Abstract
HrcA is a regulator of bacterial heat shock gene expression that binds to a cis-acting DNA element called CIRCE. It has been proposed that HrcA and CIRCE function as a repressor-operator pair. We have purified recombinant HrcA from the pathogenic bacterium Chlamydia trachomatis and have shown that it is a DNA-binding protein that functions as a negative regulator of transcription. HrcA bound specifically to the CIRCE element in a concentration-dependent manner. HrcA repressed the in vitro transcription of a chlamydial heat shock promoter, and this repression was promoter specific. HrcA-mediated repression appears to be dependent on the topological state of the promoter, as repression on a supercoiled promoter template was greater than that on a linearized template. These results provide direct support for the role of HrcA as a transcriptional repressor in bacteria. This is the first report of the in vitro reconstitution of transcriptional regulation in Chlamydia.
Chlamydia trachomatis is a gram-negative bacterium that is the cause of the most prevalent sexually transmitted disease in the United States (13). C. trachomatis is also the causative agent of trachoma, an ocular infection that is one of the leading causes of preventable blindness (13). Chlamydia spp. share an unusual developmental life cycle (4) in which infectious, but metabolically inert, elementary bodies (EBs) enter eukaryotic host cells and differentiate into metabolically active reticulate bodies (RBs). RBs divide by binary fission and later convert back to the EB form for release from the host cell to start another cycle of infection. Gene expression is clearly regulated during this unusual developmental cycle (2, 3, 7, 18), but, owing to the lack of an experimental method for genetic transformation, the mechanisms of gene regulation are not well understood.
Chlamydial pathogenesis involves the host immune response to chronic chlamydial infection, and it has been proposed that chlamydial heat shock proteins play an important role. Chlamydial GroEL (Hsp60) and GroES (Hsp10) have been implicated as primary antigens for eliciting this immune response (6). It has been proposed that chlamydial DnaK (Hsp70) is a factor for binding chlamydiae to host cells (15).
We are interested in the mechanisms that control the regulation of heat shock genes in Chlamydia. Unlike Escherichia coli, Chlamydia does not appear to use the heat shock sigma factor, σ32, to regulate heat shock transcription since the coding sequence for a σ32 homologue has not been identified in the chlamydial genome (5, 20). Instead, Chlamydia appears to contain the necessary components for an alternative mechanism that involves the derepression of heat shock gene transcription in response to a temperature upshift (10). The C. trachomatis genome contains a homologue of the heat shock repressor gene, hrcA, as well as DNA sequences that are similar to the consensus HrcA binding sequence. This DNA element is an inverted-repeat sequence called CIRCE (controlling inverted repeat of chaperone expression), and, in Chlamydia, it is located upstream of the promoters of the dnaK and groE operons (22). In several other bacterial systems, disruption of either hrcA or the CIRCE element results in derepression of heat shock protein expression (1, 17, 27). Recombinant Bacillus subtilis HrcA binds a CIRCE-containing DNA fragment, as shown by an electrophoretic mobility shift assay (EMSA) (26). The CIRCE element has been identified in over 40 eubacterial species, typically in association with a groE or dnaK heat shock operon (10). Largely because of the difficulty in overexpressing recombinant HrcA protein in E. coli, direct evidence for the activity of HrcA as a transcriptional regulator in vitro has not yet been shown.
In this report, we present a functional in vitro analysis of chlamydial HrcA by examining two of its activities: DNA binding and transcriptional repression. Our results show that HrcA binds specifically to CIRCE and that HrcA represses transcription of the dnaK promoter in a promoter-specific manner. We also present evidence that transcriptional repression by HrcA may be dependent on supercoiling.
MATERIALS AND METHODS
Reagents.
The following products were obtained from the sources given and were used according to the manufacturer's specifications: restriction enzymes, calf intestinal alkaline phosphatase, T4 DNA ligase, rRNasin, the pGEM-7Zf(+) plasmid vector, and Taq DNA polymerase, Promega Biotech (Madison, Wis.); T4 polynucleotide kinase and pRSET expression vector, Invitrogen (Carlsbad, Calif.); Sequenase kit, U.S. Biochemicals (Cleveland, Ohio); E. coli BL21(DE3), Stratagene (La Jolla, Calif.); nucleoside triphosphates, 3′-O-methylguanosine 5′-triphosphate, poly(dI-dC), HiTrap chelating column, and HiPrep Sephacryl S-200 gel filtration column, Amersham (Arlington Heights, Ill.); Centriplus columns, Millipore (Bedford, Mass.); oligonucleotide primers, Sigma Genosys (The Woodlands, Tex.); 32P-labeled nucleoside triphosphates, ICN (Costa Mesa, Calif.); Pwo DNA polymerase and mini-Quick Spin DNA columns, Roche Diagnostics (Indianapolis, Ind.); protein assay reagent, Bio-Rad (Hercules, Calif.).
DNA manipulation.
Standard recombinant methods were used for nucleic acid preparation and analysis (12). DNA was amplified by PCR as previously described (21) with an MJ Research (Incline Village, Nev.) PT-200 thermocycler. DNA sequencing was performed by the dideoxy chain termination method with a Sequenase kit on double-stranded plasmid DNA.
Cloning of chlamydial hrcA.
Plasmid pMT1133 is an expression vector that contains the entire coding region for C. trachomatis hrcA cloned into expression vector pRSET-C, which contains the coding sequence for the six-His tag. The hrcA insert (excluding the initial ATG start codon) was amplified by PCR from C. trachomatis L2 genomic DNA by using Pwo DNA polymerase. The PCR primers T122 (5′-GAAAATAGAATAGAAATGTCCCAACTGCGA) and T123 (5′-CCGGTACCTCATGATAGCTCCTTAGCGGGTAAT) were based on sequence information from the Chlamydia Genome Project (http://chlamydia-www.berkeley.edu:4231/) (20). The PCR product was digested with KpnI and cloned into pRSET-C between KpnI and blunted BamHI sites. The HrcA expression construct was sequenced, and its sequence was compared to the sequence of the C. trachomatis D genome and partially completed L2 genome.
Overexpression and purification of HrcA protein.
His6-HrcA was overexpressed in E. coli BL21(DE3) freshly transformed with pMT1133. Two liters of cells was grown at 37°C to an optical density at 600 nm of 0.5 and induced with 1 mM isopropyl-β-d-thiogalactosidase (IPTG). After 2 h, cells were pelleted, resuspended in 20 ml of buffer N (10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of pepstatin A/ml), and disrupted with a Branson Sonifier 450 (twice for 60 s each). Soluble protein was separated from cell debris by centrifugation at 10,000 × g for 10 min at 4°C (Beckman JA-17 rotor), and the lysate was loaded onto a 1-ml Ni2+-charged HiTrap chelating column. The column was washed with 10 ml of buffer N containing 50 mM imidazole followed by 10 ml of buffer N containing 100 mM imidazole. Protein was eluted with buffer N containing 250 mM imidazole, and fractions containing the highest protein concentrations were pooled. Pooled fractions were loaded onto an Amersham HiPrep 26/60 Sephacryl S-200 gel filtration column and eluted with 360 ml of buffer N. Fractions with the highest protein concentration were collected and concentrated with a Centriplus YM-30 column and spun for 65 min at 3,000 × g (Beckman JA-17 rotor). Supernatant was dialyzed against storage buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 100 μM EDTA, 10 mM 2-mercaptoethanol, 100 mM NaCl, 30% glycerol) overnight and again for 4 h. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by silver staining. Protein concentrations were assayed by the Bio-Rad protein assay. Aliquots were stored at −70°C.
Production of polyclonal anti-HrcA antibodies.
Recombinant HrcA was gel purified and used to generate rabbit polyclonal antibodies (Harlan Bioproducts for Science, Madison, Wis.).
EMSA templates.
A 110-bp restriction fragment containing the dnaK promoter region and its CIRCE element was used as a probe for gel retardation assays. This fragment was amplified by PCR with C. trachomatis serovar L2 genomic DNA and primers T155 (5′-GGGAATTCATACAAGAAAGATCCCTCCA) and T156 (5′-GGGGTACCGTGTCATTATAAGAAAACCGTCT). A nonspecific competitor fragment was produced by amplifying a portion of the 5′ untranslated region of the groES gene, which does not contain a putative CIRCE region. This fragment was amplified from L2 genomic DNA with primers T157 (5′-GGGAATTCGACTTTGCTATCGTTCTTCCTC) and T158 (5′-GGGGTACCTATTTTTATATTCTATGAGGCCTCGT). PCR products were cloned into the SmaI site of pGEM-7Zf(+) to generate plasmids pMT1134 (dnaK with CIRCE) and pMT1122 (nonspecific competitor) for use in the EMSA experiments described below. A double-stranded oligonucleotide containing CIRCE was created by reannealing primers T224 (5′-AACTAGCACTCTTTAGTTGCGAGCGCTAAAA) and T225 (5′-TTTTAGCGCTCGCAACTAAAGAGTGCTAGTT). Equimolar amounts of T224 and T225 were combined in annealing buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA), heated to 94°C in a thermocycler, and cooled to 4°C over a period of 1 h.
EMSA.
The EMSA restriction fragments were prepared by digestion of pMT1134 and pMT1122 with XbaI and EcoRI, dephosphorylation with calf intestinal alkaline phosphatase, and gel purification from a 2% agarose gel. The probe DNA was labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Free nucleotides were removed with mini-Quick Spin DNA columns, and probe activity was determined by liquid scintillation counting. The EMSA reaction mixture contained 1× binding buffer (40 mM Tris-HCl [pH 8.0], 4 mM MgCl2, 70 mM KCl, 135 μM EDTA, 100 μM dithiothreitol, 7.5% glycerol), 0.5 nM labeled probe, and various amounts of purified His6-HrcA. Some reaction mixtures also included a competitor restriction fragment, competitor double-stranded oligonucleotide, or polyclonal anti-HrcA antibodies. Reaction mixtures were incubated for 20 min at room temperature and then loaded onto a 7% polyacrylamide EMSA gel and electrophoresed at 75 V in 0.5× Tris-borate-EDTA buffer (11). After electrophoresis, the gels were dried and exposed to a phosphorimager plate. Plates were scanned with a Bio-Rad Personal FX scanner, and the data were analyzed with Quantity One software (Bio-Rad).
Purification of C. trachomatis RNA polymerase.
RNA polymerase was partially purified from C. trachomatis LGV serovar L2 at 20 h postinfection by heparin-agarose chromatography as previously described (21).
Construction of transcription plasmids.
Plasmid pMT1150 contains the promoter region of omcB (−122 to +5), which was amplified by PCR from C. trachomatis serovar L2 genomic DNA with primers T179 (5′-TTATAGACAATATTACATTATAAAATAAAAATTATATCAATTGT) and T180 (5′-AGCGAATTCTTTGAATCCGAGCTGTTTATTATTT). Plasmid pMT1161 contains the dnaK promoter (−230 to +5) amplified from plasmid pMT579 (22) with primers dnaK2 (5′-AAGTTGGTGTCATTATAGGAAAACC) and T227 (5′-AGCGAATTCCCTATAAATTGATCATTGGG). Each promoter insert was cloned upstream of a promoterless, G-less cassette transcription template in plasmid pMT1125. pMT1125 is a derivative of pMT504 (21), which was modified by removal of the rRNA promoter and control G-less cassette and introduction of a pair of PacI restriction sites into the remaining G-less cassette. The tandem PacI sites provide a convenient method for generating a transcription plasmid with a shorter G-less cassette encoding a transcript that is shorter by 28 nucleotides. This approach allows the transcripts from two plasmids in the same transcription reaction to be distinguished by electrophoresis. Thus, pMT1158, encoding a shorter omcB-specific transcript, was produced by digestion of pMT1150 with PacI, followed by ligation. Transcription of pMT1161(dnaK) resulted in a 154-nucleotide (nt) transcript, while transcription of pMT1158(omcB) produced a 126-nt transcript. For some experiments, the plasmid DNA was linearized by digestion with BamHI and gel purified from a 1% agarose gel.
In vitro transcription.
The reaction mixture contained 50 mM potassium acetate, 8.1 mM magnesium acetate, 50 mM Tris acetate (pH 8.0), 27 mM ammonium acetate, 2 mM dithiothreitol, 400 μM ATP, 400 μM UTP, 1.2 μM CTP, 0.21 μM [α-32P]CTP (800 Ci/mmol), 100 μM 3′-O-methylguanosine 5′-triphosphate Na salt, 18 U of rRNasin, 5% glycerol, 15 nM supercoiled DNA template, and various amounts of purified His6-HrcA. The reaction mixture was incubated for 20 min at room temperature. Then, 0.7 μl of heparin-agarose-purified C. trachomatis RNA was added, and the reaction mixture was incubated for a further 15 min at 37°C. The final reaction volume was 10 μl. The reaction was terminated by the addition of 10 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). A 10-μl portion of the sample was electrophoresed on an 8 M urea-6% polyacrylamide gel. After electrophoresis, the gels were dried and exposed to a phosphorimager plate. Plates were scanned with a Bio-Rad Personal FX scanner, and the data were analyzed with Bio-Rad Quantity One software.
RESULTS
Cloning, overexpression, and purification of chlamydial HrcA.
To study the function of HrcA as a candidate repressor of heat shock promoters, we purified HrcA as a six-histidine-tagged recombinant protein. We chose to overexpress and purify the chlamydial HrcA protein in E. coli, an organism that does not carry an hrcA homologue. The hrcA coding region was cloned into the E. coli protein expression vector pRSET and overexpressed in E. coli strain BL21(DE3). Initial experiments showed that over 90% of chlamydial His6-HrcA expressed in E. coli was insoluble. Rather than resorting to denaturing conditions, we chose to purify the soluble fraction of HrcA. After nickel column and gel filtration chromatography, the HrcA protein was >95% pure as assayed by silver staining (data not shown).
Chlamydial HrcA binds to the dnaK promoter region in vitro.
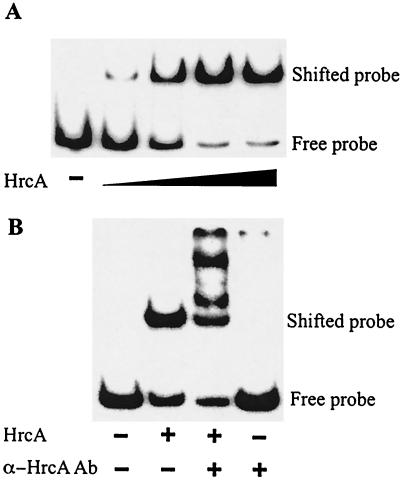
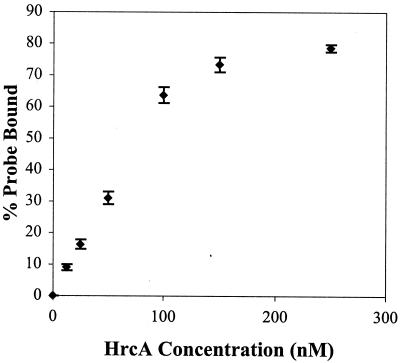
The binding of chlamydial HrcA to the dnaK promoter region was demonstrated with EMSA. HrcA bound the dnaK promoter region in a concentration-dependent manner, with saturation of binding at the highest concentrations of HrcA (Fig. 1A). Addition of polyclonal anti-HrcA antibodies produced a supershift with several slower-migrating products, indicating that the binding was HrcA specific (Fig. 1B, lane 3). The antibody alone did not cause a gel shift (Fig. 1B, lane 4). To measure the kinetics of binding, EMSA reactions were performed with six concentrations of HrcA and the results were quantified (Fig. 2). A 520:1 HrcA/DNA ratio was required for 80% DNA binding. An apparent dissociation constant (Kd) of 88 nM was obtained from a Lineweaver-Burk analysis of data points in the linear range.
FIG. 1.
EMSA with C. trachomatis dnaK CIRCE probe and recombinant HrcA. Positions of free and bound probes are marked. (A) Lane 1 (counting from the left), labeled probe alone; lanes 2 to 5, labeled probe with addition of twofold-increased concentrations of HrcA. (B) Lane 1, labeled probe alone; lane 2, addition of HrcA alone; lane 3, addition of HrcA and polyclonal anti-HrcA antibody; lane 4, addition of polyclonal anti-HrcA antibody alone. All reactions were performed with a 0.5 nM 32P-labeled dnaK CIRCE probe.
FIG. 2.
Quantification of EMSA. EMSA reactions with a 0.5 nM 32P-labeled dnaK CIRCE probe and various concentrations of recombinant HrcA were performed in triplicate and quantified by phosphorimager analysis. Error bars are standard deviations from the means.
HrcA specifically binds the CIRCE element.
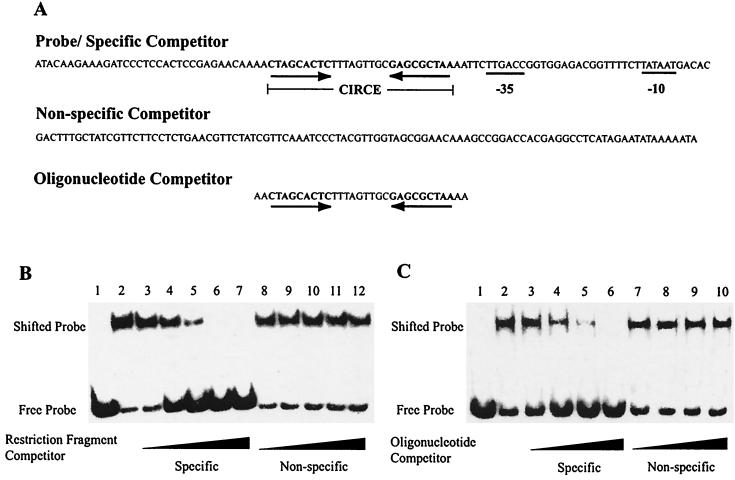
Competitive EMSA demonstrated that HrcA bound to the dnaK promoter region in a specific manner. HrcA binding to the dnaK promoter region was decreased in a concentration-dependent manner by a specific competitor consisting of the unlabeled dnaK restriction fragment (Fig. 3A). Almost complete loss of binding was observed in the presence of a 16-fold molar excess of specific competitor (Fig. 3B, lanes 3 to 7). In contrast, a nonspecific competitor that did not contain CIRCE had no effect on HrcA binding (Fig. 3B, lanes 8 to 12).
FIG. 3.
Competitive EMSA with C. trachomatis dnaK CIRCE probes and purified recombinant HrcA. (A) Representation of DNA fragments used in EMSA. CIRCE and promoter elements are marked. (B) Competition with CIRCE-containing restriction fragments. Lane 1, probe alone; lane 2, no competitor; lanes 3 to 7, molar excess of specific competitor relative to labeled probe (2-, 4-, 8-, 16-, and 32-fold); lanes 8 to 12, molar excess of nonspecific competitor (2-, 4-, 8-, 16-, and 32-fold). (C) Competition with CIRCE-containing oligonucleotides. Lane 1, probe alone; lane 2, no competitor; lanes 3 to 6, molar excess of specific competitor (4-, 8-, 16-, and 32-fold); lanes 7 to 10, molar excess of nonspecific competitor (4-, 8-, 16-, and 32-fold). All reactions, with the exception of reactions lacking HrcA, were performed with 0.5 nM labeled probe and 160 nM HrcA.
Additional competitive EMSA demonstrated that HrcA binding was dependent on the CIRCE element. A double-stranded oligonucleotide containing the dnaK CIRCE element, but not sequences from the dnaK core promoter, inhibited HrcA binding to the labeled dnaK probe in a concentration-dependent manner, with complete loss of binding in the presence of a 32-fold molar excess of specific oligonucleotide competitor (Fig. 3C, lanes 3 to 6). A control oligonucleotide containing poly(dI-dC) had no effect on HrcA binding (Fig. 3C, lanes 7 to 10).
HrcA represses the in vitro transcription of a heat shock promoter.
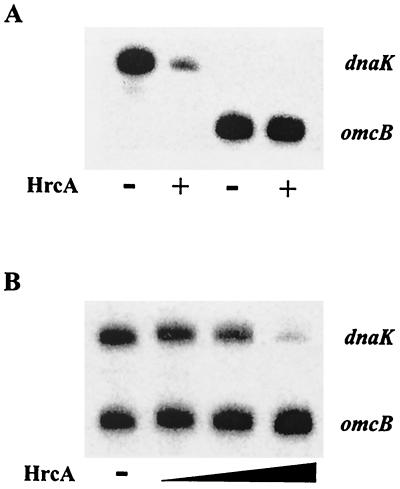
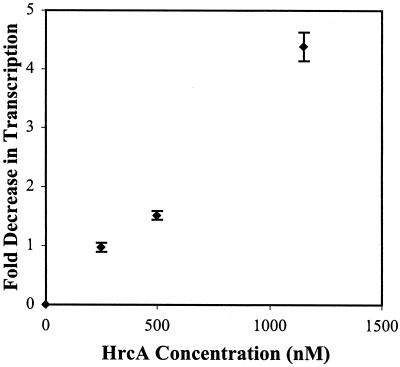
To determine if HrcA can function as a transcription factor, we measured the effect of HrcA on in vitro transcription of selected chlamydial promoters. HrcA repressed the dnaK promoter but not the omcB promoter (Fig. 4A). Repression of the dnaK promoter was concentration dependent, and a 76:1 HrcA/DNA ratio produced a fivefold decrease in transcription or 80% repression (Fig. 5). When the dnaK and omcB promoters were cotranscribed, the dnaK promoter was repressed in a concentration-dependent manner, and the omcB promoter showed a concentration-dependent increase in transcription (Fig. 4B). Transcription of the omcB promoter by itself was not affected by addition of HrcA (Fig. 4A), suggesting that the omcB promoter is not directly regulated by HrcA. These experiments showed that chlamydial HrcA specifically represses the dnaK heat shock promoter.
FIG. 4.
In vitro transcription with purified recombinant HrcA. (A) Transcription of single promoters. The upper transcript is from the dnaK promoter; the lower transcript is from the omcB promoter. Lanes 1 and 3 (counting from the left), no HrcA; lanes 2 and 4, 1,150 nM HrcA present. (B) Cotranscription of the dnaK and omcB promoters. Lane 1, no HrcA; lane 2, 230 nM HrcA; lane 3, 460 nM HrcA; lane 4; 1,150 nM HrcA.
FIG. 5.
Quantification of HrcA-mediated transcriptional repression. Data from transcription reactions performed in triplicate and quantified by phosphorimager analysis. Error bars are standard deviations from the means.
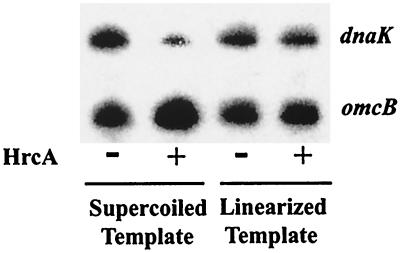
HrcA transcriptional repression depends on DNA topology.
To determine if HrcA-mediated repression is dependent on DNA topology, the effects of HrcA on transcription of linearized and supercoiled templates were compared. The dnaK and omcB plasmids were linearized by digestion with BamHI, which cuts downstream of the G-less cassette transcription template, so as not to disrupt the promoter region. Transcription of both linearized templates in the absence of HrcA was decreased slightly (8% decrease for both the dnaK and omcB promoters) as shown in Fig. 6 (compare lanes 1 and 3). Repression of the dnaK promoter by HrcA on a linearized DNA template was decreased (13% repression between lanes 3 and 4) compared to that on a supercoiled template (80% repression between lanes 1 and 2). These results demonstrate that HrcA repression of the dnaK promoter is dependent on DNA supercoiling.
FIG. 6.
In vitro transcription from supercoiled and linearized templates. Lanes 1 and 3 (counting from the left), no HrcA; lanes 2 and 4, 1,150 nM HrcA.
DISCUSSION
In this report, we show that HrcA represses in vitro transcription of heat shock genes in the pathogenic bacterium C. trachomatis. In other bacteria, HrcA has been shown to bind the cis-acting sequence CIRCE, and genetic evidence has implicated HrcA as a transcriptional repressor (16, 26). We present the first direct, in vitro evidence that HrcA acts as a transcriptional repressor in bacteria. To the best of our knowledge, this is also the first report of the in vitro reconstitution of regulated transcription in Chlamydia.
Using competitive EMSA we showed that chlamydial HrcA binds to the CIRCE element of the dnaK promoter in a sequence-specific manner. However, an excess of HrcA protein relative to DNA was required for this binding, as had been the experience with other bacterial systems (8, 9). It has been noted that the recombinant B. subtilis HrcA protein is insoluble and prone to aggregation (9), and it is possible that only a fraction of our recombinant HrcA protein was active and able to bind DNA.
Transcriptional repression by HrcA is promoter specific, as demonstrated by the lack of an effect on the control omcB promoter, which does not contain an identifiable CIRCE element. We also saw a compensatory increase in omcB transcription when the dnaK and omcB promoters were cotranscribed in the presence of HrcA, which is consistent with competitive transcription by limiting amounts of RNA polymerase. Under these conditions, HrcA binding to CIRCE would be predicted to reduce the binding of RNA polymerase to the dnaK promoter, leading to increased availability of RNA polymerase for transcription of the omcB promoter.
Our results also suggest that HrcA-mediated transcriptional repression is affected by DNA topology. We noted that an HrcA/DNA ratio that produced the maximal 80% decrease in transcription only caused 19% DNA binding and that a sevenfold-higher HrcA/DNA ratio was required for 80% DNA binding by EMSA. We found that this difference was due to the use of supercoiled templates in the transcription assay and was not due to different reaction conditions in the two assays (data not shown). The effect of DNA topology could be a result of increased HrcA binding to CIRCE on a supercoiled template or other effects on HrcA-mediated transcriptional repression.
If HrcA functions as a transcriptional repressor, derepression would provide a likely mechanism for the upregulation of dnaK and other heat shock genes in response to heat shock. In the simplest case, HrcA binding to CIRCE may be decreased with higher temperatures. In Bacillus thermoglucosidasius, the HrcA-CIRCE complex has been shown to be relatively unstable at higher-than-normal growth temperatures (24). The HrcA protein may be heat labile, or its protein conformation may be altered at higher temperatures so that its intrinsic binding to CIRCE is decreased. Alternatively, higher temperatures may affect CIRCE structure, perhaps as a consequence of changes in DNA topology. In E. coli, heat shock and cellular stress are known to produce large effects on DNA supercoiling (23, 25). Our observations suggest that, in C. trachomatis, HrcA-mediated repression of transcription is sensitive to DNA supercoiling. At this point, we do not know the supercoiling optima for DNA binding and transcriptional repression by HrcA or the effect of heat shock on DNA supercoiling in Chlamydia. However, the superhelical density in Chlamydia appears to be regulated during the chlamydial life cycle (19).
A more complicated model for derepression of the heat shock genes would involve the regulation of HrcA-CIRCE binding by an additional factor that is responsive to heat shock and cellular stress. For example, it has been proposed that the GroE chaperonin machinery modulates HrcA activity (1, 9), although this hypothesis has been disputed (8). It is also possible that the effect of an additional factor is to modulate DNA topology in response to heat shock.
The in vitro reconstitution of HrcA- and CIRCE-mediated transcriptional repression provides a useful system for studying heat shock regulation in bacteria. It provides a convenient assay for defining the mechanisms of repression and derepression by HrcA and other possible factors. The development of a regulated chlamydial transcription system will also be useful for studying gene regulation in this important human pathogen. This general approach, of reconstituting regulated transcription by the addition of a recombinant transcription factor and the cognate cis-acting DNA element to the basal transcription system, can be applied to other regulated genes in Chlamydia.
Acknowledgments
We thank Hilda Hiu Yin Yu, Chris Schaumburg, and Allen Chen for their support.
This work is supported by a grant from the NIH (AI 44198). A.C.W. was supported by a predoctoral training grant from the NIH (GMT3207311).
REFERENCES
- 1.Babst, M., H. Hennecke, and H. M. Fischer. 1996. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol. Microbiol. 19:827-839. [DOI] [PubMed] [Google Scholar]
- 2.Douglas, A. L., and T. P. Hatch. 2000. Expression of the transcripts of the sigma factors and putative sigma factor regulators of Chlamydia trachomatis L2. Gene 247:209-214. [DOI] [PubMed] [Google Scholar]
- 3.Gerard, H. C., J. A. Whittum-Hudson, and A. P. Hudson. 1997. Genes required for assembly and function of the protein synthetic system in Chlamydia trachomatis are expressed early in elementary to reticulate body transformation. Mol. Gen. Genet. 255:637-642. [DOI] [PubMed] [Google Scholar]
- 4.Hatch, T. P. 1999. Developmental biology, p. 29-68. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 5.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 6.LaVerda, D., M. V. Kalayoglu, and G. I. Byrne. 1999. Chlamydial heat shock proteins and disease pathology: new paradigms for old problems? Infect. Dis. Obstet. Gynecol. 7:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews, S. A., K. M. Volp, and P. Timms. 1999. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Lett. 458:354-358. [DOI] [PubMed] [Google Scholar]
- 8.Minder, A. C., H. M. Fischer, H. Hennecke, and F. Narberhaus. 2000. Role of HrcA and CIRCE in the heat shock regulatory network of Bradyrhizobium japonicum. J. Bacteriol. 182:14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Read, M. 1996. Electrophoretic mobility shift assay (EMSA), p. 6-11. In K. Docherty (ed.), Gene transcription: DNA binding proteins. John Wiley & Sons, Chichester, West Sussex, United Kingdom.
- 12.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Schachter, J. 1999. Infection and disease epidemiology, p. 139-169. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. American Society for Microbiology, Washington, D.C.
- 14.Schaumburg, C. S., and M. Tan. 2000. A positive cis-acting DNA element is required for high-level transcription in Chlamydia. J. Bacteriol. 182:5167-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmiel, D. H., S. T. Knight, J. E. Raulston, J. Choong, C. H. Davis, and P. B. Wyrick. 1991. Recombinant Escherichia coli clones expressing Chlamydia trachomatis gene products attach to human endometrial epithelial cells. Infect. Immun. 59:4001-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz, A., B. Tzchaschel, and W. Schumann. 1995. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol. Microbiol. 15:421-429. [DOI] [PubMed] [Google Scholar]
- 18.Shaw, E. I., C. A. Dooley, E. R. Fischer, M. A. Scidmore, K. A. Fields, and T. Hackstadt. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37:913-925. [DOI] [PubMed] [Google Scholar]
- 19.Solbrig, M. V., M. L. Wong, and R. S. Stephens. 1990. Developmental stage specific plasmid supercoiling in Chlamydia trachomatis. Mol. Microbiol. 4:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 21.Tan, M., and J. N. Engel. 1996. Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter. J. Bacteriol. 178:6975-6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan, M., B. Wong, and J. N. Engel. 1996. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J. Bacteriol. 178:6983-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse-Dinh, Y. C., H. Qi, and R. Menzel. 1997. DNA supercoiling and bacterial adaptation: thermotolerance and thermoresistance. Trends Microbiol. 5:323-326. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe, K., T. Yamamoto, and Y. Suzuki. 2001. Renaturation of Bacillus thermoglucosidasius HrcA repressor by DNA and thermostability of the HrcA-DNA complex in vitro. J. Bacteriol. 183:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein-Fischer, D., M. Elgrably-Weiss, and S. Altuvia. 2000. Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 35:1413-1420. [DOI] [PubMed] [Google Scholar]
- 26.Yuan, G., and S.-L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]