Abstract
Many multidrug transporters from gram-negative bacteria belong to the resistance-nodulation-cell division (RND) superfamily of transporters. RND-type multidrug transporters have an extremely broad substrate specificity and protect bacterial cells from the actions of antibiotics on both sides of the cytoplasmic membrane. They usually function as three-component assemblies spanning the outer and cytoplasmic membranes and the periplasmic space of gram-negative bacteria. The structural determinants of RND transporters responsible for multidrug recognition and complex assembly remain unknown. We constructed chimeric RND transporters composed of N-terminal residues of AcrB and C-terminal residues of MexB, the major RND-type transporters from Escherichia coli and Pseudomonas aeruginosa, respectively. The assembly of complexes and multidrug efflux activities of chimeric transporters were determined by coexpression of hybrid genes either with AcrA, the periplasmic component of the AcrAB transporter from E. coli, or with MexA and OprM, the accessory proteins of the MexAB-OprM pump from P. aeruginosa. We found that the specificity of interaction with the corresponding periplasmic component is encoded in the T60-V612 region of transporters. Our results also suggest that the large periplasmic loops of RND-type transporters are involved in multidrug recognition and efflux.
In Escherichia coli and Pseudomonas aeruginosa the major, constitutively expressed multidrug transporters are AcrB and MexB, respectively, which belong to the resistance-nodulation-cell division (RND) superfamily (22). Several features distinguish RND-type transporters from other multidrug pumps (MDRs). First, these transporters have a very unusual membrane topology. The predicted structure consists of 12 transmembrane α-helices (TMS) with two large hydrophilic extracytoplasmic domains between TMS 1 and 2 and TMS 7 and 8 (6). These unusually large hydrophilic domains (ED1 and ED2) make the membrane topology of RND transporters look very similar to that of receptors rather than transporters. Indeed, the RND transporters share some degree of homology with the cholesterol-sensing domain of HMG-CoA reductase. This finding suggested that these proteins evolved as machinery that senses, or senses and then extrudes, relatively apolar molecules found within the lipid bilayer of membranes (22). Second, unlike the vast majority of MDRs which mostly pump out hydrophobic cations, the RND-type transporters from gram-negative bacteria also interact with neutral, zwitterionic, and anionic compounds (10, 16). Furthermore, some RND transporters provide cell protection on both the cytosolic and the periplasmic sides of the inner membrane. In particular, AcrB and its homologs efficiently expel some β-lactam antibiotics whose target is located in the periplasmic space rather than in cytoplasm (9, 13). In addition, some transported drugs have been shown not to traverse the cytoplasmic membrane (13). These observations led to the suggestion that (i) these transporters have multiple drug-binding sites to capture drugs from different locations within the cell. Alternatively, (ii) the topology of these transporters is such that both hydrophilic and lipophilic compounds can gain access to drug-binding sites (15, 27). Presently, the structure, number, and location of the multidrug-binding sites in RND transporters remain unknown.
The amino acid sequences of AcrB and MexB are 70% identical. However, despite the high degree of structural and functional similarity, these transporters appear to possess structure variations responsible for several unique features. AcrB and MexB provide a different degree of drug resistance in E. coli and P. aeruginosa cells, respectively, and also vary in substrate specificities (8, 10). In addition, these transporters interact in a highly specific manner with at least two additional proteins: the periplasmic membrane fusion proteins (MFPs) (AcrA and MexA) and the outer membrane channels (OMFs) (TolC and OprM). These accessory proteins together with a unique RND transporter, AcrB or MexB, form complexes that span both the inner and the outer membranes of gram-negative bacteria and provide an efflux of drugs across two-membrane envelopes directly into the external medium.
In P. aeruginosa all three components of multidrug pumps are usually expressed from a single operon, for example, mexAB-oprM or mexCD-oprJ, providing the coordinated expression of each component. Genetic analysis has indicated that the periplasmic MFPs are specific for each RND transporter and cannot be interchanged between homologous pumps (23). This result implied that, despite the high degree of homology, RND transporters possess structure variations responsible for the specificity of interaction with MFPs. In contrast, a single OMF can support the function of several homologous pumps. The OprM channel encoded in the mexAB-oprM operon can also function together with MexCD (23) or MexXY (1). The versatility of OMFs is also supported by the fact that the enterobacterial MDR operons do not contain genes for these proteins. In contrast, a single protein, multifunctional channel TolC, encoded elsewhere on the chromosome is capable of supporting the transport needs of several MDRs (4) and also the export of such proteins as hemolysin and colicin V (2, 7). Interestingly TolC also supports the activity of the Pseudomonas multidrug transporters MexD and MexY when they are expressed in E. coli cells in the absence of their native OMFs (11). Based on these and other data, it was proposed that MFPs play a major role in the formation of tripartite complexes and that the inner membrane transporters (IM transporters) do not interact directly with OMFs (21).
In this study, we have taken advantage of functional differences between AcrB and MexB to search for the region(s) involved in drug transport and/or interaction with accessory proteins. Our approach is based on the construction of chimeric AcrB/MexB proteins and assaying their activity in the complexes with AcrA-TolC or MexA-OprM proteins.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani medium (10 g of Bacto-tryptone, 10 g of Bacto yeast extract, and 5 g of NaCl per liter) was used to grow all bacterial cultures. Ampicillin (100 μg/ml), kanamycin (35 μg/ml), and spectinomycin (50 μg/ml) were used for selection. All plasmids (Table 1; Fig. 1) were constructed by common cloning techniques (18) and propagated in the DH5α strain of E. coli. Nucleotides of the acrAB and mexAB-oprM loci are numbered according to the methods of Ma et al. (9) and Poole et al. (16).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| P. aeruginosa strain | ||
| PAO1 | Wild type | |
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | General cloning |
| AG100AX | ΔacrAB::kan ΔacrEF::spe | 12 |
| Plasmids | ||
| pUC18 | E. coli cloning vector, Ampr | |
| pUC151A | pUC18 vector carrying the acrAB genes | 10 |
| pBP | pUC18 vector expressing acrB under native promoter | This study |
| pOM | pUC18 vector expressing the oprM gene under Plac promoter | This study |
| pMAO | pOM derivative carrying the mexA gene | This study |
| pMABO | pMAO derivative carrying the mexB gene | This study |
| pPV20 | mexAB-oprM genes in the pAK1900 shuttle cloning vector | 16 |
FIG. 1.
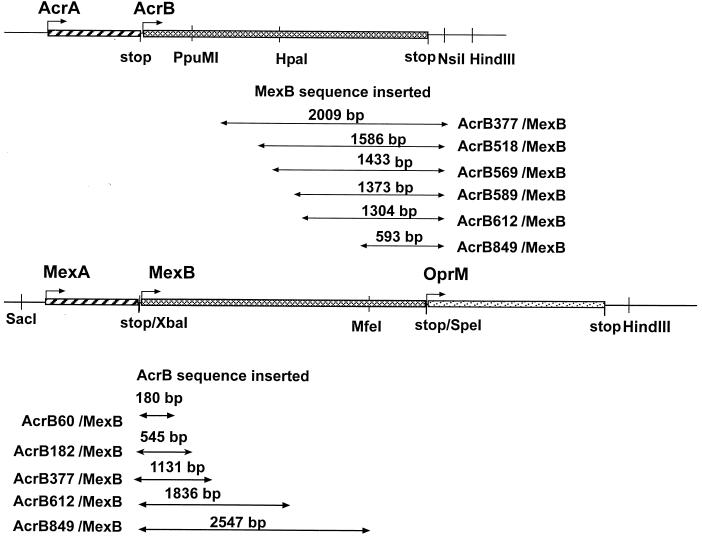
Construction of chimeric AcrB/MexB transporters. Physical maps of acrAB and mexAB-oprM operons are shown. Arrows indicate the DNA fragments corresponding to acrB or mexB present in chimeric constructs. Numbers in the names of constructs correspond to the N-terminal amino acid residues of AcrB constituting the chimeric transporters. Only restriction sites relevant to cloning are shown.
To construct pBP, pUC151A was first digested with BlpI and XhoI restriction enzymes, treated with T4 DNA polymerase, and self-ligated to produce pUC151XB. Next, a 3,554-bp SacI-NsiI fragment of pUC151XB was ligated into a SacI-PstI-treated pUC18 vector. Expression plasmids for OprM (pOM) and MexA-OprM (pMAO) were generated by PCR amplification and subcloning into XbaI-HindIII sites and SacI-XbaI sites of pUC18 and pOM, respectively. The pMABO plasmid was generated by cloning the PCR-amplified mexB gene in the XbaI and SpeI sites of pMAO.
Construction of expression plasmids for AcrB/MexB hybrid proteins.
To construct expression plasmids for AcrB/MexB hybrid proteins, we designed PCR primers complementary to the acrB and mexB regions indicated in Fig. 1 (the sequences of primers are available upon request). Primers contained added restriction sites, if required. For amplification of mexB regions, the chromosomal P. aeruginosa DNA or pPV20 was used as a template. For amplification of acrB regions, pUC151A or pBP was used as a template.
All plasmids for the expression of chimeric genes downstream of acrA are derivatives of pUC151A. Plasmid pAcrAB377 was constructed by replacement of a PpuMI-NsiI fragment of pUC151A with a PCR-amplified 2,019-bp fragment of mexB. To construct pAcrAB518, pAcrAB569, and pAcrAB612, the corresponding chimeric genes were assembled by PCR. In the expression plasmids, the chimeric genes replaced the DraIII-NsiI fragment of pUC151A. pAcrAB589 and pAcrAB849 were constructed by replacement of the PpuMI-HindIII fragment of pUC151A with acrB589/mexB and acrB849/mexB chimeric genes. acrB589/mexB and acrB849/mexB were assembled by cloning of corresponding mexB fragments into HincII-PstI sites of pUC18 to produce pBSpe and pBMfe, respectively, and subsequent subcloning of acrB fragments into XmaI-SpeI-treated pBSpe and XmaI-MfeI-treated pBMfe.
All plasmids for the expression of chimeric AcrB/MexB genes in a single operon with MexA and OprM are derivatives of pMAO plasmid. The chimeric acrB612/mexB and acrB849/mexB genes were PCR amplified using pAcrAB612 and pAcrAB849 as templates. The obtained DNA fragments were subcloned into a XbaI-SpeI-treated pMAO plasmid to yield pMexAB612 and pMexAB849. Expression plasmid pMexAB377 was constructed by the replacement of the PpuMI- SpeI fragment of pMexAB849 with the PpuMI-SpeI fragment of pMABO. To construct pMexAB60 and pMexAB182, the corresponding DNA fragments of the acrB gene were PCR amplified using pBA plasmid as a template with forward primer containing an XbaI site and the reverse primer containing sequences complementary to both acrB and mexB. The PCR products were then used as forward primers to amplify the mexB gene, with pPV20 as a template. After treatment with XbaI and SpeI, the obtained 3,150-bp DNA fragments were subcloned into the pMAO plasmid to yield pMexAB182 and pMexAB60.
When PCR was used in the construction of a plasmid, the final structure was verified by DNA sequencing. PCRs were performed with a Peltier PTC-200 thermal cycler (MJ Research) with Biolase DNA polymerase (Bioline). Restriction endonucleases, alkaline phosphatase, and T4 DNA ligase were obtained from New England BioLabs Inc. or Gibco BRL.
MIC determinations.
For the determination of MICs of various antimicrobial agents, exponentially growing cultures (optical density at 600 nm of 1.0) were inoculated at a density of 104 cells per ml into Luria-Bertani medium in the presence of twofold-increasing concentrations of the drug under investigation. Cell growth was determined after overnight incubation at 37°C.
Cross-linking, preparation of membrane fractions, and immunoblotting analysis.
Membrane fractions of the dithiobis(succinimidylpropionate) (DSP)-treated and nontreated AG100AX E. coli cells expressing different hybrids or wild-type proteins were prepared as described previously (26). Protein concentrations were determined using the Bio-Rad protein assay with bovine serum albumin as a standard. Equal protein amounts of membrane fractions containing different hybrids were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels. Membrane fractions containing wild-type AcrB and AcrB849/MexB were loaded onto gels in concentrations 5 to 10 times lower than other samples because of the higher reactivity of anti-AcrB antibody with these two proteins. SDS-polyacrylamide gel electrophoresis was performed by standard methods through 5% stacking and 8 or 10% separating gels. For immunoblotting, proteins were transferred electrophoretically to polyvinylidine difluoride membrane (Millipore) in 3-(cyclohexylamino)-1-propanesulfonic acid-NaOH (10 mM; pH 11) and methanol (10%). Protein visualization was performed according to standard protocols with anti-AcrA (24), anti-AcrB (25), and anti-OprM (a kind gift of K. Poole) antibodies and alkaline phosphatase-conjugated anti-rabbit antibodies (Sigma).
RESULTS
AcrB and MexB demonstrate distinct substrate specificities in E. coli.
Previous studies demonstrated that the MexAB-OprM multidrug efflux complex from P. aeruginosa is functional when expressed in E. coli cells (19). However, the pattern of resistance to different antimicrobials is less broad than that attributed to MexAB-OprM in P. aeruginosa. The substrate specificities of AcrAB-TolC and MexAB-OprM appear to be very similar in E. coli, consistent with the high degree of homology between corresponding components of these complexes (AcrA and MexA, 57.7% identity; AcrB and MexB, 69.8% identity).
The high degree of homology between the inner membrane components enabled us to detect the expression of MexA and MexB proteins by immunoblotting analysis with polyclonal antibody raised against AcrA and AcrB proteins. In agreement with previously reported data (19), we found that the MexAB-OprM complex is expressed in E. coli cells (Fig. 2) and is functionally active, as judged by the restoration of the drug-resistant phenotype of the AG100AX strain, which is deficient in the AcrAB and AcrEF pumps (Table 2).
FIG. 2.
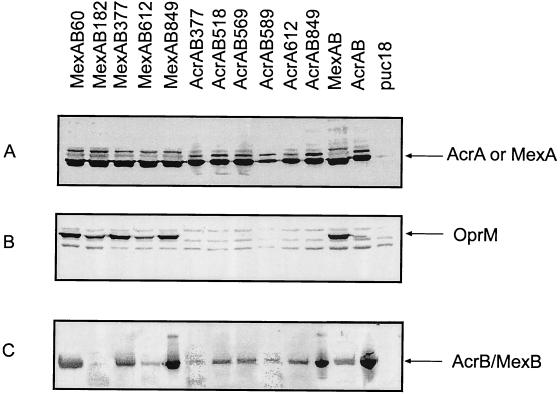
Expression of wild-type and chimeric AcrB/MexB in E. coli. Cells expressing wild-type AcrB and MexB as well as chimeric AcrB/MexB transporters in operons with either AcrA (AcrAB) or MexA/OprM (MexAB) were harvested and analyzed by Western blotting with anti-AcrA (A), anti-OprM (B), or anti-AcrB (C) antibodies. The expression of AcrA, MexA, and OprM was detected in whole cells (A and B). AcrB, MexB, and chimeric AcrB/MexB proteins were visualized in membrane fractions of corresponding cells (C). Arrows indicate the positions of expressed proteins.
TABLE 2.
Antimicrobial susceptibility of E. coli cells expressing wild-type and chimeric AcrAB and MexAB-OprM efflux pumps
| Construct | MIC (μg/ml)c
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SDS | CA | NOV | EtB | OLE | ERY | PUR | LIN | CIN | NAL | CF | PRO | TET | CTET | NOR | |
| pUC18 | 40 | 2,325 | 4 | 3.125 | 4 | 4 | 2 | 32 | 2 | 1 | 0.8 | 6.25 | 0.32 | 0.04 | 0.01 |
| AcrAB | >10,240 | >9,300 | 256 | 800 | 4,096 | 256 | 256 | 1,024 | 2 | 8 | 12.8 | 25 | 2.56 | 0.4 | 0.16 |
| MexAB-OprM | 10,240 | >9,300 | 256 | 100 | 256 | 32 | 64 | 512 | 16 | 16 | 12.8 | 25 | 2.56 | 0.8 | 0.08 |
| AcrAB849a | >10,240 | >9,300 | 256 | 200 | 512 | 256 | 256 | 256 | 8 | 8 | 6.4 | 25 | 2.56 | 0.4 | 0.08 |
| MexAB849b | 40 | 2,325 | 2 | 6.25 | 4 | 4 | 2 | 16 | 2 | 1 | 0.8 | 6.25 | 0.16 | 0.01 | 0.01 |
| AcrAB612 | >10,240 | >9,300 | 256 | 200 | 256 | 64 | 32 | 256 | 1 | 1 | 1.6 | 6.25 | 0.64 | 0.1 | 0.04 |
| MexAB612 | 40 | 2,325 | 2 | 3.125 | 2 | 4 | 2 | 16 | 2 | 1 | 0.8 | 6.25 | 0.32 | 0.02 | 0.005 |
| AcrAB589 | 40 | 2,325 | 4 | 6.25 | 2 | 4 | 1 | 32 | 2 | 1 | 0.4 | 6.25 | 0.32 | 0.01 | 0.01 |
| AcrAB569 | 40 | 2,325 | 4 | 6.25 | 2 | 4 | 2 | 32 | 2 | 1 | 0.8 | 6.25 | 0.32 | 0.01 | 0.02 |
| AcrAB518 | 40 | 2,325 | 4 | 6.25 | 2 | 4 | 2 | 32 | 2 | 1 | 0.8 | 6.25 | 0.32 | 0.01 | 0.02 |
| AcrAB377 | 40 | 2,325 | 2 | 3.125 | 2 | 2 | 2 | 32 | 2 | 1 | 0.4 | 6.25 | 0.16 | 0.01 | 0.02 |
| MexAB377 | 40 | 4,650 | 2 | 3.125 | 4 | 4 | 2 | 16 | 2 | 1 | 0.8 | 6.25 | 0.16 | 0.02 | 0.01 |
| MexAB182 | 40 | 2,325 | 2 | 3.125 | 4 | 4 | 2 | 16 | 2 | 1 | 0.8 | 3.125 | 0.32 | 0.04 | 0.04 |
| MexAB60 | 10,240 | >9,300 | 64 | 6.25 | 64 | 8 | 32 | 32 | 8 | 2 | 0.8 | 6.25 | 0.64 | 0.1 | 0.08 |
The numbers correspond to AcrB N-terminal amino acid residues present in hybrid transporters. Functional constructs and corresponding MIC values are shown in bold.
All constructs expressing chimeric pumps in a single operon with the mexA gene also contain the oprM gene.
Abbreviations: SDS, sodium dodecyl sulfate; CA, cholic acid; NOV, novobiocin; LIN, lincomycin; EtB, ethidium bromide; ERY, erythromycin; OLE, oleandomycin; NOR, norfloxacin; PUR, puromycin; NAL, nalidixic acid; CF, chloramphenicol; CIN, cinoxacin; PRO, proflavine; TET, tetracycline; CTET, chlortetracycline.
Although both AcrAB and MexAB-OprM conferred resistance to a broad spectrum of antimicrobials, the levels of resistance provided by these transporters differed substantially for a number of compounds (Table 2). In particular, E. coli cells expressing MexAB-OprM were capable of surviving in higher concentrations of cinoxacin (CIN; 8-fold difference in MICs compared to AcrAB-expressing cells). On the other hand, MexAB-OprM-containing cells were less resistant than AcrAB-expressing cells to oleandomycin (OLE; 16-fold difference), erythromycin (ERY; 8-fold difference), and ethidium bromide (EtB; 4-fold difference). It is noteworthy that the MexAB-OprM complex provides higher levels of resistance to the latter drugs when expressed in the P. aeruginosa envelope, presumably because of the difference in the permeability properties of the outer membranes of E. coli and P. aeruginosa (8).
The AcrAB pump needs, for its efficient action, the presence of the multifunctional outer membrane channel TolC (4). Although TolC- and AcrAB-deficient phenotypes are very similar, they are not identical, presumably because TolC provides the route for drugs across the outer membrane for other multidrug transporters of E. coli (20). Despite the high homology between AcrAB and MexAB, the TolC protein does not seem to interact with and support the function of MexAB. When we introduced mexAB into E. coli cells in the absence of the OprM channel, the cells remained highly susceptible to different drugs, demonstrating that all three components, MexA, MexB, and OprM, must be present to protect E. coli from multiple drugs (data not shown). Consistent with this result is the finding that the OprM protein, when produced in TolC-deficient cells, does not rescue the drug-sensitive phenotype of these cells. Thus, all three components of the Pseudomonas efflux transporter MexAB-OprM must be present in E. coli cells to provide protection against multiple drugs.
Construction and expression of chimeric AcrB/MexB molecules.
Comparative sequence analyses have indicated that the N- and C-terminal halves of RND proteins share sequence similarities, suggesting that they may have evolved via tandem gene duplication events (17). Using deletion analysis we found that neither N- nor C-terminal halves of the AcrB transporter can act autonomously (data not shown). This result suggested that both halves of multidrug transporters are required for multidrug transport or interaction with accessory proteins.
To identify regions of the RND transporters responsible for the interactions with accessory proteins and/or for the transport activities, we generated a series of hybrid AcrB/MexB molecules (Fig. 1 and 3). The hybrid acrB/mexB genes were cloned either immediately downstream of the acrA gene to assay their functionality with this protein (AcrA-AcrB/MexB) or between the mexA and oprM genes, in a manner to preserve the chromosomal arrangement of these genes in a single operon (MexA-AcrB/MexB-OprM) (Fig. 1). In these arrangements, the role of N-terminal residues in the function and assembly of multicomponent drug transporters can be evaluated by assessment of the functional activity of the AcrB/MexB hybrids in combination with AcrA and TolC, whereas the role of the C-terminal residues can be assessed in combination with MexA and OprM proteins. The native acrA promoter has provided the efficient expression of the acrA-acrB/mexB operon, but the expression of the mexA-acrB/mexB-oprM operon was dependent on the Plac promoter of the pUC18 vector.
FIG. 3.
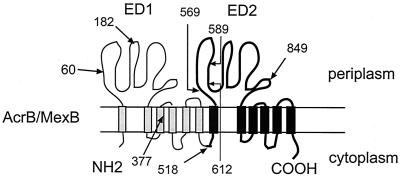
Membrane topology of AcrB and MexB. These transporters were proposed to consist of 12 TMSs with two large extracytoplasmic domains (ED1 and ED2). The N- and C-terminal halves of AcrB and MexB share sequence similarities. Arrows with numbers show the sites of fusions between AcrB and MexB amino acid sequences and the number of N-terminal amino acid residues from AcrB in chimeric AcrB/MexB transporters.
All constructs were transformed into E. coli cells deficient in AcrAB and AcrEF multidrug transporters, and their expression was assayed by immunoblotting (Fig. 2). As judged by the Western blotting of whole-cell extracts prepared from E. coli expressing either AcrA-AcrB/MexB or MexA-AcrB/MexB-OprM, all constructs produced similar amounts of MexA and AcrA (Fig. 2A). However, the levels of expression of chimeric AcrB/MexB proteins were affected by the position of junctions between AcrB and MexB (Fig. 2C). The amounts of AcrB849/MexB were comparable to that of the wild-type AcrB protein and higher than the amount of wild-type MexB, regardless of whether this hybrid was coexpressed with AcrA or with MexA/OprM proteins. All other hybrids, with the exception of AcrB589/MexB and AcrB182/MexB, were produced at levels comparable to that of the wild-type MexB. When the link between the AcrB and MexB portions was located either in the N-terminal region of ED1 or in the similar position in ED2, the hybrids were unstable, independently of the periplasmic proteins. In particular, AcrB589/MexB and AcrB182/MexB were present in membranes at low levels. The accessory proteins seem to affect the expression of the AcrB377/MexB hybrid. This protein was produced in larger amounts when expressed in combination with the MexA and OprM proteins.
The specificity of interaction with accessory proteins is encoded within the T60-V612 region of AcrB and MexB transporters.
Previous chemical cross-linking studies have shown that the periplasmic lipoprotein AcrA interacts specifically with the inner-membrane-associated AcrB transporter and that this complex is assembled even in the absence of the outer membrane component TolC (26). We used the same approach to determine which hybrid AcrB/MexB molecules can form complexes with AcrA or MexA proteins. To stabilize protein complexes, intact E. coli cells expressing different constructs were treated with cross-linker DSP (26). As previously reported, the AcrA and AcrB proteins form a stable complex in the inner membrane of E. coli (Fig. 4). Under the same conditions, MexA and MexB form complexes with molecular masses similar to those of AcrAB. In our assay we did not detect high-molecular-weight species that could indicate the presence of the tripartite MexA-MexB-OprM complex.
FIG. 4.
Western blotting analysis of AcrA (A) and MexA (B) in E. coli cells cross-linked with DSP. Positions of oligomeric AcrA and MexA are indicated as CA1 and CA2. Higher-molecular-weight complexes C1 and C2 presumably contain the periplasmic and inner membrane components of AcrAB-TolC (A) and MexAB-OprM (B) complexes, respectively. The membrane fraction of cells expressing MexA and OprM proteins without MexB (panel B, lane1) is shown as a negative control.
The high-molecular-weight complexes reacting with both anti-AcrA (Fig. 4) and anti-AcrB (data not shown) antibodies were detected in the membrane fractions of E. coli cells expressing AcrA-AcrB612/MexB, AcrA-AcrB849/MexB, and MexA-AcrB60/MexB-OprM hybrid transporters. This result is consistent with the finding that these AcrB/MexB hybrid proteins are functionally active and that the interaction between the periplasmic component and the IM transporter is required for the efflux of drugs (Table 2 and see below). The interactions between AcrB612/MexB, AcrB849/MexB, and AcrA appear to be very specific, as evidenced by the inability of AcrB612/MexB or AcrB849/MexB to associate with MexA and OprM (data not shown) and the lack of functional activity of these hybrids (Table 2). Thus, the N-terminal 612 amino acid residues of AcrB contain the structural determinants for specific association with AcrA.
Of five hybrids expressed with MexA and OprM, only AcrB60/MexB restored the drug-resistant phenotype to E. coli cells, suggesting that only the first 60 N-terminal amino acid residues of MexB can be substituted by corresponding regions of the AcrB transporter. Taken together, these results suggested that the specificity of interaction with the periplasmic MFPs is encoded in the T60-V612 region of the IM transporter.
We could not detect reproducibly the complexes between the periplasmic proteins and hybrid pumps for other constructs. The expression of AcrB182/MexB and AcrB589/MexB hybrids was substantially lower than that of the wild-type AcrB or MexB proteins. One explanation for the absence of detectable complexes is the low sensitivity of the in vivo chemical cross-linking technique. Also, the low amount of hybrid proteins in the membrane fractions could be an indication that these proteins are not properly folded within the membrane and, as a result, are vulnerable to the action of proteases.
Drug specificity of chimeric AcrB and MexB efflux transporters.
Cells expressing AcrA-AcrB612/MexB, AcrA-AcrB849/MexB, and MexA-AcrB60/MexB-OprM displayed increased resistance to a number of tested compounds compared with cells transformed with the vector alone (Table 2). However, the transport activity of these hybrid pumps differed substantially from the wild-type AcrB and MexB transporters. Since the complex formation appears to be unaffected in AcrA-AcrB612/MexB, AcrA-AcrB849/MexB, and MexA-AcrB60/MexB-OprM (see above and Fig. 4), the observed differences in substrate specificities or efficiencies of efflux among these hybrids arise most probably from the structural variations in the IM transporters that affect the multidrug recognition or multidrug transport mechanisms.
All three functional chimeras recognized and expelled the detergents SDS and cholic acid (CA) and novobiocin (NOV) with efficiencies comparable to that of the wild-type AcrAB or MexAB pumps (Table 2). This result shows that the 60 amino acid residues of the N terminus and the C-terminal amino acid residues of AcrB and MexB are interchangeable with respect to efflux of these substrates.
The wild-type AcrB and MexB transporters differ from each other in the specificity to several compounds (Table 2). Surprisingly, the chimeric AcrA-AcrB849/MexB pump, which contains only the last 200 C-terminal residues from MexB, showed the substrate specificities characteristic of both wild-type transporters. E. coli cells expressing this pump were indistinguishable from those with the wild-type AcrB transporter in susceptibility to the majority of tested compounds (Table 2). On the other hand, the specificity of the AcrA-AcrB849/MexB pump to EtB, lincomycin (LIN), and CIN was similar to that of the wild-type MexB transporter. This result suggested that the structure variations responsible for differences in transport of the latter compounds reside in the C-terminal 200 amino acid residues of the AcrB and MexB transporters. For other substrates, these residues are not sufficient to convert the specificity of the AcrB849/MexB hybrid to a MexB-like one.
The AcrA-AcrB612/MexB pump contains an additional 235 C-terminal amino acid residues from MexB. All these residues are located entirely in the extracytoplasmic domain ED2. This chimeric transporter showed, distinct from other constructs, substrate specificity. AcrB612/MexB transported LIN, EtB, ERY, OLE, and puromycin (PUR) with the efficiency of the wild-type MexB. This result suggested that the region V612-S/K849 of RND transporters might contribute to the specificity toward these drugs. The AcrB612/MexB hybrid, however, lost the ability to expel other substrates, including CIN, nalidixic acid (NAL), or proflavine (PRO), which are efficiently expelled by the wild-type pumps and AcrB849/MexB hybrid. Perhaps the binding of these compounds occurs in different site of transporter. The presence of the V612-S/K849 region from MexB could disrupt this site in the AcrB transporter.
The N-terminal 60 amino acid residues of AcrB, which are present in AcrB60/MexB, dramatically reduced the spectrum of substrates of the wild-type MexB transporter. Although AcrB60/MexB conferred resistance to PUR, CIN, SDS, CA, and NOV, this hybrid had only partial or no activity against the rest of the tested drugs.
Taken together our results suggest that such substrates as SDS, CA, and NOV differ from others in terms of recognition and transport efficiency by AcrB/MexB transporters. The partial replacements of N-terminal or C-terminal residues in AcrB/MexB transporters did not change the ability of hybrids to expel these compounds from cells. The 200 C-terminal amino acid residues of MexB are sufficient to convert the specificity of AcrB/MexB hybrids to a MexB-like specificity to EtB, OLE, and CIN. In addition, two other regions of AcrB/MexB appear to be essential for transport of various substrates. The N-terminal residues M1-T60 and amino acid residues located in the V612-S/K849 region of ED2 strongly affect the substrate specificities of hybrid transporters.
DISCUSSION
In this study we investigated the structural organization of RND transporters by constructing chimeric proteins containing the regions of two multidrug transporters, AcrB and MexB. These transporters function as the tripartite complexes AcrAB-TolC and MexAB-OprM in E. coli and P. aeruginosa, respectively, and provide an efficient protection of cells against a multitude of structurally dissimilar antimicrobial agents (8, 10). In agreement with previous studies (19), we found that the functional MexAB-OprM complex can be efficiently expressed in E. coli cells, suggesting that no other proteins are required for the functional activity of this transporter. Surprisingly, the outer membrane channel TolC did not complement the function of OprM, as judged from the drug susceptibility of TolC-containing E. coli cells expressing plasmid-borne MexA and MexB proteins. Similarly, OprM when expressed in TolC-deficient E. coli cells did not render the cells drug resistant. This finding was somewhat unexpected, since the OprM channel has been shown to be readily interchangeable between Pseudomonas MDR pumps and to function efficiently with MexCD and MexXY transporters (1, 5). Furthermore, both MexCD and MexXY proteins can function with the TolC channel when expressed in E. coli (11, 19). Thus, despite the interchangeability of TolC and OprM in some transporters, these channels interact with AcrAB and MexAB, respectively, in a highly specific manner.
Besides structural determinants responsible for the specificity of interaction, the RND transporters may possess variations defining differences in substrate specificities. The efficiency and substrate specificities of AcrAB-TolC and MexAB-OprM are difficult to compare when they are expressed in their native environments because of the well-established difference in the permeability properties of the outer membrane of E. coli and P. aeruginosa (14). However, when both pumps are expressed in E. coli their functional differences can be assessed. AcrB and MexB appeared to have very similar substrate specificities; however, they provided different levels of resistance to some compounds (Table 2). In particular, MexAB-OprM provides higher levels of resistance against CIN. On the other hand, AcrAB-TolC is more efficient in pumping out positively charged compounds such as EtB and OLE. Thus, despite the high degree of primary sequence homology between the components, these pumps do not efflux all substrates with equal efficiency.
We have taken advantage of the differences described above between AcrAB-TolC and MexAB-OprM to identify the regions of the RND transporters involved in specific interactions with the other two components of complexes and/or for regions defining substrate specificities. Among 11 hybrid pumps, only AcrA-AcrB612/MexB, AcrA-AcrB849/MexB, and MexA-AcrB60/MexB-OprM were capable of multidrug efflux, demonstrating that the complexes are assembled and that the IM hybrid transporters retained their function. We have verified the complex assembly by using an in vivo cross-linking technique (26). Although we cannot detect the tripartite complexes by using this assay, the complexes between the periplasmic components and the IM transporters were found for all functional hybrid pumps (Fig. 4). Thus, the structural determinants defining the specificity of interaction with the periplasmic components of complexes are encoded within the T60-V612 region of the IM transporters. This region spans the most of ED1, TMS 2 through TMS 7, and the 75 amino acid residues of ED2.
Since neither AcrA nor MexA proteins possess transmembrane segments, it is unlikely that TMS 2 through TMS 7 contain structure variations defining the specificity of interaction with MFPs. Thus, amino acid residues of ED1 and ED2 presumably interact with accessory MFPs. Interestingly, the hybrid transporters AcrB589/MexB and AcrB612/MexB differ by only 22 amino acid residues. However, in contrast to AcrB612/MexB, AcrB589/MexB is deficient in the complex assembly. Similarly, AcrB182/MexB, which contains an extra 122 amino acids of AcrB compared to AcrB60/MexB, is a nonfunctional construct. Both these regions are entirely located in the moderately conserved regions of ED1 or ED2. Figure 5 shows the alignment of the V590-V612 regions of RND-type multidrug transporters from bacteria belonging to the γ-Proteobacteria group. Among 22 amino acid residues of this region, 5 are highly conserved in transporters from enterobacteria and differ in the MexB transporter. The site-specific mutagenesis of these residues is under way to determine their role in drug efflux and complex assembly.
FIG. 5.
Alignment of the V590-V612 region of AcrB homologs from bacteria belonging to the γ-Proteobacteria division and MexB from P. aeruginosa. The conserved residues in AcrB and its close homologs that differ in MexB are in boldface and highlighted.
The comparison of functional activities of AcrB612/MexB, AcrB849/MexB, and AcrB60/MexB (Table 2) suggested that the recognition of negatively charged compounds, such as the detergents SDS and CA and NOV, is distinct from other compounds. These substrates are expelled by all three hybrids with an efficiency similar to that in the wild-type transporters AcrB and MexB. However, hybrid pumps differed significantly in the efflux of other tested compounds.
The AcrA-AcrB849/MexB chimeric pump displayed a MexB-like specificity to the substrates CIN, EtB, OLE, and LIN (Table 2). In this hybrid, only the C-terminal 200 amino acid residues are from MexB. Thus, the structural variations responsible for differences in substrate specificities of AcrB and MexB to these substrates appear to localize in the C-terminal domains of RND transporters. Alignment of these regions in AcrB and MexB indicated that these domains are highly conserved (data not shown). The only variable regions in this domain, which might be responsible for the observed differences between AcrB and MexB, are the cytoplasmic loop connecting TMS 10 and TMS 11 and the C-terminal cytoplasmic tail.
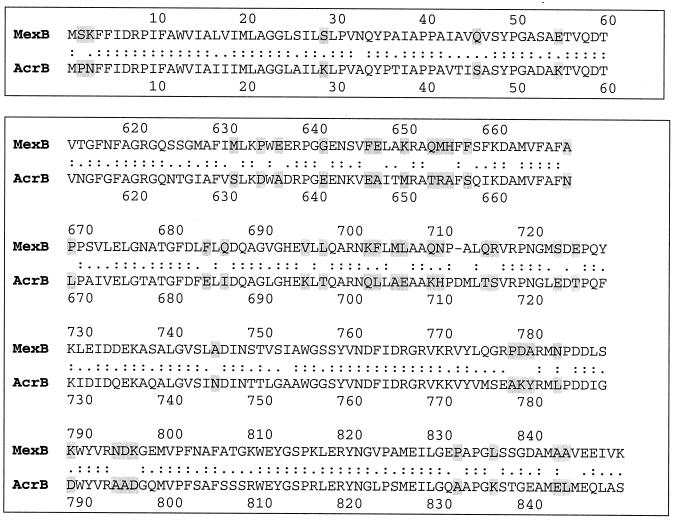
On the other hand, AcrA-AcrB849/MexB was similar to the wild-type AcrB in recognition of ERY and PUR, suggesting that other regions of AcrB are also involved in interactions with substrates. AcrA-AcrB612/MexB, containing an additional 235 C-terminal residues from MexB, transported these substrates with an efficiency comparable to that of the wild-type MexB transporter. This result suggested that the residues V612-S/K849 located in the extracytoplasmic domain ED2 play an important role in transport of these substrates. The alignment of the V612-S/K849 regions in AcrB and MexB is shown in Fig. 6. These regions appear to differ substantially in the distribution of charged amino acids, with the obviously high density of negative charges in the MexB protein. On the contrary, AcrB contains an extra number of positively charged amino acids. The reverse distribution of charges in AcrB612/MexB and AcrB849/MexB could explain the inability of AcrB612/MexB to pump out quinolones (CIN and NAL) and other substrates (chloramphenicol [CF], PRO, and tetracycline [TET]). The important role of extracytoplasmic domains in the drug specificities of RND transporters has also been demonstrated in domain replacement studies of the AcrB and AcrD transporters (3).
FIG. 6.
Alignment of AcrB and MexB sequences in functionally important regions. Amino acid residues that differ in these two transporters are highlighted.
It is noteworthy that MexA-AcrB60/MexB-OprM appears to be functionally similar to AcrB612/MexB. This similarity is obvious in the specificities to NAL, PUR, CF, PRO, tetracycline, and chlortetracycline (CTET). The AcrB60/MexB construct contains only 60 N-terminal amino acid residues from AcrB. Since structural variations in the M1-T60 region of AcrB/MexB profoundly affect the specificity toward such a broad spectrum of substrates, this result suggests that all these substrates might interact with the same drug-binding region of AcrB/MexB. Perhaps this drug-binding site comprises amino acid residues from both the N- and C-terminal domains of RND transporters. Indeed, the N-terminal residues of all our constructs originate from the AcrB transporter. The shortest region of AcrB required for the function with AcrA was found to be 612 N-terminal residues (AcrB612/MexB). Still, similar to MexA-AcrB60/MexB-OprM, the AcrA-AcrB612/MexB hybrid pump appears to be defective in transport of several substrates. On the other hand, cells expressing AcrA-AcrB849/MexB were resistant to all tested compounds. Thus, amino acid residues from both the N- and C-terminal domains could contribute to the substrate specificity of RND transporters. However, we cannot exclude the possibility that the defects in transport of particular substrates are caused by partial misfolding of chimeric transporters.
Acknowledgments
This work was supported by a Scientist Development grant from the American Heart Association to H.I.Z.
This study was begun in the lab of H. Nikaido. We are indebted to Hiroshi Nikaido for his continuing support and critical reading of the manuscript. We thank K. Poole for providing polyclonal anti-OprM antibody.
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan, S. K. 2001. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci. 26:3-6. [DOI] [PubMed] [Google Scholar]
- 3.Elkins, C. A., and H. Nikaido. 2002. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrB of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 184:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotoh, N., H. Tsujimoto, A. Nomura, K. Okamoto, M. Tsuda, and T. Nishino. 1998. Functional replacement of OprJ by OprM in the MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 165:21-27. [DOI] [PubMed] [Google Scholar]
- 6.Guan, L., M. Ehrmann, H. Yoneyama, and T. Nakae. 1999. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonas aeruginosa. J. Biol. Chem. 274:10517-10522. [DOI] [PubMed] [Google Scholar]
- 7.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 8.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 10.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinolones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikaido, H., M. Basina, V. Nguyen, and E. Y. Rosenberg. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido, H., and H. I. Zgurskaya. 2001. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:215-218. [PubMed] [Google Scholar]
- 16.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saier, M. H., Jr., I. T. Paulsen, M. K. Sliwinski, S. S. Pao, R. A. Skurray, and H. Nikaido. 1998. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12:265-274. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulavik, M. C., C. Houseweart, C. Cramer, N. Jiwani, N. Murgolo, J. Greene, B. DiDomenico, K. J. Shaw, G. H. Miller, R. Hare, and G. Shimer. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thanabalu, T., E. Koronakis, C. Hughes, and V. Koronakis. 1998. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 17:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng, T. T., K. S. Gratwick, J. Kollman, D. Park, D. H. Nies, A. Goffeau, and M. H. Saier, Jr. 1999. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1:107-125. [PubMed] [Google Scholar]
- 23.Yoneyama, H., A. Ocaktan, N. Gotoh, T. Nishino, and T. Nakae. 1998. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 244:898-902. [DOI] [PubMed] [Google Scholar]
- 24.Zgurskaya, H. I., and H. Nikaido. 1999. AcrA is a highly asymmetric protein capable of spanning the periplasm. J. Mol. Biol. 285:409-420. [DOI] [PubMed] [Google Scholar]
- 25.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]