Abstract
Many proteobacteria use acyl-homoserine lactones as quorum-sensing signals. Traditionally, biological detection systems have been used to identify bacteria that produce acyl-homoserine lactones, although the specificities of these detection systems can limit discovery. We used a sensitive approach that did not require a bioassay to detect production of long-acyl-chain homoserine lactone production by Rhodobacter capsulatus and Paracoccus denitrificans. These long-chain acyl-homoserine lactones are not readily detected by standard bioassays. The most abundant acyl-homoserine lactone was N-hexadecanoyl-homoserine lactone. The long-chain acyl-homoserine lactones were concentrated in cells but were also found in the culture fluid. An R. capsulatus gene responsible for long-chain acyl-homoserine lactone synthesis was identified. A mutation in this gene, which we named gtaI, resulted in decreased production of the R. capsulatus gene transfer agent, and gene transfer agent production was restored by exogenous addition of N-hexadecanoyl-homoserine lactone. Thus, long-chain acyl-homoserine lactones serve as quorum-sensing signals to enhance genetic exchange in R. capsulatus.
Many proteobacteria use acyl-homoserine lactone (acyl-HSL) signals in cell density-dependent gene regulation (9, 11, 45). Acyl-HSLs act as intercellular signals that allow bacterial species to monitor their population density and activate specific sets of genes at high cell densities. This type of cell density-dependent gene regulation, also called quorum sensing, was first described in the marine bacterium Vibrio fischeri (6, 21), which uses an acyl-HSL to activate luminescence gene expression. The V. fischeri quorum-sensing regulatory elements are LuxI and LuxR (8). The LuxI protein is the acyl-HSL synthase responsible for production of the N-3-oxohexanoyl-HSL. LuxR is a transcription factor that activates luminescence gene expression when bound by the acyl-HSL signal (8, 9, 11, 12, 23).
Acyl-HSL signaling controls a number of bacterial processes, including virulence factor production, secondary metabolite production, and biofilm development in Pseudomonas aeruginosa (25, 27, 46) and conjugal transfer in Agrobacterium tumefaciens (10, 29, 49). Generally, LuxR and LuxI homologs serve as signal receptors and signal generators, respectively. Depending on the system, the signal varies in acyl group length and substitution (11), and these acyl side chain differences confer signal specificity (7, 35). Differences in acyl chain lengths are also a factor in signal permeability. Short-chain acyl-HSLs, like N-3-oxohexanoyl-HSL (3OC6-HSL) and butanoyl HSL, can diffuse freely through the cell membrane (14, 26). While still diffusible, long-chain acyl-HSLs like the P. aeruginosa signal N-3-oxododecanoyl-HSL (3OC12-HSL) appear to partition to the cell membrane. The MexAB-OprD efflux pump and perhaps other efflux pumps can aid in 3OC12-HSL export (26).
Rhodobacter capsulatus and Paracoccus denitrificans are members of the α group of the Proteobacteria. These free-living organisms are closely related and have been well studied for reasons related to their physiological flexibility (1, 20). We assessed their ability to synthesize acyl-HSLs by using a previously described radiotracer technique (3, 34, 38). This allowed us to discover that R. capsulatus and P. denitrificans made a long-chain acyl-HSL that was not readily detected by any available bioassay. We show that the most abundant acyl-HSL produced by these bacteria is N-hexadecanoyl-HSL (C16-HSL).
Because the R. capsulatus genomic sequence is available, we studied acyl-HSL signaling in this organism further. We identified luxI and luxR homologs in the R. capsulatus genome. We demonstrate that cells concentrate this hydrophobic acyl-HSL. We also show it activates production of the gene transfer agent when added to an R. capsulatus acyl-HSL synthesis mutant. Gene transfer agent is a bacteriophage-like particle that transfers random 4.5-kb fragments of host genomic DNA between R. capsulatus cells (16, 40, 47). This type of genetic exchange is a biological process that we show is related to quorum sensing.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The plasmids and strains used are listed in Table 1. R. capsulatus cultures were grown aerobically or phototrophically in either yeast extract-peptone-salts (YPS) (44) or RCV minimal medium (2) at 30°C. P. denitrificans was grown aerobically in Sistrom's succinic acid minimal medium A (SIS) (4) or Luria broth (LB) (33) at 30°C. V. fischeri was grown at 23°C in morpholinepropanesulfonic acid (MOPS) minimal medium (22) modified to contain 0.3% (vol/vol) glycerol and 75% (vol/vol) artificial seawater. E. coli was grown at 37°C in LB. Antibiotic concentrations used for R. capsulatus were as follows: rifampin, 100 μg/ml; kanamycin, 5 μg/ml; spectinomycin, 10 μg/ml; and tetracycline, 0.5 μg/ml. For E. coli, antibiotic concentrations were kanamycin, 30 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 12.5 μg/ml; and ampicillin, 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| R. capsulatus | ||
| SB1003 | Parental strain for gtaI mutant, Rifr | 48 |
| B10 | Wild-type GTA indicator strain, Rifs | 19 |
| ALS1 | gtaI::ΩSpr derivative of SB1003, Rifr Spr | This work |
| P. denitrificans ATCC 177441 | Wild type | |
| V. fischeri MJ-1 | Wild type | 32 |
| E. coli S17-1λpir | Donor in interspecies conjugations | 28 |
| Plasmids | ||
| pGtaI-1 | pCR2.1 containing TA-cloned 2.7-kb RCC03805 PCR fragment, Apr | This work |
| pHP45ΩSp | ΩSpr cassette source | 30 |
| pGtaIKO | pGtaI-1 with ΩSpr cassette inserted in RCC03805 ORF, Apr Spr | This work |
| pJP5603 | Mobilizable suicide vector, Kmr | 28 |
| pJPGtaIKO | 4.7-kb MfeI-XbaI fragment of pGtaIKO cloned into pJP5603, Kmr Spr | This work |
| pBBRMCS-2 | Broad-host-range cloning vector, Kmr | 15 |
| pBBRGtaI | RRC03805 cloned in pBBRMCS-2, Kmr | This work |
| pYP | GTA gene fusion orfg2::lacZ, Tcr | 18 |
| pYNP | pYP construct lacking promoter, Tcr | 18 |
GTA, gene transfer agent.
Detection and identification of R. capsulatus and P. denitrificans acyl-HSLs.
Previously, discovery of acyl-HSLs has relied on bioassays, which are limited by signal specificity constraints (36). Novel acyl-HSLs might not be detected by any of the available bioassays. We adapted an assay that detects the incorporation of 14C label into acyl-HSLs (3, 34, 38) to screen R. capsulatus, P. denitrificans, and other bacteria for acyl-HSL production regardless of whether the molecules could be readily detected in any bioassay. Late-logarithmic-phase cultures (5-ml volume) grown in methionine-free medium (RCV or SIS) were labeled for 30 to 60 min with 5 μCi of carboxy[14C]methionine, 55 mCi/mmol (American Radiochemical Company, St. Louis, Mo.). Unless indicated, total cultures were extracted twice with an equal volume of acidified ethyl acetate (100 μl of glacial acetic acid per liter). Extracts were combined and dried under a stream of N2 gas. The residue was suspended in 200 μl of methanol and separated by C18 reverse-phase high-performance liquid chromatography (HPLC) in a 20 to 100% methanol-in-water gradient (27). Four milliliters of scintillation cocktail 3a70b (Research Products Inc., Mount Prospect, Ill.) was added to each of the 70 1-ml fractions collected, and radioactivity was determined by scintillation counting.
To determine C16-HSL concentrations, R. capsulatus ALS1 harboring pYP (18), a plasmid that produces β-galactosidase in response to added C16-HSL (see Results), was used. The procedure was identical to that described for the 3OC12-HSL bioassay described elsewhere (27, 36) except for the different reporter and that cultures were grown aerobically in YPS medium for 22 h. Synthetic C16-HSL was used to generate a standard curve.
To identify acyl-HSLs, we extracted late-logarithmic-phase culture fluid with acidified ethyl acetate. The acyl-HSLs were purified from extracts of LB-grown cultures by C18 reverse-phase HPLC as described above. The purified material was analyzed by chemical ionization mass spectrometry (CI-MS) with a VG Trio-1 quadropole mass spectrometer with methane as the reagent gas. We restricted our isolation of material for mass spectrometry to material from the culture fluid. As described in the Results, this represented about half of the total for each of the acyl-HSLs. This facilitated purification because the bulk of the cellular lipids were not included in the extract. Alternatively, identification was done by coelution of radiolabeled material from whole cultures (cells plus culture fluid) with chemically synthesized acyl-HSLs.
Acyl-HSL association experiments.
To determine if the long-chain acyl-HSLs are associated with cells, we compared the amount of radiolabeled acyl-HSL associated with the cell-free culture fluid and cell pellet for R. capsulatus, P. denitrificans, and, as a control, V. fischeri. Radiolabeling was performed as described above except that prior to extraction, the cultures were centrifuged (10,000 × g, 4°C), and the culture fluid was separated from the cell pellet. The cell pellet was resuspended in 5 ml of fresh medium (the original culture volume). Acyl-HSLs from the culture fluid and suspended cell pellet were extracted and separated by HPLC as described above. The total amounts of [14C]3OC6-HSL (fraction 10), [14C]C16-HSL (fractions 65 to 67), and [14C]C14-HSL (fractions 60 to 62) in the culture fluid and cell pellet were determined by scintillation counting.
Acyl-HSLs.
C16-HSL was synthesized in a manner similar to that described elsewhere (5). Hexadecanoic acid (Sigma Chemical Company, St. Louis, Mo.) was mixed with l-homoserine lactone hydrochloride in dry dichloromethane with activation by dicyclohexylcarbodiimide and 1-hydroxybenzotriazol. The crude synthetic product was washed with 5% NaHCO3, and the C16-HSL was purified by C18 reverse-phase HPLC. As shown by CI-MS, the product showed the expected molecular mass. All other synthetic acyl-HSLs were purchased from Aurora Biosciences.
Identification and genetic analysis of R. capsulatus acyl-HSL synthase.
We performed a Blast search of the R. capsulatus genome (http://ergo.integratedgenomics.com/ERGO; Integrated Genomics, Chicago, Ill.) for translation products showing similarity to the Rhodobacter sphaeroides CerI protein (accession number AAC46022). One putative R. capsulatus gene, open reading frame (ORF) RRC03805, coded for a polypeptide that showed significant similarity to CerI and was adjacent to a gene coding for a probable LuxR-type regulatory protein, ORF RRC03806. A 2.7-kb fragment of R. capsulatus SB1003 chromosomal DNA containing ORF RRC03805 was amplified with the Expand long-template PCR system (Boehringer Mannheim) and the following primers: forward, 5′-CAATTGGGCTACCGCCGTCTGAACCG-3′, and reverse, 5′-TCTAGACGGGTCCGATCCGCGGACGG-3′.
The PCR fragment was used to create pGtaI-1 with the Original TA cloning kit (Invitrogen, Carlsbad, Calif.). A 2-kb SmaI fragment of pHP45ΩSp (containing the spectinomycin resistance [Spr] cassette) was cloned into a unique PshAI site of pGtaI-1, disrupting RRC03805. This construct was designated pGtaIKO. A pGtaIKO MfeI-XbaI fragment containing the inactivated RRC03805 gene was ligated to EcoRI- and XbaI-digested pJP5603 to create pJPGtaIKO, which contains the Spr cassette 178-bp downstream of the predicted translational start codon of RRC03805. The Spr cassette in pJPGtaIKO is flanked by 1.4 kb of upstream and 1.3 kb of downstream R. capsulatus DNA. E. coli S17-1 λpir was used to mobilize pJPGtaIKO into R. capsulatus SB1003 (28). We selected spectinomycin-resistant colonies and screened for a kanamycin-sensitive mutant. One mutant, ALS1, contained an Spr cassette insertion in RRC03805, as shown by Southern blot analysis with RRC03805 and Spr cassette probes.
For complementation studies, pBBRGtaI was constructed by cloning the RRC03805 ORF into the SalI and EcoRI sites of pBBRMCS-2, a broad-host-range plasmid (15). The resulting plasmid was introduced into R. capsulatus ALS1 by conjugation as described above. A kanamycin-resistant exconjugant was selected for subsequent studies.
Assessment of gene transfer agent transcription and production.
Plasmid pYP, which contains the gene transfer agent promoter, orfg1, and an in-frame translational fusion of the gene transfer agent structural gene orfg2 to lacZ (18), was introduced into R. capsulatus strains SB1003 and ALS1 by conjugation. Exconjugants were grown phototrophically in YPS at 30°C, and β-galactosidase activity was measured in stationary-phase cells (≈22 h) (46). Where indicated, synthetic C16-HSL (2 μM) was added to cultures. As a control, similar experiments were performed with pYNP, which contains a promoterless orfg2::lacZ fusion (18). With pYNP, β-galactosidase activity was less than 0.1 U.
Transducing particle production was measured as gene transfer agent-mediated transfer of a rifampin resistance (Rifr) marker from the gene transfer agent donor strains (SB1003 and ALS1) to the Rifs gene transfer agent recipient strain (B10) as described previously (18). R. capsulatus SB1003 and ALS1 were grown as described above, and synthetic C16-HSL (2 μM) was added to cultures where indicated.
RESULTS
R. capsulatus and P. denitrificans synthesize acyl-HSLs.
Acyl-HSL discovery has relied on detection with bioassays employing heterologous reporter constructs (36). These bioassays have certain limitations: they are labor intensive, require construction of an appropriate indicator strain, and are biased towards detection of acyl-HSLs close enough in structure to the natural signal to be recognized by specific LuxR homologs. The development of a radiotracer assay to detect acyl-HSLs overcomes these limitations. We have developed such an assay and used it with success to monitor the relative abundance of multiple acyl-HSLs produced by P. aeruginosa (34, 38). The radiotracer assay depends upon incorporation of radiolabel from carboxyl-[14C]methionine into acyl-HSLs. Radiolabeled acyl-HSL can be separated from methionine or S-adenosylmethionine by solvent extraction.
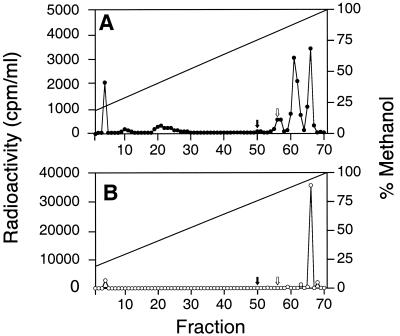
We screened R. capsulatus and P. denitrificans for acyl-HSL synthesis with the radiotracer assay. These were both organisms in which we had failed to detect acyl-HSLs with a variety of bioassays (31; unpublished results). P. denitrificans incorporated 14C label into a product that was ethyl acetate extractable and was eluted with approximately 95% methanol in our C18 reverse-phase HPLC (Fig. 1B). R. capsulatus appeared to make two acyl-HSLs, one that coeluted with the P. denitrificans product and an additional product that was eluted with 90% methanol in water (Fig. 1A). These acyl-HSLs were more hydrophobic than any previously described naturally occurring acyl-HSLs. Our results demonstrate one significant advantage of the nonbiological detection approach, that acyl-HSLs exhibiting little or no activity in existing bioassays can be detected.
FIG. 1.
HPLC analysis of radiolabeled acyl-HSLs from (A) R. capsulatus and (B) P. denitrificans culture extracts. Acyl-HSLs from P. aeruginosa (3OC12-HSL) and R. sphaeroides (7,8-cis-C14-HSL) eluted at fractions 50 and 56, respectively, as shown by the solid and open arrows, respectively. C14-HSL eluted at fraction 61 and C16-HSL at fractions 65 and 66. These are indicated as references. The void volume was in fractions 1 to 5. The radioactivity in the void volume likely represents a small amount of methionine in the ethyl acetate. The methanol gradient is shown by the solid line. No radioactive material was eluted after fraction 70.
Identification of R. capsulatus and P. denitrificans acyl-HSLs.
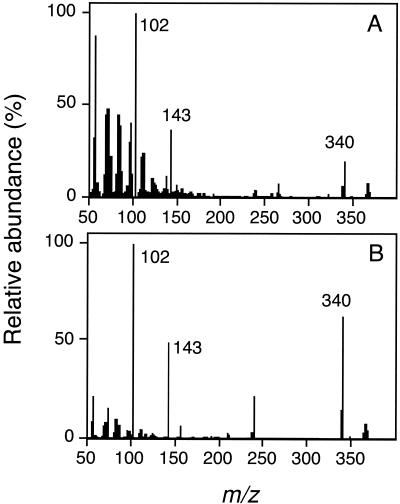
The R. capsulatus product from fraction 60 coeluted with synthetic tetradecanoyl-HSL (C14-HSL), but the identities of the more hydrophobic compounds produced by R. capsulatus and P. denitrificans (fractions 65 and 66) were unknown. The material in fractions 65 and 66 was purified from P. denitrificans culture fluid as described in Materials and Methods and analyzed by CI-MS, which showed a quasimolecular (M + H)+ ion with an m/z of 340 (Fig. 2A). There were also peaks at 102 and 143 m/z, corresponding to homoserine lactone and the homoserine lactone ring with two of the acyl carbons, respectively, which are characteristic peaks found in CI-MS spectra of acyl-HSLs (36). The spectrum was consistent with the conclusion that the compound in fractions 65 and 66 was C16-HSL. As a confirmation, we chemically synthesized C16-HSL. Synthetic C16-HSL showed an elution profile identical to that of the P. denitrificans material and had a CI-MS spectrum similar to that of the P. denitrificans material (Fig. 2B). It was not possible to obtain sufficiently pure material from R. capsulatus for further analysis, but we conclude that the R. capsulatus material that coeluted with C16-HSL in HPLC is also C16-HSL.
FIG. 2.
Chemical ionization mass spectra of acyl-HSLs. (A) Spectrum of material purified from a P. denitrificans culture and (B) spectrum of synthetic C16-HSL.
R. capsulatus acyl-HSL synthase gene.
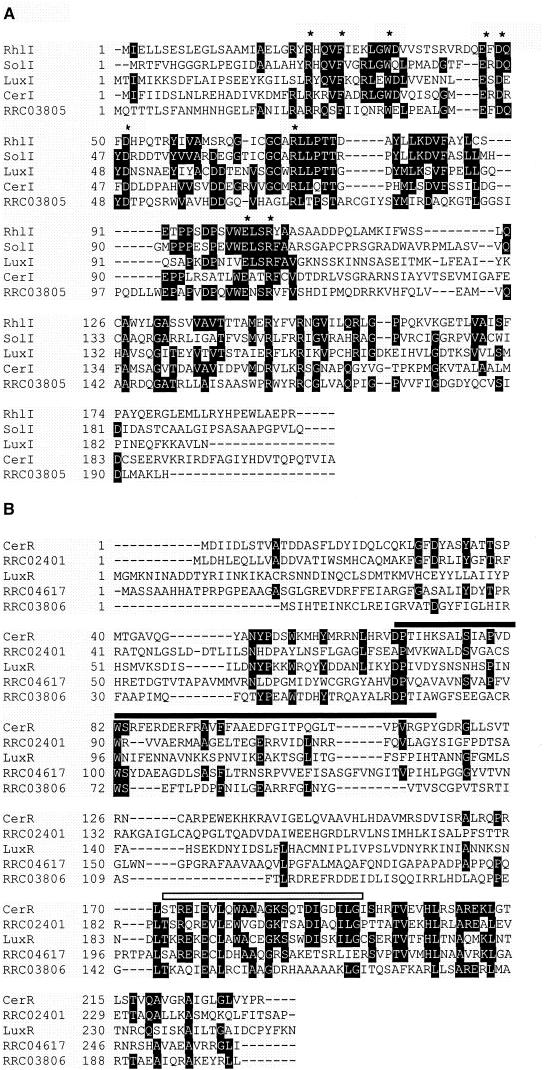
We chose to study C16-HSL production further in R. capsulatus, an organism for which the genome has been sequenced. We identified an ORF, RRC03805, that encoded a product that showed 26% identity (43% similarity) to the R. sphaeroides CerI and contained each of the nine completely conserved amino acid residues found in acyl-HSL synthases (Fig. 3A) (24).
FIG. 3.
Multiple alignments of several LuxI and LuxR family members with homologs found in the R. capsulatus genome. (A) Representative LuxI family members. The asterisks indicate residues conserved among most LuxI family members (24). Each of these nine residues occurs in the aligned sequence of RRC03805. ClustalW (43) was used to align the sequences, and Boxshade (0.6 setting) was used to determine the degree of residue shading. (B) Representative LuxR family members. The solid bar above the residues corresponding to LuxR amino acids 79 to 127 represents the conserved signal-binding region (13, 37, 39), and the open bar above the residues corresponding to LuxR amino acids 184 to 210 indicates the conserved helix-turn-helix-containing region in the DNA-binding domain. The sequences used in the alignments were R. sphaeroides CerI (accession number AAC46022) and CerR (AAC47021), V. fischeri LuxI (CAA68562) and LuxR (B33538), Ralstonia solanacearum SolI (030920), and P. aeruginosa RhlI (AAC44037).
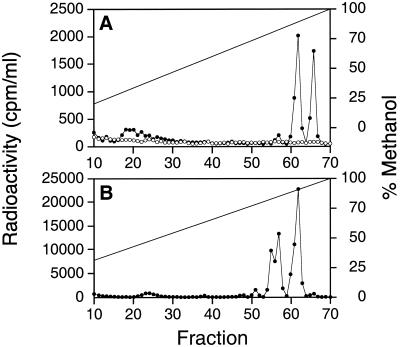
To verify that RRC03805 coded for an acyl-HSL synthase, we created an RR03805 ΩSPr insertion mutant, R. capsulatus ALS1 (Table 1). Strain ALS1 did not synthesize C16-HSL or C14-HSL (Fig. 4A), and we detected no other acyl-HSLs. This suggested that the RRC03805 gene codes for an acyl-HSL synthase. Unlike the R. sphaeroides cerI mutant, which overproduces exopolysaccharide (31), R. capsulatus ALS1 had no obvious phenotype. When the RRC03805 mutation in strain ALS1 was complemented with pBBRGtaI, acyl-HSL synthesis was restored (Fig. 4A). Interestingly, E. coli containing pBBRGtaI synthesized C14-HSL, additional putative acyl-HSLs (fractions 57 to 58), but not C16-HSL (Fig. 4B). To our knowledge, this appears to be the first acyl-HSL synthase that fails to direct the synthesis of all cognate acyl-HSL molecules in E. coli.
FIG. 4.
HPLC analysis of radiolabeled acyl-HSLs from culture extracts. (A) HPLC profile of extract of R. capsulatus ALS1 (○) and ALS1 containing pBBRGtaI (•). (B) HPLC profile of extract of E. coli containing pBBRGtaI.
A Blast search of the R. capsulatus translation products with the R. sphaeroides CerR (AAC47021) revealed three related polypeptides, RRC03806, RRC04617, and RRC02401. These translation products were 26%, 21%, and 23% identical (37%, 31%, and 36% similar) to CerR, respectively. Two of the three, RRC03806 and RRC04617, contained all seven of the most highly conserved residues found among LuxR family members (Fig. 3B) (42). RRC02401 possessed only three of the seven conserved residues, but did show similarity to other LuxR-related proteins along what is thought to be the acyl-HSL binding region (Fig. 3B). RRC03806 was 53 bp upstream of the acyl-HSL synthase, and the two genes were in the same orientation. The ORFs encoding the other two LuxR-like proteins were unlinked.
C16-HSL induction of R. capsulatus gene transfer agent.
The R. capsulatus genome harbors a 15-kb cluster of genes that code for production of a phage-like particle known as the gene transfer agent (17, 18). Each gene transfer agent particle can deliver approximately 4.5 kb of randomly packaged genomic DNA to a recipient cell, where recombination may occur (40, 47). Because gene transfer agent production is maximal in stationary phase (18, 41), when cell densities are high, and because a gene transfer system in another bacterium has been reported to be controlled by quorum sensing (10, 49), we tested whether the gene transfer agent is regulated by C16-HSL.
First, we examined transcription of a gene transfer agent structural gene. Plasmid pYP (Table 1), which carries the gene transfer agent gene orfg2 fused to lacZ (18), was introduced into the wild-type and acyl-HSL synthase mutant R. capsulatus strains. Measurements of lacZ-encoded β-galactosidase activity indicated that orfg2 expression was reduced sevenfold in the acyl-HSL synthase mutant compared to the wild-type strain (Table 2). Addition of C16-HSL restored wild-type β-galactosidase activity to the mutant (Table 2). This sevenfold reduction in β-galactosidase activity was observed in early, mid-, and late logarithmic phase and in stationary phase (data not shown). Second, we assessed the number of gene transfer agent particles produced by the wild-type and acyl-HSL synthase mutant R. capsulatus strains. In agreement with the lacZ reporter data, gene transfer agent production was fivefold less in the acyl-HSL-deficient strain, and the number of transductants was increased by addition of C16-HSL (Table 2). These data indicate that gene transfer agent production is controlled by C16-HSL.
TABLE 2.
Effect of hexadecanoyl-HSL on gene transfer agent transcription and particle production
| Strain | β-Galactosidase activity (U) from pYPa | No. of gene transfer agent transductantsb |
|---|---|---|
| Wild type | 69.2 | 28.0 |
| ALS1 | 9.9 | 5.3 |
| ALS1 plus 2 μM C16-HSL | 61.2 | 37.3 |
Average of two samples.
Average of three samples.
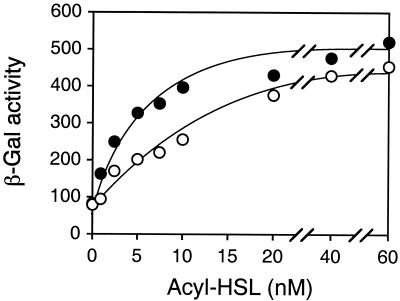
Both C16-HSL and C14-HSL activated gene transfer agent gene expression in a concentration-dependent manner (Fig. 5). The half-maximal response occurred at 2.5 to 5 nM C16-HSL and 7.5 to 10 nM C14-HSL. This is consistent with information about other acyl-HSL systems, in which responses generally require nanomolar concentrations of signals (14, 27, 49). C16-HSL appeared to be the preferred signal for gene transfer agent gene expression (Fig. 5). It is clear from the available R. capsulatus genomic sequencing data (assembled into nine contigs) that the acyl-HSL synthase gene and the gta genes are not tightly linked; the minimum distance between them is 570 kbp (www.integratedgenomics.com).
FIG. 5.
Dose-response curves for activity of C16-HSL (•) and C14-HSL (○), measured as β-galactosidase (β-gal) activity in R. capsulatus ALS1(pYP).
Concentrations and locations of long-chain acyl-HSLs in R. capsulatus cultures.
One limitation of the 14C-labeled acyl-HSL assay is that it does not provide information on how much of an acyl-HSL accumulates during culture growth (34). Thus, we needed to perform bioassays to determine the absolute level of C16-HSL. Our discovery that a gene transfer agent structural gene-lacZ fusion is expressed in response to C16-HSL (Fig. 5) provided us with a bioassay tool (see Materials and Methods). Cell-free late-logarithmic-phase R. capsulatus and P. denitrificans culture fluids contained approximately 325 to 390 nM and 350 to 400 nM C16-HSL, respectively. Unlike the case with V. fischeri, for example, where almost all of the 3OC6-HSL is found in the culture supernatant fluid (14), a significant fraction of C14 and C16-HSL remained with the cells.
Ethyl acetate extracts of P. denitrificans cell pellets contained 48 to 50% of the total 14C-labeled C16-HSL. Extracts of R. capsulatus cells contained 33 to 34% and 49 to 52% of the total 14C-labeled C16-HSL and putative 14C-labeled C14-HSL signals, respectively. For comparison, we measured the fraction of 3OC6-HSL in V. fischeri cells. With V. fischeri, 2 to 4% of the total 14C-labeled 3OC6-HSL remained with cells, consistent with a previous report (14). These data suggest that the long-chain acyl-HSLs produced by P. denitrificans and R. capsulatus are concentrated in cells. Presumably, they partition with the membranes, where they may be unavailable as signals.
DISCUSSION
We have used a radiotracer technique rather than bioassays to show that R. capsulatus and P. denitrificans synthesize the long-chain C16-HSL (both organisms) and material that coelutes with C14-HSL in HPLC (R. capsulatus). This extends the range of known acyl-HSL signals. Other bacteria also produce relatively long-chain acyl-HSLs, for example, R. sphaeroides, which produces 7,8-cis-3-tetradecenoyl-HSL (31). It is interesting that, as a rule, long-chain acyl-HSLs are produced by alpha-proteobacteria. The hydrophobic nature of the R. capsulatus and P. denitrificans long-chain acyl-HSLs may explain why about half of the total acyl-HSLs in cultures were associated with cells. Because the cells constitute only a small percentage of the total culture volume, the cellular concentrations of these acyl-HSLs were estimated to be quite high, on the order of 50 times higher than the extracellular concentrations.
We assume that the bulk of the cellular acyl-HSLs was associated with the membranes. The implications of this are that although there would be significant levels of acyl-HSLs in membranes, the cytoplasmic concentrations might be similar to the environmental concentrations. Thus, the acyl-HSLs could still serve a signaling function. Short-chain acyl-HSLs, like 3OC6-HSL and butanoyl HSL, are freely diffusible and not concentrated in cells (14, 26). Longer-chain signals like 3OC12-HSL in P. aeruginosa are concentrated in cells, and an efflux pump has been shown to aid 3OC12-HSL export (26).
We studied acyl-HSL production by R. capsulatus further because of the availability of the genome sequence for this organism. We identified a single ORF that showed significant sequence similarity with LuxI family members, and we showed that this ORF directed the synthesis of acyl-HSLs. Insertion of an Spr cassette in this ORF generated a strain that did not produce acyl-HSLs. Introduction of a wild-type copy of this gene into the R. capsulatus ALS1 mutant or into E. coli resulted in acyl-HSL synthesis. In the complemented R. capsulatus mutant, the acyl-HSLs produced were identical to those produced by the wild-type strain, but E. coli did not produce C16-HSL. To our knowledge, this is the first example of an I gene-containing recombinant E. coli that does not produce the native acyl-HSL. Adjacent to this acyl-HSL synthase gene is a gene coding for a LuxR homolog. However, there are other LuxR homologs in the R. capsulatus genome database, and we do not know if the adjacent gene is a transcription factor that responds to C16-HSL.
Although we have not identified the receptor experimentally, we have shown that C16-HSL can serve as an environmental signal for activation of gene transfer agent structural genes in R. capsulatus. This demonstrates that even though C16-HSL is concentrated in cells, it is a signal for gene expression when added externally. Of course, it is possible that long-chain acyl-HSLs play an additional role in R. capsulatus and P. denitrificans membranes.
The R. capsulatus long-chain acyl-HSL induced gene transfer agent gene expression five- to sevenfold, whereas expression of gene transfer agent and flagellar genes was reduced 10-fold or more in cckA and ctrA mutants (16). It will be interesting to evaluate the possibility of connections between the cckA/ctrA system and the acyl-HSL-dependent regulation described here. Perhaps there are independent but overlapping signals that cooperatively induce maximal production of gene transfer agent.
ADDENDUM IN PROOF
After acceptance of our paper, Marketon et al. (M. M. Marketon, M. R. Gronquist, A. Eberhard, and J. E. Gonzalez, J. Bacteriol. 184:5686-5695, 2002) showed with radiotracers that Sinorhizobium meliloti produces long-chain acyl-HSLs (14- to 18-carbon acyl groups).
Acknowledgments
We thank R. Haselkorn for sharing R. capsulatus genomic sequence data and A. S. Lang for comments.
This work was supported by grants from the W. M. Keck Foundation, the National Institutes of Health (GM59026), the National Science Foundation (MCB 9808308), and the Natural Sciences and Engineering Research Council (Canada). A.L.S. was supported by U.S. Public Health Service Training Grant T32-AI07343.
REFERENCES
- 1.Baker, S. C., S. J. Ferguson, B. Ludwig, M. D. Page, O. M. Richter, and R. J. V. Spanning. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62:1046-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty, J. T., and H. Gest. 1981. Generation of succinyl-coenzyme A in photosynthetic bacteria. Arch. Microbiol. 129:335-340. [Google Scholar]
- 3.Blosser-Middleton, R. D., and K. M. Gray. 2001. Multiple N-acyl homoserine lactone signals of Rhizobium leguminosarum are synthesized in a distinct temporal pattern. J. Bacteriol. 183:6771-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen-Bazire, G., W. R. Sistrom, and R. Y. Stanier. 1956. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Comp. Phys. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 5.Dekhane, M., K. T. Douglas, and P. Gilbert. 1996. A novel convenient route to the naturally occurring 3-oxoacyl-l-homoserine lactones and related bacterial autoinducers. Tetrahedron Lett. 37:1883-1884. [Google Scholar]
- 6.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 7.Eberhard, A., C. A. Widrig, P. McBath, and J. B. Schineller. 1986. Analogs of the autoinducers of bioluminescence in Vibrio fischeri. Arch. Microbiol. 146:35-40. [DOI] [PubMed] [Google Scholar]
- 8.Engebrecht, J., K. H. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of the functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Gen. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, W. C., and S. C. Winans. 1993. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 16.Lang, A., and J. Beatty. 2002. Bacterial signal transduction system simultaneously control genetic exchange and motility. J. Bacteriol. 184:913-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang, A. S., and J. T. Beatty. 2001. The gene transfer agent of Rhodobacter capsulatus and “constitutive transduction” in prokaryotes. Arch. Microbiol. 175:241-249. [DOI] [PubMed] [Google Scholar]
- 18.Lang, A. S., and J. T. Beatty. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 97:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrs, B. L. 1974. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 71:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEwan, A. G. 1994. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur phototrophic bacteria. Antonie Van Leeuwenhooek 66:151-164. [DOI] [PubMed] [Google Scholar]
- 21.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescence system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsek, M. R., A. L. Schaefer, and E. P. Greenberg. 1997. Analysis of random and site-directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 26:301-310. [DOI] [PubMed] [Google Scholar]
- 25.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 26.Pearson, J. P., C. V. Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 29.Piper, K. R., S. Beck v. Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature (London) 362:21670-21676. [DOI] [PubMed] [Google Scholar]
- 30.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 31.Puskas, A., E. P. Greenberg, S. Kaplan, and A. L. Schaefer. 1997. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 179:7530-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruby, E. G., and K. H. Nealson. 1976. Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica L, a model of symbiosis based on bacterial studies. Biol. Bull. 151:574-596. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 34.Schaefer, A. L., E. P. Greenberg, and M. R. Parsek. 2001. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Methods Enzymol. 336:41-47. [DOI] [PubMed] [Google Scholar]
- 35.Schaefer, A. L., B. L. Hanzelka, A. Eberhard, and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 178:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer, A. L., B. L. Hanzelka, M. R. Parsek, and E. P. Greenberg. 2000. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 305:288-301. [DOI] [PubMed] [Google Scholar]
- 37.Shadel, G. S., R. Young, and T. O. Baldwin. 1990. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J. Bacteriol. 172:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 39.Slock, J., D. VanRiet, D. Kolibachuk, and E. P. Greenberg. 1990. Critical regions of the Vibrio fischeri LuxR protein defined by mutational analysis. J. Bacteriol. 172:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solioz, M., and B. Marrs. 1977. The gene transfer agent of Rhodopseudomonas capsulata: purification and characterization of its nucleic acid. Arch. Biochem. Biophys. 181:300-307. [DOI] [PubMed] [Google Scholar]
- 41.Solioz, M., H. C. Yen, and B. Marrs. 1975. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 123:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens, A. M., and E. P. Greenberg. 1999. Transcriptional activation by LuxR, p. 231-242. In G. Dunny and S. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wall, J. D., P. F. Weaver, and H. Gest. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch. Microbiol. 105:217-224. [DOI] [PubMed] [Google Scholar]
- 45.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
- 48.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulatus. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, L., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-homoserine lactones. Nature (London) 362:446-448. [DOI] [PubMed] [Google Scholar]