTHis6 after IPTG induction; WT, purified PilT-His6; K149Q, purified PilT K149Q. The positions of molecular size markers (lane M) are indicated.
In addition to the Walker A and B sequence motifs, PilT shares aspartate and histidine-containing sequence motifs (Fig. 1A) (18) with a large group of putative nucleotide-binding proteins involved in bacterial type II and IV secretion, type IV pilus assembly, natural competence, and tight adherence (27). Several such proteins from type IV secretion systems have been shown to possess nucleotide triphosphate (NTP)-hydrolyzing activity in vitro. These include VirB11, TrwD, TrbB, and HP0525 (7, 18, 28). Recently, putative secretion ATPases for type IV pilus assembly and tight adherence were characterized biochemically and also shown to have ATPase activity (2, 29). PilT motility proteins have not been investigated in vitro.
In order to better understand the mechanism of type IV pilus retraction, we sought to purify PilT and characterize its biochemical properties. This effort was aided by the fact that the thermophilic eubacterium Aquifex aeolicus encodes within its genome a PilT (10) which is 51% identical to the P. aeruginosa and N. gonorrhoeae PilTs at the amino acid level (Fig. 1A). Because of the expected thermostability of the protein and because of high sequence homology to mesophilic PilTs, including motifs seen in twitching motility proteins that are not seen in other secretion ATPases (for example, the DFSF motif at amino acid 83, the AIRNLIRE sequence at amino acid 301, and the Q-rich region near the C terminus; Fig. 1A), we chose A. aeolicus PilT as an excellent model to undertake biochemical and structural studies on this class of proteins. We report here that purified A. aeolicus PilT is a heat-stable oligomeric NTPase.
MATERIALS AND METHODS
Cloning.
A. aeolicus pilT was cloned from genomic DNA (a gift from R. Huber, Universität Regensburg) by PCR and inserted within the multicloning region of pBAD18 (15) for arabinose-inducible and glucose-repressible expression in Escherichia coli, creating pBAD-AapilT. A. aeolicus pilT was subcloned into two T7 RNA polymerase-based expression plasmids, pET15b and pET23a(+) (33) (Novagen). pET15bHis6AapilT allows expression of PilT with a 20-amino-acid N-terminal tag containing hexahistidine. Creation of pET23a(+)AapilTHis6 introduced codons for Leu and Glu after the last PilT codon and before the coding region for the C-terminal hexahistidine tag.
With pET23a(+)AapilTHis6 as a template, the codon for Lys149 was changed to a codon for Gln by full-circle mutagenesis (Stratagene) according to the manufacturer's protocol with 4-min extension times, forward primer 5′-TTA CAG GTC CTA CGG GGT CGG GTC AGT CTA CAA-3′, and reverse primer 5′-TTG TAG ACT GAC CCG ACC CCG TAG GAC CTG TAA-3′ (mutagenic bases italicized) to create pET23a(+)AapilTHis6-K149Q. The pilT coding region in each plasmid described was verified by automated DNA sequencing.
Expression and purification.
pET23a(+)AapilTHis6 was transformed into Escherichia coli BL21(DE3) cells (Novagen), which were used to inoculate a 3-ml Luria broth-ampicillin (100 μg/ml) (LB-ampicillin) culture. After 6 h, 125 μl was spread on each of 48 LB-ampicillin plates. After overnight growth, these cells were washed and used to inoculate 6-liters of LB-ampicillin containing 2% ethanol. After 1 h, the liquid culture was induced with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). At 4 h postinduction, cells were harvested by centrifugation and frozen overnight. Cells resuspended in 40 ml of lysis buffer (25 mM Tris-Cl [pH 7.9] at room temperature, 10 mM imidazole [pH 8.0] at room temperature, 300 mM KCl, 5% glycerol) were lysed by three passes through a French pressure cell operating at 16,000 lb/in2 and cleared by ultracentrifugation (100,000 × g for 1.5 h). Two milliliters of nickel-nitrilotriacetic acid-agarose (Qiagen) was added to the supernatant with stirring and incubated for 1 h.
A column (0.8 by 4 cm) was packed with the loaded resin and washed with wash buffer (lysis buffer with 30 mM imidazole and 750 mM KCl) until the A280 fell below 0.005. PilT was eluted with elution buffer (lysis buffer with 200 mM imidazole and 150 mM KCl) at a flow rate of 0.03 ml/min. Protein from peak elution fractions was heated to 80°C for 30 min, and precipitated protein was removed by centrifugation for 30 min at 14,000 × g in a benchtop microcentrifuge (Beckman). Purity was judged by chemifluorescence of a SYPRO Ruby (Molecular Probes)-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel scanned on a Storm 860 phosphorimager and analyzed with ImageQuant software (Molecular Dynamics). Protein concentrations were determined with the Bradford assay reagent (Bio-Rad) with bovine serum albumin (Sigma) as the standard.
ATPase end point assay.
ATPase reactions were run in 5% glycerol-150 mM KCl-5 mM MgCl2-150 mM buffer (pH 5 to 6.5 morpholineethanesulfonic acid [MES], pH 7.0 to 7.5 morpholinepropanesulfonic acid [MOPS], pH 8 to 8.5 Tris, based on pH measured at room temperature with calculated correction for temperature point depression). Protein diluted in assay buffer to 1.2 to 1.4 mg/ml final concentration (28 to 33 mM protein, with 17 to 40 mM residual imidazole and 2 to 5 mM residual Tris) was equilibrated at 80°C for 15 min, following which ATP was added to a 2.5 mM final concentration and the reaction was separated into three aliquots for 10, 15, and 20 min time points. Reactions were stopped by freezing in liquid N2. The resulting final ADP concentration was determined by a modification of a standard real-time coupled enzyme assay (35). One unit of activity is defined at the amount of protein required to hydrolyze 1 nmol of ATP per min.
NTPase assay.
NTP hydrolysis reactions were performed as described above except that the protein concentration was 0.1 to 0.2 mg/ml and the time points were 4, 8, and 12 min. The final NTP concentration was 2.5 mM, and the ADP concentration, when included, was 250 μM. The resulting inorganic phosphate concentration was determined with a colorimetric malachite green assay (12). Briefly, the color-developing mix containing 3 volumes of 0.045% malachite green HCl in H2O, 1 volume of 4.2% (NH4)2MoO4 in 4N HCl, and 1/50th volume of 1% Triton X-100 was freshly made and filtered. Then 800 μl of the color-developing mix was added immediately to 100 μl of the NTP hydrolysis reaction, 100 μl of 34% citrate was added after 1 min, and the absorbance was measured at 660 nm within 5 min. Background inorganic phosphate concentration measurements with elution buffer replacing protein were subtracted from the protein time points. A standard curve was generated every time the color-developing mix was made.
Size exclusion chromatography.
A Superdex 200 HR10/30 prepacked column (Pharmacia) was run at 0.8 ml/min in previously described elution buffer with 100 mM imidazole on an Äkta protein purification system (Pharmacia). A total of 1 mg of each of six standards (thyroglobulin, 669 kDa; ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa; albumin, 67 kDa; and ovalbumin, 43 kDa) was loaded in a 100-μl volume via a 100-μl sample loop. Then 1.4 mg of PilT was also loaded in 100 μl. Then 1-ml fractions were collected, and the specific activity of these fractions was measured if the protein concentration was sufficient.
Sedimentation equilibrium studies.
Protein was exhaustively dialyzed against 5% (vol/vol) glycerol-150 mM KCl-25 mM Tris-Cl (pH 7.9) at room temperature-100 mM imidazole (pH 8.0) at room temperature. The final dialysate was used to dilute protein samples to 12, 26, and 39 μM. Double-sector charcoal-filled Epon centerpieces were used with 100 μl of sample and 105 μl of the final dialysate as the reference in a Beckman Optima XL-A analytical ultracentrifuge. Concentration gradients were recorded at 280 nm every 2 to 4 h until gradients became superimposable; in some instances after equilibrium was established, gradients were monitored for up to 12 h to further confirm the equilibrium state. Equilibrium at 40°C was established at speeds of 5,900, 6,300, 7,400, 8,600, and 10,000 rpm. A nonsedimenting baseline absorbance was measured at the end of the run by high-speed depletion of the protein.
The polypeptide molecular weight and partial specific volume were calculated from the sequence as 42,252 and 0.746 ml/g, respectively. The extinction coefficient for aqueous media was calculated as 14,900 M−1cm−1 (24). The density of the final dialysate was measured at 40°C as 1.01292 g/ml with an Anton Paar DMA 5000 density meter.
Data collected at three initial concentrations and various speeds were simultaneously fit to various models with a program written for Igor Pro (Wavemetrics Inc., Lake Oswego, Oreg.) by D. R. McCaslin. Models evaluated included single species, two and three species in equilibrium, and two independent noninteracting species. For the present analysis, parameters to account for nonsedimenting absorbances were constrained to the values measured after high-speed depletion of the protein. The reduced molecular weight, M(1 − 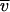 ρ), where M is the molecular weight,
ρ), where M is the molecular weight, 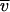 is the partial specific volume, and ρ is the solvent density, was used as the actual model parameter and was converted to a molecular weight only after the fit was completed. This approach eliminated biases to the fit propagated by possible errors in partial specific volume and density.
is the partial specific volume, and ρ is the solvent density, was used as the actual model parameter and was converted to a molecular weight only after the fit was completed. This approach eliminated biases to the fit propagated by possible errors in partial specific volume and density.
Dynamic light scattering.
Purified PilT well above the oligomerization threshold concentration was used in a Protein Solutions Dynapro dynamic light-scattering instrument. Data were collected at successively increasing temperatures from 10°C to 70°C after a 10-min equilibration at each temperature. At least 40 readings were taken at each temperature, and data were analyzed with the manufacturer's software, DYNALS.
RESULTS
Cloning, overexpression, and purification of A. aeolicus PilT.
pilT was cloned from the hyperthermophile A. aeolicus. The protein was overexpressed as a C-terminal hexahistidine fusion and purified by Ni2+ affinity chromatography. Taking advantage of the thermostability of the A. aeolicus protein compared to potential contaminating E. coli proteins, a 30-min, 80°C heat shock served as a final purification step. The overall yield from each 6-liter culture was 10 to 13 mg of approximately 96% pure PilT (Fig. 1B). (Similar results were obtained for an N-terminal hexahistidine fusion; data not shown.) The major protein band was shown by N-terminal sequence analysis to correspond to PilT (MFEKQEVEQK), as was a slightly higher molecular weight contaminant. Matrix-assisted laser desorption/time-of-flight mass spectrometry results were in agreement with the predicted mass of 42.2 kDa for the full-length PilT-Leu-Glu-His6 protein.
PilT is an ATPase.
Based on the highly conserved nucleoside triphosphate binding motif found in the PilT amino acid sequence (Fig. 1A), it has long been hypothesized that PilT has the ability to bind and hydrolyze ATP (23, 38). In order to test this hypothesis, the specific ATPase activity of purified PilT was measured in a standard coupled enzyme assay under a variety of conditions. Negligible ATPase activity was detected with the standard real-time assay up to the upper temperature limit of 55°C, at which point coupling enzymes began to precipitate. The standard colorimetric assay was therefore amended so that PilT and ATP were coincubated for the indicated times at the indicated temperatures (up to 80°C). Then, coupling enzymes were added and end point values were determined (see Materials and Methods).
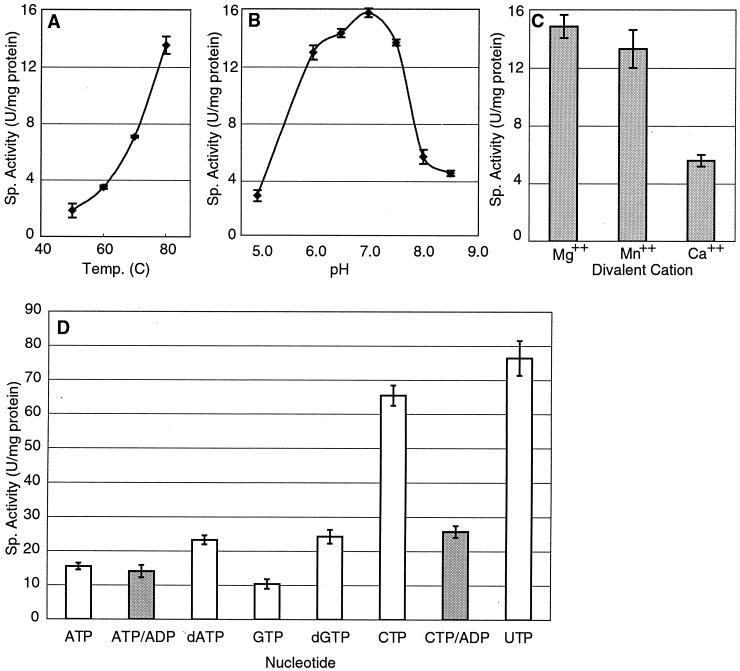
The PilT protein is a thermostable ATPase with a specific activity of 15.7 ± 0.9 U/mg of protein (mean ± standard deviation) at pH 7.0 in the presence of Mg2+ (Fig. 2). The same specific activity was measured for N-terminally histidine-tagged and purified PilT (data not shown). The activity was negligible at temperatures below 50°C and increased up to the maximum temperature testable, 80°C (Fig. 2A). Optimum pH was determined to be between 6.5 and 7.0 (Fig. 2B). Substitution of Mn2+ for Mg2+ did not significantly affect the specific activity of the enzyme (Fig. 2C), although it did consistently lead to decreased stability at high temperature. Substitution of Ca2+ for Mg2+ did decrease the maximal activity to 5 U/mg. The specific activity was linear with protein concentration from 0.6 to 2.4 mg/ml. Thus, A. aeolicus PilT is a well-behaved enzyme in vitro, with low but highly reproducible ATPase activity.
FIG. 2.
NTPase activity of A. aeolicus PilT. Specific activity (in nanomoles of ATP per minute per milligram of protein) is reported as a function of (A) temperature (pH constant at 7.5; metal ion Mg2+), (B) pH (temperature constant at 80°C; metal ion Mg2+), and (C) metal ion identity (temperature and pH constant at 80°C and 7.0, respectively). (D) Specific activity (in nanomoles of NTP per minute per milligram of protein) as a function of substrate, with (shaded bars) or without (open bars) excess ADP. ATPase assays (A, B, and C) were done with the coupled enzyme assay, while NTPase assays (D) were done with the malachite green assay, both described in Materials and Methods. Each point includes three replicates.
PilT is promiscuous with respect to substrate in vitro.
A sensitive inorganic phosphate release assay (12) was used to assess the substrate specificity of the PilT enzyme (Fig. 2D). The highest activity in vitro was observed with the pyrimidines CTP and UTP, which yielded specific activities of 65.5 ± 3.0 and 76.5 ± 5.1 U/mg of protein, respectively. The nucleotides and deoxynucleotides tested increased in efficiency in the following order: GTP < ATP < dGTP ≈ dATP ≪ CTP < UTP. At an ADP concentration of 1/10th the concentration of NTP, pyrimidine hydrolysis was substantially inhibited but ATP hydrolysis was unaffected (Fig. 2D).
Lys149Gln PilT loses NTPase activity.
To investigate the role of the proposed nucleotide-binding motif in the in vitro NTPase activity of PilT, a protein was created in which the lysine of the invariant GxxxxGKT/S motif was replaced with glutamine, a structurally conservative change which is nonetheless expected to cause substantially decreased ability to hydrolyze or indeed to bind nucleoside triphosphates. The overexpression and purification of this variant PilT were done exactly as for the wild-type PilT, with similar yields (Fig. 1B).
Circular dichroism measurements also yielded virtually indistinguishable results for the wild-type and Lys149Gln proteins and indicated an alpha-helical content of ≈30% (data not shown). The ATPase and CTPase activities of the altered protein were below the sensitivity of our assays, verifying the importance of the lysine residue and the P-loop motif in the in vitro activity of PilT and confirming that the measured NTPase activity of the wild-type protein was due to PilT rather than a minor contaminating host enzyme.
PilT behaves as an oligomer.
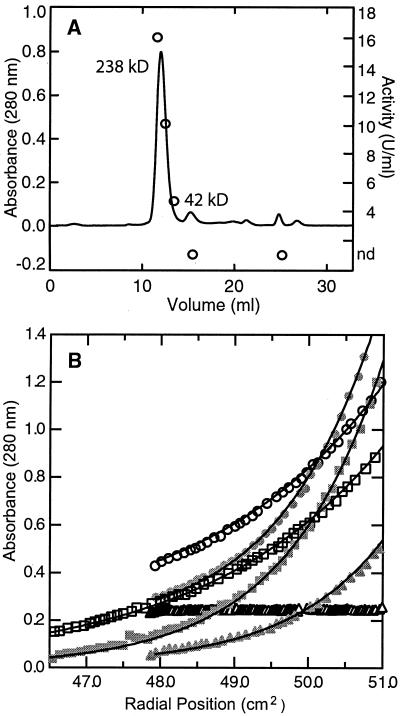
We investigated the oligomeric state of purified PilT by several biophysical methods (Fig. 3), including equilibrium sedimentation, size exclusion chromatography, and dynamic light scattering. Based on size exclusion chromatography at room temperature, PilT had an estimated molecular mass of 239 kDa (Fig. 3A), corresponding to a pentamer or hexamer of the 42-kDa polypeptide.
FIG. 3.
Oligomeric state of PilT. (A) Size exclusion chromatogram including specific ATPase activities in units per milliliter of protein-containing fractions (open circles). Main peaks on the trace are labeled with calculated molecular masses. (B) Equilibrium sedimentation. Only the 6,300 rpm (open symbols) and 8,600 rpm (solid symbols) data at 40°C for the three initial concentrations, 12 μM (triangles), 26 μM (squares), and 39 μM (circles), are shown. Every fourth data point is shown with the exception of the 12 μM, 6,300 rpm data, for which every data point is shown. The solid lines were determined from the global fit of the complete data set (11 sets of data) to a model consisting of a single species with a molecular weight of 204,000. The 12 μM sample did not form a gradient at speeds below 8,600 rpm, nor did the 26 μM sample at 5,900 rpm. At 40°C and an initial 39 μM concentration of protein, PilT generated gradients at all speeds examined.
Sedimentation equilibrium provides a thermodynamically rigorous measurement of the weight average molecular weight of a purified protein as a function of its concentration. Data exhibiting concentration gradients of PilT at 40°C were analyzed as a single species with a reasonably good fit to a weight average molecular weight of 203,800 (Fig. 3B), corresponding to a pentamer of PilT subunits. Fits to this model exhibited small systematic deviations which became apparent when we examined the fit residuals, suggesting that it may not be a complete description of the underlying molecular events. Results of analysis of a system of two oligomeric species in concentration-dependent equilibrium have better overall statistics; however, if the fitted curve for this model were plotted in Fig. 3B, the differences would be very hard to discern. Whether these observations arose from the presence of a single species or two or more species in a concentration-dependent equilibrium will require further study.
At lower concentrations and speeds, PilT did not form a gradient in these 40°C experiments, suggesting that it remained monomeric (Fig. 3B). To date, we have found no conditions in which a significant population of monomer could be observed in the presence of the oligomer, indicating that the conversion to a larger aggregate is highly cooperative. At temperatures of 30°C and below and at all loading concentrations and speeds, oligomer was present. Thus, the association to the oligomer is also highly temperature dependent, being stronger at lower temperatures.
Equilibrium sedimentation studies on K149Q PilT showed all of the properties described above (data not shown).
Dynamic light scattering was also used to assess the homogeneity of the PilT oligomer. PilT above the oligomerization concentration at temperatures between 10°C and 40°C had polydispersity of 10 to 15%, well within the recommended range for protein crystallization (1). At temperatures of up to 70°C, the polydispersity increased to 22%, although the predicted radius of the predominant species did not change significantly (data not shown). The radius is compatible with oligomeric PilT as determined by sedimentation equilibrium and chromatography.
DISCUSSION
To the best of our knowledge, this is the first published demonstration of NTPase activity for a PilT protein. This result is consistent with the hypothesis that PilT family members may be part of a pilus-dependent surface motility retraction motor, although it does not rule out additional regulatory or structural roles for PilT, nor does it prove that A. aeolicus PilT is directly involved in motility in vivo.
The substrate promiscuity of A. aeolicus PilT in vitro does not allow definitive identification of its in vivo substrate. Because the concentration of free ATP in the cell is likely to be substantially higher than that of free pyrimidine triphosphate, and because the presence of ADP inhibits use of CTP as a substrate (Fig. 2D), higher in vitro specific activity is not necessarily indicative of a pyrimidine triphosphate as the physiological substrate. Our observation that PilT has a higher specific activity with deoxypurine triphosphates than with the corresponding purine triphosphates (Fig. 2D) is again difficult to correlate with in vivo substrate use, in part because deoxynucleoside triphosphate concentrations in vivo are much lower than the corresponding NTP concentrations. The TrbB secretion NTPase uses dATP 10 times more efficiently than ATP in vitro, although ATP inhibits this activity (18). Thus, while our data are compatible with ATP as the in vivo substrate for A. aeolicus PilT, final determination awaits further kinetic studies under as yet poorly understood physiological conditions.
If PilT is directly responsible for pilus retraction via NTP hydrolysis, as our data allow (Fig. 2), how does it work? Oster and Kaiser have proposed a modified version of the F1-ATPase rotary motor to explain retraction (17). There are several reasons to explore this model. First, the ability to operate in forward or reverse is a common feature of both the F1F 0-ATPase and the pilus biogenesis-retraction system. F1F0-ATPase can function with α3β3 subunit rotation driven by the proton motive force and coupled to ATP synthesis or with ATP hydrolysis leading to rotation in the opposite direction and an increase in proton motive force. In a similarly reversible way, type IV pili can be assembled from pilin monomers or retracted, concomitant with or followed by disassembly of filaments to a pool of monomers. Possibly in the case of PilT-mediated retraction, once the bottommost pilin subunit has been moved into the membrane environment, disassembly is spontaneous as the hydrophobic α-helix becomes solubilized in the lipid bilayer (23). The reversibility requires two NTPase proteins: PilT would interact with the same pilus base proteins as the putative secretion ATPase required for pilus assembly, PilB (named PilQ in R64 thin pilus biogenesis, where ATPase activity has been demonstrated [29]) but with the opposing outcome. PilT and PilB could be present at distinct times or form a mixed multimer at the pilus base. Second, the F1-ATPase γ subunits which form the shaft within the α3β3 stator barrel are a helical coiled coil (32), structurally similar to the pilin monomer N-terminal α-helices thought to be exposed at the bacterial membrane end of the pilus filament (11, 13, 25). Additionally, F1 is the soluble portion of the membrane-bound ATP synthase complex and is connected to the membrane by protein-protein interactions. This is likely to be the case for PilT, which has no predicted transmembrane segments and is found both in the cytoplasm and associated with the membrane (6; our unpublished data).
Another appropriate rotary motor analogy may be the flagellar motor, which similarly has the ability to rotate clockwise (for tumbling) or counterclockwise (for swimming). Twitching motility in N. gonorrhoeae and P. aeruginosa is regulated by multiple single-domain response regulator proteins (9), analogous to the prototype response regulator CheY, which binds directly to the base of the flagellar motor to switch the direction of rotation for chemotaxis (31). In Synechocystis spp., PilT2 is required for accurate phototaxis and may be regulated directly by response regulator domains (4). Perhaps pilus-mediated movement is a consequence of ATP-dependent force-generating retraction by a rotary motor, and the decision to extend or retract is controlled by a direct interaction of a response regulator protein with PilT.
A second, mechanistically quite distinct possibility is that PilT acts as an antichaperone in facilitating the disassembly of pilin monomers from the base of the homopolymeric pilus filament (23). The PilT oligomer observed in this study (Fig. 3) is likely to have an internal cavity, as suggested by electron micrographs of type II and IV secretion ATPases showing ringed oligomers in vitro (19) and as seen in the crystal structure of the type IV secretion ATPase HP0525 (44). That structure suggested a chaperone-like function for the ATP-dependent secretion of partially folded substrates across the inner membrane (44). Likewise, NTP hydrolysis by PilT would catalyze disassociation of pilin monomers from the stable, hydrophobic assembly of the pilus filament.
PilT and bacterial type II and IV secretion ATPases are distant members of the AAA+ family of often hexameric proteins that mediate remodeling and disassembly of macromolecular complexes (17, 36). Vale has suggested that protein unfolding is facilitated within these oligomers because subunits can alternately tense and relax (presumably upon binding ATP and release of ADP) while remaining bound to the substrate protein complex, thus generating concerted conformational changes or, in this case, disassembly (36). In this model, CheY-like response regulator proteins would interact with PilT to coordinate the appropriate location and timing for pilus assembly, and pilus retraction would be a constitutive rather than a regulated function of normal pili in the presence of active PilT. In N. gonorrhoeae, PilT does modulate pilus biogenesis (42, 43); in Myxococcus xanthus, the location of pilus biogenesis is controlled by the frz chemosensory system (34, 39); and in Synechocystis spp., the single-domain response regulator TaxY1 has been proposed to mediate pilus function via PilT1 (4).
Comparison of PilT to type II and IV secretion ATPases reveals that when specific ATPase activity has been measured, it is similar in magnitude to that measured here, 15 nmol/min/mg for TadA (2), 4.5 nmol/min/mg for TrwD (28), and only 1 nmol/min/mg for PilQ (29), in comparison to 15.7 nmol/min/mg for PilT (Fig. 2). For PilT, this means a steady-state rate of ≈0.65 ATP molecule hydrolyzed per PilT monomer per minute. Given that pilus retraction rates have been calculated to be approximately 1,500 pilin subunit monomers per second (21), it seems likely that if PilT acts as a retraction motor in vivo, it must be stimulated by protein-protein, protein-membrane, or protein-substrate interactions or posttranslational modifications. Little is known about the in vivo interactions of proteins in the pilus assembly-disassembly pathway. Alternatively, in an antichaperone disassembly model for retraction, NTP binding or hydrolysis may be required to allow an open-gated conformation of the PilT oligomer for retraction. In this case, there may not be a requirement for NTP hydrolysis with every pilin monomer that disassociates from the base of the pilus.
These and other models can be experimentally addressed now that active purified PilT is available. Current questions include what components, if any, in addition to PilT are required to transmit the energy of NTP hydrolysis to the pilus filament, the role of PilT in retraction (direct or indirect), whether it is indeed the retraction motor, and the molecular mechanism of retraction. Further biochemical and genetic analyses, high-resolution microscopy of purified components and/or living cells, and structural biology will be critical for answering these important questions.
Acknowledgments
We thank R. Schwartz for initial cloning of the A. aeolicus pilT gene and G. Worzalla for technical assistance.
This work was supported by the NIH (GM59721, K.T.F.) and a W. M. Keck Foundation Distinguished Young Investigator Award (K.T.F.). The UW Biophysics Instrumentation Facility is supported by the NSF (BIR-9512577).
REFERENCES
- 1.Bernstein, B. E., P. A. M. Michels, H., Kim, P. H., Petra, S., and W. G. J. Hol. 1998. The importance of dynamic light scattering in obtaining multiple crystal forms of Trypanosoma brucei PGK. Protein Sci. 7:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee, M. K., S. C. Kachlany, D. H. Fine, and D. H. Figurski. 2001. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 183:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaya, D., N. R. Bianco, D. Bryant, and A. R. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941-951. [DOI] [PubMed] [Google Scholar]
- 4.Bhaya, D., A. Takahashi, and A. R. Grossman. 2001. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA 98:7540-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 6.Brossay, L., G. Paradis, R. Fox, M. Koomey, and J. Hebert. 1994. Identification, localization, and distribution of the PilT protein in Neisseria gonorrhoeae. Infect. Immun. 62:2302-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie, P. J., J. E. Ward, Jr., M. P. Gordon, and E. W. Nester. 1989. A gene required for transfer of T-DNA to plants encodes an ATPase with autophosphorylating activity. Proc. Natl. Acad. Sci. USA 86:9677-9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comolli, J., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darzins, A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol. Microbiol. 11:137-153. [DOI] [PubMed] [Google Scholar]
- 10.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olson, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 11.Dupuy, B., M. K. Taha, A. P. Pugsley, and C. Marchal. 1991. Neisseria gonorrhoeae prepilin export studied in Escherichia coli. J. Bacteriol. 173:7589-7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichelberg, K., C. C. Ginocchio, and J. E. Galan. 1994. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J. Bacteriol. 176:4501-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forest, K. T., and J. A. Tainer. 1997. Type-4 pilus structure: outside to inside and top to bottom — a minireview. Gene 192:165-169. [DOI] [PubMed] [Google Scholar]
- 14.Graupner, S., N. Weger, M. Sohni, and W. Wackernagel. 2001. Requirement of novel competence genes pilT and pilU of Pseudomonas stutzeri for natural transformation and suppression of pilT deficiency by a hexahistidine tag on the type IV pilus protein PilAI. J. Bacteriol. 183:4694-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose P-BAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgkin, J., and D. Kaiser. 1979. Genetics of gliding motility in Myxococcus xanthus (Myxobacteriales): two gene systems control movement. Mol. Gen. Genet. 171:177-191. [Google Scholar]
- 17.Kaiser, D. 2000. Bacterial motility: how do pili pull? Curr. Biol. 10:R777-R780. [DOI] [PubMed] [Google Scholar]
- 18.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause, S., M. Barcena, W. Pansegrau, R. Lurz, J. M. Carazo, and E. Lanka. 2000. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA 97:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 21.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 22.Merz, A. J., and M. So. 2000. Interactions of pathogenic Neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 23.Merz, A. J., and K. T. Forest. 2002. Bacterial surface motility: slime trails, grappling hooks, and nozzles. Curr. Biol. 12:R297-R303. [DOI] [PubMed] [Google Scholar]
- 24.Pace, N., and F. X. Schmid. 1997. Protein structure: a practical approach. IRL Press, Oxford, UK.
- 25.Parge, H. E., K. T. Forest, M. J. Hickey, D. A. Christensen, E. D. Getzoff, and J. A. Tainer. 1995. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378:32-38. [DOI] [PubMed] [Google Scholar]
- 26.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 27.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivas, S., S. Bolland, E. Cabezon, F. M. Goniand, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272:25583-25590. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, D., T. Horiuchi, and T. Komano. 2001. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 276:17968-17975. [DOI] [PubMed] [Google Scholar]
- 30.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 32.Stock, D., A. G. Leslie, and J. E. Walker. 1999. Molecular architecture of the rotary motor in ATP synthase. Science 286:1700-1705. [DOI] [PubMed] [Google Scholar]
- 33.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 34.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 35.Tchen, T. T. 1958. Mevalonic kinase: purification and properties. J. Biol. Chem. 233:1100-1103. [PubMed] [Google Scholar]
- 36.Vale, R. D. 2000. AAA proteins. Lords of the ring. J. Cell Biol. 150:F13-F19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 39.Ward, M. J., and D. R. Zusman. 1997. Regulation of directed motility in Myxococcus xanthus. Mol. Microbiol. 24:885-893. [DOI] [PubMed] [Google Scholar]
- 40.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 41.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert and, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 42.Wolfgang, M., H. S. Park, S. F. Hayes, J. P. van Putten, and M. Koomey. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95:14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]