Abstract
In order to determine whether ClpXP-mediated proteolysis is a common mechanism used to regulate the chemotaxis machinery during the cell cycle of Caulobacter crescentus, we have characterized a soluble cytoplasmic chemoreceptor, McpB. The mcpB gene lies adjacent to the major chemotaxis operon, which encodes 12 chemotaxis proteins, including the membrane chemoreceptor McpA. Like McpA, McpB possesses a C-terminal CheBR docking motif and three potential methylation sites, which we suggest are methylated. The McpB protein is degraded via a ClpX-dependent pathway during the swarmer-to-stalked cell transition, and a motif, which is 3 amino acids N-terminal to the McpB CheBR docking site, is required for proteolysis. Analysis of the degradation signal in McpB and McpA reveals a common motif present in the other four chemoreceptors that possess CheBR docking sites. A green fluorescent protein (GFP) fusion bearing 58 amino acids from the C terminus of McpA, which contains this motif, is degraded, suggesting that the C-terminal sequence is sufficient to confer ClpXP protease susceptibility.
Temporal and spatial control of proteins is of fundamental importance in cell differentiation and organization. Caulobacter crescentus's unique life cycle comprising an asymmetric cell division and the obligate differentiation of a motile swarmer cell to a nonmotile stalked cell makes it an attractive system to study these processes (10). The differentiation of the swarmer cell is accompanied by the degradation of key components of the flagellar and chemotaxis apparatus (4, 9, 14, 19). Once the stalked cell has differentiated into the predivisional cell, most of the components for the flagellar and chemotaxis apparatus are synthesized and targeted to the nascent swarmer portion of the cell (13). When cell division occurs, the swarmer cell has all the components necessary for motility. The swarmer cell swims by rotating its single polar flagellum in a clockwise direction (22), while the short intermittent reversals in rotation of the flagellum enable reorientation in a three-dimensional space, resulting in swimming towards attractants or away from repellents. The receptors for the chemotactic response, the chemoreceptors, or alternatively referred to as methyl-accepting chemotaxis proteins (MCPs), are typically integral cytoplasmic membrane proteins which form clusters at the pole of the cell (3, 12, 15, 25). Membrane spanning MCPs are composed of an extracytoplasmic substrate-binding domain and a cytoplasmic signaling domain. The signaling domain interacts with the sensor kinase CheA, the linker protein CheW, and the CheR and CheB proteins, which methylate the chemoreceptors and are required for the adaptation response (7). Both CheB and CheR compete for the same docking site on the receptor, which is located at the extreme C terminus of the chemoreceptor (5). In Escherichia coli the CheBR binding pentapeptide sequence is typically NWE(T/S)F (28), and that in C. crescentus is typically XWEEF (27). The CheBR docking site is found in two out of the five E. coli chemoreceptors and in six out of the eighteen chemoreceptors in C. crescentus.
The integral membrane McpA chemoreceptor was the first protein recognized to undergo specific cell-cycle-regulated proteolysis in C. crescentus (4). It is synthesized in the predivisional cell, where it is targeted to the cell pole of the nascent swarmer compartment (3). When the swarmer cell differentiates into a stalked cell, the McpA is degraded via a ClpX-dependent pathway (33). ClpX is an ATP-dependent chaperone that interacts with the ClpP protease and, upon unfolding of substrate proteins, delivers them to the ClpP proteolytic chamber (21, 29). With a few exceptions ClpX typically recognizes hydrophobic residues at the extreme C terminus of its substrates (24), for example the C. crescentus response regulator CtrA requires hydrophobic amino acids at its extreme C terminus for its ClpX-dependent degradation (9, 18). Although the degradation of McpA is also ClpX dependent, it does not require hydrophobic amino acid residues at the extreme C terminus but rather those located immediately upstream of the C-terminal CheBR binding site (33). To test whether this unusual location for a ClpX-dependent degradation signal is specific only to McpA or whether it is a common theme present in the other MCPs containing the C-terminal CheRB docking site, we have characterized a soluble cytoplasmic chemoreceptor, McpB.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were cultured at 37°C in Terrific Broth for liquid media or Luria-Bertani for solid media supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), or tetracycline (15 μg/ml) as required. C. crescentus strains were grown in PYE medium (0.2% [wt/vol] Bacto Peptone [Difco], 0.1% [wt/vol] yeast extract [Difco], 1 mM MgSO4, 0.5 mM CaCl2) and incubated at 28°C or at room temperature supplemented with chloramphenicol (1 μg/ml), tetracycline (1 μg/ml), nalidixic acid (20 μg/ml), or kanamycin (5 μg/ml) as necessary and 20 mM xylose (PYEX) or 10 mM glucose (PYEG), where indicated. Plasmids were mobilized into C. crescentus from the E. coli strain S17-1 (Table 1).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH10B | F−mrcA Δ(mrr-hsdRMS-mcrBC) ΔlacX74 φ80dlacZΔM15 deoR recA1 endA1 araΔ139 Δ(ara leu)7697 rpsL galU galK nupG | Gibco BRL |
| S17-1 | M294::RP4-2 (tet::Mu) (Kan::Tn7) | 30 |
| C. crescentus | ||
| CB15 | Wild type | ATCC 19089 |
| MRKA949 | ΔmcpA3 | This study |
| MRKA952 | ΔmcpB1 | This study |
| MRKA990 | ΔmcpA3 ΔmcpB1 | This study |
| MRKA1005 | ΔmcpA3 ΔcheRI1 | This study |
| MRKA1006 | ΔmcpA3 ΔcheB12 | This study |
| MRKA993 | MRKA949(pJMB) | This study |
| MRKA1020 | MRKA1005(pJMB) | This study |
| MRKA1021 | MRKA1006(pJMB) | This study |
| MRKA994 | MRKA949(pGMB) | This study |
| MRKA1111 | MRKA949(pJS14) | This study |
| MRKA1110 | MRKA949(pJMBD) | This study |
| MRKA1255 | CB15ATCC19089 mcpBΩpXMB | This study |
| MRKA1292 | MRKA1255(pMR20) | This study |
| MRKA1266 | MRKA1255(pMO88) | This study |
| MRKA1245 | NA1000 clpXΩaadA xylXΩpUJ175 Δ(cagAI-cheYIII)che17 | This study |
| MRKA1280 | MRKA949 xylXpΩpGEG1 | This study |
| MRKA1281 | MRKA949 xylXpΩpGEG1M | This study |
Plasmid constructions.
To generate an in-frame mcpA deletion strain, we constructed pDMPA1213 (Table 2) by cloning the 5′ and 3′ flanking DNA of mcpA in pNPTS128 (1). For the construction we used four primers to amplify the flanking regions from the mcpA locus: KOmcpA-5Spe, CGC TTT GGT GAC ACT AGT TGT GCG GCG AGG; KOmcpA-5Nco, GC CGG ATC CGT TCC ATG GCA AGG TCC CCT C; KOmcpA-3Nco, TGG GA GGA ATT CCC ATG GTT GCC GCG ATC T; and KOmcpA-3Xba, CTT GGA ATT CCG TCT AG AGG AGC GCC CCT G. Both KOmcpA-5Spe-KOmcpA-5Nco and KOmcpA-3Nco-KOmcpA-3Xba PCR products were cloned consecutively into the plasmid pNPTS128. For the mcpB deletion construct the following primer pairs were used to generate the construct pDMPB1415 (Table 2) with the 5′ and 3′ mcpB flanking DNA fused in frame: KOmcpB-5Spe, CGC TGG AGG CCG ACT AGT CCA AGC TGT TCG; KOmcpB-Hin-R, TGG CGG TCC CCA AAG CTT CGC TTG GGG AAA; KOmcpB-Hin-F, AGG AAG AGT GGG AAG CTT TCT AGC GCG CGG; and KOmcpB-3Xba, CGC AGG TGA TGC TCT AGA GCA ACG CTG ACG. To create deletions we used the previously described procedure (1). The deletion mutants were verified by PCR and immunoblotting. To delete the cheBI and cheRI genes we followed the procedure outlined previously (1).
TABLE 2.
Plasmids used in this study
| Plasmid | Description and/or use | Source |
|---|---|---|
| pJS14 | Chlr pBBR1-derived medium-copy-number broad-host-range vector of 10 to 20 copies per cell in C. crescentus (31) | J. Skerker |
| pJMB | 2.6-kb C. crescentus DNA fragment containing the mcpB gene and its promoter cloned into pJS14 | This study |
| pJMBD | pJMB A527D V528D A529I | This study |
| pGL10 | Mini RK2 replicon containing oriT and Kanr | D. Helinski |
| pGMB | 2.6-kb C. crescentus DNA fragment containing the mcpB gene and its promoter in pGL10 | This study |
| pMR20 | pMR4-based Tetr mini-RK2 replicon with multiple-cloning site | R. Roberts and C. Mohr |
| pMO88 | clpX with mutation in ATP binding site under the control of xylXp cloned in pMR20 as a KpnI-BglII fragment | M. Østerås and U. Jenal, unpublished data |
| pXSU31 | Narrow-host-range vector with 103-bp xylXp fragment upstream of multiple-cloning sites | This study |
| pXMB | 600-bp PCR fragment of the mcpB gene cloned into XbaI-SphI sites of pXSU31 | This study |
| pGEG1 | Last 58 amino acids of McpA fused to the C terminus of GFP | This study |
| pGEG1M | GFP fusion to the last 58 amino acids of McpA with ELD mutation in the degradation signal (33) | This study |
To clone the mcpB gene and its promoter region we isolated the 5.8-kb BamHI fragment from the mcpA operon complementing cosmid pR6 (2) and ligated it into pBluescript KSII(+) to generate pBMKS6 (Table 2). We then deleted the StuI-SpeI fragment to generate pBMKS6dSS. The 2.6-kb XbaI-SalI fragment from this plasmid, which carries the mcpB gene and its promoter, was cloned into pGL10 to generate pGMB and into pJS14 to generate pJMB (Table 2). To generate the amino acid substitutions A527D, V528D, and A529I in McpB, we used inverse PCR to amplify the pJMB plasmid with the primers MBDDIF, G GGC GCT ACG GCG GAC GAT ATC AAG GAA GAG TGG G, and MBDDIR, C CCA CTC TTC CTT GAT ATC GTC CGC CGT AGC GCC C. The construct was tested for presence of an EcoRV site and was sequenced to confirm the mutation. The xylose inducible mcpB construct pXMB was constructed by amplifying a 600 bp fragment of the mcpB gene with the primers McpBsphF, CC CCA AGC GAA AGC ATG CGG ACC GCC ATG AAC C, and McpBXbaR, TCG CTG CAG ACC TCA CTC TAG ATC CTC GCC AGC, and by cloning it into the SphI-XbaI sites of pXSU31 downstream of the xylX promoter.
To generate the green fluorescent protein (GFP) fusion to the last 58 amino acids of McpA, we used the following primers: 3XmcpA, G AGA TCG CGG CAA CCT CTA GAA TTC CTC CC, and 5BFmcpA, GC TTC CAG GTG GGA TCC GGT TCG TCG TCC TA. The PCR product was digested with BamHI and XbaI and ligated into the BglII and SpeI sites of pJUG1, which has gfp from pEGFP-C1 (Clontech) downstream of xylXp, to generate the construct pGEG1 (Table 2). To generate the stabilized derivative of this fusion the same primers were used but the template DNA was pXCP3ELD (33) with the residues A660, A661, and L662 replaced with the amino acids E, L, and D, respectively, and the resulting plasmid was named pGEG1M (Table 2).
RESULTS
A chemoreceptor homologue is encoded by a gene upstream of the major chemotaxis operon.
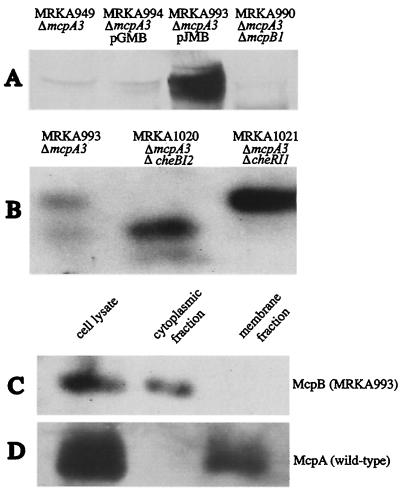
An open reading frame encoding a chemoreceptor homologue was identified upstream of the major chemotaxis operon and named mcpB (27). Although the monocistron mcpB is transcribed in the same direction as the mcpA operon, it is separated by the cagAI gene, which is transcribed in the opposite direction from both mcpB and mcpA cistrons. The methylation and signaling domains of McpB and McpA are highly conserved. However, the N-terminal regions involved in ligand binding lack any homology, suggesting that McpB responds to different environmental cues. To test whether mcpB was expressed, we used antisera raised against McpA. We prevented cross-reaction with McpA by creating an in-frame mcpA deletion strain, MRKA949, while an mcpA mcpB double deletion strain, MRKA990, was created as a negative control (Table 1). We were able to detect McpB in cell extracts from the strain MRKA949 and found that the protein was expressed at very low levels (Fig. 1A). Since no chemotaxis defects have been observed by placing chemoreceptors on multicopy plasmids (8, 17), we decided to clone the mcpB gene, including its promoter, onto a low-copy-number plasmid (pGMB) and a medium-copy-number plasmid (pJMB) to improve its detection. Both plasmids were conjugated into the mcpA mcpB deletion strain, MRKA990, and the best signal was achieved with the strain bearing the medium-copy-number plasmid pJMB (Fig. 1A). The increased expression of mcpB observed in MRKA993 did not affect swarming of this strain (data not shown).
FIG. 1.
Analysis of McpB. (A) Detection of McpB. All cell extracts were prepared from cultures with an mcpA deletion to reduce cross-reaction with the McpA antisera. Samples were ΔmcpA3 (MRKA949), pGMB (MRKA994 expressing McpB from a low-copy-number plasmid), pJMB (MRKA993 expressing McpB from a medium-copy-number plasmid) and ΔmcpA3 ΔmcpB1(MRKA990) and were subjected to SDS-8% PAGE and electroblotted to nitrocellulose (32). The immunoblots were incubated with the antisera raised against McpA (3). The secondary antibody was goat anti-rabbit immunoglobulin G-horseradish peroxidase. (B) McpB methylation. All strains bear the plasmid pJMB, ΔmcpA3 (MRKA993), ΔmcpA3 ΔcheBI2 (MRKA1021), and ΔmcpA3 ΔcheRI1 (MRKA1020). The separation was achieved on an SDS-6% polyacrylamide gel (16 cm) run overnight at 5 mA. Extracts of MRKA993 (C) and CB15 ATCC 19089 (D) were separated into membrane and cytoplasmic fractions as described previously for McpA (3). Equal amounts of protein were analyzed by SDS-8% PAGE and immunoblots were performed with McpA antisera.
McpB is a methylated cytoplasmic chemoreceptor.
The mcpB deletion strain, MRKA952, did not show any obvious defects in semisolid agar with the following carbon sources: xylose, glucose, sucrose, tryptone, and Casamino Acids. Therefore, we tested whether McpB could be methylated as this would indicate its involvement in chemotaxis. The electrophoretic mobility of a chemoreceptor in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is affected by its methylation status (6). We have shown previously that the methylation status of McpA is modulated by the CheBI methylesterase and CheRI methyltransferase (1); therefore, we introduced a cheBI or cheRI deletion into the mcpA deletion strain MRKA949, generating strains MRKA1006 and MRKA1005. The plasmid pJMB was then conjugated into the cheBI mcpA strain MRKA1006 and the cheRI mcpA strain MRKA1005. Extracts of the resulting strains MRKA1020 and MRKA1021 were analyzed by SDS-6% PAGE, and two bands, each representing a different methylation status, were detected (Fig. 1B). In the cheBI mutant strain, only the faster migrating band could be observed, while in the cheRI mutant strain, only the upper band was detectable, suggesting that McpB is methylated. Sequence analysis of the deduced mcpB sequence revealed that this protein lacks the transmembrane regions typically found in chemoreceptors. To verify the cellular localization of the McpB protein we fractionated MRKA993 cell extracts into a membrane and a cytoplasmic fraction. As a control we also fractionated cell extracts from the wild-type CB15 strain. McpB was detected only in the cytoplasmic fraction (Fig. 1C), while McpA was detected in the membrane fraction of wild-type cell extracts (Fig. 1D).
A hydrophobic degradation signal and the ClpX ATPase are required for the cell-cycle-dependent degradation of McpB.
In order to test whether McpB is degraded differentially during the cell cycle, we first determined the expression pattern of the mcpB gene. By using an mcpB-lacZ transcriptional fusion, we found that the cell-cycle expression pattern for mcpB was much like that of mcpA in that it was transcribed mainly in predivisional cells (data not shown). This has also been observed recently when the cell-cycle transcription profile of the mcpB gene was determined in the DNA microarray analysis of synchronous C. crescentus cell populations (23).
To test whether McpB protein might be degraded during the C. crescentus cell cycle we performed immunoblots on cell extracts of synchronized cells of strain MRKA993. The levels of McpB dropped drastically during the swarmer to stalked cell transition (Fig. 2B), suggesting that McpB is degraded during this phase of the cell cycle. McpB levels increased concomitantly with the onset of mcpB promoter activity in the predivisional cell (Fig. 2B). To show that this modulation in McpB levels is due to regulated proteolysis we constructed the strain MRKA1255, which has the mcpB gene under the control of a xylose inducible promoter (Fig. 3A). Swarmer cells from strain MRKA1255 were isolated and allowed to progress through the cell cycle in PYEX. Although, the xylX promoter is constitutively active throughout the cell cycle (26), we noticed that the McpB protein levels dropped as the swarmer cells differentiated into a stalked cell and later increased in predivisional cells (Fig. 3B). As an additional control we repeated the experiment shown in Fig. 3B but grew the swarmer cells in PYEG. Under these conditions, McpB was not synthesized and the protein levels dropped very quickly, resulting in the complete disappearance of McpB after the swarmer cells differentiated into stalked cells (Fig. 3C). These results suggest that the proteolysis of McpB is modulated during the cell cycle.
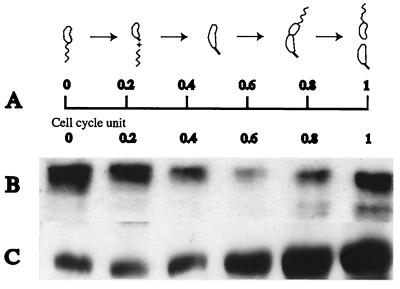
FIG. 2.
The McpB protein levels are modulated during the cell cycle by proteolysis and C-terminal residues are required for this modulation. (A) Schematic diagram of C. crescentus cell cycle. (B) Swarmer cells were isolated by a modification of the Evinger and Agabian method (11) using Percoll (Pharmacia) instead of Ludox (33). Cell extracts from equal culture volumes of a synchronous population of strain MRKA993, which bears the plasmid with the mcpB gene under the control of its own promoter, were analyzed for the presence of McpB by immunoblotting with McpA antisera. One cell cycle unit was 150 min. (C) Swarmer cells from MRKA1110 bearing the plasmid encoding the mutated mcpB (A527D V528D A529I) gene were isolated and allowed to proceed through the cell cycle in PYE medium. Equal volumes of cells were analyzed for the presence of McpB by immunoblotting with McpA antisera. One cell cycle unit was 150 min.
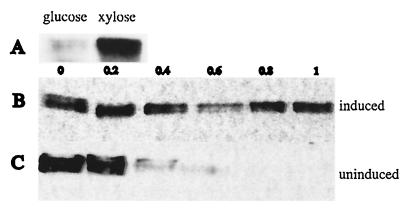
FIG. 3.
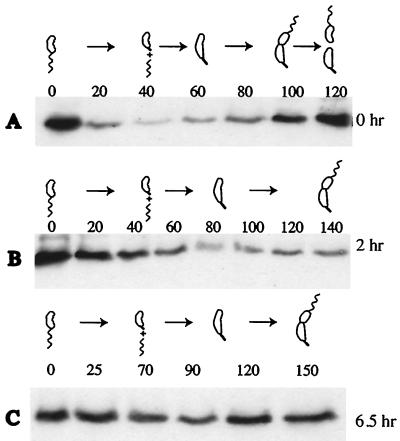
The McpB protein is degraded during the swarmer-to-stalked-cell transition. (A) Strain MRKA1374 bearing the xylose inducible mcpB gene was grown in PYE medium supplemented with glucose or xylose, and extracts from equal amount of cell material as calculated by A600 were analyzed for presence of McpB by immunoblotting with McpA antisera. (B) Strain MRKA1374 was grown in PYEX before cells were synchronized and allowed to proceed through the cell cycle in the same medium. Extract of equal amounts of cell material were analyzed by immunoblotting with McpA antisera. One cell cycle unit was 100 min. (C) Strain MRKA1374 was grown in PYEX before synchronization and was then allowed to proceed through the cell cycle in PYEG. Cell extracts from equal culture volumes of a synchronous population of MRKA1374 were analyzed for the presence of McpB by immunoblotting with McpA antisera. One cell cycle unit was 100 min.
We have shown earlier that the integral membrane protein McpA is degraded by a ClpX dependent pathway (33). To test a possible requirement of ClpX for the degradation of McpB, we used a novel conditional mutant system to rapidly turn off the activity of the essential ClpX ATPase in C. crescentus. Plasmid pMO88, which contains a mutated clpX copy under the control of the xylX promoter, was introduced into the C. crescentus MRKA1255 strain, which contains a single chromosomal mcpB gene under the control of the xylX promoter. The mutant derivative of ClpX has a mutated Walker A box in the ATP binding domain, and upon xylose induction displays a strong dominant negative effect, which leads to the rapid loss of ClpX activity (M. Østerås and U. Jenal, unpublished data). The degradation of McpA and CtrA was affected after only 1 h of induction (data not shown). Strain MRKA1255 bearing pMO88 was grown in PYE medium except for 1 h prior to synchronization, when both the clpX ATP mutant allele and mcpB were induced by the addition of 20 mM xylose. Swarmer cells were then isolated and allowed to progress through the cell cycle in PYEG to prevent any further expression of McpB and the ClpX mutant. While the degradation of the McpB protein was not affected in a strain bearing the parental plasmid pMR20 (Fig. 4A), it was drastically reduced in the strain bearing pMO88 (Fig. 4B). The flagellar motor protein FliF is degraded by the protease ClpAP (Østerås and Jenal, unpublished). Since both ClpX and ClpA share the same protease component, we tested whether the induction of a ClpX mutant might affect ClpA dependent degradation. As shown in Fig. 4C, the ClpAP dependent degradation of FliF is not affected. As a further control for our novel system we compared the degradation of McpB in a clpX conditional strain. Since the original clpX conditional strain UJ200 (18) has an intact copy of the mcpA gene, we transduced the Δche17 chemotaxis deletion (34) into UJ200 to create the strain MRKA1245. The plasmid pJMB, which bears mcpB, was conjugated into the MRKA1245 strain. This strain was grown in PYE medium supplemented with 20 mM glucose for 0, 2, and 6.5 h to deplete ClpX and then swarmer cells were isolated to test the effect on the degradation of McpB. As expected, McpB was still degraded after zero hours of ClpX depletion (Fig. 5A). However, after 2 h of ClpX depletion, which corresponds to a single generation time, the degradation of McpB was affected (Fig. 5B). This effect was more pronounced after 6.5 h of ClpX depletion, which corresponds to just under three generation times (Fig. 5C). These data support our findings from the ClpX mutant inductions experiments, which demonstrated that the degradation of the soluble chemoreceptor McpB was dependent on ClpX.
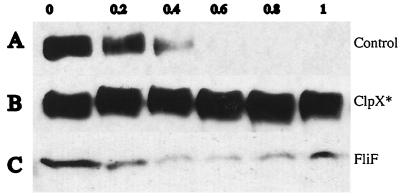
FIG. 4.
Induction of an ATPase ClpX mutant inhibits McpB degradation. (A) MRKA1292 cells bearing the control plasmid pMR20 and a chromosomal copy of the xylose inducible mcpB gene were induced in PYEX for 1 h immediately before they were synchronized and allowed to proceed through the cell cycle in PYEG. Extracts from equal volumes of cells were analyzed for the presence of McpB by immunoblotting with McpA antisera. One cell cycle unit was 120 min. (B) MRKA1266 cells bearing the pMO88 plasmid with the xylose inducible ClpX mutant allele were induced in PYEX for 1 h before they were synchronized and allowed to proceed through the cell cycle in PYEG. Extracts from equal volumes were analyzed for the presence of McpB by immunoblotting with McpA antisera and (C) with FliF antisera (19). One cell cycle unit was 150 min.
FIG. 5.
ClpX depletion affects McpB degradation. (A) MRKA1245 cells bearing the plasmid pJMB were grown to an A660 of 0.5 in PYEX, and swarmer cells were isolated and allowed to go through the cell cycle in PYEG. The numbers above the panel correspond to the time in minutes. The cell cycle was 120 min. (B) MRKA1245 cells bearing the plasmid pJMB were grown to an A660 of 0.5 in PYEX. The cells were washed thrice with PYEG and diluted to an A660 of 0.258 and then they were allowed to grow in PYEG for 2 h to an A660 of 0.583. Swarmer cells were then isolated and allowed to go through the cell cycle in PYEG media. The numbers above the panel correspond to the time in minutes. At 140 min the majority of cells were at the predivisional cell stage. (C) MRKA1245 cells bearing the plasmid pJMB were grown to an A660 of 0.5 in PYEX. The cells were washed thrice with PYEG and diluted to an A660 of 0.092 and then they were allowed to grow in PYEG for 6.5 h to an A660 of 0.781. Swarmer cells were then isolated and allowed to go through the cell cycle in PYEG media. The numbers above the panel correspond to the time in minutes. At 150 min the majority of cells were at the predivisional cell stage. Equal amounts of cells as determined by A660 were analyzed for the presence of McpB by immunoblotting with McpA antisera.
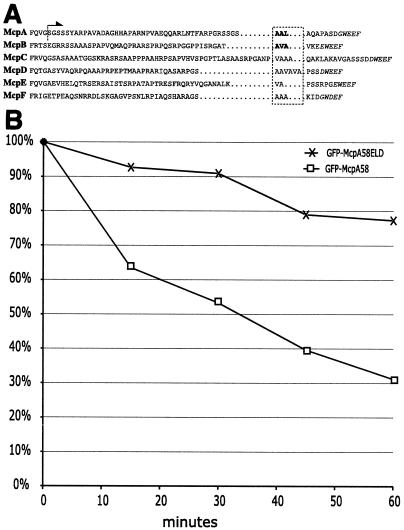
ClpX substrates have been shown to be recognized via hydrophobic residues at their extreme C termini (24). Both McpA and McpB have docking sites for CheR and CheB proteins at this position. We have previously shown that hydrophobic residues just upstream of the XWEEF docking motif are required for McpA proteolysis (33). Close inspection of the C-terminal amino acid sequence of the McpB protein revealed the presence of hydrophobic residues at a similar position (Fig. 6A). To test the requirement of these amino acids in proteolysis we have substituted the residues A527, V528, and A529 in McpB with D, D, and I, generating the plasmid pJMBD, and conjugated it into the mcpA deletion strain, creating MRKA1110. When strain MRKA1110, which expresses this mutant version of McpB, was analyzed in the context of the cell cycle, the characteristic fluctuation of the protein level as observed for the wild-type protein was not seen (Fig. 2C). The observed increase in McpB levels late in the cell cycle are due to renewed synthesis in the predivisional cells. Thus, the substitution of these hydrophobic residues for negatively charged amino acids abolished McpB degradation during the cell cycle.
FIG. 6.
Chemoreceptor degradation signal. (A) Amino acid sequences from the exposed C terminus of all C. crescentus MCPs bearing a CheBR binding site were taken from McpA (Q00986), McpB (AAK22415), McpC (AAK22330), McpD (AAK23633), McpE (AAK24252), and McpF (AAK24656). The italicized amino acids indicate the putative pentapeptide CheBR binding site. The boxed hydrophobic amino acids are the putative sequences required for proteolysis. In the McpA and McpB sequences, the bold letters denote hydrophobic residues required for proteolysis as demonstrated by in vitro mutagenesis. The arrow indicates that the amino acid sequence C terminal of the dotted line is present in the GFP-McpA58 fusion. (B) MRKA1280 (GFP-McpA58) and MRKA1281 (GFP-McpA58ELD) cells bearing the xylose inducible GFP fusions were grown in PYE medium to an A660 of 0.4 and then the GFP fusions were induced for 2 h by adding 20 mM xylose. After 2 h the cells were centrifuged for 5 min at 5,000 × g and the cells were then resuspended in PYE medium containing 20 mM glucose. This was repeated three times to ensure removal of xylose. Cells from MRKA1280 (GFP-McpA58) and MRKA1281 (GFP-McpA58ELD) were resuspended to an A660 of 0.3 in PYE medium containing 20 mM glucose and grown for 60 min. Equal volumes of cells were taken every 15 min. The cell extracts were analyzed by immunoblotting with GFP antisera (AbCam) and anti-goat horseradish peroxidase (AbCam). The signal generated by Lumi Light Western blotting substrate (Roche) was captured by XLS film (Kodak), which was scanned and quantified using ImageJ (NIH). Each time point was compared with time zero, which was given a value of 100%.
The McpA degradation sequence is sufficient to confer susceptibility to proteolysis onto GFP.
Comparison of McpA and McpB with the three-dimensional structure of the cytoplasmic domain from the homologous E. coli chemoreceptor Tsr (20) revealed that the residues required for proteolysis form part of an exposed and flexible C terminus. This observation led us to attempt to transfer the degradation signal onto a different protein, GFP. We chose to use the McpA sequence instead of McpB, because it is the archetypal C. crescentus chemoreceptor. The last 58 amino acids of McpA (Fig. 6A) were fused to the C terminus of GFP to generate the plasmid pGEG1 (GFP-McpA58). The protein fusion in this construct is under the control of the xylose inducible promoter xylXp. As a control we also generated a stabilized derivative, pGEG1M, where the hydrophobic residues A, A, and L are substituted for E, L, and D, respectively (33). Both plasmids pGEG1 and pGEG1M were introduced into the mcpA deletion strain to generate strains MRKA1280 and MRKA1281. We grew both strains in PYE medium until they reached an A660 of 0.4 when xylose was added to induce expression of the GFP fusions. After 2 h the cells were centrifuged and the cells were resuspended in PYE medium with 20 mM glucose to repress expression of the GFP fusions. The level of the wild-type GFP fusion in MRKA1280 decreased significantly and by 60 min it had dropped to 31% of the level at time zero (Fig. 6B). While the level of the GFP fusion with the mutated degradation signal had decreased only slightly by 60 min, it had only dropped to 77% of the level at time zero (Fig. 6B). This suggests that the extreme C terminus of McpA possesses a degradation signal, which is sufficient to confer susceptibility to degradation onto a noncognate protein.
DISCUSSION
In this study we have shown that McpB is a soluble cytoplasmic chemoreceptor. Analysis of the C. crescentus genome predicts a total of eighteen chemoreceptor genes (27). The absence of any obvious chemotaxis phenotype in the mcpA (I Potocka and M. R. K. Alley, unpublished data) and mcpB deletion strains suggests that many of these chemoreceptors are functional and perform redundant tasks. Analysis of the N-terminal substrate specificity domain in the soluble cytoplasmic chemoreceptor McpB points to a possible role in oxygen sensing. McpB appears to be a direct homologue of the myoglobin-like aerotaxis transducers found in Halobacterium salinarum and Bacillus subtilis (17). The residues that are conserved between the HemAT proteins in both organisms are also conserved in McpB. A second HemAT homologue, McpM (AAK24288), is present in C. crescentus (27). McpM has already been shown to bind heme when overexpressed in E. coli (16). The presence of a second HemAT homologue in C. crescentus may explain the absence of a chemotaxis phenotype in the mcpB deletion strain. Although B. subtilis has only one HemAT homologue, a phenotype could only be observed in a strain that had all ten chemoreceptors deleted (17). Therefore it is likely that a large proportion of the eighteen chemoreceptors need to be deleted before an obvious phenotype is observed in C. crescentus. We have recently found that a strain deleted for McpA, McpB, McpC and McpD has a chemotaxis phenotype in swarm plates (L. C. Edwards and M. R. K. Alley, unpublished data).
In addition to the two HemAT transducers, there are five additional chemoreceptors that can be classified as energy sensors. McpD, McpH (AAK25311), and McpR (AAK24774) have PAS or PAC domains and are likely to be soluble cytoplasmic chemoreceptors. The other potential energy sensing chemoreceptors, McpI (AAK24811) and McpP (AAK23380), are predicted to have six transmembrane domains and are homologous to the HtrVIII chemoreceptor from H. salinarum. The N-terminal substrate specificity domain of HtrVIII is homologous to cytochrome c oxidase (8). The large number of chemoreceptors devoted to energy sensing would suggest that it is an important component of C. crescentus chemotaxis.
We have demonstrated that the soluble McpB chemoreceptor is degraded during the cell cycle. The fact that both C. crescentus chemoreceptors characterized to date are removed by ClpX-dependent proteolysis points to a more general mechanism by which C. crescentus rids itself of chemotactic proteins it does not require after differentiation into a nonmotile stalked cell. Further analysis of the sequences of the remaining chemoreceptor genes appears to support this hypothesis. The remaining chemoreceptors that have a CheBR docking site at their C-termini (McpC, McpD, McpE, and McpF) have hydrophobic residues immediately upstream of this site, suggesting a common degradation signal for these methylation-dominant chemoreceptors (Fig. 6). Alignments of the sequences of the cytoplasmic MCP domains with that of the E. coli Tsr protein, for which the three dimensional structure is known (20), suggest that the hydrophobic residues and the CheBR binding pentapeptide are not part of any secondary structure, but rather form a flexible tail. The ability of this sequence in McpA to confer degradation onto the GFP protein strongly suggests that these hydrophobic residues form an exposed degradation signal. Eight of the remaining twelve MCPs that lack the CheBR binding site have hydrophobic residues at their extreme C termini that are commonly observed in proteins subject to ClpX-mediated proteolysis (24). These residues can also be found in CheYI, which has already been reported to be unstable (14). Further analysis of the cell cycle modulation of other chemotactic proteins will reveal whether C. crescentus has indeed evolved a mechanism to actively remove the motility machinery by targeted proteolysis as it differentiates into a nonmotile cell.
Acknowledgments
We thank Misha Rahman for helpful advice.
We thank the Imperial College for funding I.P. M.R.K.A. is supported by a Royal Society University Research Fellowship. The preliminary findings in this study were funded by a Wellcome Trust project grant (044761) to MRKA.
REFERENCES
- 1.Alley, M. R. K. 2001. The highly conserved domain of the Caulobacter McpA chemoreceptor is required for its polar localization. Mol. Microbiol. 40:1335-1343. [DOI] [PubMed] [Google Scholar]
- 2.Alley, M. R. K., S. L. Gomes, W. Alexander, and L. Shapiro. 1991. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics 129:333-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alley, M. R. K., J. R. Maddock, and L. Shapiro. 1992. Polar localization of a bacterial chemoreceptor. Genes Dev. 6:825-836. [DOI] [PubMed] [Google Scholar]
- 4.Alley, M. R. K., J. R. Maddock, and L. Shapiro. 1993. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science 259:1754-1757. [DOI] [PubMed] [Google Scholar]
- 5.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 96:10667-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, A., and M. I. Simon. 1980. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J. Bacteriol. 143:809-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooun, A., J. Bell, T. Freitas, R. W. Larsen, and M. Alam. 1998. An archaeal aerotaxis transducer combines subunit I core structures of eukaryotic cytochrome c oxidase and eubacterial methyl-accepting chemotaxis proteins. J. Bacteriol. 180:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 10.Domian, I. J., K. C. Quon, and L. Shapiro. 1996. The control of temporal and spatial organization during the Caulobacter cell cycle. Curr. Opin. Genet. Dev. 6:538-544. [DOI] [PubMed] [Google Scholar]
- 11.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gestwicki, J. E., A. C. Lamanna, R. M. Harshey, L. L. McCarter, L. L. Kiessling, and J. Adler. 2000. Evolutionary conservation of methyl-accepting chemotaxis protein location in Bacteria and Archaea. J. Bacteriol. 182:6499-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gober, J. W., M. R. K. Alley, and L. Shapiro. 1991. Positional information during Caulobacter cell differentiation. Curr. Opin. Genet. Dev. 1:324-329. [DOI] [PubMed] [Google Scholar]
- 14.Grunenfelder, B., G. Rummel, J. Vohradsky, D. Roder, H. Langen, and U. Jenal. 2001. Proteomic analysis of the bacterial cell cycle. Proc. Natl. Acad. Sci. USA 98:4681-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison, D. M., J. Skidmore, J. P. Armitage, and J. R. Maddock. 1999. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol. Microbiol. 31:885-892. [DOI] [PubMed] [Google Scholar]
- 16.Hou, S., T. Freitas, R. W. Larsen, M. Piatibratov, V. Sivozhelezov, A. Yamamoto, E. A. Meleshkevitch, M. Zimmer, G. W. Ordal, and M. Alam. 2001. Globin-coupled sensors: A class of heme-containing sensors in Archaea and Bacteria. Proc. Natl. Acad. Sci. USA 98:9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou, S., R. W. Larsen, D. Boudko, C. W. Riley, E. Karatan, M. Zimmer, G. W. Ordal, and M. Alam. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540-544. [DOI] [PubMed] [Google Scholar]
- 18.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 15:2393-2406. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, K. K., H. Yokota, and S. H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 21.Kim, Y. I., R. E. Burton, B. M. Burton, R. T. Sauer, and T. A. Baker. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5:639-648. [DOI] [PubMed] [Google Scholar]
- 22.Koyasu, S., and Y. Shirakihara. 1984. Caulobacter crescentus flagellar filament has a right-handed helical form. J. Mol. Biol. 173:125-130. [DOI] [PubMed] [Google Scholar]
- 23.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 24.Levchenko, I., C. K. Smith, N. P. Walsh, R. T. Sauer, and T. A. Baker. 1997. PDZ-like domains mediate binding specificity in the Clp/Hsp100 family of chaperones and protease regulatory subunits. Cell 91:939-947. [DOI] [PubMed] [Google Scholar]
- 25.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 26.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okumura, H., S. Nishiyama, A. Sasaki, M. Homma, and I. Kawagishi. 1998. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J. Bacteriol. 180:1862-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega, J., S. K. Singh, T. Ishikawa, M. R. Maurizi, and A. C. Steven. 2000. Visualization of substrate binding and translocation by the ATP-dependent protease, ClpXP. Mol. Cell 6:1515-1521. [DOI] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Stephens, C., C. Mohr, C. Boyd, J. Maddock, J. Gober, and L. Shapiro. 1997. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J. Bacteriol. 179:5355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai, J. W., and M. R. K. Alley. 2001. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J. Bacteriol. 183:5001-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai, J. W., and M. R. K. Alley. 2000. Proteolysis of the McpA chemoreceptor does not require the Caulobacter major chemotaxis operon. J. Bacteriol. 182:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]