Abstract
We showed previously that transcription in Escherichia coli promotes C · G-to-T · A transitions due to increased deamination of cytosines to uracils in the nontranscribed but not the transcribed strand (A. Beletskii and A. S. Bhagwat, Proc. Natl. Acad. Sci. USA 93:13919-13924, 1996). To study mutations other than that of C to T, we developed a new genetic assay that selects only base substitution mutations and additionally excludes C · G to T · A transitions. This novel genetic reversion system is based on mutations in a termination codon and involves positive selection for resistance to bleomycin or kanamycin. Using this genetic system, we show here that transcription from a strong promoter increases the level of non-C-to-T as well as C-to-T mutations. We find that high-level transcription increases the level of non-C-to-T mutations in DNA repair-proficient cells in three different sequence contexts in two genes and that the rate of mutation is higher by a factor of 2 to 4 under these conditions. These increases are not caused by a growth advantage for the revertants and are restricted to genes that are induced for transcription. In particular, high levels of transcription do not create a general mutator phenotype in E. coli. Sequence analysis of the revertants revealed that the frequency of several different base substitutions increased upon transcription of the bleomycin resistance gene and that G · C-to-T · A transversions dominated the spectrum in cells transcribing the gene. These results suggest that high levels of transcription promote many different spontaneous base substitutions in E. coli.
Transcription is an inherently asymmetric process that separates transiently the two strands of DNA and copies one strand as RNA. One DNA strand (the transcribed or template strand [TS]) is paired with 8 to 9 nucleotides of RNA in the transcription bubble and is enveloped by the RNA polymerase. The other DNA strand (the nontranscribed or nontemplate strand [NTS]) is unpaired and is thought to lie on the outside of the RNA polymerase (9). This asymmetry creates differential sensitivities of the two DNA strands within the bubble for chemical probes such as hydroxyl radicals and permanganate (8, 11).
Beletskii and Bhagwat have shown that there is also asymmetry in the susceptibility of cytosines in the two strands to deamination (3). Cytosines in the NTS are up to 10 times more likely to deaminate to uracil than those in the TS (1, 3), and in a strain of Escherichia coli defective in uracil excision (genotype ung), this causes a strand-dependent increase in C-to-T mutations. We refer to instances of this phenomenon as transcription-induced mutations (TIM). The extent of this susceptibility of cytosines in the NTS to deamination is roughly proportional to the frequency of transcription of the gene (1). This phenomenon has been seen with plasmid-borne as well as chromosomal genes (2) and with genes transcribed by the T7 RNA polymerase (4). Further, the frequency of cytosine deamination is directly related to the length of time the transcription bubble stays open (4). Recently, Mokkapati and Bhagwat showed that transcription-induced cytosine deaminations occur in the absence of protein synthesis (16), and this eliminates the possibility that TIM may result from indirect effects of high transcription such as interference with the synthesis of DNA repair proteins. Further, it was also found that TIM is present in cells defective in homologous recombination, mismatch repair, or nucleotide excision repair (A. Johnson, J. Klapacz, and A. S. Bhagwat, unpublished results), eliminating the possibility that TIM is caused by strand-biased repair of DNA.
Because cytosine deaminations are thought to be caused by the attack of the base by water or hydroxyl ions (20), we wondered whether other endogenous chemicals could also react preferentially with NTS and cause mutations. To study this, we have constructed a new genetic system that can be used to assay all base substitutions other than C to T. We describe below a study of TIM using this genetic selection system.
MATERIALS AND METHODS
Bacterial strains.
The parent strain was AB1157 [λ− thr-1 leuB6 hisG4 argE3 rpsL31 supE44 lacY1 ara-14 galK2 tsx-33 Δ(gpt-proA) racO rfbD1 kdgK5 xyl-5 mtl-1 thi-1 qsr′ mgl-51]. Strains BH205 (= AB1157 ung::Tn10), BH196 (= BH205 tyrT35), and BH198 (= BH205 tyrU20) were constructed by transducing the ung, tyrT, and tyrU markers from strains BW504, AB2216, and AB2577, respectively, into AB1157 or BH205 by using P1 transduction. tyrT35 and tyrU20 code for ochre suppressors SupM and SupC, respectively, which insert tyrosine in response to a TAA stop codon. ung::Tn10 transductants were selected on Luria broth (LB) plates supplemented with 15 μg of tetracycline/ml, and the ochre-suppressing mutations were selected on minimal plates supplemented with 40 μg each of threonine, leucine, and proline per ml. We had originally hoped that in the strains BH196 and BH198, a TGA-to-TAA mutation within the opal mutations of ble and kan alleles (see below) would be suppressed and scored as ble+ or kan+. However, the mutation spectrum of the revertants of these alleles suggested that this was not the case. This was confirmed by replacing the TGA within ble with ochre (TAA) mutations and testing these plasmids in BH196 and BH198 for ochre suppression. No suppression of TAA was observed (phenotype, Ble−; data not shown). Thus, the opal mutations in ble used in this work cannot revert by a G · C-to-A · T change in any of the strains used.
ble mutants.
A unique-restriction-site elimination procedure (21) was used to introduce the stop codon TGA at codon 39 or 75 within the bleomycin resistance gene in pUP21 (1). The mutagenic primer used to change codon 39 was 5′-GGCTGGATGATCCTCTGACGTGGGGATCTCATGC and the primer for codon 75 was 5′-GGAGTTCTACCGGTGATGCAAATCCGTCG. The underlined sequences are the recognition sites for BtrI and BsaBI, respectively. The selection primer was 5′-GACTTGGTTGAGTACTCACCAGTCAC. All primers were synthesized by GIBCO/BRL (Gaithersburg, Md.) and purified from a DNA sequencing gel prior to use.
The success of the mutagenesis procedure was confirmed with appropriate restriction digestions and by using DNA sequencing. The primer for DNA sequencing was 5′-GCTTCCTCGTGCTTTACGG and was synthesized by Sigma-Genosys (The Woodlands, Tex.). DNA sequencing was done at the DNA Sequencing Facility of Wayne State University. The plasmids resulting from mutagenesis at codons 39 and 75 are referred to as pUP21-op39 and pUP21-op75, respectively.
Reversion assays.
For preliminary reversion tests, pUP21-op39 and pUP21-op75 plasmids were introduced into host strains and three or more independent colonies were picked from LB plates with carbenicillin (50 μg/ml). Cells were grown in LB liquid medium with the same antibiotic, and when the turbidity of the culture reached 0.3, samples from all the cultures were diluted 100-fold to a final volume of 5 ml (for pUP21-op39) or 25 ml (for pUP21-op75) of LB with carbenicillin. Two such cultures were prepared, one with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) and the other without, and both cultures were grown until turbidity reached 0.3. Dilutions of the cultures were spread on LB plates with carbenicillin to determine the total number of viable cells, and the rest of each culture was concentrated and resuspended in 1 ml of LB. Revertants were selected by plating these cells on LB plates containing phleomycin (an analog of bleomycin; 5 μg/ml for 5-ml cultures and 6 μg/ml for 25-ml cultures) (Cayla, Toulouse, France). After incubation overnight at 37°C, colonies were picked for further analysis (see below).
In experiments in which both kanamycin-resistant (Kanr) and phleomycin-resistant (Phlr) revertants were studied, the plasmids were introduced in appropriate hosts (strain BH196 or BH198 for pUP21-op39 and strain BH205 for pUP21-op75) and cultures were grown from a colony until they reached a turbidity of ∼0.3. A volume from each culture was diluted 100-fold to start a pair of 26-ml cultures in LB with carbenicillin. IPTG was present in one culture of each pair, and all of the cultures were grown until turbidity reached 0.3. Dilutions of cultures were spread on LB plates with carbenicillin to determine the number of viable cells, 1 ml of each culture was spread on LB plates with kanamycin (50 μg/ml) (Sigma-Aldrich, St. Louis, Mo.), and the remaining portion of each culture was concentrated by centrifugation and spread on LB plates with phleomycin (6 μg/ml). Revertant frequency was calculated as the ratio of the total number of Phlr or Kanr colonies to the total number of viable cells.
To determine mutation rates, AB1157 cultures containing the appropriate plasmid were prepared as described above and Phlr reversion assays were performed. The number of colonies on LB plates with carbenicillin was used to determine the average number of cells in the cultures (C), and the number of cultures without Phlr revertants was used to determine p0 (number of cultures without revertants/total number of cultures). The mutation rate (μ) was calculated using the equation μ = −ln p0/C (18).
Characterization of ble+ revertants.
To study independent revertants, one colony per independent culture was picked from an LB plate containing phleomycin and the cells were grown overnight in LB medium containing the antibiotic. Plasmid DNAs were prepared from these cultures by using the Perfectprep Plasmid Mini kit (VWR, Brisbane, Calif.). These DNAs were used for restriction analysis using restriction enzyme BtrI or BsaBI.
In early experiments, the plasmid DNAs were retransformed into AB1157 and colonies were selected on LB plates with phleomycin. This procedure separated any nonrevertant plasmid copies within the population from the copies containing the revertants. Once again, single Phlr colonies were picked following retransformation and plasmid DNAs were prepared. This DNA was used for DNA sequence analysis.
Subsequently, we found that it was not necessary to retransform the pool of plasmids obtained from the original Phlr revertant colonies. When the plasmids from cultures prepared from these colonies were directly used for DNA sequencing and sequencing chromatograms were viewed with Chromas software (Technelysium Pty. Ltd., Australia), two overlapping peaks were clearly seen on the chromatograms at the site of reversion. One of these peaks always corresponded to the original base and the other peak was assumed to represent the reversion mutation.
Growth rate determination.
AB1157 cells containing pUP21, pUP21-op75, or one of the revertants were grown in LB liquid medium containing 50 μg of carbenicillin/ml until turbidity reached 0.3. Each culture was then diluted 100-fold into two 5-ml cultures, one of which was additionally supplemented with IPTG to 1 mM. At various times during the growth in the exponential phase (optical density at 550 nm of 0.05 to 0.5), dilutions of the cultures were spread on plates with carbenicillin to determine the total number of viable cells. After overnight incubation at 37°C, colonies were counted and linear regression was performed on a plot of log10 (total number of cells) versus time of growth (h). The slope of the plot was the growth rate for each culture, and the doubling time (in minutes) for each culture was calculated using the following equation: doubling time = (0.301 × 60)/(growth rate).
Construction of kan-op218 and reversion assays.
A kan+ derivative of pUP31 plasmid (1) was used to introduce the TGA stop codon at position 218 within the kan gene. The mutagenic primer (5′-GGTGTGGCGGACCGGTGACAGGACATAGCG) was utilized in a unique-restriction-site elimination procedure (21) for this purpose. (The opal codon within the primer is presented in bold, the recognition sequence for the restriction enzyme AgeI is underlined, and the recognition sequence for the restriction enzyme HphI is in italics.) Success of the mutagenesis was confirmed by using restriction digestions with AgeI and HphI and by DNA sequencing. The resulting plasmid was named pUP31-op218. This plasmid was introduced into AB1157, and the reversion assays were done in a manner similar to that with pUP21-op39 or pUP21-op75, with the following changes. When IPTG was included in the growth medium, it was used at a final concentration of 150 μM (1), and in all cases the revertants were selected on plates containing neomycin (60 μg/ml; Sigma-Aldrich). The reversion frequency was analyzed using the Lea-Coulson method of the median (18).
Rifampin resistance assay.
AB1157 cells carrying pUP21-op75 plasmid were grown in the presence of carbenicillin until the culture reached a turbidity of ∼0.25. It was diluted 10,000-fold to start 6 to 12 new 50-ml cultures. Half of the cultures in each set contained IPTG at 1 mM, while the remaining cultures lacked the inducer. Each culture was grown to a turbidity of ∼0.3 (∼6.5 h), and a 106-fold dilution of each culture was plated on LB with carbenicillin. The remaining cultures were concentrated to 1 ml, and 0.5-ml samples of the concentrated cultures were plated on LB with phleomycin (6 μg/ml) or rifampin (100 μg/ml). The Phlr revertant frequency was defined as described above, and the rate of accumulation of rifampin-resistant (Rifr) mutants in the culture was calculated by the Lea-Coulson method of the median (18).
RESULTS
Rationale behind a genetic system specific for non-C-to-T mutations.
The three termination codons cannot revert to sense codons through C-to-T (same as G to A) changes. The ochre codon (TAA) does not contain a C · G pair, and the amber (TAG) and the opal (TGA) codons change to an ochre codon when they suffer C-to-T mutations. Consequently, when a genetic selection requires that a nonsense codon be replaced with a sense codon, the only way in which this can occur is by non-C-to-T base substitutions or addition and/or deletion mutations. We used this property of nonsense codons to develop a new genetic reversion assay that is specific for all base substitutions except that of C to T.
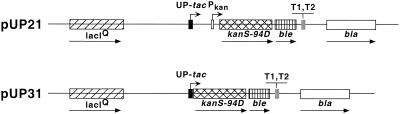
The codons 39 and 75 in the gene for bleomycin resistance (ble) were mutated to TGA in separate constructs, and the sequence surrounding these opal codons was changed to introduce new restriction sites (Table 1). In the plasmid pUP21, the ble gene is downstream from an inactive kanamycin resistance gene (kan) and the two genes are transcribed from two promoters, the natural promoter of kan-ble genes, Pkan, and a synthetic promoter called UP-tac (Fig. 1). While Pkan is a weak constitutive promoter, UP-tac is a very strong promoter that is repressed by the lac repressor (1). Both of the opal mutations inactivate the ble gene, and cells containing these plasmids (pUP21-op39 and pUP21-op-75) are sensitive to as little as 3 μg of phleomycin (an analog of bleomycin) per ml.
TABLE 1.
Sequence contexts of mutations in ble and kan
| Plasmid | Codon | Allele name | Mutation | Unique restriction sitea |
|---|---|---|---|---|
| pUP21-op39 | 39 | ble-op39 | CAG to TGA | BtrI (TGACTGGGG) |
| pUP21-op75 | 75 | ble-op75 | CAG to TGA | BsaBI (TGATGCAAATCC) |
| pUP31-op218 | 218 | kan-op218 | TAT to TGA | HphI (CCGTGACAG) |
Termination codons are in bold and the restriction sites are underlined.
FIG. 1.
Structures of pUP21 and pUP31 plasmids. The plasmids pUP21 and pUP31 are shown in linear form, and relevant genetic elements within the plasmid are shown. Genes: lacIQ, lac super repressor; kanS94D, kanamycin resistance gene (kan) from Tn5 in which codon 94 has been mutated, inactivating the gene; ble, bleomycin resistance gene; bla, β-lactamase gene. UP-tac and Pkan are promoters that transcribe the kan and ble genes, and T1 and T2 are transcription terminators.
Although we expected that the op39 and op75 alleles would, as a result of base substitution mutations, revert to the state in which they confer phleomycin resistance (phenotype Phlr), we were concerned that some of the revertants might have arisen from intragenic second-site mutations, addition and/or deletion mutations, or nonsense suppression. To assess whether this had occurred, we isolated independent spontaneous Phlr revertants from cultures containing pUP21-op39 or pUP21-op75 and characterized the isolates. Plasmids were isolated from these cultures, the DNA was retransformed into E. coli, and the transformants were spread on plates with phleomycin. Following retransformation, all the plasmids conferred resistance to phleomycin, eliminating the possibility that the original Phlr colonies resulted from nonsense suppression.
Next, we digested the plasmid DNAs with BtrI (for pUP21-op39) or BsaBI (for pUP21-op75) and separated the products on agarose gels. Because the restriction sites include only two of the three bases of the opal codons (Table 1), random base substitutions within the opal codons should result in the loss of the overlapping restriction site in two out of three revertants. However, we found that only 2 out of 8 revertants from pUP21-op39 and 6 out of 13 from pUP21-op75 had lost the corresponding restriction site (data not shown). While this showed that a significant number of revertants contained mutations in (or near) the termination codon, it raised the possibility that some of the mutations were at sites other than those of the termination codons.
To clarify this point, several revertants were sequenced and were found to contain the expected sequence changes. None had suffered frameshift mutations or had deleted the nonsense codon. The revertants that had lost the relevant restriction site had acquired substitutions in the second or the third position of the opal codons. Further, the revertants that retained the restriction site had a substitution at the first position (data not shown). These and other results described below show that reversion of ble-op39 or ble-op75 occurs only as a result of base substitutions within one of the three positions of the opal codon.
Both C-to-T and non-C-to-T mutations increase in frequency as a result of transcription.
Wyszynski et al. showed previously that the kanS-94D allele present in pUP21-op39 and pUP21-op75 reverts to kan+ only as a result of a C-to-T change within codon 94 of the gene (reference 23 and unpublished results). Consequently, simultaneous scoring of revertants of the kan and ble alleles within these plasmids may allow one to separately monitor C-to-T and non-C-to-T mutations. Thus, pUP21-op39 and pUP21-op75 plasmids can potentially be used to monitor all base substitution mutations.
To demonstrate this use, these plasmids were introduced into an ung strain (BH205). In this host, uracils generated as a result of cytosine deamination are not repaired and C-to-T mutations promoted by transcription are easier to score (3). Multiple independent E. coli cultures containing pUP21-op39 or pUP21-op75 were grown to mid-log phase, and each culture was diluted and split into two cultures. IPTG was added to one of each pair of cultures, and all of the cultures were grown for 3 more hours before the spreading of appropriate dilutions on LB plates containing carbenicillin (to determine the total cell count), kanamycin, or phleomycin was performed. The frequency of Kanr and Phlr revertants for each culture is presented in Tables 2 and 3.
TABLE 2.
Transcription-induced mutations in pUP21-op39a
| Culture | Frequency of Kanr mutation in culture
|
Frequency of Phlr mutation in culture
|
||||
|---|---|---|---|---|---|---|
| IPTG | No IPTG | Ratiob | IPTG | No IPTG | Ratio | |
| 1 | 6.1 × 10−6 | 1.6 × 10−6 | 3.8 | 7.8 × 10−9 | <5.1 × 10−9c | >1.5 |
| 2 | 6.2 × 10−6 | 2.1 × 10−6 | 2.9 | 1.7 × 10−8 | <6.3 × 10−9c | >2.7 |
| 3 | 9.5 × 10−6 | 2.2 × 10−6 | 4.3 | 7.1 × 10−9 | 9.1 × 10−9 | 0.8 |
| 4 | 4.8 × 10−6 | 1.4 × 10−6 | 3.4 | 1.8 × 10−8 | 1.1 × 10−8 | 1.6 |
| 5 | 5.7 × 10−6 | 2.0 × 10−6 | 2.8 | 1.5 × 10−8 | <6.5 × 10−9c | >2.3 |
| 6 | 5.2 × 10−6 | 1.4 × 10−6 | 3.8 | 3.6 × 10−8 | 6.6 × 10−9 | 5.4 |
The host strain was BH198.
The ratios were calculated by using the following equation: (frequency of revertants in culture with IPTG)/(frequency of revertants in culture without IPTG).
No Phlr colonies.
TABLE 3.
Transcription-induced mutations in pUP21-op75a
| Culture | Frequency of Kanr mutation in culture
|
Frequency of Phlr mutation in culture
|
||||
|---|---|---|---|---|---|---|
| IPTG | No IPTG | Ratiob | IPTG | No IPTG | Ratio | |
| 1 | 1.3 × 10−6 | 1.8 × 10−7 | 7.2 | 9.3 × 10−9 | 1.0 × 10−9 | 9.0 |
| 2 | 7.9 × 10−7 | 2.9 × 10−7 | 2.8 | 1.0 × 10−8 | 4.0 × 10−9 | 2.6 |
| 3 | 9.8 × 10−7 | 2.9 × 10−7 | 3.4 | 1.6 × 10−8 | 9.2 × 10−9 | 1.7 |
| 4 | 7.9 × 10−7 | 2.7 × 10−7 | 2.9 | 8.3 × 10−9 | 2.0 × 10−9 | 4.2 |
| 5 | 1.0 × 10−6 | 2.6 × 10−7 | 3.9 | 9.3 × 10−9 | 2.0 × 10−9 | 4.7 |
| 6 | 8.0 × 10−7 | 2.0 × 10−7 | 4.0 | 5.1 × 10−9 | <2.4 × 10−10c | >21.4 |
The host strain was BH205.
The ratios were calculated by using the following equation: (frequency of revertants in culture with IPTG)/(frequency of revertants in culture without IPTG).
No Phlr colonies.
The frequencies of Kanr revertants were higher by a factor of ∼3 to 7 in cultures grown in the presence of IPTG than in paired cultures lacking the inducer (Tables 2 and 3). The magnitudes of the reversion frequencies were higher with pUP21-op39 than with pUP21-op75, probably because the experiments with these plasmids used different host strains (see Materials and Methods). Regardless, these data confirm the previous results of Beletskii and Bhagwat (1, 3), which showed that C-to-T mutations resulting from cytosine deamination increase in frequency as a result of transcription.
In the same experiments, reversion to Phlr also occurred at a higher frequency in nearly all the cultures containing IPTG than in the uninduced cultures. In one culture only (op39 culture 3), the revertant frequency was slightly lower in the presence of IPTG in the culture. In some cultures, no Phlr revertants were recovered in the uninduced cultures (op39 cultures 1, 2, and 5 and op75 culture 6); hence, only a maximum possible revertant frequency could be estimated in these cultures. Overall, these data show a clear trend of increasing frequencies of Phlr revertants following the induction of the Up-tac promoter, but it is difficult to estimate the exact magnitude of the increase due to the low mutation frequencies and variability within the data.
To determine the effects of transcription on Phlr more accurately, we used a large number of independent paired cultures containing pUP21-op75 and measured the revertant frequencies within these cultures. The experiments were done using two different E. coli strains, and the data were analyzed using the P0 method of Luria and Delbrück (14, 18). The results are summarized in Table 4.
TABLE 4.
Summary of the fluctuation test data
| Plasmid | IPTG | Total no. of cultures | No. of cells per culture | P0 | Mutation rated | Ratioc |
|---|---|---|---|---|---|---|
| pUP21-op39a | + | 30 | 7.2 × 108 | 0.10 | 2.2 × 10−9 | 2.3 |
| − | 30 | 9.6 × 108 | 0.27 | 9.6 × 10−10 | ||
| pUP21-op75b | + | 84 | 2.2 × 109 | 0.21 | 4.7 × 10−10 | 3.5 |
| − | 84 | 2.5 × 109 | 0.61 | 1.3 × 10−10 |
The host strain was AB1157 Sup−.
The host strain was AB1157.
Calculated as (mutation rate in cultures with IPTG)/(mutation rate in cultures without IPTG).
Number of mutations per cell per generation. Mutation rate was calculated as −ln p0/C (see Materials and Methods).
In both sets of data, the presence of IPTG in the growth medium caused a significant increase in the frequency of mutations. Consistent with what was seen in the small-scale experiments (Tables 2 and 3), many more cultures grown without IPTG were devoid of Phlr revertants compared to the induced cultures, and consequently, the mutation rate was two- to fourfold higher in the latter cultures (Table 4). Based on these results, we conclude that in exponentially growing E. coli cells proficient in DNA repair, transcription from a strong promoter induces non-C-to-T in addition to C-to-T mutations.
High transcription does not create a general mutator phenotype.
A possible mechanism by which base substitutions could increase during high transcription is the creation of a general mutator phenotype. For example, this could happen if the transcription from the UP-tac promoter were to inhibit the synthesis of a critical DNA repair enzyme or the proof-reading subunit of the DNA polymerase III. To eliminate this possibility, we simultaneously monitored rates of Phlr and Rifr mutants.
As described above, the rate of accumulation of Phlr revertants was higher in induced than in uninduced cultures (Table 5). It is probable that the rate of Phlr reversion is greater in these cultures than that shown in Table 4 because of differences in the experimental procedures for the two sets of experiments. While the cells in the former experiment were grown for 6.5 h following dilution, they were grown for only 3 h in the experiments involving the Delbrück-Luria fluctuation test (Table 4). In contrast to the findings for Phlr revertants, the rate of accumulation of Rifr mutants was slightly lower in cultures containing IPTG. Therefore, it is unlikely that the high level of transcription of the kan-ble genes in pUP21 plasmids creates a mutator phenotype in the cells.
TABLE 5.
Mutation rates of Phlr and Rifr mutants
| IPTG | No. of cells per culture | Phleomycin resistancea
|
Rifampin resistancea
|
||||
|---|---|---|---|---|---|---|---|
| No. of mutations per culture | Mutation rateb | Ratioc | No. of mutations per culture | Mutation rate | Ratio | ||
| + | 4.3 × 109 | 19.5 | 3.2 × 10−9 | 2.6 | 12.9 | 2.1 × 10−9 | 0.67 |
| − | 3.5 × 109 | 6.0 | 1.2 × 10−9 | 15.4 | 3.1 × 10−9 | ||
Based on 11 independent cultures.
Number of mutations per cell per generation, based on the Lea-Coulson method of the median (18). Mutation rate = (number of mutations per culture)/(1.44 × number of cells per culture).
Calculated as (mutation rate in cultures with IPTG)/(mutation rate in cultures without IPTG).
Spectrum of TIM.
To determine the spectrum of non-C-to-T mutations caused by transcription, pUP21-op75 was introduced into a E. coli strain that is proficient in all known DNA repair pathways (strain AB1157). Plasmid DNA was isolated from independent Phlr colonies obtained from induced and control cultures, and the ble genes within these plasmids were sequenced. These data are summarized in Table 6.
TABLE 6.
Spectrum of mutations in ble-op75 revertants
| Mutation | Independent mutation(s) in induced culture
|
Independent mutations in uninduced culture
|
Amino acida | ||
|---|---|---|---|---|---|
| No. | % of total | No. | % of total | ||
| 5′ T | |||||
| T to A | 22 | 19.6 | 13 | 14.8 | Arg |
| T to C | 24 | 21.4 | 27 | 30.7 | Arg |
| T to G | 14 | 12.5 | 8 | 9.1 | Gly |
| G | |||||
| G to C | 1 | 0.9 | 0 | 0.0 | Ser |
| G to T | 29 | 25.9 | 18 | 20.5 | Leu |
| A 3′ | |||||
| A to C | 7 | 6.3 | 8 | 9.1 | Cys |
| A to G | 12 | 10.7 | 12 | 13.6 | Trp |
| A to T | 3 | 2.7 | 2 | 2.3 | Cys |
| Total | 112 | 100 | 88 | 100 | |
Amino acid at position 75 as a result of reversion.
No addition and/or deletion mutations were detected among the revertants, and in every case, only non-C-to-T base substitutions were found. The detection of all eight non-C-to-T base substitutions among the revertants (Table 6) shows that the genetic selection does not exclude any specific substitutions. While most of the base substitutions were obtained on multiple occasions, G · C to C · G transversion was obtained only once. To confirm that this substitution can indeed create Phlr, the DNA from the revertant was retransformed into E. coli, Phlr revertants were selected, and plasmid DNA from one of the revertants was resequenced to confirm the identity of the mutation. The likely reason that the original amino acid at position 75, Gln, can be replaced with any of six other amino acids is that this residue is not critically involved in the binding of the antibiotic (15).
The overall distribution of mutations was roughly the same in cultures grown with or without IPTG, and G · C to C · G was the rarest of base substitutions in both cases. However, because the overall rate of mutations was higher by a factor of 3.5-fold upon the induction of transcription (Table 4), these data show that the frequencies of many types of base substitution detected by the assay increased as a result of transcription. For example, although T · A-to-C · G transitions decreased from ∼31% (uninduced) to ∼21% (induced) of the total, there was still a ∼2.4-fold (= 3.5 × 21/31) increase in these mutations upon transcription. Consequently, these data suggest that transcription promotes an increase in many types of non-C-to-T base substitutions.
The only significant differences in the sets of mutational data with and without IPTG concerned the relative frequencies of G · C-to-T · A and T · A-to-C · G mutations. While these base substitutions were the two most frequent mutations in both data sets, the highest percentage of mutations in the presence of IPTG was that of G · C to T · A, while without IPTG it was that of T · A to C · G (Table 6). Therefore, it is possible that there are some subtle changes in the spectrum of mutations upon transcription of ble gene.
Growth rates of mutants and revertants.
We considered the possibility that although phleomycin was not included in the growth medium, there is some growth advantage for the ble+ revertants compared to the opal mutants under conditions of high levels of transcription and that this advantage may translate into a higher Phlr reversion rate. The existence of such a growth rate advantage for the wild-type ble+ gene has been suggested in some previous reports (5, 6). To evaluate this possibility, we determined the growth rates of cells containing wild-type ble+ and ble-op75 and three different revertants containing AGA (amino acid, Arg), CGA (Arg), or TTA (Leu) at codon 75 of ble. The revertants chosen for this work were among the most frequent revertants and included 66 and 67% of the revertants in cultures grown without and with IPTG, respectively (Table 6). Cultures containing each of the plasmids were grown under identical conditions, and their doubling times, based on four or more cultures, are reported in Table 7.
TABLE 7.
Doubling time of culturesa
| Codonb (mutation) | Doubling time (min) in:
|
|
|---|---|---|
| No IPTG | IPTG | |
| CAG (Gln) | 19.6 ± 3.1 | 21.4 ± 7.6 |
| TGA (opal) | 19.9 ± 7.5 | 22.2 ± 7.4 |
| CGA (Arg) | 19.6 ± 5.6 | 23.1 ± 6.8 |
| AGA (Arg) | 18.6 ± 6.9 | 21.7 ± 6.6 |
| TTA (Leu) | 21.5 ± 4.6 | 25.3 ± 14.1 |
Results are from four or more independent experiments; values represent means ± standard deviations.
Codon 75 in ble.
All the cultures grew with a doubling time of ∼20 min in the absence of IPTG, and including the inducer in the growth medium increased the doubling time of all the cultures by 10 to 20% (Table 7). Further, no statistically significant growth advantage was apparent for any of the revertants compared to that for the opal mutant. These results strongly suggest that a two- to fourfold increase in the rate of mutations seen in the IPTG-induced cultures is unlikely to be explained by a transcription-dependent growth advantage for the revertants.
Transcription-induced non-C-to-T mutations in kan genes.
We wondered whether high levels of transcription can promote non-C-to-T mutations in genes other than ble. To investigate this issue, we converted codon 218 of kan+ (TAT, Tyr) to TGA in the plasmid pUP31 (Fig. 1) (1, 2) and used this plasmid (pUP31-op218) in reversion assays. Independent cultures were grown in pairs, with one culture in each pair containing IPTG in its growth medium. Following growth, these cultures were plated on carbenicillin or neomycin plates to determine the rates of accumulation of neomycin-resistant (Neor) revertants.
The Neor revertants of the kan-op218 allele arose in cultures containing IPTG at about four times the rate seen with the uninduced cultures (Table 8). This increase in the mutation rate due to transcription is similar to the increase in the mutation rate of Phlr revertants seen with the ble-op39 and ble-op75 alleles (Table 4). Consequently, the transcription-dependent increase in non-C-to-T mutations seen with the ble allele is unlikely to result from features that are unique to that gene or its protein product and is expected to be a general property of transcription in E. coli.
TABLE 8.
Reversion rates of kan-op218 allelea
| IPTG | No. of cells per culture | No. of mutations per culture | Mutation rateb | Ratioc |
|---|---|---|---|---|
| + | 3.8 × 108 | 4.0 | 7.3 × 10−9 | 3.8 |
| − | 6.1 × 108 | 1.7 | 1.9 × 10−9 |
Based on 19 independent cultures.
No. of Neor mutations per cell per generation based on the Lea-Coulson method of the median (18). Mutation rate = (number of mutations per culture)/(1.44 × number of cells per culture).
Calculated as (mutation rate in cultures with IPTG)/(mutation rate in cultures without IPTG).
DISCUSSION
We have designed a novel genetic reversion system by using the antibiotic resistance genes kan and ble that provides a positive selection for base substitutions and scores all such mutations except C to T (G to A). We placed one kan and two ble alleles under the control of a strong regulated promoter and used them to show that induction of transcription promotes non-C-to-T mutations. Transcription-dependent increases in mutations were seen with all the alleles, suggesting that this phenomenon is not strongly dependent on the sequence context of the mutation. Qualitatively, the spectra of mutations obtained with or without induction of transcription in the op75 allele were roughly similar, but the overall rate of mutations increased by a factor of 2 to 4. We also showed that the observed increase in the revertants is not caused by a growth advantage for the revertants over the ble mutant during growth under conditions of high transcription. Additionally, we found that cells in which the UP-tac promoter was induced did not display a general mutator phenotype. This suggests that the increases in the mutation rate were restricted to the ble or kan gene and were directly caused by the high transcription level. These increases in the level of mutations were seen in DNA repair-proficient cells, suggesting that even larger increases may be seen in cells defective in DNA repair pathways or upon treatment with certain mutagens. When coupled with the previous results of Beletskii and Bhagwat, which show that the frequency of C-to-T mutations increases upon transcription (3), these results suggest that several different classes of base substitution increase as a result of transcription.
A qualitatively similar spectrum of mutations was seen among the much smaller number of independent revertants sequenced using the op39 allele of ble (data not shown). Among the revertants obtained from IPTG-induced cultures, G · C-to-T · A transversions dominated (5 out of 15) while levels of T · A-to-C · G transitions were also high (4 out of 15). However, levels of G · C-to-C · G transversions were also high (3 out of 15), suggesting that there may be some differences in the spectra of mutations induced by transcription at the two sites.
We found no growth advantage for the wild-type ble+ or the revertants compared to that for the op-75 mutant (Table 7), and this appears to contradict the reported growth advantage of the ble+ gene over ble in chemostat experiments (5). We would like to suggest that this advantage is realized only at high cell densities or after very long growth periods. In the experiments described by Blot et al. (5, 6) cells were maintained at a turbidity of 1.0 for 10 to 200 h. Blot et al. attribute the growth advantage of cells with ble+ to a decreased rate of cell death (5). However, no statistically significant differences were apparent after 4 h of growth for three out four strains used, and clear differences emerged only at 18 h of growth (5). In our experiments, cells were not grown beyond a turbidity of 0.3 and the time period of induction was typically 3 h. Consequently, the type of growth advantage for the ble+ revertants over ble observed by Blot et al. is not relevant to the experiments reported here.
Traditionally, mutation spectra have been studied using forward mutation assays involving acquisition of resistance to rifampin or mutations in lacI. The Phlr reversion assay described here is as convenient as any assay involving antibiotic resistance but has one significant advantage over the other assays for studying spontaneous base substitutions. In most forward mutation assays, insertion and/or deletion mutations or C-to-T substitutions dominate the spectrum. For example, in one study of spontaneous Rifr mutants (17) G · C-to-A · T transitions comprised 72% of the obtained mutations. In a study of lacId mutations (19), 29% of the mutations were insertions or deletions, and of the base-substitution mutations 47% were G · C-to-A · T changes. Such dominance of frame-shifting mutations and C-to-T mutations in the spectra means that other base substitutions such as G · C to T · A are rarely seen. In the Rifr and lacId mutational studies mentioned above, G-to-T transversions were seen in only 6.5 and 5.6%, respectively, of all the mutations (17, 19). In contrast, in our study G-to-T mutations constituted ∼21% of the mutations obtained in the absence of IPTG in the growth medium (Table 6). Thus, the genetic system described in this paper has certain advantages for the study of less-frequent base substitutions in comparison to assays based on forward mutations.
The pattern of mutations summarized in Table 6 shows a clear preference for mutations at the first nucleotide in the opal codon, T. This is true regardless of whether the ble gene is heavily transcribed (54% of total) or not (55%). However, these mutations could result from damage either to the thymine in the nontranscribed strand or to the adenine in the transcribed strand. Remarkably, the frequencies of mutations at the third position within the opal codon (A) are not similar to those of the complementary mutations in the first position (T). For example, A-to-G changes in the third position (14% without IPTG and 11% with) are significantly lower than the equivalent T-to-C changes at the first position (31 and 21%). Whether this reflects a sequence context or strand bias in DNA damage and/or repair is yet unclear.
The most frequent base change observed in the presence of IPTG in the growth media is G to T, which is the signature mutation of oxidative damage to a guanine (7) as well as of bypass of the abasic site generated as a result of hydrolysis of the glycosidic linkage within a deoxyguanosine (10). Therefore, the 4.4-fold increase [=3.5 × (25.9/20.5)] in G-to-T mutations as a result of transcription could be due to an increase in the chemical susceptibility either of the guanosines in the nontranscribed strand or of their glycosidic linkage. It is known that the rate of depurination is four times higher in single-stranded than in double-stranded DNA (12, 13). It is also known that free DNA bases react with hydroxyl radicals 8 to 15 times more readily than those in double-stranded DNA (22). It would be useful to know whether these differences in the chemical susceptibilities of single- and double-stranded DNA are indeed the underlying causes of the observed increases in non-C-to-T mutations upon transcription.
Acknowledgments
The work presented here was supported by National Institutes of Health grant GM 57200.
The idea for a termination codon-based reversion assay originated in a discussion of A.S.B. with R. Sinden (Texas A&M University System Health Sciences Center, Houston, Texas).
REFERENCES
- 1.Beletskii, A., and A. S. Bhagwat. 1998. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem. 379:549-551. [PubMed] [Google Scholar]
- 2.Beletskii, A., and A. S. Bhagwat. 2001. Transcription-induced cytosine-to-thymine mutations are not dependent on sequence context of the target cytosine. J. Bacteriol. 183:6491-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beletskii, A., and A. S. Bhagwat. 1996. Transcription-induced mutations: increase in C to T mutations in the non-transcribed strand during transcription in Escherichia coli. Proc. Natl. Acad. Sci. USA 93:13919-13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beletskii, A., A. Grigoriev, S. Joyce, and A. S. Bhagwat. 2000. Mutations induced by bacteriophage T7 RNA polymerase and their effects on the composition of the T7 genome. J. Mol. Biol. 300:1057-1065. [DOI] [PubMed] [Google Scholar]
- 5.Blot, M., B. Hauer, and G. Monnet. 1994. The Tn5 bleomycin resistance gene confers improved survival and growth advantage on Escherichia coli. Mol. Gen. Genet. 242:595-601. [DOI] [PubMed] [Google Scholar]
- 6.Blot, M., J. Meyer, and W. Arber. 1991. Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage to Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 88:9112-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, K. C., D. S. Cahill, H. Kasai, S. Nishimura, and L. A. Loeb. 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J. Biol. Chem. 267:166-172. [PubMed] [Google Scholar]
- 8.Kainz, M., and J. Roberts. 1992. Structure of transcription elongation complexes in vivo. Science 255:838-841. [DOI] [PubMed] [Google Scholar]
- 9.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel, T. A. 1984. Mutational specificity of depurination. Proc. Natl. Acad. Sci. USA 81:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, D. N., and R. Landick. 1992. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J. Mol. Biol. 228:759-777. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl, T., and B. Nyberg. 1972. Rate of depurination of native deoxyribonucleic acid. Biochemistry 11:3610-3618. [DOI] [PubMed] [Google Scholar]
- 14.Luria, S. E., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama, M., T. Kumagai, Y. Matoba, M. Hayashida, T. Fujii, Y. Hata, and M. Sugiyama. 2001. Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin. J. Biol. Chem. 276:9992-9999. [DOI] [PubMed] [Google Scholar]
- 16.Mokkapati, S. K., and A. S. Bhagwat. Lack of dependence of transcription-induced cytosine deaminations on protein synthesis. Mutat. Res., in press. [DOI] [PubMed]
- 17.Otterlei, M., B. Kavli, R. Standal, C. Skjelbred, S. Bharati, and H. E. Krokan. 2000. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 19:5542-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaaper, R. M., and R. L. Dunn. 1991. Spontaneous mutation in the Escherichia coli lacI gene. Genetics 129:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro, R. 1981. Damage to DNA caused by hydrolysis, p. 3-18. In E. Seeberg and K. Kleppe (ed.), Chromosome damage and repair. Plenum Press, New York, N.Y.
- 21.Trower, M. K. (ed.). 1996. In vitro mutagenesis protocols, vol. 57. Humana Press Inc., Totowa, N.J.
- 22.von Sonntag, C. 1987. The chemical basis of radiation biology. Taylor & Francis, London, United Kingdom.
- 23.Wyszynski, M., S. Gabbara, and A. S. Bhagwat. 1994. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot-spots at sites of cytosine methylation in E. coli. Proc. Natl. Acad. Sci. USA 91:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]