Abstract
The Rhizobium-legume symbiosis involves the formation of a novel plant organ, the nodule, in which intracellular bacteria reduce molecular dinitrogen in exchange for plant photosynthates. Nodule development requires a bacterial signal referred to as Nod factor, which in Sinorhizobium meliloti is a β-(1,4)-linked tetramer of N-acetylglucosamine containing N-acyl and O-acetyl modifications at the nonreducing end and a critical 6-O-sulfate at the reducing end. This sulfate modification requires the action of three gene products: nodH, which catalyzes the sulfonyl transfer, and nodPQ, which produce the activated form of sulfate, 3′-phosphoadenosine-5′-phosphosulfate. It was previously reported that S. meliloti cell surface polysaccharides are also covalently modified by sulfate in a reaction dependent on NodPQ. We have further characterized this unique form of bacterial carbohydrate modification. Our studies have determined that one of the nodPQ mutant strains used in the initial study of sulfation of cell surface harbored a second unlinked mutation. We cloned the gene affected by this mutation (referred to as lps-212) and found it to be an allele of lpsL, a gene previously predicted to encode a UDP-glucuronic acid epimerase. We demonstrated that lpsL encoded a UDP-glucuronic acid epimerase activity that was reduced in the lps-212 mutant. The lps-212 mutation resulted in an altered lipopolysaccharide structure that was reduced in sulfate modification in vitro and in vivo. Finally, we determined that the lps-212 mutation resulted in a reduced ability to elicit the formation of plant nodules and by altered infection thread structures that aborted prematurely.
In order to acquire reduced nitrogen, many leguminous plants enter into symbiotic associations with bacteria, culminating in the formation of a novel plant organ, the nodule, in which intracellular bacteria reduce molecular dinitrogen to ammonia. In a compatible symbiotic interaction, the bacteria trigger an alteration in the growth of the plant root hairs, resulting in a curled structure that entraps a microcolony of the bacteria. This curled root hair is the site for a plant cell wall-encapsulated ingrowth of the root hair, referred to as an infection thread. The infection thread, filled with proliferating bacteria, extends to the base of the root hair cell and then penetrates the root, allowing bacterial entry into the plant. Concurrent with the development of the infection thread, the cells in the root cortex dedifferentiate, leading to new cell division and the consequent formation of the nodule. The infection thread branches and penetrates the developing nodule, delivering the bacteria, which are then released into the plant cytoplasm. These intracellular bacteria undergo a series of developmental changes and signal transduction events to induce the expression of nitrogenase, which catalyzes the reduction of molecular dinitrogen to ammonia (4, 5, 18, 20, 31, 41).
Symbiotic nitrogen-fixing relationships between legumes and the genera Rhizobium, Bradyrhizobium, Mesorhizobium, and Sinorhizobium (collectively called rhizobia) are mediated by chemical signals exchanged between the symbiotic partners (21, 32). The plant produces a chemical signal (usually in the form of flavonoid molecules) that is perceived by the bacterium and activates transcription of the nod genes. Most of the known nod gene products catalyze synthesis of an oligosaccharide signal, referred to as Nod factor. All known Nod factors consist of β-(1,4)-linked N-acetylglucosamine residues which are N-acylated at the nonreducing end (9-11, 15, 17, 38). To this Nod factor structure are added host-specific modifications, which in Sinorhizobium meliloti consist of a 16:2 N-acyl group and 6-O-acetyl group at the nonreducing end of the molecule and a 6-O-sulfate modification at the reducing end (24).
The presence of the sulfate modification on Nod factor is absolutely required for the establishment of the symbiosis on alfalfa and is dependent on the products of three genes, nodH, nodP, and nodQ. The nodH gene product catalyzes the sulfonyl transfer to the Nod factor backbone (13, 33), while the nodP and nodQ gene products form a sulfate-activating complex which catalyzes the conversion of sulfate to 3′-phosphoadenosine-5′-phosphosulfate (34, 35), an activated form of sulfate used by all known carbohydrate sulfotransferases. Two copies of nodPQ exist in S. meliloti (36). One copy (referred to nodP1Q1) is present on pSymA, a large (1.35-Mb) symbiotic plasmid which carries the majority of the nod genes. An additional copy of nodPQ (referred to as nodP2Q2) is present on pSymB, a distinct large (1.68-Mb) symbiotic megaplasmid. The genes nodP1Q1 and nodP2Q2 are functionally redundant in that both copies have to be inactivated to impair nodule formation on alfalfa (36).
In addition to sulfate modification of the S. meliloti Nod factor, sulfuryl modifications are also carried on polysaccharides that constitute the S. meliloti cell surface (6). Sulfate modification of cell surface polysaccharides is increased in the presence of plant inducers of nod gene transcription (19) and is dependent upon NodPQ (6). To understand the mechanism and symbiotic function of this rare form of sulfate modification, we investigated the role of nodPQ in the sulfation of cell surface polysaccharide. During these studies, we identified an unlinked mutation in the nodP1Q1 mutant background used in the previous studies of cell surface sulfation (6). We characterized this unlinked mutation, lps-212, and found it to be an allele of the gene lpsL. We demonstrate that the lpsL gene encodes a UDP-glucuronic epimerase activity that is reduced in the lps-212 mutant. The lps-212 mutation alters the structure of S. meliloti lipopolysaccharide (LPS) and reduces its sulfation in vitro and in vivo. Finally, the lps-212 mutant is symbiotically compromised, exhibiting at least a 10-fold reduction in nitrogen fixation.
MATERIALS AND METHODS
Bacterial strains and media.
All strains used were derivatives of Sinorhizobium meliloti 1021 and are described in Table 1. All strains were grown in Luria-Bertani (LB) (8) or M9 medium (26) with the antibiotic concentrations described previously (28).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Rm1021 | SU47 Smr | 27 |
| JT402 | Tn5 located between nodE and nodG | 39 |
| JSS12 | nodP1Q1::Tn5-233 lps-212 | 36 |
| JSS12Tr | nodP1Q1::Tn5-233 | 36 |
| JSS14 | nodP2Q2::Tn5 | 36 |
| DKR1 | lps-212, Tn5 located in noncoding region between nodE and nodG | This study |
| DKR58 | lps-212 pMS03 | This study |
| DKR59 | lps-212 pDKR59 | This study |
| DKR60 | lps-212 pDKR60 | This study |
| DKR61 | lps-212 pDKR61 | This study |
| DKR62 | lpsL::pDKR62 | This study |
| DKR63 | rkpK::pDKR63 | This study |
| JT210 | nodH::Tn5 | 39 |
| Plasmids | ||
| pMB393 | Broad-host-range plasmid pBBS containing pBluescript multicloning site | 2 |
| pMS03 | Broad-host-range plasmid pMB393 containing constitutive trp promoter | This study |
| pDKR59 | pMS03/lpsL | This study |
| pDKR60 | pMS03/lpsL212 | This study |
| pDKR61 | pMS03/rkpK | This study |
| pDKR62 | pVO155/lpsL | This study |
| pDKR63 | pVO155/rkpK | This study |
| pXLGD4 | pRK290/hemA::lacZ | 1 |
| pVO155 | Integrational vector | 28 |
| pTB93G | pMB393/GFP | 16 |
Strain construction.
Strain DKR1 was constructed by using N3 phage grown on strain JT402 (39) to transduce strain JSS12 to neomycin resistance. Neomycin-resistant transductants which were spectinomycin sensitive (due to replacement of the nodP1Q1::Tn5-233 with wild-type sequence) were screened for altered sulfation and LPS structure.
Plasmid construction.
Plasmid pDKR59 was constructed by PCR amplification of lpsL from strain Rm1021 with primers 5′-TCTGCGAAAGCTTCCCGACCCTGGA-3′ and 5′-TGCAATTGGGTACCGAAGCACGCGC-3′. The PCR product was cloned into the Topo2.1 plasmid with the TopoTA cloning kit (Invitrogen). The resulting plasmid was then digested with KpnI and ClaI, and the insert was isolated from an agarose gel with a gel extraction kit (Qiagen) and ligated with plasmid pMS03 digested with the same enzymes. Plasmid pDKR60 was constructed in the same manner as pDKR59, but chromosomal DNA prepared from strain DKR1 was used to amplify lpsL.
Plasmid pDKR61 was constructed by PCR amplification of rkpK with primers 5′-TGTGCGAAGCTTGTGCTTCCGCACC-3′ and 5′-GGCAGAGGGATCCCCGTGCAGCTTC-3′. The PCR product was then cloned into pCR2.1 as described above. The plasmid was then digested with KpnI and ClaI, and the insert was isolated and cloned into pMS03 digested with the same enzymes. Plasmid pMS03 was constructed by digestion of plasmid pTE3 with HindIII. The insert containing the trp promoter was isolated as described above and ligated into pMB393 (3) digested with the same enzymes.
Plasmid pDKR62 was constructed by PCR amplification of an internal fragment of lpsL from strain Rm1021 with primers 5′-TATGTCGACGCGAACCTCGT-3′ and 5′-ATTTCTAGAGCGCCATGTCCGGCCG-3′. The PCR product was cloned into pCR2.1 as described above. The plasmid was then digested with SalI and XbaI, and the insert was isolated and ligated with pVO155 (28) digested with the same enzymes. Plasmid pDKR63 was constructed by PCR amplification of an internal fragment of rkpK with primers 5′-AGAAAGTCGGTACCAATGTGCAGGA-3′ and 5′-TACTTGGATCCGCGGATATCGCCGC-3′. The PCR product was cloned into pCR2.1 as described above. The plasmid was then digested with SalI and BamHI, and the insert was isolated and then ligated into pVO155 digested with the same enzymes.
Preparation of extracts for LPS analysis.
Extracts were prepared according to Reuhs et al. (30) with the following modifications. The cells from 1.5 ml of log-phase (optical density at 600 nm, 0.5) culture were centrifuged at 8,000 × g and resuspended in 1 ml of water. The cells were again centrifuged at 8,000 × g, and the pellet was resuspended in 0.15 ml of solution A (0.05 M Na2HPO4, 0.005 M EDTA, pH 7). To the cell suspension was added 0.15 ml of 90% phenol, and the sample was vortexed and then incubated at 65°C for 15 min, followed by incubation on ice for 10 min. Samples were then centrifuged at 8,000 × g for 10 min, and the aqueous phase was removed and dried under vacuum. The pellet was resuspended in sample loading buffer and fractionated by deoxycholate-polyacrylamide gel electrophoresis (PAGE) as described previously (30). The polysaccharides were then visualized by silver staining (Bio-Rad) or phosphorimaging (Bio-Rad).
Preparation of cell surface protein extracts.
Cultures of 100 ml of cells were grown in LB medium until the cultures reached stationary phase, and the cells were then collected by centrifugation at 600 × g. The cell pellets were resuspended in 3 ml of buffer A (25 mM Tris-HCl [pH 7.5] containing 5 mM 2-mercaptoethanol and 10% glycerol) and disrupted by two passes through a Bionebulizer (Glasco), and the cell debris was removed by centrifugation at 6,000 × g. A cell surface extract was then prepared by centrifugation at 100,000 × g for 30 min in a tabletop ultracentrifuge (Beckman). The resulting pellet was resuspended in 100 μl of buffer A, and the protein concentration was determined by a modified Bradford assay (Bio-Rad).
In vitro cell surface sulfation assay.
From 2.5 to 10 μg of membrane extract was combined with 5 μCi of 35SO4-labeled 3′-phosphoadenosine-5′-phosphosulfate (prepared as described previously [13, 25, 34]), and 2 μl of 5× buffer B (50 mM Tris-HCl [pH 8], 30 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol) in a total reaction volume of 10 μl. The mixture was then allowed to incubate for 30 min at 30°C, and the reaction was stopped by incubation for 2 min at 100°C. Sodium dodecyl sulfate (SDS) sample buffer, prepared as described above was then added, and the samples were heated at 95°C for 5 min and fractionated on an SDS-12.5% PAGE gel. The gel was dried, and the incorporation of 35SO4 into LPS was then determined by quantitation of the radioactivity in the gel with a Bio-Rad phosphorimager.
UDP-glucuronic acid epimerase assay.
[U-14C]uridine 5′-diphosphoglucuronic acid (10 μCi of 225 mCi/mmol; ICN) was incubated with 2 to 10 μg of clarified soluble cell extracts that had been desalted by passage through a G-25 spin column (Pharmacia). The reaction was allowed to proceed for 30 min at 37°C. The extracts were then spotted on fluorescent polyethyleneimine-cellulose plates (Baker) and developed in 0.3 M lithium chloride. The migration of the labeled material was monitored by autoradiography or phosphorimaging, (Bio-Rad). The migration of UDP-glucose, UDP-glucuronic acid, and UDP-galacturonic acid was determined by comparison to unlabeled standards (Sigma) that were visualized by long-wave UV light.
UDP-glucose dehydrogenase assay.
UDP-glucose dehydrogenase was assayed spectrophotometrically as described previously (19).
Nodulation assay.
The ability of wild-type and lps-212 mutants to initiate formation of nodules on alfalfa was assayed as described previously (12). Briefly, alfalfa seeds were sterilized by shaking in 70% ethanol for 45 min, followed by shaking for 45 min in 20% hypochlorite. The seeds were then washed four times with sterile H2O, allowed to imbibe H2O overnight, and germinated on an inverted petri dish in the dark. The seedlings were then transferred to slant agar tubes containing BNM agar (12). The plants were allowed to grow in the tubes for 4 days and then were inoculated with bacterial strains which had been cultured to log phase (optical density at 600 nm, 0.5) in TY medium and then diluted 1:200 in BNM liquid medium. A total of 5 ml of medium was poured over the plants and then removed from the tube. At various times, inoculated plants were observed under a dissecting scope, and the number of nodules was counted. Each experiment was performed with replicate tubes.
Nitrogenase assay.
Nitrogen fixation was quantitated in plants 28 days after inoculation via the acetylene reduction assay (40). Ethylene-acetylene separation and quantitation were carried out on a Shimadzu GC-8A1F gas chromatograph with a Porapak N column and flame ionization detector. The amount of ethylene produced was calculated by integration of the peak and converted to nanomoles of ethylene formed per plant by comparison to a standard curve developed from injected standard amounts of ethylene.
LacZ staining of bacteria in infection threads.
The progress of infection was monitored by visualization of the bacteria within the infection thread as described previously (1, 3). Briefly, wild-type and lps-212 mutant strains were engineered to express β-galactosidase by introduction of plasmid pXLGD4 containing lacZ under the control of the constitutive hemA promoter. The plants were inoculated with the mutant strains as described above, and the infection was allowed to proceed for either 7 days or 28 days. Sections of the plant root containing nodules were then removed and fixed with 1.25% glutaraldehyde in 200 mM sodium cacodylic acid (pH 7.2) under vacuum for 30 min, followed by incubation under normal atmospheric pressure for an additional hour. The plants were washed four times for 15 min with 200 mM sodium cacodylic acid buffer (pH 7.2) and then placed in 1 ml of 200 mM sodium cacodylate (pH 7.2)-5 mM potassium ferrocyanide-5 mM sodium ferricyanide-0.8% X-gal (5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside). The samples were allowed to stain overnight in the dark and were then washed twice with 200 mM sodium cacodylate (pH 7.2), followed by treatment for 5 min with 20% hypochlorite. Finally, the plants were washed two times with 200 mM sodium cacodylate (pH 7.2) and then photographed under visible light with a Nikon Optiphot microscope with a 20× objective.
RESULTS
Isolation of S. meliloti mutant with altered cell surface sulfation.
A previous study reported that carbohydrate residues on the S. meliloti cell surface are covalently modified by sulfate in a reaction dependent on NodPQ (6). To further characterize this modification in vivo, we examined the sulfate modification of LPS in strains having null mutations in either nodP1Q1 or nodP2Q2. Our results were consistent with the previous studies (6), demonstrating that NodPQ are required for sulfation of cell surface carbohydrates (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data).
However, we observed that LPS purified from exponentially growing strain JSS12 (nodP1Q1::Tn5-233 lps-212), fractionated by deoxycholate-PAGE and visualized by silver staining, was structurally distinct from wild-type LPS in that the LPS core region migrated at a higher relative mobility than wild-type LPS core (Fig. 1A). We found that exponentially growing S. meliloti produces primarily rough LPS (comprised of lipid A and LPS core) and lacks detectable O-antigen. Insertions that inactivated nodP2Q2 did not produce this altered form of LPS (Fig. 1A). Consistent with the observed changes in LPS structure, strain JSS12 was also found to be sensitive to the detergent deoxycholate (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data), a common characteristic of mutants altered in LPS biosynthesis (19, 22). These results indicated that strain JSS12 produced an altered form of LPS.
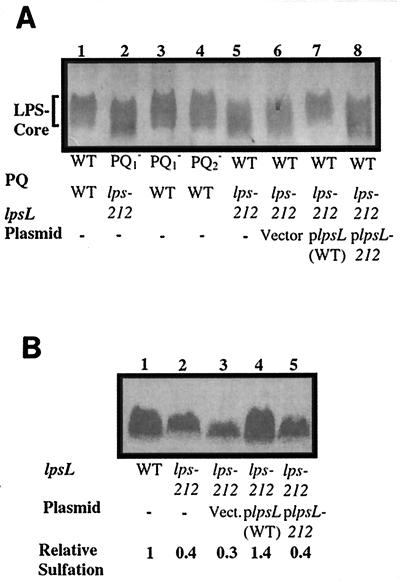
FIG. 1.
Analysis of LPS from wild-type and mutant strains. (A) Silver-stained deoxycholate-PAGE gel of purified LPS core from S. meliloti. Lane 1, strain Rm1021 (wild type [WT]); lane 2, strain JSS12 (nodP1Q1::Tn5-233 lps-212); lane 3, JSS12Tr (nodP1Q1::Tn5-233); lane 4, JSS14 (nodP2Q2::Tn5); lane 5, DKR1 (lps-212); lane 6, DKR58 (lps-212 pVector); lane 7, DKR59 (lps-212 plpsL); lane 8, DKR60 (lps-212 plpsL212). (B) In vivo sulfate labeling of S. meliloti LPS core. Wild-type and lps-212 strains were cultured in the presence of 10 μCi of Na235SO4; the LPS was extracted, fractionated by deoxycholate-PAGE, and analyzed by phosphorimaging as described in Materials and Methods. Lane 1, Rm1021 (wild type [WT]); lane 2, DKR1 (lps-212); lane 3, DKR58 (lps-212 pVector); lane 4, DKR59 (lps-212 plpsL); lane 5, DKR60 (lps-212 plpsL212).
Altered form of LPS found in JSS12 is genetically separable from the nodP1Q1::Tn5-233 insertion.
Transduction via phage N3 of the nodP1Q1::Tn5-233 insertion from strain JSS12 into strain Rm1021 yielded strain JSS12Tr, which no longer showed the structural alteration of LPS seen in JSS12 (Fig. 1A). In order to demonstrate conclusively that the mutation leading to the altered LPS was unlinked to nodP1Q1::Tn5-233, we used strain JT402, which harbors a Tn5 tightly linked to nodP1Q1 (39). Strain JT402 was used as a donor in phage N3-mediated transduction, selecting for the antibiotic resistance of the insertion (in a noncoding position adjacent to nodP1Q1), and the resulting transductants were then scored for retention or loss of the nodP1Q1::Tn5-233 insertion.
A significant portion (24%) no longer harbored the nodP1Q1::Tn5-233 insertion and were presumed to result from replacement of the nodP1Q1::Tn5-233 region with the wild-type sequence. The transductants that no longer contained the nodP1Q1::Tn5-233 (and were wild type for nodP1Q1) retained the altered LPS phenotype, as judged by silver-stained PAGE of LPS preparations (Fig. 1A) and by deoxycholate sensitivity (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data). One of these strains, DKR1, was chosen for use in subsequent experiments. These data demonstrate that strain JSS12 harbors two mutations, nodP1Q1::Tn5-233 and an additional mutation (referred to as lps-212) that affects the structure of LPS.
lps-212 mutant is deficient in sulfate modification of LPS.
Our finding that strain JSS12 contained two distinct mutations brought into question whether the decreased sulfation seen in strain JSS12 (6) resulted from the nodP1Q1::Tn5-233 insertion or from the presence of the lps-212 mutation. Strain JSS12Tr (which contains nodP1Q1::Tn5-233 in an otherwise wild-type background) showed a reduced amount of sulfation in vivo (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data). However, strain DKR1 (which is wild type for nodPQ) also exhibited a reduction in the relative amount of sulfation when analyzed on deoxycholate-PAGE (Fig. 1B).
lps-212 mutant is deficient in cell surface sulfation in vitro.
To understand the role of the lps-212 mutation in LPS sulfate modification, we developed an in vitro sulfation assay. This assay uses previously established protocols to prepare 3′-phosphoadenosine-5′-phosphosulfate labeled with 35S at high specific activity (13, 25, 34). This radiolabeled 3′-phosphoadenosine-5′-phosphosulfate was then used as the donor for a recently discovered sulfate transfer activity (D. H. Keating and S. R. Long, unpublished results), which is membrane associated and modifies carbohydrates present in the S. meliloti cell surface fraction.
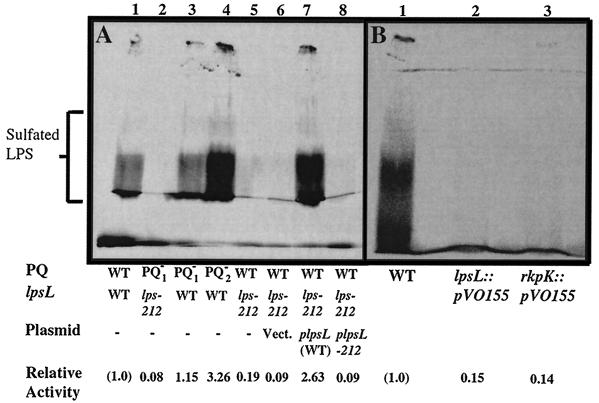
When wild-type Rm1021 was assayed, an abundant sulfation activity was observed (Fig. 2). Strains JSS12Tr (nodP1Q1::Tn5-233) and JSS14 (nodP2Q2::Tn5) were also found to be wild type for this activity, demonstrating that NodPQ (which produces 3′-phosphoadenosine-5′-phosphosulfate) is not required for in vitro sulfation when 3′-phosphoadenosine-5′-phosphosulfate is provided (Fig. 2A). However, strains JSS12 and DKR1 were found to be strongly deficient in this activity (Fig. 2A). JSS12 and DKR1 have in common the alteration in LPS structure, implying that the mutant form of LPS associated with lps-212 is unable to undergo sulfation in vitro.
FIG. 2.
In vitro LPS sulfation of wild-type S. meliloti and lps-212 mutants. (A) In vitro sulfation activity of wild-type and mutant extracts. Extracts were prepared and assayed for in vitro polysaccharide sulfotransferase activity as described in Materials and Methods. Lane 1, strain Rm1021 (wild type [WT]); lane 2, JSS12 (nodP1Q1::Tn5-233 lps-212); lane 3, JSS12Tr (nodP1Q1::Tn5-233); lane 4, JSS14 (nodP2Q2::Tn5); lane 5, DKR1 (lps-212); lane 6, DKR58 (lps-212 pVector); lane 7, DKR59 (lps-212 plpsL); lane 8, DKR60 (lps-212 plpsL212). (B) In vitro sulfation of mutants lacking either rkpK or lpsL. Lane 1, Rm1021 (wild type [WT]); lane 2, DKR62 (lpsL::pVO155); lane 3, DKR63 (rkpK::pVO155).
Cloning of lps-212.
The deoxycholate-sensitive phenotype of strain DKR1 was used to clone the lps-212 gene by complementation. A medium containing 0.6 mg of deoxycholate per ml impaired the growth of the lps-212 mutant (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data). A cosmid library containing ca. 20-kb pieces of the Rm1021 genome was mated into DKR1, and tetracycline-resistant exconjugants were selected on deoxycholate-containing medium. Colonies which grew on this medium were then screened for complementation, as judged by silver-stained PAGE of cell surface extracts, and in vitro carbohydrate sulfation activity. Surprisingly, none of the colonies initially selected on this medium (20 colonies screened) were found to complement the altered LPS phenotype of DKR1. Replating all of the colonies initially selected for growth on deoxycholate-containing medium resulted in only about 1% survival during subsequent challenge with deoxycholate. Of these, the majority (9 of 10 colonies tested) retained deoxycholate resistance in subsequent replating experiments and displayed wild-type LPS (as judged by silver-stained PAGE of polysaccharide preparations) and wild-type sulfation activity in vitro (D. H. Keating, M. G. Willits, and S. R. Long, unpublished data). Three of these colonies were kept for further analysis.
lps-212 is an allele of lpsL.
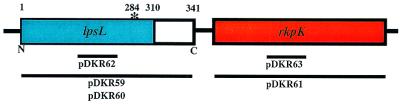
The nucleotide sequence of the complementing clones was determined. Two of the sequenced clones carried identical inserts, whereas the third clone had a distinct but overlapping insert. Comparison with the GenBank database showed that the nucleotide sequence of the inserts had >90% nucleotide identity to lpsL and rkpK of S. meliloti strain Rm41 (Fig. 3). The S. meliloti strain Rm41 rkpK gene was previously reported to encode UDP-glucose dehydrogenase, which converts UDP-glucose to UDP-glucuronic acid (19). The upstream gene, lpsL, had significant sequence similarity to nucleotide-sugar epimerases (19). Sequencing of PCR-amplified lpsL from the wild-type and DKR1 strains demonstrated a single nucleotide difference between the genes isolated from the two different sources, a deletion of a guanosine nucleotide near the predicted C terminus (nucleotide 847), which resulted in a shift of the reading frame and premature truncation of the predicted protein (Fig. 3).
FIG. 3.
S. meliloti genomic region containing lpsL and rkpK. The site of the frameshift mutation is denoted with an asterisk. The unshaded region corresponds to the part of LpsL truncated as the result of the premature termination resulting from the frameshift mutation. The numbers correspond to the amino acid positions of the open reading frame. The lines placed below the open reading frame depiction correspond to the regions of lpsL and rkpK carried on the plasmids used for complementation and gene inactivation.
The primary sequence information therefore suggested that lps-212 was an allele of lpsL. To confirm these data, minimal clones containing either lpsL or rkpK were constructed and inserted into plasmid pMS03 downstream of the Salmonella enterica serovar Typhimurium trp promoter, which allows constitutive expression in S. meliloti. The plasmids were conjugated into DKR1, and complementation of the LPS defect was scored by assaying deoxycholate sensitivity, in vitro polysaccharide sulfation of plasmid-bearing extracts, and LPS structures.
A minimal clone containing the lpsL gene derived from the wild-type strain complemented DKR1, as judged by recovery of wild-type polysaccharide structure (Fig. 1A), in vivo LPS sulfation (Fig. 1B), and in vitro sulfation activity (Fig. 2A), suggesting that the mutated gene in strain lps-212 was an allele of lpsL. A minimal clone containing lpsL isolated from strain DKR1 was unable to complement the alteration of polysaccharide structure (Fig. 1A), LPS sulfation in vivo (Fig. 1B), or in vitro sulfation (Fig. 2A), implying that the mutation in the lpsL gene of strain DKR1 eliminated its ability to restore wild-type phenotypes to strains containing the original lps-212 mutation. Minimal rkpK clones from wild-type S. meliloti were unable to complement any of the phenotypes of strain DKR1 (D. H. Keating, M. G. Willits, and S. R. Long, unpublished results).
To demonstrate that lps-212 mutant phenotypes arose from an altered allele of lpsL, we constructed null mutations in either lpsL or rkpK with the insertional mutagenic plasmid pVO155 (28). Strain DKR62, containing an insertion in lpsL, and DKR63, which contains an insertion in rkpK, were both found to be affected in LPS biosynthesis, as judged by deoxycholate sensitivity (D. H. Keating, M. G. Willits, and S. R. Long, unpublished results) and in vitro sulfation (Fig. 2B). This suggested that both lpsL and rkpK are required for both LPS synthesis and its modification with sulfate.
lpsL is required for UDP-glucuronic acid epimerase activity.
As mentioned above, lpsL isolated from strain Rm41 has been reported previously to bear a high degree of similarity to sugar epimerases (19). The gene lpsL from S. meliloti strain Rm1021 is also highly similar to genes encoding sugar epimerases (51% amino acid identity to Escherichia coli galE). Previous studies of lpsL from strain Rm41 predicted that UDP-glucose is converted to UDP-glucuronic acid via RkpK, which is then epimerized to UDP-galacturonic acid by LpsL (19).
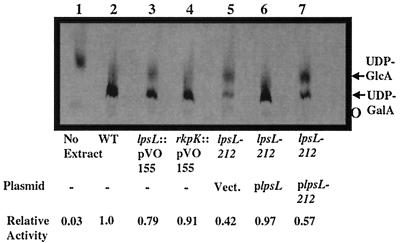
To determine if lpsL encodes an epimerase activity capable of converting UDP-glucuronic acid to UDP-galacturonic acid, we used a thin-layer chromatography system which separates UDP-galacturonic acid from UDP-glucuronic acid. Radiochemically pure UDP-glucuronic acid was incubated with clarified S. meliloti cell extracts and then analyzed by thin-layer chromatography (Fig. 4). Wild-type Rm1021 extract was able to convert a detectable amount of UDP-glucuronic acid to UDP-galacturonic acid (as judged by migration with respect to unlabeled standards). However, the extract from strain DKR1 showed a reduced ability to carry out this reaction in vitro. The extract from strain DKR62, which carries an insertionally inactivated lpsL, also had a reduced amount of epimerase activity (Fig. 4), although greater than that of the strain DKR1 extract. The activity in strain DKR1 could be restored by addition of wild-type lpsL on a multicopy plasmid but not by a multicopy plasmid carrying the lpsL212 allele, indicating that the product of the wild-type lpsL gene is required for nucleotide sugar epimerase activity in vitro. Strain DKR63, containing an insertionally inactivated rkpK, was found to have nucleotide sugar epimerase activity similar to that of the wild type (Fig. 4).
FIG. 4.
Assay of UDP-glucuronic acid epimerase activity in the wild type and lpsL mutants. Wild-type and mutant extracts were assayed for UDP-glucuronic acid epimerase as described in Materials and Methods. Lane 1, no extract; lane 2, strain Rm1021 (wild type [WT]); lane 3, strain DKR62 (lpsL::pVO155); lane 4, DKR63 (rkpK::pVO155); lane 5, DKR58 (lps-212 pVector); lane 6, DKR59 (lps-212 plpsL); lane 7, DKR60 (lps-212 plpsL212). UDP-GlcA, UDP-glucuronic acid; UDP-GalA, UDP-galacturonic acid; O, origin. Radioactive species were identified by migration in relation to unlabeled standards. Similar results were seen in four assays performed with independently prepared extracts.
Strains Rm1021 and DKR1 and strains carrying insertions that inactivated rkpK or lpsL were also assayed for UDP-glucose dehydrogenase activity. Extracts prepared from the wild-type and DKR1 strains retained wild-type UDP-glucose dehydrogenase activity (Table 2), whereas extracts prepared from strain DKR63 (rkpK::pVO155) lacked detectable UDP-glucose dehydrogenase activity (Table 2). Interestingly, extracts prepared from strain DKR62 (lpsL::pVO155) were found to have a ca. 3-fold reduction in dehydrogenase activity. Because extracts from strain DKR1 had wild-type activity, the reduction in UDP-glucose dehydrogenase activity seen in DKR62 may result from a polar effect of the lpsL::pVO155 insertion on the transcription of rkpK (which is downstream of lpsL).
TABLE 2.
UDP-glucose dehydrogenase activity of wild-type and mutant strains
| Strain | Genotype | UDP-glucuronic acid dehydrogenase activity (nmol/min/mg of protein) | Relative activitya |
|---|---|---|---|
| None | <0.23 | <0.003 | |
| Rm1021 | Wild type | 74 | 1.0 |
| DKR1 | lpsL212 | 66 | 0.89 |
| DKR62 | lpsL::pDKR62 | 23 | 0.31 |
| DKR63 | rkpK::pDKR63 | <0.23 | <0.003 |
The activity of strain Rm1021 was set at 1.
lps-212 mutant is deficient in nodulation.
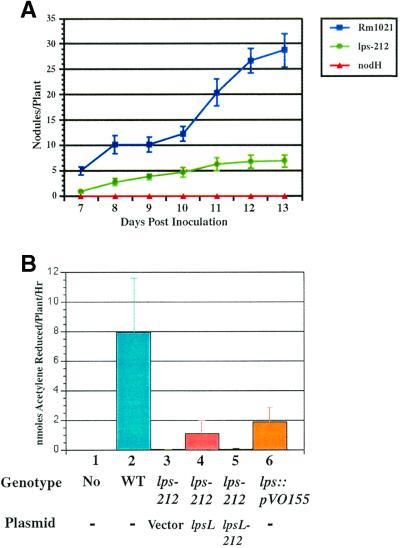
We assayed the ability of strain DKR1 to enter into a nitrogen-fixing symbiosis with alfalfa and found that DKR1 was deficient in several symbiotic phenotypes. First, the number of nodules produced by DKR1 was consistently lower than that formed by the wild type (Fig. 5A). Second, the DKR1-induced nodules had undetectable levels of nitrogenase activity (Fig. 5B). In fact, the rates of nitrogen fixation were the same as those of plants inoculated with an S. meliloti nodH mutant, which is incapable of producing nodules on alfalfa (Fig. 5A). Strain DKR1 harboring a wild-type copy of lpsL on a multicopy plasmid showed a significant but not wild-type level of nitrogen fixation. Strain DKR1 containing a plasmid with the lpsL212 allele showed no detectable nitrogen fixation.
FIG. 5.
Symbiotic assay of Rm1021 (wild type) and DKR1 mutant on alfalfa hosts. (A) Nodulation kinetics of Rm1021 (wild type), DKR1 (lps-212), and JT210 (nodH::Tn5). Plants were grown on agar slants in the absence of added nitrogen and inoculated with bacterial strains, and the number of nodules was quantitated and plotted as a function of time as described in Materials and Methods. Error bars denote standard error in nodule number. (B) Nitrogen fixation assay of alfalfa nodules produced by wild-type and mutant S. meliloti. Plants were inoculated with S. meliloti and assayed for nitrogen fixation 21 days after inoculation as described in Materials and Methods. Lane 1, mock inoculation; lane 2, Rm1021 (wild-type [WT]); lane 3, DKR58 (lps-212 pVector); lane 4, DKR59 (lps-212 plpsL); lane 5, DKR60 (lps-212 plpsL212); lane 6, DKR62 (lpsL::pVO155). Error bars denote standard error.
We believe that the inability of plasmid-borne wild-type lpsL to restore normal levels of nitrogen fixation is due to loss of the plasmid under the nonselective conditions present in planta. The pTB93G plasmid, which is based on the same vector as pMS03, has been reported previously to be unstable in planta (7). Strain DKR62 containing the lpsL::pVO155 mutation also produced nodules with a reduced amount of nitrogen fixation, but this activity was significantly higher than the level seen in plants inoculated with strain DKR1 (Fig. 5B).
Strain DKR1 is altered in nodule invasion.
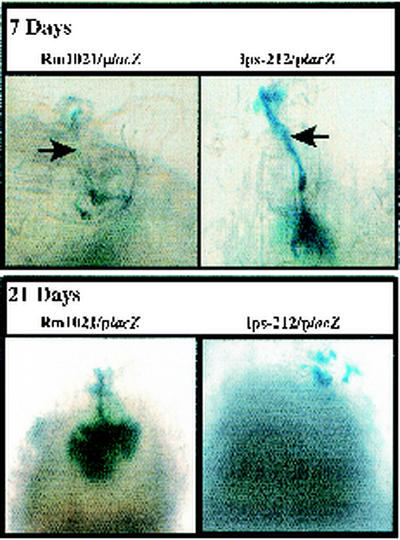
To further characterize the nature of the symbiotic deficiency associated with strain DKR1, we compared infection with the wild-type and DKR1 strains. using a chromogenic reporter to monitor the bacteria in planta. The wild-type and DKR1 strains were transformed with plasmid pXLGD4, which contains lacZ under the control of the hemA promoter. The hemA promoter allows strong constitutive expression of β-galactosidase in bacterial cells inhabiting the infection thread and plant cytoplasm (1, 23). The β-galactosidase activity encoded by lacZ could be visualized by staining the fixed nodule tissue with X-Gal, allowing observation of bacteria within the infection thread and nodule.
Analysis of the infection by wild-type S. meliloti demonstrated typical infection threads that rapidly penetrated the epidermal and cortical layers of the root (Fig. 6). Strain DKR1 showed a very different infection phenotype. Strain DKR1 was able to enter the plant (as judged by the ability of the X-Gal-staining cells to survive treatment with bleach). However, the infection threads differed from those of the wild type in that they had a thicker appearance and were unable to efficiently penetrate the developing nodule (Fig. 6). Instead, the bacteria formed darkly staining clusters, suggesting that the mutant bacteria were able to replicate extensively and were viable but were unable to enter the developing nodule.
FIG. 6.
Visualization of infection threads elicited by wild-type and lps-212 mutant strains. Plasmid pXLGD4 containing a hemA::lacZ transcriptional fusion was mobilized into strain Rm1021 (wild type) and DKR1 (lps-212) by conjugation. The transconjugants were inoculated onto plants, and the progress of infection was examined at either 7 days or 28 days postinoculation by fixing and staining the plants for β-galactosidase activity as described in Materials and Methods. The arrows point to infection threads.
DISCUSSION
A hallmark of the legume nitrogen-fixing symbiosis is the limited number of plant symbiotic partners that are responsive to a given species of bacteria. Chemical modifications of the Nod factors produced by the bacteria are essential for this host specificity (9-11, 15, 17, 38). However, several lines of evidence suggest that additional determinants of host specificity must exist. This was implied by experiments with heterologous rhizobia engineered to produce the S. meliloti Nod factor, which could elicit nodule formation on alfalfa but could not establish a nitrogen-fixing symbiosis (14).
Consistent with the requirement for additional types of host-specific signals, three classes of mutants which are able to elicit the formation of nodules but cannot establish an effective symbiosis have been isolated. Mutants of S. meliloti strain 1021 which lack a specific class of exopolysaccharide (known as succinoglycan), mutants blocked in synthesis of capsular polysaccharides (K-antigens) in S. meliloti strain Rm41, and mutations that disrupt the synthesis of LPS in Rhizobium leguminosarum all elicit the formation of nodules that are unable to fix nitrogen (7, 22, 29). Each of these mutants have in common defects in the synthesis of carbohydrates associated with the cell surface. The findings presented here with the lpsL212 mutant further implicate cell surface carbohydrate species in the establishment of the S. meliloti-alfalfa symbiosis.
The lpsL212 mutation present in strain DKR1 arose spontaneously in strain JSS12, a nodP1Q1 mutant with a reduced ability to produce 3′-phosphoadenosine-5′-phosphosulfate, the form of sulfate used as a substrate in all carbohydrate sulfotransfer reactions. Despite the intriguing connection between nodP1Q1 and lpsL, we have been unable to assign a role for the lpsL212 mutation in this genetic context. The growth rates of JSS12 (nodP1Q1 lpsL212) and JSS12Tr (nodP1Q1 in an otherwise wild-type background) were similar, and no difference in viability was seen under a variety of conditions, including stationary phase and sulfate deprivation (D. H. Keating, M. G. Willits, and S. R. Long, unpublished results).
The mutation in strain DKR1 is an allele of lpsL, previously predicted to encode a UDP-sugar epimerase, based on substantial sequence identity to epimerases which convert UDP-glucose and UDP-galactose (19). Based on the phenotypes of lpsL and rkpK insertion mutations and their genomic proximity, lpsL was predicted to encode an epimerase that converts UDP-glucuronic acid (produced by RkpK) to UDP-galacturonic acid (19). In this report we demonstrated that lpsL is clearly required for UDP-glucuronic acid epimerase activity in vitro. Our studies with Rm1021 demonstrated that both rkpK and lpsL mutants are altered in cell surface, as judged by deoxycholate sensitivity and LPS analysis. rkpK mutants of strain Rm41were also found to have defects in the synthesis of K-antigen; this has not been tested in strain Rm1021, which has a K-antigen of distinct but as yet unknown polysaccharide composition (B. Reuhs, personal communication). Mutations in lpsL did not affect the synthesis of K-antigen in strain Rm41 (19), but this has not been tested in strain Rm1021.
Although the lpsL212 mutation present in strain DKR1 shows phenotypic similarities with the lpsL::pVO155 insertion in strain DKR62, several differences also exist between the two different alleles. The lpsL::pVO155 insertion mutant was better able to fix nitrogen than the lpsL212 mutant. This increased ability to fix nitrogen may in part result from loss of the lpsL::pVO155 insertion in planta, which would restore a wild-type gene and was shown to occur in ca. 1% of the bacteria isolated from the nodules of lpsL::pVO155-infected plants. However, the lpsL::Tn5 insertion in strain Rm41 was also found to be capable of nitrogen fixation (19). The lpsL studies in strain Rm41 lacked a quantitative measure of nitrogen fixation and likely would have missed a subtle phenotype, such as was seen with the lpsL::pVO155 insertion. Thus, although the differences between the Rm41 and Rm1021 alleles of lpsL could simply be the result of differences between the two strain backgrounds, it is also possible that they reflect differences between the insertionally inactivated allele of lpsL and the lpsL212 allele.
The differences seen between the lpsL212 and lpsL::pVO155 alleles in symbiotic assays reflect those seen in biochemical assays of UDP-glucuronic epimerase activity. Extracts prepared from the lpsL212 mutant consistently showed a reduced ability to produce UDP-galacturonic acid in vitro compared to extracts from the lpsL::pVO155 mutant. Although we do not yet understand the basis for the discrepancy in biochemical behavior between the lpsL212 and lpsL::pVO155 alleles in this assay, S. meliloti contains five open reading frames that show substantial sequence identity to genes encoding UDP-sugar epimerases. In the absence of LpsL activity, proteins encoded by these additional open reading frames may contribute to the low level of UDP-galacturonic acid synthesis seen in the lpsL::pVO155 mutant. The lpsL212 allele, on the other hand, may prevent conversion of UDP-glucuronic acid to UDP-galacturonic acid by these alternative epimerases. The lpsL212 allele is a frameshift mutation that results in a polypeptide which is truncated by only 31 amino acids (ca. 9% of the open reading frame). This truncated open reading frame may retain an ability to bind UDP-glucuronic acid or otherwise interfere with UDP-sugar metabolism.
Although we currently do not understand the biochemical mechanism by which lpsL212 differs from lpsL::pVO155, it is clear that that lpsL212 renders strain DKR1 incapable of entering into a nitrogen-fixing symbiosis. There are two potential explanations for the inability of strain DKR1 to establish an effective symbiosis. First, the abnormal infection thread structures seen in the chromatically tagged DKR1 strains could arise from a simple lack of viability within the infection thread microenvironment. While this remains a formal possibility, two lines of evidence are inconsistent with such a model. First, DKR1 can be recovered and efficiently cultured from infected nodules, indicating that viable bacteria are present. However, this assay is not quantitative, and subtle variations in viability would not have been detectable.
Second, although the infection thread structures in DKR1-infected nodules were clearly abnormal, they appeared to be filled with bacteria, as judged by expression of reporter fusions. It should be pointed out that the chromatic visualization technique used in these experiments relies on a protein molecule that is relatively stable within bacterial cells. Therefore, it is possible that the majority of cells within the infection thread are no longer viable and that the β-galactosidase activity seen in nodules arose from accumulation prior to the loss of bacterial viability. Future studies with fluorescent and chromatic tags with shorter half-lives are required to eliminate this possibility.
Despite the lack of quantitative measures for infection thread viability, qualitative assessment clearly showed that bacteria were present within the infection threads elicited by strain DKR1. This argues against a model postulating simple lack of viability of strain DKR1 in planta. In this respect, the lpsL212 mutant is similar to exo mutants, which are unable to produce the acidic exopolysaccharide succinoglycan. Such exo mutants have been reported to elicit the formation of altered infection thread structures containing clusters of bacteria (7, 29), leading to the hypothesis that exopolysaccharides act as chemical signals between the bacteria and plant.
Based on the similarity of the phenotypes of DKR1 and exo mutants, it is possible that the inability of DKR1 to invade nodules stems from a similar lack of a signal required for infection thread growth. Strain DKR1 produces wild-type quantities of succinoglycan (D. H. Keating, M. G. Willits, and S. R. Long, unpublished results), and thus we propose that either the structure of the LPS on the S. meliloti cell surface or possibly its modification by sulfate is a likely candidate for such a signal. Studies of additional S. meliloti mutations affecting the synthesis of LPS and K-antigen as well as the identification of mutants specifically blocked in the sulfate modification of cell surface are required to test this hypothesis.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and Department of Energy grant DE-FG03-90ER20010.
We thank Bradley Reuhs and members of our laboratory for helpful discussions. We also thank Monica Stein for construction of plasmid pMS03.
REFERENCES
- 1.Ardourel, M., N. Demont, F. Debellé, F. Maillet, F. de Billy, J. C. Promé, J. Dénarié, and G. Truchet. 1994. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic responses. Plant Cell 6:1357-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., V. Oke, and S. R. Long. 2000. New genetic tools for use in the Rhizobiaceae and other bacteria. BioTechniques 29:240-242, 244-245. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, C., S. Camut, C. A. Malpica, G. Truchet, and C. Rosenberg. 1990. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewin, N. J. 1991. Development of legume root nodules. Annu. Rev. Cell Biol. 7:191-226. [DOI] [PubMed] [Google Scholar]
- 5.Brewin, N. J., J. A. Downie, and J. Young. 1992. Nodule formation in legumes, p. 239-248. In J. Lederberg (ed.), Encyclopedia of microbiology. Academic Press, San Diego, Calif.
- 6.Cedergren, R. A., J. Lee, K. L. Ross, and R. I. Hollingsworth. 1995. Common links in the structure and cellular localization of Rhizobium chitolipooligosaccharides and general rhizobium membrane phospholipid and glycolipid components. Biochemistry 34:4467-4477. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.de Bruijn, F. J., and J. A. Downie. 1991. Biochemical and molecular studies: symbiotic nitrogen fixation. Curr. Opin. Biotechnol. 2:184-192. [DOI] [PubMed] [Google Scholar]
- 10.Dénarié, J., F. Debellé, and C. Rosenberg. 1993. Signaling and host range variation in nodulation. Annu. Rev. Microbiol. 46:497-531. [DOI] [PubMed] [Google Scholar]
- 11.Downie, J. A. 1994. Signaling strategies for nodulation of legumes by rhizobia. Trends Microbiol. 2:318-324. [DOI] [PubMed] [Google Scholar]
- 12.Ehrhardt, D. W., E. M. Atkinson, and S. R. Long. 1992. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256:998-1000. [DOI] [PubMed] [Google Scholar]
- 13.Ehrhardt, D. W., E. M. Atkinson, K. F. Faull, D. I. Freedberg, D. P. Sutherlin, R. Armstrong, and S. R. Long. 1995. In vitro sulfotransferase activity of NodH, a nodulation protein of Rhizobium meliloti required for host-specific nodulation. J. Bacteriol. 177:6237-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faucher, C., F. Maillet, J. Vasse, C. Rosenberg, A. A. van Brussel, G. Truchet, and J. Dénarié. 1988. The nodH and nodQ host range genes of Rhizobium meliloti behave as avirulence genes in Rhizobium leguminosarum bv. viciae and determine changes in the production of plant specific extracellular signals. Mol. Plant-Microbe Interact. 2:291-300. [Google Scholar]
- 15.Fisher, R. F., and S. R. Long. 1992. Rhizobium-plant signal exchange. Nature 357:655-660. [DOI] [PubMed] [Google Scholar]
- 16.Gage, D., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higashi, S. 1993. (Brady)Rhizobium-plant communications involved in infection and nodulation. J. Plant Res. 106:201-211. [Google Scholar]
- 18.Hirsch, A. M. 1992. Developmental biology of legume nodulation. New Phytol. 122:211-237. [DOI] [PubMed] [Google Scholar]
- 19.Kereszt, A., E. Kiss, B. L. Reuhs, R. W. Carlson, K. A. Kondorosi, and P. Putnoky. 1998. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and invasion of the symbiotic nodule: the rkp gene encodes a UDP-glucose dehydrogenase. J. Bacteriol. 180:5426-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kijne, J. W., R. Bakhuizen, A. A. N. van Brussel, H. C. J. CanterCremers, C. L. Diaz, B. S. de Pater, G. Smit, H. P. Spaink, S. Swart, C. A. Wiffelman, and B. J. J. Lugtenberg. 1992. The Rhizobium trap: root hair curling in root-nodule symbiosis. Soc. Exp. Biol. Semin. Ser. 48:267-284. [Google Scholar]
- 21.Kondorosi, A. 1992. Regulation of nodulation genes in rhizobia, p. 325-340. In D. P. S. Verma (ed.), Molecular signals in plant-Microbe communication. CRC Press, Boca Raton, Fla.
- 22.Lagares, A., G. Caetano-Anollés, K. Niehaus, J. Lorenzen, H. D. Ljunggren, A. Pühler, and G. Favelukes. 1992. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation in alfalfa. J. Bacteriol. 174:5941-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leong, S. A., P. Williams, and G. S. Ditta. 1985. Analysis of the 5′ regulatory region of the gene for γ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 13:5965-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lerouge, P., P. Roche, C. Faucher, F. Maillet, G. Truchet, J.-C. Promé, and J. Dénarié. 1990. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulfated and acylated glucosamine oligosaccharide signal. Nature 344:781-784. [DOI] [PubMed] [Google Scholar]
- 25.Leyh, T. S., J. C. Taylor, and G. D. Markham. 1988. The sulfate activation locus of Escherichia coli K12: cloning, genetic, and enzymatic characterization. J. Biol. Chem. 274:355-360. [PubMed] [Google Scholar]
- 26.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 29.Pellock, B. J., H.-P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kumar Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and Sinorhizobium meliloti produce structurally conserved lipopolysaccharides and strain-specific K-antigens. Appl. Environ. Microbiol. 64:4030-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridge, R. 1992. A model of legume root hair growth and rhizobium infection. Symbiosis. 14:359-373. [Google Scholar]
- 32.Schlaman, H. R. M., R. J. H. Okker, and B. J. J. Lugtenberg. 1992. Regulation of nodulation gene expression by NodD in rhizobia. J. Bacteriol. 174:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultze, M., C. Staehelin, H. Röhrig, M. John, J. Schmidt, E. Kondorosi, J. Schell, and A. Kondorosi. 1995. In vitro sulfotransferase activity of Rhizobium meliloti NodH protein:lipochitooligosaccharide nodulation signals are sulfated after synthesis of the core structure. Proc. Natl. Acad. Sci. USA 92:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwedock, J., and S. R. Long. 1990. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature 348:644-647. [DOI] [PubMed] [Google Scholar]
- 35.Schwedock, J. S., S. Liu, T. S. Leyh, and S. R. Long. 1994. Rhizobium meliloti NodP and NodQ form a multifunctional sulfate-activating complex requiring GTP for activity. J. Bacteriol. 176:7055-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwedock, J. S., and S. R. Long. 1992. Rhizobium meliloti genes involved in sulfate activation: the two copies of nodPQ and a new locus, saa. Genetics 132:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sindhu, S. S., N. J. Brewin, and E. L. Kannenberg. 1990. Immunochemical analysis of lipopolysaccharides from free-living and endosymbiotic forms of Rhizobium leguminosarum. J. Bacteriol. 172:1804-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaink, H. P. 1995. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu. Rev. Phytopathol. 33:345-368. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, J. A., J. K. Tu, J. Ogawa, R. Sanga, R. F. Fisher, and S. R. Long. 1987. Extended region of nodulation genes in Rhizobium meliloti 1021. I. Phenotypes of Tn5 insertion mutants. Genetics 117:181-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner, G. L., and A. H. Gibson. 1980. Measurement of nitrogen fixation by indirect means, p. 111-138. In F. J. Bergersen (ed.), Methods for evaluating biological nitrogen fixation. John Wiley and Sons, Chichester, United Kingdom.
- 41.Vijne, I., L. das Neves, A. van Kammen, H. Franssen, and T. Bisseling. 1993. Nod factors and nodulation in plants. Science 260:1764-1765. [DOI] [PubMed] [Google Scholar]