Abstract
Mechanisms involved in maintaining cytoplasmic metal ion homeostasis play a central role in the adaptation of Helicobacter pylori to the changing gastric environment. An investigation of the global regulatory responses to copper ions by using RNA profiling with a threshold factor of 4.0 revealed that copper induces transcription of 19 H. pylori genes and that only the ferritin gene pfr is repressed. The 57-fold copper induction identified the HP1326 gene encoding an H. pylori-specific protein as a candidate for a novel copper resistance determinant. The HP1326 gene is expressed as a monocistronic unit, and two small HP1326 mRNAs are copper induced. The HP1326 protein is secreted and is required for copper resistance maintained by cytoplasmic copper homeostasis, as H. pylori HP1326 mutants were copper sensitive and displayed increased copper induction of HP1326 transcription as well as elevated copper repression of ferritin synthesis. The clear copper-sensitive phenotype displayed by H. pylori HP1327 and HP1328 mutants provides strong evidence that the HP1326 protein, together with the signal peptide site of the H. pylori-specific protein HP1327, whose gene is located downstream from that encoding HP1326, and the CzcB and CzcA metal efflux system component homologs HP1328 and HP1329, constitutes a novel type of copper efflux pump, as discussed below. The HP1329 gene could not be inactivated, but the 14-fold transcriptional copper induction determined by RNA profiling points towards a function of the encoded CzcA homolog in copper resistance. In summary, results from RNA profiling identified the novel H. pylori-specific copper resistance determinants CrdA (HP1326) and CrdB (HP1327), which are required for adaptation to copper-rich environmental conditions.
The bacterial pathogen Helicobacter pylori (14) colonizes the human gastric mucosa. Once H. pylori infects a host, the organism remains for the remainder of life and causes various disorders of the upper gastrointestinal tract, including gastric adenocarcinoma and lymphoma (3). The persistence in the hostile gastric niche necessitates molecular mechanisms which enable H. pylori to effectively adapt to changes in the environmental ionic composition (52). Recent investigations have shown that H. pylori metal ion homeostasis is of extraordinary importance for gastric adaptation (52), as proteins involved in iron uptake (55) and iron storage (7, 56), as well as the iron-dependent superoxide dismutase (45), are required for the establishment of the H. pylori infection in animal models. Furthermore, CorA-mediated magnesium ion uptake was recently shown to be essential for H. pylori viability in vitro (39), and nickel homeostasis is required for maintaining correct regulation (53, 54) and activity of the central H. pylori pathogenicity factor urease (27, 52), which is required for colonization of the gastric niche. Besides magnesium, iron, and nickel, copper plays a substantial role in metabolism, as it is a cofactor for electron transport, oxidases, and hydroxylases (26, 28). Under physiological conditions, copper ions participate in electron transfer reactions and their importance for H. pylori is underlined by the presence of copper in the cb-type cytochrome oxidase, the terminal oxidase in the respiratory chain (33, 50). On the other hand, the copper-mediated generation of toxic hydroxyl radicals, which is analogous to the iron-catalyzed Fenton reaction (21, 29), necessitates mechanisms that keep the concentration of free copper ions in the cytoplasm below toxic levels (36). H. pylori controls the cytoplasmic copper concentration by efflux via the P-type ATPase CopA (6, 18, 19) and responds actively to changes in the external copper concentration, as significant changes in gene expression have been reported for the ferritin gene pfr (encoding the major iron storage protein of H. pylori), which is repressed by the presence of copper (7). Pfr synthesis is also repressed by nickel and by zinc, and this response is thought to secure the availability of free iron ions in the cytoplasm when other metals are present at increasing concentrations (7). The absence of metal repression of Pfr synthesis in an H. pylori mutant lacking the ferric uptake regulator Fur suggests that the Fur protein is substantially involved in copper repression of Pfr synthesis (7). At the present time, other proteins involved in H. pylori copper metabolism have not been investigated in detail. The second H. pylori P-type ATPase, CadA (23), exporting cobalt, cadmium, and zinc, and the nickel-storage protein Hpn (31) do not contribute to copper resistance, as shown by mutational analysis. The annotation of the H. pylori genome sequence (2, 48) revealed the presence of additional open reading frames (ORFs) encoding the P-type ATPase homolog HP1503 and two CzcA homologs (designated HP0969 and HP1329) which are possibly involved in metal efflux. In Ralstonia sp. (formerly Alcaligenes eutrophus), the Czc metal efflux system is composed of the inner membrane, periplasmic, and outer membrane proteins CzcA, CzcB, and CzcC, respectively, which mediate heavy metal resistance via proton-driven export of cobalt, zinc, and cadmium (35, 36, 41). A CzcC homolog is absent in the H. pylori genome, whereas genes for two CzcB homologs (annotated as HP0970 and HP1328) are located directly upstream of the corresponding CzcA homologs (48). Copper efflux by Czc systems has been reported for the Cus (YbdE) copper-silver resistance determinant of Escherichia coli (17, 38), which is encoded in an copper-induced operon together with the neighboring cusB, ylcC, and cusC genes (Fig. 1). Copper induction of cus transcription, starting from a promoter upstream of cusC, is mediated by the CusR/CusS two-component regulatory system (32). In H. pylori, the multiplicity and the genetic organization of metal efflux systems in distinct genomic loci underline the importance of cytoplasmic ion homeostasis in the gastric environment and suggest that H. pylori is able to exactly modulate its cytoplasmic metal ion content in response to a wide variety of environmental conditions (52). In the present study, we used whole-genome analysis for identification of copper-regulated H. pylori genes by using RNA profiling. The ORF HP1326, encoding a methionine-rich 13.8-kDa H. pylori-specific protein, was found to be strongly copper induced at the transcriptional level. The HP1326 mutant displayed increased copper sensitivity, elevated copper induction of HP1326 transcription, and copper repression of Pfr synthesis, indicating that HP1326 is substantially involved in maintaining cytoplasmic copper homeostasis. Similar phenotypes were observed when the neighboring genes HP1327 and HP1328 (coding for an H. pylori-specific protein and a CzcB homolog, respectively) were inactivated. Together with the protein's molecular properties, the copper regulation of HP1326 and HP1329 expression, the phenotypes displayed by H. pylori HP1326, -27, and -28 mutants support the model that HP1326 and HP1327, together with the CzcB and CzcA homologs, constitute a novel type of copper efflux pump. According to their role as copper resistance determinants, we propose the designation of the H. pylori HP1326 and HP1327 genes as crdA and crdB, respectively.
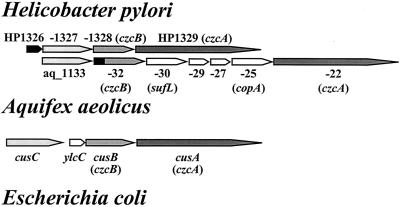
FIG. 1.
Comparative view of orthologs of HP1326 to HP1329 in H. pylori, A. aeolicus, and E. coli. Genes represented by arrows indicating the transcriptional direction are numbered according to the annotated genome sequences of H. pylori strain 26695 (48), A. aeolicus (12), and E. coli (17, 38). Clear homologies of H. pylori HP1328 and HP1329, A. aeolicus aq_1122 and aq_1132, and E. coli cusB and cusA to the Ralstonia genes czcB and czcA, respectively, are indicated. The orthologous gene pairs HP1327/aq_1133/cusC, HP1328/aq_1132/cusB, and HP1329/aq_1122/cusA are indicated with boxes shaded light gray, medium gray, and dark gray, respectively. The HP1326 gene and the homologous DNA region in the A. aeolicus aq_1132 gene are shown with black boxes. A. aeolicus and E. coli genes (shown with white boxes) share no homology with the H. pylori gene cluster HP1326 to -29. They encode A. aeolicus orthologs of the periplasmic cell division protein SufL, the cation-exporting P-type ATPase CopA, two putative proteins of unknown function, and the E. coli YlcC protein, as indicated. The sequences and annotations were obtained from the TIGR microbial database (http://www.tigr.org/tdb/mdb/mdbcomplete.html).
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The bacterial strains used in this study are listed in Table 1. H. pylori was routinely cultivated on Dent blood agar in a microaerobic atmosphere, as described previously (53). Growth inhibition experiments with copper were performed in brucella broth with 5% fetal calf serum (BBF). The total copper content of BBF medium is 0.5 μM, as determined previously by atomic absorption mass spectrometry (7). Metal-enriched conditions were established by supplementation of BBF with copper chloride (catalog no. C6641; Sigma). For growth inhibition experiments with copper ions, H. pylori wild-type (wt) and mutant strains were precultured in BBF medium to an optical density at 600 nm (OD600) of 1.0 and diluted 1:100 in test medium supplemented with copper or sodium chloride. Subsequently, the influence of metals on bacterial growth was determined after growth for 48 h by photometrical determination of the OD600 values of the cultures. The growth inhibition experiments were performed in triplicate and were repeated at least three times. Control cultures were supplemented with sodium chloride at the highest metal concentrations to exclude the influence of osmotic stress and chloride ions on copper regulation or resistance. E. coli was grown in Luria-Bertani medium. When appropriate, growth media were supplemented with 20 mg of kanamycin (Km) or 20 mg of chloramphenicol (Cm)/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source and/or reference(s) |
|---|---|---|
| Strains | ||
| H. pylori | ||
| 1061 | wt but lacking the entire cag PAI | 13, 53, 54 |
| 26695 | wt but containing the entire cag PAI | 48 |
| 26695-1326 | 26695 ΔHP1326::cat Cmr | This study |
| 26695-1327 | 26695 HP1327::Pcat Cmr | This study |
| 26695-1328 | 26695 HP1328::Pcat Cmr | This study |
| NCTC11638 | wt but containing the entire cag PAI | NCTC,b7 |
| P1 | wt | 37 |
| E. coli | ||
| MC4100 | araD139 Δ(argF-lac)169 flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 | 10 |
| Plasmids | ||
| pZERO-2 | Cloning vector; MCS in lacZ′ neo Kmr | Invitrogen |
| p1326-CAT | pZERO-2 ΔHP1326::cat Cmr Kmr | This study |
| p1327-PCAT | pZERO-2 HP1327::Pcat Cmr Kmr | This study |
| p1328-PCAT | pZERO-2 HP1328::Pcat Cmr Kmr | This study |
| p1329-PCAT | pZERO-2 HP1329::Pcat Cmr Kmr | This study |
cat, cat gene without its own promoter; Pcat, cat gene with its own promoter; MCS, multiple cloning site; PAI, pathogenicity island.
NCTC, National Collection of Type Cultures.
DNA techniques and mutagenesis of H. pylori.
Restriction and modifying enzymes (Roche Diagnostics, Mannheim, Germany) were used according to the manufacturer's instructions. DNA cloning was performed with E. coli according to standard protocols (4). Plasmids were isolated by using a kit from Qiagen. Sequences of the Cm-acetyl-transferase gene cat with (Pcat) and without (cat) its own promoter were amplified by PCR with primers CATS1 and CATS2, respectively, in combination with the primer CATAS1. Pcat or cat genes were fused to upstream and downstream DNA regions of mutagenized genes (Fig. 2A) by using a modified version of the megaprimer PCR protocol (43), as described previously (39). Briefly, DNA regions flanking cat or Pcat were amplified by PCR from DNA of H. pylori strain 26695 by using primers carrying 5′ extensions complementary to the 5′ and 3′ ends of the Pcat and cat cassettes, respectively (Table 2). The resulting PCR products were purified with a kit from Qiagen and subsequently mixed with PCR-amplified Pcat or cat cassettes for use as megaprimers in a second PCR using only the flanking primers. The resulting PCR products, carrying cat or Pcat inserted into the HP1326, HP1327, HP1328, or HP1329 gene, were cloned into plasmid pZERO-2 (Invitrogen). The resulting plasmids, pHP1326-CAT, pHP1327-PCAT, pHP1328-PCAT, and pHP1329-PCAT (Table 1), were used for mutagenesis of the corresponding genes in the H. pylori chromosome (Fig. 2A). Correct construction of the plasmids was confirmed by sequencing or by restriction analysis with appropriate enzymes. Marker exchange mutagenesis of H. pylori was performed by using electroporation according to standard procedures (20). H. pylori mutants, carrying the cat gene inserted into the chromosome, were selected by growth on Dent blood agar containing Cm at concentrations of 20 mg/liter. Correct insertion of cat into the HP1326, -27, and -28 genes (Fig. 2A) was verified by PCR analysis with the appropriate primers (Table 2 and data not shown).
FIG. 2.
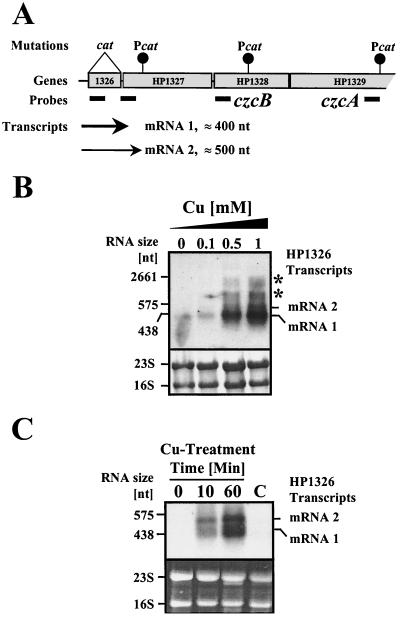
Transcriptional analysis of the HP1326 gene in H. pylori strain 26695. (A) Schematic overview on transcriptional and mutational analysis of the gene cluster HP1326 to -29. Genes numbered according to the annotated genome sequence of H. pylori strain 26695 (48) are indicated by grey boxes. The insertion sites of cat resistance cassettes are marked by black circles. The positions of antisense RNA probes are marked by black bars. The transcripts detected by Northern blot analysis are indicated by arrows. The thickness of the arrows reflects the abundance of the transcripts upon exposure to copper. (B) Influence of copper on transcription of the HP1326 gene. The upper panel shows total RNA (15 μg) isolated from H. pylori strain 26695 grown in BBF (lane 0) and in BBF supplemented with copper at the micromolar concentrations indicated on the top of each lane (0.1 to 1) and hybridized with the DIG-labeled HP1326-specific antisense RNA probe (panel A). The lower panel shows methylene blue-stained RNA samples on the nylon membrane prior to hybridization. Transcripts are marked on the right (mRNAs 1 and 2). Transcripts which accumulated in the 16S and 23S bands are marked by asterisks. The sizes of marker RNAs are indicated on the left. (C) Time kinetics of copper-mediated HP1326 mRNA accumulation. Exponentially growing cells of H. pylori strain 26695 were treated with 1 mM copper chloride for 10 and 60 min, as indicated at the top of the lanes. Samples taken prior to copper treatment (lane 0) and after 60 min without copper treatment (lane C) served as controls. Total RNA samples were hybridized with DIG-labeled HP1326 antisense mRNA (upper panel). The lower panel shows ethidium bromide-stained RNA samples in the agarose gel prior to transfer and hybridization. The HP1326 transcripts which accumulated in presence of copper are indicated on the right (mRNAs 1 and 2). Sizes of marker RNAs are indicated on the left.
TABLE 2.
Oligonucleotides used in this study
| Genea | Application | Primer or probe | Sequence (5′→3′)b |
|---|---|---|---|
| HP1326 | Mutagenesis, upstream | FUMC-L1 | CCCGCCTATGGCTAATTCTC |
| CAT1326-R1 | 1-TTACATTAAGATAACAAGAA | ||
| Mutagenesis, downstream | CAT1326-L1 | 3-GATTTATAAGAGCATGCTA | |
| 1327-R1 | TTTCTTCATCAAGCCTGGCG | ||
| Antisense RNA probe | 1326-L1 | GGGTTATGGGTTTAAACGCA | |
| 1326-R1T7 | T7-TTATAAATCCAGGCTTGTTT | ||
| HP1327 | Mutagenesis, upstream | 1327-L1 | TTATAAGCGCGTTTGATAAA |
| PCAT1327-R1 | 2-GCGCTTCTTCTAAATTGTTT | ||
| Mutagenesis, downstream | CAT1327-L1 | TCTATGGGTGAATTGGCTTT | |
| 1327-R1 | TTGTAAAGACAAATAAGTGC | ||
| Antisense RNA probe | 1327-L1 | TTATAAGCGCGTTTGATAAA | |
| 1327-R1T7 | T7-GCGCTTCTTCTAAATTGTTT | ||
| HP1328 | Mutagenesis, upstream | 1328-L1 | TACAACGCTTATTACAACGC |
| PCAT1328-R1 | 2-AATAATGCCGTTGAAGTGAG | ||
| Mutagenesis, downstream | CAT1328-L1 | 3-CAAATCATAGATTTAAGCC | |
| 1328-R1 | CTTAAATCCGGCAAAGCGTC | ||
| HP1329 | Mutagenesis, upstream | I329-L1 | ACCAGCGTGTATGACAGGAG |
| PCAT1329-R1 | 2-CAAGCTCACGCCATAAATTT | ||
| Mutagenesis, downstream | CAT1329-L1 | 3-TTAATAGCAAGCGTCCTGGG | |
| 1329-R1 | ATTTAATCGCATCTAACACG | ||
| Antisense RNA probe | 1329-L1 | ACCAGCGTGTATGACAGGAG | |
| 1329-R1T7 | T7-CAAGCTCACGCCATAAATTT | ||
| cat | cat gene without promoter | CATS2 | TCCGAGATTTTCAGGAG |
| CATAS1 | TTACGCCCCGCCCTGCCA | ||
| Pcat | cat gene with promoter | CATS1 | TCCGGTTTTTGTTAATCCGCC |
| CATAS1 | TTACGCCCCGCCCTGCCA |
Gene numbers refer to the H. pylori 26695 genome sequence (48).
The 5′ extensions used for fusion of PCR products to the cat gene by megaprimer PCR are labeled as follows: 1 (5′-CTCCTGAAAATCTCGGA), complementary to the 5′ region of the cat gene without promoter; 2 (5′-GGCGGATTAACAAAAACCGGA), complementary to the 5′ region of the cat gene with promoter; 3 (5′-TGGCAGGGCGGGGCGTAA), complementary to the 3′ end of the cat gene; and T7 (5′-CTAATACGACTCACTATAGGGAGA), adds a T7 promoter sequence for creation of DIG-labeled antisense RNA.
Transcriptome analysis using DNA array hybridization.
For RNA profiling analysis, H. pylori organisms were grown in brain heart infusion (BHI) medium (supplemented with 10% fetal calf serum at 37°C) with moderate shaking (120 rpm). Anaerocult C kits (Merck) were used to generate microaerobic conditions. In the main experiment, 24-h-old precultures of H. pylori strain P1 (Table 1) were diluted to an OD600 of 0.1 in brain heart infusion medium containing 500 μM copper chloride and were further incubated for 3 h. Control cultures were treated identically except for the addition of copper chloride. Cells were collected by centrifugation and lysed using peqGold RNAPure (PeqLab). Total RNA was isolated by using RNeasy Mini kits (Qiagen), followed by DNase I (Roche) treatment for 60 min at 37°C and an additional purification step with RNeasy Mini kits. The array preparation and hybridization data are to be published elsewhere (I. Ivanov et al., unpublished data). Briefly, ORFs were prepared from 10 ng of genomic DNA by PCR amplification in two rounds of amplification using 30 cycles (94°C for 10 s and 65°C for 5 min). The first round of amplification was performed directly from genomic DNA, whereas in the second round, DNA was reamplified from the product of the first round (diluted 10,000-fold, according to a standard procedure [42]). After amplification, the PCR products were directly repipetted to the polystyrene 384-well microtiter plates (Genetix, New Milton, Hampshire, United Kingdom) and further used for spotting, as described previously (15). All H. pylori ORFs were spotted in duplicates on an array area of 7 by 10 cm. Total RNA, which was isolated from exponentially growing H. pylori cells with and without treatment for 3 h with 1 mM copper chloride according to standard protocols (4), was labeled by using a random primer protocol in the presence of 50 μCi of [33P]dCTP (corresponding to approximately 0.5 μM). The labeled probes were purified by gel filtration, followed by denaturation in 0.5 M NaOH at 65°C for 20 min and neutralization with 1 M Na-phosphate buffer at pH 7.5. For each hybridization, three arrays derived from different batches of production were used. The arrays were prehybridized for at least 2 h and then hybridized for 20 h at 50°C in 10 ml of buffer containing 50% formamide, 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 7% sodium dodecyl sulfate (SDS), and 50 mM Na-phosphate (pH 7.5) and washed twice in 2× SSC-0.5% SDS and 0.1× SSC-0.5% SDS buffers. Subsequently, the arrays were exposed for 100 h to Fuji BAS-IP MS 2025 plates and scanned at 25-μm resolution on a Fuji BAS-5000 Phosphorimager (Raytest, Straubenhardt, Germany). The acquired images were then analyzed by using a BioChipExplorer image analysis package from GPC Biotech (Munich, Germany), generating expression values for each gene. After normalization and scaling, the data were analyzed with an Expressionist software package (GeneData, Basel, Switzerland). The RNA profiling experiments were repeated a minimum of three times.
Northern blot hybridization.
Isolation of total RNA and detection of mRNA by hybridization with digoxigenin (DIG)-labeled antisense RNA probes were performed according to a standard protocol (4), as described previously (7, 16, 53). PCR products, carrying parts of the genes HP1326, HP1327, and HP1329 (Fig. 1 and 2A), were amplified with primers listed in Table 2 and were used for the production of antisense RNA probes labeled with DIG by in vitro transcription using T7 RNA polymerase (Roche). Northern hybridization and stringency washes were performed at 68°C, and the bound probe was visualized with the DIG Detection kit (Roche) by using the chemiluminescent substrate CPD-Star from Amersham Pharmacia. The sizes of the HP1326 mRNAs were determined by using the digoxigenylated RNA length standards from Roche (set 1; catalog no. 1526529).
Protein analysis.
H. pylori cultures grown in broth to an OD600 of 1.0 to 1.2 were harvested by centrifugation for 10 min at 4,000 × g at 4°C. Determination of protein concentrations, SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed as described previously (7). The H. pylori ferritin Pfr was detected with the specific antiserum AK198 (Max-von-Pettenkofer-Institute, LMU Munich, Germany). Bound rabbit antibodies were detected with a protein A-alkaline-phosphatase conjugate, followed by incubation with nitroblue tetrazolium as the substrate. The Cat protein was quantitated by enzyme-linked immunosorbent assay (ELISA) using a commercial system from Roche Diagnostics (CAT ELISA; catalog no. 1 363 727) according to the manufacturer's instructions as described previously (7). The amount of Cat protein was calculated from a standard curve prepared with purified E. coli Cat protein and normalized to the total protein content.
Bioinformatic analysis.
Genome sequence database searches were performed by using National Center for Biotechnology Information (NCBI) BLAST software (www.ncbi.nlm.nih.gov/BLAST/). Operon analysis was done with a Phylosopher program (Genedata, Basel, Switzerland; www.genedata.com). The current version (version 4) contains 67 completely sequenced bacterial genomes, as published at GenBank (NCBI; see above). The Phylosopher program allows identification of bacterial protein families by using a proprietary cluster algorithm. Putative transcription terminators behind the HP1326 gene were identified by using the Terminator program in a GCG software package (8), a computer algorithm for testing potential prokaryotic transcriptional terminator structures. For this identification procedure, the corresponding genomic nucleotide sequence, spanning ORFs HP1326 to HP1329, was extracted from the H. pylori 26695 genome (nucleotides [nt] 1385783 to 1392554) and analyzed using the default parameters. Database analysis and protein sequence alignments were performed by using the BLAST software of the NCBI server (http://www.ncbi.nlm.nih.gov/[last accessed August 2002]). Protein signal sequences and functional domain motifs were identified by using SMART (Simple Modular Architecture Research Tool; 44) as provided by the European Molecular Biology Laboratory (http://smart.embl-heidelberg.de/[last accessed August 2002]).
RESULTS
Identification of H. pylori genes differentially regulated upon copper treatment.
To identify new H. pylori genes involved in copper resistance or homeostasis, we performed a global survey of copper-regulated genes, with cDNA populations generated from total RNA of H. pylori with and without copper treatment, by using transcriptome analysis and DNA array hybridization. Computer-assisted analysis of the differentially expressed genes revealed that 20 H. pylori ORFs respond to copper, as indicated by results with induction and repression values over the significance threshold of 4.0 (Table 3). While 19 ORFs were induced, only the ferritin gene pfr (HP0653) was repressed. The ORFs HP1326 and HP1428, encoding an H. pylori-specific protein and a conserved hypothetical protein whose function is thus far unknown, respectively, showed the highest copper induction levels (approximately 57-fold) (Fig. 1; Table 3). ORF HP1329, encoding a CzcA metal efflux pump orthologue and located downstream of the HP1326 gene (Fig. 1), was also copper induced but at a lower level (14-fold; Table 3). The strong copper induction of the HP1326 and HP1329 genes, as well as their localization (Fig. 1) near the CzcB metal efflux system homolog HP1328 (48), provided strong evidence for a function in copper metabolism. This was further supported by results from database searches, which revealed that the C-terminal part (amino acids 125 to 444) of the HP1326 amino acid sequence shares 30% identity and 47% similarity with the N-terminal part of the Aquifex aeolicus (12) protein aq_1132 (TIGR locus name, NT01AA0901), which is orthologous to those of the H. pylori HP1328 protein and to other CzcB homologs (Fig. 1). ORF aq_1132 is part of an operon-like structure located downstream from the HP1327 homolog aq_1133, which contains genes for homologs of the putative copper exporter CopA (aq_1125) and CzcA (aq_1127) further downstream (Fig. 1). In this context, it seems interesting that the HP1327 protein shares some homologous regions with the E. coli CusC protein, which is encoded by a gene positioned upstream from those of the HP1328/CzcB homolog CusB and the E. coli copper-silver resistance determinant CusA (Fig. 1). These properties highlighted HP1326 and the neighboring genes as excellent candidates for a novel type of H. pylori copper resistance determinant. Thus, we studied these genes in more detail.
TABLE 3.
Expression values of copper-regulated H. pylori genes identified by RNA profilinga
| ORF | Description | Expression value
|
Factor | |
|---|---|---|---|---|
| −Cu | +Cu | |||
| HP1326 | H. pylori predicted coding region HP1326 | 529 | 30,250 | 57.2 |
| HP1428 | Conserved hypothetical protein | 90 | 5,044 | 55.9 |
| HP1329 | Cation efflux system protein (czcA) | 38 | 531 | 14.0 |
| HP1355 | Nicotinate-nucleotide pyrophosphorylase (nadC) | 187 | 1,812 | 9.7 |
| HP1446 | Biopolymer transport protein (exbD) | 308 | 2,346 | 7.6 |
| HP1340 | Biopolymer transport protein (exbD) | 61 | 379 | 6.3 |
| HP1158 | Pyrroline-5-carboxylate reductase (proC) | 72 | 447 | 6.2 |
| HP0994 | H. pylori predicted coding region HP0994 | 157 | 917 | 5.9 |
| HP0886 | Cysteinyl-tRNA synthetase (cysS) | 586 | 3,193 | 5.5 |
| HP0768 | Molybdenum cofactor biosynthesis protein A (moaA) | 13 | 69 | 5.2 |
| HP1516 | H. pylori predicted coding region HP1516 | 36 | 174 | 4.8 |
| HP1208 | Ulcer-associated adenine-specific DNA methyltransferase (hpyIM) | 55 | 263 | 4.8 |
| HP1533 | Conserved hypothetical protein | 165 | 765 | 4.6 |
| HP1349 | H. pylori predicted coding region HP1349 | 309 | 1,390 | 4.5 |
| HP0753 | Flagellar protein (fliS) | 11 | 48 | 4.5 |
| HP0733 | H. pylori predicted coding region HP0733 | 10 | 46 | 4.4 |
| HP1225 | Conserved hypothetical integral membrane protein | 34 | 146 | 4.3 |
| HP1534 | IS605 transposase (tnpB) | 114 | 486 | 4.3 |
| HP1277 | Tryptophan synthase, alpha subunit (trpA) | 53 | 221 | 4.2 |
| HP0653 | Nonheme iron containing ferritin (pfr) | 664 | 93 | −7.1 |
Genes with induction or repression factors of at least 4.0 are listed. Descriptions refer to the complete genome sequence of H. pylori strain 26695 (48). The data represent mean values from three hybridizations. Standard deviations were in the range of 10%.
Transcriptional analysis of the HP1326 gene region.
The expression values determined by RNA profiling provided strong evidence that the HP1326 gene is separately transcribed from the downstream genes HP1327, -28, and -29. The basal expression levels, as well as the copper induction values, of the HP1326 and HP1329 genes differ considerably (Table 3). The enclosed ORFs HP1327 and HP1328 displayed basal expression levels of 148 and 181 and were only threefold and twofold induced, respectively, upon copper treatment. Transcriptional analysis by Northern blot hybridization confirmed that the HP1326 gene is expressed as a monocistronic unit (Fig. 2). Hybridization of total RNA isolated from the H. pylori strain 26695 with an HP1326-specific antisense RNA probe (Fig. 2A) detected a single transcript (mRNA 1; Fig. 2B), which was associated with about 400 nt and was thus not long enough to contain other genes besides HP1326. The signal strength indicated that in the absence of copper, the HP1326 mRNA levels are near the lower detection limit (Fig. 2B and C). Supplementation of the growth medium with 0.5 and 1 mM copper increased the HP1326 mRNA levels drastically, as indicated by accumulation of the 400-nt transcript and by appearance of a second transcript, which was approximately 500 nt in size (mRNA 2; Fig. 2B). Lower copper concentrations (0.1 mM) did not influence the HP1326 transcript levels (Fig. 2B). A time kinetics of copper-induced HP1326 transcription revealed that the copper regulation occurs within minutes (Fig. 2C). The transcript levels showed a clear increase 10 min after copper treatment and reached the levels detected in cells grown with copper for 48 h (Fig. 2B) 50 min later (Fig. 2C). To investigate the possibility that longer transcripts occur as a result of copper-induced readthrough, the transcription of the HP1326 downstream genes was analyzed with antisense RNA probes specific for the ORFs HP1327 and HP1329 (Fig. 2A). The results obtained by hybridizations performed with a probe located at the beginning of the HP1327 gene (Fig. 2A; see also Fig. 5) were identical to those obtained with the HP1326-specific probe (data not shown), confirming that both HP1326 mRNA 1 and 2 (Fig. 2B and C) contain the N-terminal part of the HP1327 gene (see Fig. 5). Hybridization experiments performed with probes specific for HP1328 and HP1329 (Fig. 2A) did not detect any transcript, even when the bacteria were grown in the presence of 1 mM copper (data not shown). This demonstrated that copper does not induce longer HP1326 transcripts and that the HP1328 and HP1329 transcript levels are below the detection limit of the hybridization approach. This is in agreement with the expression values determined by RNA profiling (see above and Table 3), which show that even in the presence of copper, the levels of the HP1327, -28 (see above), and -29 (Table 3) mRNAs only reach levels comparable to that of basal HP1326 expression, which is near the lower detection limit for Northern blotting (Fig. 2B and C). However, the smears observed near the 16S and 23S rRNA bands (Fig. 2B) as the result of the presence of copper in the medium may have been caused by the accumulation of longer transcripts covering the HP1326, -27, and -28 genes, which may have been due to instability not detectable in separate bands. The analysis of HP1326 transcription in the H. pylori reference strains 1061 and NCTC11638 (Table 1) gave comparable results, demonstrating that the regulation by copper of HP1326 transcription is not strain specific (data not shown).
FIG. 5.
Localization of DNA sequence motifs in the HP1326 gene region. The binding site for primer 1327-L1 (Table 2), which was used to generate the HP1327-specific antisense RNA probe, is marked by a horizontal arrow. Sequence motifs with significant homologies to −35 and −10 RNA polymerase binding sites and to ribosome binding sites (RBS) are underlined. The two IVRs behind the HP1326 gene were predicted by using the Terminator program (8) in a GCG software package (IVR1 and IVR2). The signal peptide cleavage site of the downstream-encoded HP1327 protein, as predicted by using Signal P software, is marked by a vertical arrow.
Mutational analysis of the HP1326, -27, -28, and -29 genes and their role in copper homeostasis.
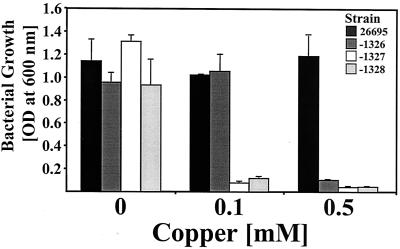
To study the functions of HP1326, -27, -28, and -29 in copper resistance, we tried to inactivate the corresponding genes in the chromosome of the H. pylori reference strain 26695 (Table 1; Fig. 2A) by marker exchange mutagenesis. To minimize possible polar effects on the downstream genes, the HP1326 gene was replaced by a cat resistance cassette with a ribosome binding site but lacking both promoter and terminator sequences (Fig. 2A). To secure expression of the downstream genes, HP1327 and HP1328 were inactivated by insertion of a cat gene with a promoter (Fig. 2A). Repeated transformation of the H. pylori wt strain 26695 with a plasmid, pHP1326-CAT, pHP1327-PCAT, pHP1328-PCAT, or pHP1329-PCAT (Table 1), carrying cat insertions in the corresponding genes (Fig. 2A), revealed that HP1326, -27, and -28, but not HP1329, could be inactivated. This provides evidence that an intact version of HP1329 is required for H. pylori viability in vitro, as even 10 repetitions of transformation of strain 26695 with plasmid pHP1329-PCAT (Table 1; Fig. 2A) did not result in mutants. Subsequently, the copper resistance functions of HP1326, -27, and -28 were investigated by growth inhibition experiments with the mutants and the wt strain in BBF medium supplemented with copper at increasing concentrations (Fig. 3). In unsupplemented BBF, growth of the strain 26695-1326, -1327, and -1328 mutants was comparable to that of the wt strain, indicating that the mutations do not generally limit bacterial fitness (Fig. 3). The growth inhibition caused by copper (Fig. 3) showed that all three mutants were clearly copper sensitive compared to the wt strain, indicating that all three genes are required for copper resistance (Fig. 3). Sodium chloride at the highest copper concentration had no influence on growth (data not shown), indicating that the metal sensitivity of the mutants was not caused by osmotic stress.
FIG. 3.
The role of the HP1326, HP1327, and HP1328 genes in H. pylori copper resistance. The growth of H. pylori strain 26695 (black) and of the isogenic mutants 26695-1326 (dark gray), -1327 (white), and -1328 (light gray) was determined by measuring the OD600. The medium was supplemented with copper at increasing concentrations, as indicated on the x axis. The data represent mean values from three independent determinations. Standard deviations are indicated.
Influence of the HP1326, -27, and -28 mutations on cytoplasmic copper homeostasis.
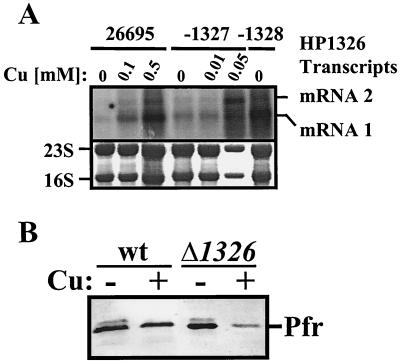
To test whether the copper sensitivity of HP1326, -27, and -28 mutants is caused by increased cytoplasmic copper levels resulting from a lack of copper efflux mediated by the corresponding proteins, we studied copper induction of HP1326 transcription in the mutants. The influence of the HP1326 mutation on copper induction of the HP1326 gene was analyzed by using the transcriptional HP1326::cat fusion in the 26695-1326 mutant (Fig. 2A; Table 1) as a reporter. Therefore, strains 26695 and 26695-1326 were grown in BBF medium without and with 0.1 mM copper and the Cat protein concentration in the cells was determined with a commercial ELISA. The Cat levels of 1.89 ± 0.87 and 51.7 ± 14 (in nanograms of Cat per microgram of total protein), as detected in cells grown without (n = 4) and with (n = 4), copper, respectively, provided strong evidence that the HP1326 mutation results in increased cytoplasmic copper availability, as a similar copper concentration did not increase HP1326 transcript levels in the H. pylori wt strain (Fig. 2B). The investigation of copper-induced HP1326 transcription by using Northern blot analysis with the HP1326-specific probe (Fig. 2A) in Hp1327 and HP1328 mutants gave similar results (Fig. 4A). The accumulation of HP1326 mRNAs in the HP1327 mutant grown with 0.05 mM copper and in the HP1328 mutant grown without additional copper (Fig. 4A) provides further evidence for increased internal copper levels. To support these findings, we used data resulting from the recent discovery of copper- and Fur-mediated repression of Pfr synthesis (7) to design biosensor experiments to detect the increased cytoplasmic copper availability in the HP1326 mutant. Because the Fur protein senses the concentration of metals in the cytoplasm, an increase in cytoplasmic copper availability should result in hyperrepression of Pfr synthesis in the presence of copper. Immunoblot analysis of Pfr in the wt strain and in the HP1326 mutant grown in the absence and presence of copper with a Pfr-specific antiserum (Fig. 4B) revealed that repression by copper of Pfr synthesis is indeed strongly increased in the HP1326 mutant, confirming that the HP1326 protein is required for maintaining cytoplasmic copper homeostasis.
FIG. 4.
Influence of the HP1326, HP1327, and HP1328 mutations on cytoplasmic copper homeostasis. (A) Copper-induced HP1326 transcription in HP1327 and HP1328 mutants. The upper panel shows total RNA (15 μg) isolated from the H. pylori wt strain 26695 and from the HP1327 (26695-1327) and HP1328 (26695-1328) mutants grown in BBF without copper (0) and in BBF supplemented with copper (Cu) at the micromolar concentrations indicated on the top of each lane and hybridized with the DIG-labeled HP1326-specific antisense RNA probe (Fig. 2A). The lower panel shows methylene blue-stained RNA samples on the nylon membrane prior to hybridization. Transcripts are marked on the right (mRNAs 1 and 2). (B) Influence of the HP1326 mutation on copper repression of Pfr synthesis. The H. pylori wt strain 26695 and the HP1326 mutant 26695-1326 (Δ1326::cat) were grown in BBF medium without (−) or with (+) 0.1 mM copper (Cu), and the ferritin Pfr (marked on the right) was detected by immunoblot analysis with the specific antiserum AK198 (7, 56).
DISCUSSION
In the gastric mucosa, metal ions released from food by proteolytic and acidic degradation can be complexed by the cation-chelating activity of gastric mucus components (40) and by binding to host proteins such as lactoferrin (34). Thus, metal starvation as well as metal-rich conditions may occur in relatively short time intervals. Given the average daily copper intake (in the range of 1 mg) (5), the copper content of drinking water (in the range of 10 to 50 μM), and the copper content of copper-rich foods like liver (up to 200 mg/kg of meat), the estimate that H. pylori is exposed to copper ions in the micromolar range is justified. Together with the important role of copper in maintaining basic metabolic functions, this suggests that H. pylori copper homeostasis plays a crucial role in gastric adaptation. In the present study, we employed genome-wide RNA profiling for the study of copper-mediated gene regulation and for the identification of the novel H. pylori copper resistance determinants CrdA (HP1326), CrdB (HP1327), and HP1328 (CzcB). This approach was validated by the clearly evident sevenfold repression of pfr transcription (Table 3), which is well in agreement with Fur-dependent copper repression of Pfr at the protein level (7). This response might secure cytoplasmic availability of other biometal cofactors in the presence of increased copper concentrations (see Fig. 6). The same might hold true for the copper induction of the H. pylori exbD and moaA genes, which are thought to be involved in iron uptake (52) and molybdenum cofactor synthesis (48), respectively. The copper induction of the H. pylori nadC, proC, cysS, and trpA genes (Table 3), encoding homologs of NAD and amino-acid metabolism enzymes, could be protective against copper-mediated enzyme inhibition (22, 49) and protein auto- oxidation (24, 46). The copper induction of the H. pylori fliS gene, which is required for the production of intact flagella (1), might enable H. pylori to escape from copper toxicity by alterations in motility. Taken together, these regulatory responses (see Fig. 6) indicate that for limitation of copper toxicity, H. pylori elicits a SOS-like stress response. This idea is further supported by the copper induction of genes for transposase TnpB and the DNA methylase HpyIIM (Table 3), which might help to overcome copper-mediated DNA damage. Other H. pylori metal export genes, namely, the P-type ATPases CadA (HP0791), CopA (HP1072), and CopA2 (HP1503), as well as the second CzcA homolog HP0969, did not appear to be significantly copper regulated (data not shown). The rapid copper induction of the two small monocistronic HP1326 mRNAs enables H. pylori to respond rapidly to increased copper concentrations. The small sizes of the HP1326 transcripts (Fig. 2A and B) and their cross-hybridization with the HP1327-specific probe (Fig. 2A and 5) allow the calculation that the transcriptional start site is located 50 bp in front of the HP1326 start codon. This fits with the position of a well-conserved consensus sequence for a RNA polymerase binding site (Fig. 5). Two inverted repeats (IVRs) behind the HP1326 stop codon might be involved in mediating transcriptional termination (Fig. 5). In view of the differential copper induction and the mRNA sizes, this genetic arrangement suggests that in the absence of copper, transcription terminates at IVR1, while copper exposure causes a partial readthrough to IVR2 (Fig. 5). The protein(s) involved in copper regulation remains to be identified in ongoing studies. Homologs of known copper regulator proteins such as E. coli CueR (47), Pseudomonas CopR/S (30), and Ralstonia CzcRS (51) are absent in the H. pylori genome (2, 48). The strong copper induction of HP1326, and its genetic organization with the czcB and czcA gene homologs, suggested that the whole HP1326 to -29 gene cluster might be involved in copper export. This idea was supported by the copper sensitivity displayed by HP1326, -27, and -28 mutants (Fig. 3) and by the copper induction of the czcA gene HP1329. However, the role of HP1329 in copper resistance could not be experimentally confirmed, because it was not possible to inactivate the gene. Copper export functions of HP1326, -27, and -28 were demonstrated indirectly by the accumulation of free copper ions in the corresponding mutants, as indicated by increased copper induction of HP1326 mRNA levels (Fig. 4A) and elevated copper repression of Pfr synthesis (Fig. 4B). In accordance with the Ralstonia CzcABC architecture (41), the homologies and molecular properties of the corresponding H. pylori proteins lead to the integrative model that HP1329 and HP1327 transport metal ions over the inner and outer membranes, respectively, while HP1328 and HP1326 act as periplasmic binding (and possibly storage) proteins (Fig. 6). In the changing gastric environment, periplasmic copper sequestration would enable H. pylori to store the essential copper ions under metal-rich conditions for subsequent access if environmental copper were to become scarce (Fig. 6). Analysis using SMART software (44) confirmed the presence of signal sequence cleavage sites in the deduced HP1326, -27, -28, and -29 protein sequences. Secretion of the HP1326 protein was experimentally confirmed by transposon TnBla mutagenesis in an earlier study (37). The corresponding clone pMu72 carries TnBla inserted behind the predicted HP1326 signal sequence (44, 48) at codon 25 (S. Odenbreit and R. Haas, personal communication). A methionine-rich motif (FMMPEMPGMPAMKEMA) in the HP1326 protein sequence could be involved in metal binding. Such motifs are also present in the A. aeolicus aq_1132 protein (Fig. 1), in the Pseudomonas periplasmic copper resistance protein CopC (QFSGAKLMMTAMPGMAAHSPMPMPAKVS) (11, 25), and in the E. coli Pco copper efflux proteins (9). The role of the HP1327 protein in copper export is further supported by the presence of an outer membrane efflux protein family domain motif, which is located in the C-terminal region (48).
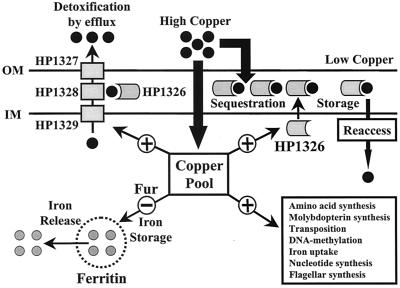
FIG. 6.
Schematic overview of H. pylori responses to increased copper concentrations. Copper influx is indicated by the arrows. Copper is detoxified by efflux via CzcAB and by periplasmic sequestration via the HP1326 protein. A possible role of HP1326 as a CzcAB helper protein is indicated. Induction (+) and repression (−) of gene expression mediated by the increased copper pool are indicated by arrows. Genes involved in metabolic pathways and various biosynthetic pathways, iron uptake, and flagellar synthesis are boxed. Regulation of H. pylori ferritin synthesis by Fur under copper-rich conditions, as well as release of iron from ferritin, is indicated. The putative architecture of the Czc system of H. pylori HP1326 to HP1329 was deduced from the Ralstonia Czc system, as published previously (41).
In summary, results from RNA profiling allowed us to study copper regulation in H. pylori and to identify the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328), which usefully complete copper resistance mediated by the H. pylori metal export pump CopA (6, 18, 19), especially under copper-rich conditions. Finally, a possible interplay between the H. pylori-specific CrdA and CrdB proteins with the CzcB and CzcA homologs provides evidence for a novel type of metal efflux pump, which might effectively combine copper efflux and sequestration in a manner similar to that of the Pseudomonas Cop system (36).
Acknowledgments
This study was supported by a grant from ALTANA Pharma, Konstanz, Germany, to S.B. Parts of this work were funded by a grant from the Deutsche Forschungsgemeinschaft (Ki201/9-1) to M.K.
We thank S. Odenbreit and R. Haas (Max-von-Pettenkofer-Institute, LMU Munich, Germany) for providing the experimental data on TnBla mutagenesis of the HP1326 gene and for the Pfr antiserum AK198.
REFERENCES
- 1.Allan, E., N. Dorrell, S. Foynes, M. Anyim, and B. W. Wren. 2000. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J. Bacteriol. 182:5274-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Asaka, M., M. Kudo, M. Kato, T. Sugiyama, and H. Takeda. 1998. Long-term Helicobacter pylori infection—from gastritis to gastric cancer. Aliment. Pharmacol. Ther. 12(Suppl. 1):9-15. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Barceloux, D. G. 1999. Copper. J. Toxicol. Clin. Toxicol. 37:217-230. [DOI] [PubMed] [Google Scholar]
- 6.Bayle, D., S. Wangler, T. Weitzenegger, W. Steinhilber, J. Volz, M. Przybylski, K. P. Schäfer, G. Sachs, and K. Melchers. 1998. Properties of the P-type ATPases encoded by the copAP operons of Helicobacter pylori and Helicobacter felis. J. Bacteriol. 180:317-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bereswill, S., S. Greiner, A. H. M. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brendel, V., and E. N. Trifonov. 1984. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 12:4411-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, N. L., S. R. Barrett, J. Camakaris, B. T. Lee, and D. A. Rouch. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004. Mol. Microbiol. 17:1153-1166. [DOI] [PubMed] [Google Scholar]
- 10.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 11.Cha, J. S., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. USA 88:8915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 13.de Jonge, R., J. G. Kusters, M. S. Timmer, V. Gimmel, B. J. Appelmelk, S. Bereswill, A. H. M. van Vliet, S. G. Meuwissen, M. Kist, C. M. J. E. Vandenbroucke-Grauls, and E. J. Kuipers. 2001. The role of Helicobacter pylori virulence factors in interleukin production by monocytic cells. FEMS Microbiol. Lett. 196:235-238. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eickhoff, H., J. Schuchhardt, I. Ivanov, S. Meier-Ewert, J. O'Brien, A. Malik, N. Tandon, E. W. Wolski, E. Rohlfs, L. Nyarsik, R. Reinhardt, W. Nietfeld, and H. Lehrach. 2000. Tissue gene expression analysis using arrayed normalized cDNA libraries. Genome Res. 10:1230-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fassbinder, F., A. H. M. van Vliet, V. Gimmel, J. G. Kusters, M. Kist, and S. Bereswill. 2000. Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp). FEMS Microbiol. Lett. 184:225-229. [DOI] [PubMed] [Google Scholar]
- 17.Franke, S., G. Grass, and D. H. Nies. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965-972. [DOI] [PubMed] [Google Scholar]
- 18.Ge, Z., K. Hiratsuka, and D. E. Taylor. 1995. Nucleotide sequence and mutational analysis indicate that two Helicobacter pylori genes encode a P-type ATPase and a cation-binding protein associated with copper transport. Mol. Microbiol. 15:97-106. [DOI] [PubMed] [Google Scholar]
- 19.Ge, Z., and D. E. Taylor. 1996. Helicobacter pylori genes hpcopA and hpcopP constitute a cop operon involved in copper export. FEMS Microbiol. Lett. 145:181-188. [DOI] [PubMed] [Google Scholar]
- 20.Ge, Z., and D. E. Taylor. 1997. Helicobacter pylori DNA transformation by natural competence and electroporation, p. 145-152. In C. L. Clayton and H. L. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 21.Goldstein, S., and G. Czapski. 1986. The role and mechanism of metal ions and their complexes in enhancing damage in biological systems or in protecting these systems from the toxicity of O2−. J. Free Radic. Biol. Med. 2:3-11. [DOI] [PubMed] [Google Scholar]
- 22.Heron, P., K. Cousins, C. Boyd, and S. Daya. 2001. Paradoxical effects of copper and manganese on brain mitochondrial function. Life Sci. 68:1575-1583. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann, L., D. Schwan, R. Garner, H. L. Mobley, R. Haas, K. P. Schäfer, and K. Melchers. 1999. Helicobacter pylori cadA encodes an essential Cd(II)-Zn(II)-Co(II) resistance factor influencing urease activity. Mol. Microbiol. 33:524-536. [DOI] [PubMed] [Google Scholar]
- 24.Kachur, A. V., C. J. Koch, and J. E. Biaglow. 1999. Mechanism of copper-catalyzed autoxidation of cysteine. Free Radic. Res. 31:23-34. [DOI] [PubMed] [Google Scholar]
- 25.Lim, C. K., and D. A. Cooksey. 1993. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J. Bacteriol. 175:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malmstrom, B. G., and J. Leckner. 1998. The chemical biology of copper. Curr. Opin. Chem. Biol. 2:286-292. [DOI] [PubMed] [Google Scholar]
- 27.McGee, D. J., and H. L. Mobley. 1999. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr. Top. Microbiol. Immunol. 241:155-180. [DOI] [PubMed] [Google Scholar]
- 28.McGuirl, M. A., and D. M. Dooley. 1999. Copper-containing oxidases. Curr. Opin. Chem. Biol. 3:138-144. [DOI] [PubMed] [Google Scholar]
- 29.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills, S. D., C. K. Lim, and D. A. Cooksey. 1994. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol. Gen. Genet. 244:341-351. [DOI] [PubMed] [Google Scholar]
- 31.Mobley, H. L., R. M. Garner, G. R. Chippendale, J. V. Gilbert, A. V. Kane, and A. G. Plaut. 1999. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter 4:162-169. [DOI] [PubMed] [Google Scholar]
- 32.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagata, K., S. Tsukita, T. Tamura, and N. Sone. 1996. A cb-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology 142:1757-1763. [DOI] [PubMed] [Google Scholar]
- 34.Nakao, K., I. Imoto, N. Ikemura, T. Shibata, S. Takaji, Y. Taguchi, M. Misaki, K. Yamauchi, and N. Yamazaki. 1997. Relation of lactoferrin levels in gastric mucosa with Helicobacter pylori infection and with the degree of gastric inflammation. Am. J. Gastroenterol. 92:1005-1011. [PubMed] [Google Scholar]
- 35.Nies, D. H. 1995. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 177:2707-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 37.Odenbreit, S., M. Till, and R. Haas. 1996. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol. Microbiol. 20:361-373. [DOI] [PubMed] [Google Scholar]
- 38.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 39.Pfeiffer, J., J. Guhl, B. Waidner, E. Bisse, M. Kist, and S. Bereswill. 2002. Magnesium transport mediated by CorA is essential for viability of Helicobacter pylori. Infect. Immun. 70:3930-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Powell, J. J., R. Jugdaohsingh, and R. P. Thompson. 1999. The regulation of mineral absorption in the gastrointestinal tract. Proc. Nutr. Soc. 58: 147-153. [DOI] [PubMed] [Google Scholar]
- 41.Rensing, C., T. Pribyl, and D. H. Nies. 1997. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 179:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 44.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seyler, R. W., Jr., J. W. Olson, and R. J. Maier. 2001. Superoxide dismutase-deficient mutants of Helicobacter pylori are hypersensitive to oxidative stress and defective in host colonization. Infect. Immun. 69:4034-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stadtman, E. R. 1990. Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic. Biol. Med. 9:315-325. [DOI] [PubMed] [Google Scholar]
- 47.Stoyanov, J. V., J. L. Hobman, and N. L. Brown. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39:502-511. [DOI] [PubMed] [Google Scholar]
- 48.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, McKenney, L. M. Fitzgerald, N. Lee, M. D. Adams, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 49.Tsekova, K., D. Dentchev, and D. Todorova. 2000. Effect of cadmium and copper on the production of citric acid by Aspergillus niger. Folia Microbiol. 45:331-334. [DOI] [PubMed] [Google Scholar]
- 50.Tsukita, S., S. Koyanagi, K. Nagata, H. Koizuka, H. Akashi, T. Shimoyama, T. Tamura, and N. Sone. 1999. Characterization of a cb-type cytochrome c oxidase from Helicobacter pylori. J. Biochem. 125:194-201. [DOI] [PubMed] [Google Scholar]
- 51.van der Lelie, D., T. Schwuchow, U. Schwidetzky, S. Wuertz, W. Baeyens, M. Mergeay, and D. H. Nies. 1997. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol. 23:493-503. [DOI] [PubMed] [Google Scholar]
- 52.van Vliet, A. H. M., S. Bereswill, and J. G. Kusters. 2001. Ion metabolism and transport, p. 193-206. In H. L. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 53.van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Vliet, A. H. M., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2001. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 56.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stähler, J. Guhl, E. Bisse, A. H. M. van Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of the ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]