Abstract
Surface layers (S-layers), which form the outermost layers of many Bacteria and Archaea, consist of protein molecules arranged in two-dimensional crystalline arrays. Bacillus anthracis, a gram-positive, spore-forming bacterium, responsible for anthrax, synthesizes two abundant surface proteins: Sap and EA1. Regulatory studies showed that EA1 and Sap appear sequentially at the surface of the parental strain. Sap and EA1 can form arrays. The structural parameters of S-layers from mutant strains (EA1− and Sap−) were determined by computer image processing of electron micrographs of negatively stained regular S-layer fragments or deflated whole bacteria. Sap and EA1 projection maps were calculated on a p1 symmetry basis. The unit cell parameters of EA1 were a = 69 Å, b = 83 Å, and γ = 106°, while those of Sap were a = 184 Å, b = 81 Å, and γ = 84°. Freeze-etching experiments and the analysis of the peripheral regions of the cell suggested that the two S-layers have different settings. We characterized the settings of each network at different growth phases. Our data indicated that the scattered emergence of EA1 destabilizes the Sap S-layer.
S-layers are ubiquitous proteinaceous paracrystalline sheaths, present on the surfaces of many Archaea and Bacteria (23, 27). S-layers are of no value as taxonomic markers, as their presence is often strain dependent. As S-layers are usually the outermost component of the cell envelope, they might be important in the interaction between the cell and its environment (28). The proposed functions of S-layers include shape maintenance, molecular sieving, and phage fixation. S-layers may act as virulence factors by protecting pathogenic bacteria from complement, facilitating their binding to host molecules, or enhancing their ability to associate with macrophages (for a review see reference 23). S-layers are derived from the noncovalent, entropy-driven assembly of a (glyco)protein protomer(s) on the bacterial surface. Most arrays consist of a single protein species, with one well-documented exception, the S-layer of Clostridium difficile, which consists of two different proteins derived from the same precursor (2, 3, 31). In some species, more than one S-layer protein may be present at the surface of the bacterium. The S-layer proteins may be synthesized simultaneously (e.g., in Brevibacillus brevis and Aquaspirillum serpens) or sequentially through chromosomal rearrangements (e.g., in Campylobacter fetus and Bacillus stearothermophilus) (for a review see reference 23).
Bacillus anthracis, a gram-positive, spore-forming bacterium, is the etiological agent of anthrax, a disease involving toxemia and septicemia. Fully virulent bacilli produce two toxins, encoded by pX01, and a poly-γ-d-glutamic acid capsule, encoded by pXO2 (22). In the absence of the capsule, the cell wall is covered by an S-layer (9, 12). Two abundant proteins (EA1 and Sap) are colocalized at the surface of the bacilli (7, 18). Both components interact with the underlying cell wall via noncovalent bonds between their S-layer homolog (SLH) domains and a polysaccharide (15, 16). The capacity of each component to form an array has been studied by use of a genetic approach involving the deletion of one or both of the corresponding genes in various B. anthracis backgrounds. The surfaces of single and double mutants were then analyzed. EA1 is an S-layer component in both capsulated and noncapsulated strains, whereas the Sap S-layer has been detected only in capsulated cells (17, 18). Although both proteins have been simultaneously observed at the surface of the parental strain, the lattice is very similar to that made by EA1 alone. These data raised several questions concerning the structural organization of the B. anthracis S-layer. In particular, it has remained unclear whether B. anthracis synthesizes a single array consisting of two protomers or whether two distinct S-layers are simultaneously present.
The recent study of the regulation of EA1 and Sap synthesis showed that these proteins are sequentially synthesized in a growth phase-dependent manner, with synthesis of Sap preceding that of EA1 (20). This is of interest because in B. anthracis the mechanism involved in the sequential synthesis of S-layer proteins is due to reversible transcriptional regulation rather than to DNA rearrangements as in other species. Furthermore, this explains why an EA1-like S-layer at the surface of parental bacteria grown to stationary phase was observed. It is noteworthy that, in the Bacillus cereus group, to which B. anthracis belongs, when an S-layer is present (S-layers are mostly limited to pathogenic bacteria from a monophyletic group), the simultaneous occurrence of two S-layer genes seems a general situation (19). We therefore chose the B. anthracis S-layer as an interesting model to study the setting of multiple S-layer components.
Our main aim was to determine whether the Sap protein can form a regular layer in the absence of the capsule. We used image processing to calculate projection map networks from electron micrographs for each array. Finally, as the synthesis of Sap and EA1 is sequential, we analyzed the dynamics of the surface structure of a parental strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All B. anthracis strains used were isogenic and derived from the plasmidless strain 9131 (7). The Sap mutant RBA91 only produces EA1. The EA1 mutant SM91 produces only Sap (18). Cells were grown in SPY medium (7), brain heart infusion (BHI) medium (Difco Laboratories), or Luria (L) broth (21). Cells were precultured on BHI medium or L agar plates. SPY was directly inoculated with spores.
Preparation and electron microscopy of negatively stained array fragments.
Samples were prepared and negatively stained as described previously (18). EA1 images were recorded at a magnification of ×45,000 on a Philips CM12 electron microscope operating at 80 kV. Sap images were recorded on a Jeol 1010 electron microscope operating at the same voltage, but at ×40,000 magnification.
Freeze fracture and freeze-etching.
Bacterial suspensions were washed with 50 mM Tris-HCl, pH 7.5, buffer and harvested by centrifugation. The pellet (15 μg) was placed between a thin copper holder and a thin copper plate before quenching in liquid propane, as described by Aggerbeck and Gulik-Krzywicki (1). The frozen sample was fractured at −125°C in a vacuum of about 10−7 torr by removing the upper plate with a knife that had been cooled in liquid propane in a Balzers 301 freeze-etching unit. The fractured sample was etched at −100°C for 3 to 5 min and then replicated with a 1.5-nm-thick platinum-carbon deposit and backed with chromic acid before being washed with distilled water.
Whole-mount bacterium immunolabeling and negative staining.
The efficient use of negative staining on whole-mount bacteria requires the induction of leakage of the cytoplasmic material; thus, growth parameters were analyzed. B. anthracis cells were resistant to osmotic shock in all of the media except for L broth. The parental strain was grown in L broth to optical densities at 600 nm of 0.8 and 2.5. The cells were then harvested by centrifugation and resuspended in 1/100 volume of L broth supplemented with 10 mM MgSO4. To avoid cell lysis during the immunolabeling, all remaining steps were carried out in the same medium. The bacteria were then directly adsorbed onto carbon collodion-coated glow-discharged nickel grids. The bacteria were then immunolabeled with mouse anti-Sap antibodies and rabbit anti-EA1 antibodies, as previously described (18). Binding was revealed with 10-nm gold-conjugated anti-mouse antibodies and 15-nm gold-conjugated anti-rabbit antibodies (British Biocell International). The grids were then rinsed in 10 mM Tris-HCl, pH 8.0-10 mM MgSO4 to produce osmotic shock, allowing cell leakage. The negative staining was carried out in uranyl acetate.
Image processing of negatively stained two-dimensional arrays.
Seven images of EA1 negatively stained two-dimensional arrays were selected for image processing. Areas of 1,024 by 1,024 pixels, corresponding to 227 by 227 nm on the specimen, were digitized (pixel size = 10 μm) with a Leafscan 45 charge-coupled device array microdensitometer. Four homogeneous Sap images were selected. Areas of 1,024 by 1,024 pixels were digitized (pixel size = 20 μm) with the Leafscan 45 microdensitometer. Within each of those areas, a 512- by 512-pixel area was further selected with Ximdisp (29). Thus, the areas finally analyzed measured 256 by 256 nm. The Fourier transforms of all areas were indexed with Spectra (25). Subsequent processing, including lattice corrections (11), was done with the Medical Research Council image analysis package (4). Space group determinations were performed with the Allspace program (32). Projection maps were generated with the CCP4 program suite (Collaborative Computational Project; 1994 edition).
Image processing of shadowed freeze fractured and freeze-etched two-dimensional arrays.
Freeze fracture electron micrographs were processed by the widely used cross-correlation averaging technique (8, 24). The areas that appeared to be the most ordered (domains) were selected by use of optical diffraction and digitized. The two-dimensional lattice parameters, a, b, and γ, were determined by least-squares refinement from the Fourier transform by using the Spectra program (25). The image was then Fourier filtered; large masking holes were used to reduce the noise but keep any lattice distortions. The cross-correlation map was calculated between a reference area extracted from the Fourier-filtered image and the image itself. An averaged motif was calculated by adding image regions centered on the best correlation peak positions by using an automated procedure specially designed for freeze fracture images (5). For unidirectional shadowing, the surface profile of the replica of the fracture face was estimated from the averaged motif by using a deconvolution process based on a simple modeling of the shadowing process, which leads to a phase correction of the Fourier coefficients (30, 33). Such a procedure has been shown to yield motifs that closely resemble the one obtained with rotary shadowing (6). The images used for comparisons were finally obtained by merging arrayed motifs (for rotary shadowing) or surface profiles (for unidirectional shadowing) calculated from different freeze fracture images.
RESULTS
Electron-microscopic visualization of Sap and EA1 S-layers.
Before studying the surface of the parental strain, we investigated a putative array formed by each single protein by use of single mutants SM91 (Sap+ EA1−) and RBA91 (Sap− EA1+). We negatively stained array fragments to enable us to observe the network of RBA91 with the aim of calculating a projection map of the EA1 S-layer (18). The layer was the most visible after detachment from the cell surface. This also improved the resolution in terms of electron microscopy. Yet, for the SM91 strain, we never obtained isolated array fragments (18). Given that Sap forms an S-layer on capsulated strains (17), we hypothesized that it would also make an S-layer on noncapsulated strains and that only technical problems had hampered its characterization. Therefore we studied negatively stained deflated SM91 bacteria. This was carried out by means of a leakage step, by the use of osmotic shock, to maintain the integrity of the S-layer. This treatment made electron microscopy analysis possible. An ordered structure at the surface of SM91 was always observed, indicating that, even in the absence of a capsule, Sap is able to form an S-layer. This S-layer appeared to form a very homogeneous and ordered network.
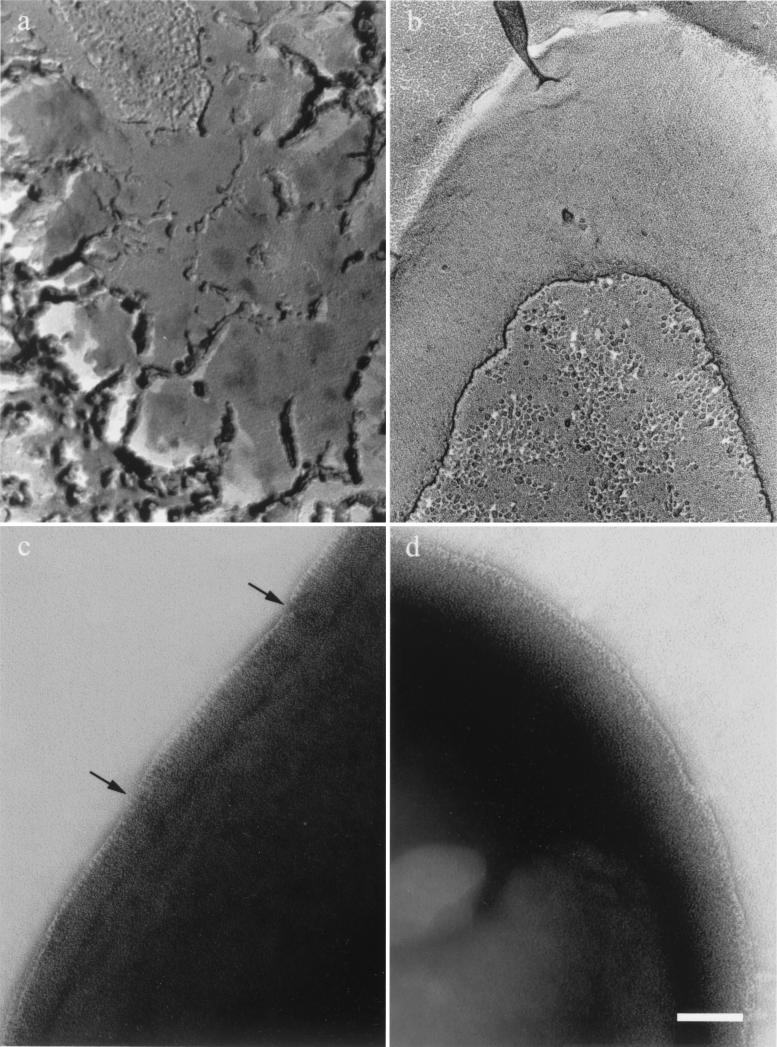
We then used a freeze fracture and freeze-etching technique to observe entire surfaces of each single mutant (Fig. 1a and b). The S-layer was visible on both strains. However, the Sap network was continuous (Fig. 1b), whereas the EA1 crystal formed relatively random patches in a mosaic-like organization (Fig. 1a). To gain further insight into these organizations, we analyzed the periphery of negatively stained whole-mount bacteria. The cells were very dense with regard to the electron beam, but we could see a thin structure surrounding the bacteria corresponding to the “edge” of the S-layer (Fig. 1c and 1d). An unbroken layer surrounded SM91 (Sap+) bacteria (Fig. 1d). In contrast, RBA91 (EA1+) bacteria were surrounded by a broken, sometimes overlapping layer (Fig. 1c). These data correlate with those obtained by freeze-etching and emphasize the different organizations of the two S-layers.
FIG. 1.
Difference in S-layer organization in RBA91 and SM91. RBA91 (EA1+) (a and c) and SM91 (Sap+) (b and d) bacteria were analyzed by freeze-etching (a and b) and by negative staining of the periphery of intact cells (c and d). Arrows indicate breakage observed in the EA1 edge. Scale bar, 100 nm.
Therefore, the electron microscopy analyses indicate that both Sap and EA1 can independently form an S-layer, albeit with mechanistic differences.
Structural analysis of each S-layer.
We calculated projection maps for both crystals by analyzing computer-filtered images of negatively stained specimens; the structural analysis was complemented with an independent freeze fracture and freeze-etching analysis.
(i) Projection map of the EA1 array.
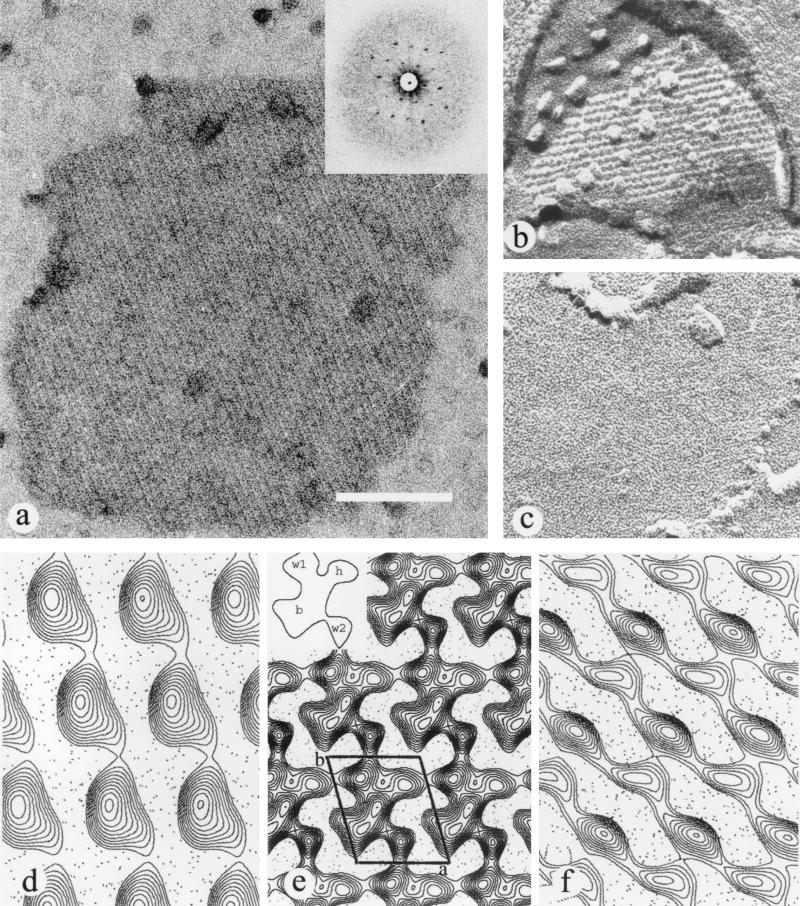
Projection maps were calculated by use of electron micrographs of negatively stained expelled fragments from the single mutant RBA91 (Fig. 2a). The final EA1 map was obtained by merging seven independent images. The final resolution used to calculate the projection map was 20 Å. The projection map shows that the EA1 protein crystallizes as one molecule per unit cell and thus belongs to the p1 plane group (Fig. 2e). The unit cell parameters were as follows: a = 69 Å, b = 83 Å, and γ = 106° (Fig. 2b). The projection map shows that the mass distribution of the protein consists of four main domains: three small domains and one very large domain (Fig. 2e, inset). The largest domain has a central body (b domain) around which the three smaller domains are distributed as head (h domain) and wings (w1 and w2 domains), the whole having a bird shape (Fig. 2e). The strongest intermolecule contacts, which ensure the natural linear crystallization of EA1, are formed by the w1 and w2 domains. The lateral association is made by the h domain of one molecule linking to the top wing (w1) of the next lateral molecule.
FIG. 2.
Structural analysis of the EA1 S-layer. (a) Negatively stained expelled fragment of an EA1 array; inset, diffraction pattern. (b and c) Freeze-etched unidirectional shadowed specimen showing both the corrugated (b) and smooth (c) sides. (e) Projection map of a negatively stained EA1 array; inset, schematic representation of the protein mass as a flying bird. (d and f) Projection map of the corrugated (d) and smooth (f) sides of the EA1 array. Scale bar, 100 nm.
The unidirectional shadowed EA1 arrays obtained on freeze fractured and freeze-etched specimens clearly showed two different sides because the fracture yielded both outside-out and inside-out fragments. A smooth side and a corrugated side were observed, in agreement with the results obtained by using different S-layers (Fig. 2b and c) (13, 14). The surface projections calculated for each side presented different characteristics. The corrugated side, thought to face inwards, showed only one large domain, with a globular shape (Fig. 2d), like those that could be observed directly on the replicas. The smooth side, which is usually thought to face outwards, showed two attached small elongated domains (Fig. 2f). Values of the mean unit cell parameters obtained for the replicas were a = 67 Å, b = 84 Å, and γ = 105° for the smooth side and a = 68 Å, b = 81 Å, and γ = 106° for the corrugated side. Images used for comparison were finally obtained by merging the surface profiles computed from different images of freeze fractured and freeze-etched samples (three for the smooth side and two for the rough side). However, for the sake of comparison, the final cell dimensions used to calculate the projection of the replicas corresponded to negatively stained images. The resolutions of the projections were around 25 Å.
(ii) Projection map for the Sap array.
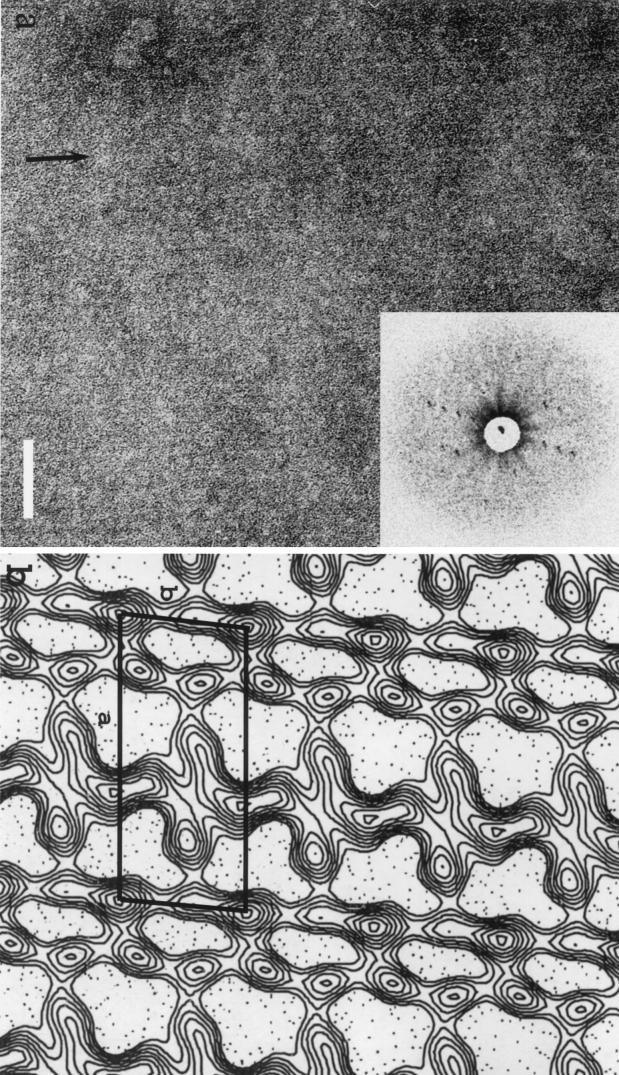
Electron micrographs were recorded by using negatively stained deflated bacteria of the single mutant SM91 (Fig. 3a). Four independent images were selected, processed, merged, and used to construct the final projection map. The resolution used to calculate the projection was 30 Å. The projection map was calculated with no symmetry applied. Values of the unit cell parameters of this crystal cell were a = 184 Å, b = 81 Å, and γ = 84° (Fig. 3b). The projection map revealed six or seven domains that repeat themselves along the two axes of the crystal. However, the molecular boundaries of each individual protein could not be unambiguously defined. The layout of the seven domains is indicative of a rather lean or linear network.
FIG. 3.
Structural analysis of the Sap S-layer. (a) Negatively stained array from a deflated SM91 (Sap+) bacterium (the lattice is best seen along the arrow); inset, diffraction pattern. (b) Projection map of the Sap array; a cell unit has been drawn. Scale bar, 100 nm.
Images were also recorded on rotationally shadowed Sap arrays obtained from freeze fractured and freeze-etched specimens. Networks, very similar to that observed with the negatively stained deflated bacteria were visible in all cases (data not shown).
Dynamic emergence of two independent S-layers on parental B. anthracis cells.
Sap and EA1 appear sequentially on the surface of the bacilli (20). However, at any given time, both proteins are present. Initially Sap is the most abundant protein, and then it is replaced by EA1. This suggests that two S-layers appear sequentially. We therefore tried to visualize the successive networks on the surface of the parental strain and to identify their protein compositions simultaneously (Sap or EA1). Thus, we immunolabeled bacteria and then used the “deflated-bacterium” technique.
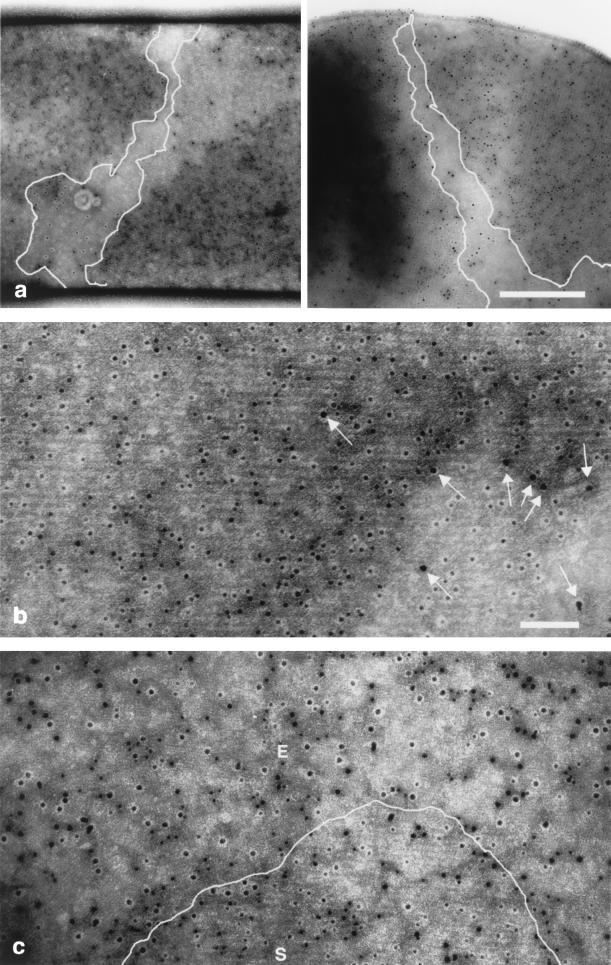
We observed the parental strain, 9131 (EA1+ Sap+), during the exponential phase and during stationary phase. During exponential phase, immunolabeling revealed an average of 88% 10-nm gold particles per surface unit, which indicates that the largest network structure is that made of Sap (Fig. 4a and b). This network seemed to wrap around the bacteria. In contrast, at the same time point, EA1 labeling was punctate and scattered and did not seem to be organized (Fig. 4b). During stationary phase, we found 50% 15-nm gold particles per surface unit, indicating that the Sap/EA1 ratio was changing. EA1 array fragments were visible, but some Sap array fragments persisted (Fig. 4c). Thus, in early stationary phase, patches of two S-layers coexist. The EA1 crystalline lattices are less visible than those of Sap, as the lattice size is smaller. They are composed of patches oriented in different directions, which is consistent with the freeze-etching images.
FIG. 4.
Alternation of the S-layer networks on the parental strain according to the growth phase. The parental 9131 strain (Sap+ EA1+) was subjected to double-immunogold labeling during the exponential (a and b) and stationary (c) phases. Deflated bacteria were incubated with mouse anti-Sap and rabbit anti-EA1 antibodies. Binding was revealed with 10-nm gold-conjugated anti-mouse antibodies and 15-nm gold-conjugated anti-rabbit antibodies. Scale bars, 500 (a) and 100 nm (b and c). The Sap S-layer can be seen to wind around the axis of the bacterium in two independent images. The areas in panel a not covered by the Sap array are delineated, and the labeled Sap array is visible in the magnified figure (b). The few 15-nm particles revealing EA1 protomers are indicated by arrows in panel b. (c) During stationary phase, the emergence of the labeled EA1 array (E) destabilizes the labeled Sap array (S).
DISCUSSION
To our knowledge, this is the first study where mutant strains have been used to elucidate the S-layer structural organization of a bacterium producing two S-layer proteins. EA1 S-layer fragments were easily visible because they detach from the surface of EA1+ bacteria during preparation for electron microscopy analysis. In contrast, Sap S-layer fragments were recovered only from a capsulated strain, suggesting that they need to be stabilized by the capsule (17). It was, therefore, important to establish whether, even in the absence of a putative stabilizing element (e.g., the capsule), Sap protomers could constitute an S-layer. Our results show that this is indeed the case. Both freeze-etching and negative staining of deflated bacteria detected a Sap S-layer at the surface of a noncapsulated Sap+ EA1− strain. EA1 and Sap arrays are clearly organized differently, suggesting that they have different settings. The Sap-forming layer is homogeneously distributed on the cell surface, and the EA1 layer displays a crazy-paving aspect. We constructed projection maps for each array by using single mutants. We used freeze fracture combined with freeze-etching to visualize the surface of the bacteria and negative staining to study the two-dimensional crystal. The combination of these two techniques applied to RBA91 helped to elucidate the spatial organization of the EA1 S-layer. The projection map of EA1, obtained from negatively stained samples, for which the resolution was 20 Å, revealed a compact globular shape, reminiscent of a bird. The results obtained from each side, by freeze-etching experiments, were strikingly different. Despite these differences, the sum of the surface projections of both sides (rough and smooth) is consistent with the projection map obtained from the negatively stained sample. Comparison of the three projection maps obtained (Fig. 2d to f) suggested that the globular domain, found on the rough side (Fig. 2d), corresponds to the central domain of the negative-staining map (Fig. 2e, inset, domain b). Conversely, one of the two elongated domains found on the smooth side may correspond to the w1 and h domains and the other may correspond to w2 (Fig. 2e). This indicates that the head and wings (w1, h, and w2; Fig. 2e, inset), seen on the smooth side, are located on the outer surface. Conversely, the globular domain (Fig. 2e, inset), seen on the rough side, is located on the inner surface and thus faces the peptidoglycan (Fig. 2d) (13, 14). The fact that the body faces inwards suggests that the SLH domain, which is anchored in the cell wall (15), can account for the density.
Unfortunately, we could not unambiguously define the molecular boundaries of Sap due to the poor resolution. This low resolution could be the result of taking images on deflated bacteria. Those bacteria may have retained internal organites which would render the surface inhomogeneously planed. For those reasons, the possible p2 symmetry observed on the map was not further explored at this stage of the structural study. It will be discussed in a separate study based on forthcoming data. Our present data suggest, however, that the Sap S-layer presents somehow a fibril-like layout compared to the EA1 layer. Moreover, a fine edge was observed with Sap, whereas a thicker edge was observed with EA1. Note also that the maps of the Sap S-layer obtained on freeze-etched samples closely resembled those obtained with negatively stained deflated Sap+ bacteria (data not shown). Finally, the apparent linear and lean nature of the Sap lattice may provide flexibility to the S-layer during the exponential phase. Indeed, the Sap edge is continuous, whereas that of EA1 has breakages (Fig. 1d).
During the exponential phase the parental strain is mainly covered by Sap proteins, organized in a noncontinuous S-layer that seems to wrap itself along the length of the bacteria (Fig. 4a). In fact, in gram-positive bacteria, the S-layer lattice primarily consists of multiple bands arranged helically on the cylindrical part of the cell (10, 26). Because the Sap protein is synthesized during the exponential phase (20), the ribbon-like appearance could also be due to the fact that this S-layer builds up during the exponential phase, during which the length of the bacterium increases. We also demonstrated that during stationary phase the Sap S-layer is progressively replaced by the EA1 S-layer, consistent with the hypothesis of Mignot et al. (20). This raises questions about where and how the EA1 subunits appear on the surface. The EA1 nucleation points first appear mainly in the free spaces between Sap S-layer covered zones. The Sap array is then destabilized, and the bacterial surface is progressively colonized by EA1, as revealed by immunolabeling. During the stationary phase, the surface displays an EA1 S-layer with a patchwork pattern. This aspect correlates with the random emergence of these protomers. Interestingly, the random insertion of new subunits has mostly been found in gram-negative species (26). The cohesion of the EA1 array fragments is probably sufficient to allow the crystallization process to propagate around the initial nucleation sites, thus disrupting the Sap array. This is reflected by the mosaic pattern found on the parental strain during stationary phase and by the broken edging observed on this strain, which is due to fractures along boundaries of the crystalline patches. Furthermore, the prefragmentation of the EA1 S-layer may ease fragment detachment. The same findings were observed with RBA91, suggesting that, even in the absence of Sap, EA1 molecules appear and nucleate randomly. As for SM91, the S-layer homogeneously covers the whole bacterial surface. This indicates that, in absence of EA1, Sap can cover the whole surface.
The differences between the lattices suggest differences in pore size or in mechanical constraints. This could be important because nutriment requirements vary during different growth phases. Each array may also maintain diverse bacterial molecules in the vicinity of the cell. It should be noted that, in vivo, B. anthracis cells are covered, as the outermost structure, by a capsule. When capsule synthesis is induced in vitro by the addition of bicarbonate, the S-layer forms under the capsule filaments. The structural interaction between each array and the capsule is therefore of importance. There may be differences in the capsule setting depending on the underlying S-layer, and we are currently investigating this hypothesis.
Acknowledgments
We are grateful to M. Mock, in whose laboratory part of this work was carried out, for her continuing interest and to T. Gulik-Krzywicki for the accessibility to the freeze-etching unit. We thank G. Pehau-Arnaudet for his help in the preparation of the figures.
T. M. and S. M. were funded by the Ministère de l'Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Aggerbeck, L. P., and T. Gulik-Krzywicki. 1986. Studies of lipoproteins by freeze-fracture and etching electron microscopy. Methods Enzymol. 128:457-472. [DOI] [PubMed] [Google Scholar]
- 2.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187-1199. [DOI] [PubMed] [Google Scholar]
- 3.Cerquetti, M., A. Molinari, A. Sebastianelli, M. Diociaiuti, R. Petruzzelli, C. Capo, and P. Mastrantonio. 2000. Characterization of surface layer proteins from different Clostridium difficile clinical isolates. Microb. Pathog. 28:363-372. [DOI] [PubMed] [Google Scholar]
- 4.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 5.Delacroix, H., T. Gulik-Krzywicki, P. Mariani, and V. Luzzati. 1993. Freeze-fracture electron microscope study of lipid systems: the cubic phase of space group Pm3n. J. Mol. Biol. 229:526-539. [DOI] [PubMed] [Google Scholar]
- 6.Delacroix, H., T. Gulik-Krzywicki, P. Mariani, and J. L. Risler. 1993. Freeze-fracture electron microscopy of lyotropic lipid systems: quantitative analysis of cubic phases of space group Ia3d (Q230). Liq. Crystals 15:605-625. [Google Scholar]
- 7.Etienne-Toumelin, I., J.-C. Sirard, E. Duflot, M. Mock, and A. Fouet. 1995. Characterization of the Bacillus anthracis S-layer: cloning and sequencing of the structural gene. J. Bacteriol. 177:614-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank, J., W. Goldfarb, D. Eisenberg, and T. S. Baker. 1978. Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy 3:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhardt, P. 1967. Cytology of Bacillus anthracis Fed. Proc. 26:1504-1517. [PubMed] [Google Scholar]
- 10.Gruber, K., and U. B. Sleytr. 1988. Localized insertion of new S-layer during growth of Bacillus stearothermophilus strains. Arch. Microbiol. 149:485-491. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, R., J. M. Baldwin, T. A. Ceska, F. Zemlin, E. Beckmann, and K. H. Downing. 1990. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213:899-929. [DOI] [PubMed] [Google Scholar]
- 12.Holt, S. C., and E. R. Leadbetter. 1969. Comparative ultrastructure of selected aerobic spore-forming bacteria: a freeze-etching study. Bacteriol. Rev. 33:346-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovmöller, S., A. Sjörden, and D. Wang. 1988. The structure of crystalline bacterial surface layers. Prog. Biophys. Mol. Biol. 51:131-163. [DOI] [PubMed] [Google Scholar]
- 14.Luckevich, M. D., and T. J. Beveridge. 1989. Characterization of a dynamic S-layer on Bacillus thuringiensis. J. Bacteriol. 171:6656-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesnage, S., T. Fontaine, T. Mignot, M. Delepierre, M. Mock, and A. Fouet. 2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mesnage, S., E. Tosi-Couture, and A. Fouet. 1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927-936. [DOI] [PubMed] [Google Scholar]
- 17.Mesnage, S., E. Tosi-Couture, P. Gounon, M. Mock, and A. Fouet. 1998. The capsule and S-layer: two independent and yet compatible macromolecular structures in Bacillus anthracis. J. Bacteriol. 180:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesnage, S., E. Tosi-Couture, M. Mock, P. Gounon, and A. Fouet. 1997. Molecular characterization of the Bacillus anthracis main S-layer component: evidence that it is the major cell-associated antigen. Mol. Microbiol. 23:1147-1155. [DOI] [PubMed] [Google Scholar]
- 19.Mignot, T., B. Denis, E. Couture-Tosi, A. B. Kolstø, M. Mock, and A. Fouet. 2001. Distribution of S-layers on the surface of. Bacillus cereus strains: phylogenetic origin and ecological pressure. Environ. Microbiol. 3:493-501. [DOI] [PubMed] [Google Scholar]
- 20.Mignot, T., S. Mesnage, E. Couture-Tosi, M. Mock, and A. Fouet. 2002. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol. Microbiol. 43:1615-1628. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 23.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxton, W. O., and J. Frank. 1977. Motif detection in quantum noise-limited electron micrographs by cross-correlation. Ultramicroscopy 2:19-27. [DOI] [PubMed] [Google Scholar]
- 25.Schmid, M. F., R. Dargahi, and M. W. Tam. 1993. SPECTRA: a system for processing electron images of crystals. Ultramicroscopy 48:251-264. [DOI] [PubMed] [Google Scholar]
- 26.Sleytr, U. B., and T. J. Beveridge. 1999. Bacterial S-layers. Trends Microbiol. 7:253-260. [DOI] [PubMed] [Google Scholar]
- 27.Sleytr, U. B., and P. Messner. 1983. Crystalline surface layers on bacteria. Annu. Rev. Microbiol. 37:311-339. [DOI] [PubMed] [Google Scholar]
- 28.Sleytr, U. B., P. Messner, D. Pum, and M. Sára. 1996. Occurrence, location, ultrastructure and morphogenesis of S-layers, p. 5-33. In U. B. Sleytr, P. Messner, D. Pum, and M. Sára (ed.), Crystalline bacterial cell surface proteins. R. G. Landes Company, Austin, Tex.
- 29.Smith, J. M. 1999. Ximdisp—a visualization tool to aid structure determination from electron microscope images. J. Struct. Biol. 125:223-228. [DOI] [PubMed] [Google Scholar]
- 30.Smith, P. R., and J. Kistler. 1977. Surface reliefs computed from micrographs of isolated heavy metal shadowed specimens. J. Ultrastruct. Res. 61:124.. [DOI] [PubMed] [Google Scholar]
- 31.Takeoka, A., K. Takumi, T. Koga, and T. Kawata. 1991. Purification and characterization of S layer proteins from Clostridium difficile GAI 0714. J. Gen. Microbiol. 137:261-267. [DOI] [PubMed] [Google Scholar]
- 32.Valpuesta, J. M., J. L. Carrascosa, and R. Henderson. 1994. Analysis of electron microscope images and electron diffraction patterns of thin crystals of phi 29 connectors in ice. J. Mol. Biol. 240:281-287. [DOI] [PubMed] [Google Scholar]
- 33.Winkler, H., and H. Gross. 1988. Image interpretation of metal shadowed specimens with the aid of Monte Carlo simulation of the shadowing process. Image and signal processing in electron microscopy. Scanning Microsc. 2(Suppl.):379-386. [PubMed] [Google Scholar]