Abstract
NAD is an indispensable redox cofactor in all organisms. Most of the genes required for NAD biosynthesis in various species are known. Ribosylnicotinamide kinase (RNK) was among the few unknown (missing) genes involved with NAD salvage and recycling pathways. Using a comparative genome analysis involving reconstruction of NAD metabolism from genomic data, we predicted and experimentally verified that bacterial RNK is encoded within the 3′ region of the nadR gene. Based on these results and previous data, the full-size multifunctional NadR protein (as in Escherichia coli) is composed of (i) an N-terminal DNA-binding domain involved in the transcriptional regulation of NAD biosynthesis, (ii) a central nicotinamide mononucleotide adenylyltransferase (NMNAT) domain, and (iii) a C-terminal RNK domain. The RNK and NMNAT enzymatic activities of recombinant NadR proteins from Salmonella enterica serovar Typhimurium and Haemophilus influenzae were quantitatively characterized. We propose a model for the complete salvage pathway from exogenous N-ribosylnicotinamide to NAD which involves the concerted action of the PnuC transporter and NRK, followed by the NMNAT activity of the NadR protein. Both the pnuC and nadR genes were proven to be essential for the growth and survival of H. influenzae, thus implicating them as potential narrow-spectrum drug targets.
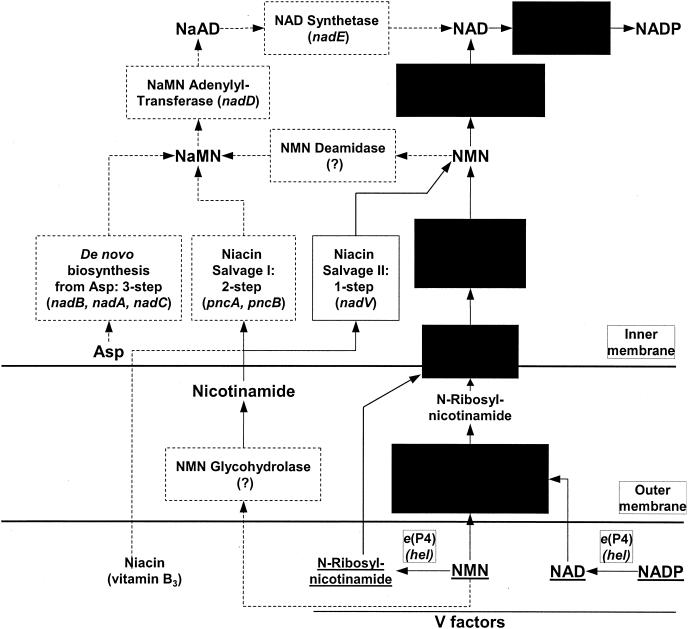
NAD and its phosphorylated analog (NADP) are indispensable cofactors for numerous oxidoreductases in all living organisms (34). Systematic studies of the bacterial NAD biosynthetic machinery in Escherichia coli and Salmonella enterica serovar Typhimurium (41) revealed a complex network of de novo, salvage, and recycling pathways. Not all of these pathways are present in other bacteria; e.g., Haemophilus influenzae is incapable of synthesizing NAD de novo or of salvaging niacin (nicotinamide or nicotinic acid; vitamin B3) from the growth medium (Fig. 1).
FIG. 1.
Biochemical transformations and pathways in the biosynthesis of NAD(P) in Enterobacteriaceae and Pasteurellaceae. Pathways and individual enzymatic steps that occur only in E. coli and not in H. influenzae are outlined by dashed boxes and arrows. Common components between E. coli and H. influenzae are shown on a black background. Gene names are given as for E. coli except with nadN (characterized for H. influenzae), hel (characterized for H. influenzae, not present in E. coli), and nadV (characterized for H. ducreyi, not present in either E. coli or H. influenzae). Missing genes are labeled with “(?).”
The genes for all major NAD or NADP biosynthetic steps in bacteria have been experimentally identified (for the latest review, see reference 6). Two previously missing bacterial genes, β-nicotinic acid mononucleotide (NaMN) adenylyltransferase (NaMNAT; EC 2.7.7.18) and NAD kinase (EC 2.7.1.23), were recently identified in E. coli (24, 33). Genes encoding some of the enzymes of the NAD salvage and recycling pathways, such as β-nicotinamide mononucleotide (NMN) glycohydrolase (EC 3.2.2.14), NMN deamidase (EC 3.5.1.42), and ribosylnicotinamide kinase (RNK; EC 2.7.1.22), remained unknown in spite of convincing genetic and biochemical data indicating the presence of corresponding enzymes in some bacteria (19, 28). RNK enzymatic activity was detected in a number of prokaryotes and eukaryotes, including humans, where it has been implicated in the activation of some clinically important nucleoside analogs (46). Previous biochemical data suggest that RNK must be of central importance for H. influenzae (13, 23), providing an obligate step of NAD biosynthesis. We used comparative genome analysis to search for missing RNK gene candidates, focusing primarily on H. influenzae and related microorganisms.
Genes encoding enzymes hydrolyzing exogenous V factors (NADP, NAD, and NMN) to N-ribosylnicotinamide, the ultimate NAD precursor required for the growth of H. influenzae, were recently identified (25, 45, 48). However, genes involved with the transport of N-ribosylnicotinamide and its conversion to NAD remained unknown. Projection of NAD biosynthetic pathways onto genomic data allow us to propose that in H. influenzae these steps are performed by the homologs of the PnuC transporter and NadR proteins previously described for E. coli and S. enterica serovar Typhimurium. The multifunctional NadR protein was previously implicated in the transcriptional regulation of NAD biosynthesis (20, 55) and PnuC transporter-mediated salvage of exogenous NMN (21, 57). Based on sequence similarity with archaeal NMN adenylyltransferase (NMNAT) (43), the NadR protein was also predicted to have NMNAT (EC 2.7.7.18) activity (28, 35), which was later confirmed for NadR from E. coli (ecNadR) (44).
A comparative analysis of the domain organizations of various NadR homologs led us to predict and experimentally verify that RNK activity resides within the C-terminal domain of the NadR protein. We propose a dual functional role for this domain in assisting PnuC-mediated transport of N-ribosylnicotinamide and providing the ultimate precursor (NMN) for NAD biosynthesis. Here we present a comparative characterization of RNK and NMNAT activities of NadR homologs from H. influenzae (hiNadR) and S. enterica serovar Typhimurium (stNadR) and the results of gene inactivation experiments supporting our model for a PnuC-NadR salvage pathway. The experimentally determined essentiality of the pnuC and nadR genes in H. influenzae suggests that the corresponding proteins may be considered narrow-spectrum targets for anti-infective agents.
MATERIALS AND METHODS
Genome comparative analysis, metabolic reconstruction, and identification of missing genes.
Comparative analysis of microbial genomes was performed using the ERGO database (available by subscription from Integrated Genomics, Inc., Chicago, Ill.) as previously described (14, 22). Additional public resources for gene and protein analysis used in this study were GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html), PSI-BLAST (http://www.ncbi.nlm.nih.gov/BLAST), and PFAM (http://pfam.wustl.edu). Genomic sequences used in this study were E. coli K-12 MG1655 (GenBank accession no. U00096; http://gib.genes.nig.ac.jp/) (7), H. influenzae Rd (GenBank accession no. L42023; http://www.tigr.org) (17), S. enterica serovar Typhimurium LT2 (accession no. AE006468), Pseudomonas aeruginosa PAO1 (GenBank accession no. AE004091; http://www.pseudomonas.com/) (52), Mycobacterium tuberculosis H37Rv (GenBank accession no. AL123456; http://www.sanger.ac.uk/) (12), Nostoc punctiforme ATCC 29133 (GenBank accession no. NC_002791; http://www.jgi.doe.gov/JGI_microbial/html/nostoc/), Lactococcus lactis IL-1403 (GenBank accession no. AE005176; http://spock.jouy.inra.fr/) (8), Haemophilus ducreyi (GenBank accession no. NC_002940; http://www.microbial-pathogenesis.org/H.ducreyi/), Actinobacillus actinomycetemcomitans HK1651 (GenBank accesssion no. NC_002924; http://www.genome.ou.edu/act.html), and Pasteurella multocida Pm70 (GenBank accession no. AE004439; http://www.cbc.umn.edu/ResearchProjects/AGAC/Pm/pmhome.html) (31).
NAD and NADP metabolism was reconstructed from genomic data (Table 1) by the approach described in reference 49. We tentatively projected all the relevant pathways over the set of genomes listed above, based on the presence or absence of orthologous genes implicated in these pathways. Missing genes are revealed with respect to functional roles in asserted pathways for which a corresponding gene is either unknown for any species or known for some species but cannot be projected to other species by sequence similarity. A combination of computational techniques, including analysis of chromosomal clustering described in references 14 and 39 and protein fusion events (16, 29), was used to infer functional coupling and to identify candidate open reading frames (ORFs) for these missing genes. Candidate ORFs were further prioritized with additional criteria such as occurrence (or phylogenetic) profiles (40) and conserved sequence motifs (using PFAM and similar resources).
TABLE 1.
Functional reconstruction of NAD(P) biosynthetic pathways from genomic dataa
| NAD(P) biosynthesis | EC no. | Genomic data for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E. colib | N. puncti- forme | L. lactis | P. aeru- ginosa | M. tuber- culosis | H. influ- enzae | H. ducreyi | A. actinomyce- temcomitans | P. multo- cida | ||
| De novo pathway: aspartate to NaMN | Yes | Yes | No | Yes | Yes | No | No | No | No | |
| Aspartate oxidase | 1.4.3.16 | nadB | + | − | + | + | − | − | − | − |
| Quinolinate synthase | nadA | + | − | + | + | − | − | − | − | |
| Quinolinate phosphoribosyltransferase | 2.4.2.19 | nadC | + | − | + | + | − | − | − | − |
| Salvage I: niacin to NaMN | Yes | Yes | Yes | Yes | ?c | No | No | No | No | |
| Nicotinamide deamidase | 3.5.1.19 | pncAd | + | ? | + | + | − | − | − | − |
| Nicotinate phosphoribosyltransferase | 2.4.2.11 | pncB | + | + | + | + | − | − | − | − |
| Salvage II: niacin to NMN | No | No | No | No | No | No | Yes | Yes | Yes | |
| Nicotinamide phosphoribosyltransferase | 2.4.2.12 | − | − | − | − | − | − | nadV | + | + |
| Salvage III: NMN/ N-ribosylnicotinamide to NAD | Yes | Yes | Yes | No | No | Yes | Yes | Yes | Yes | |
| NMN phosphohydrolase (periplasmic) | 3.1.3.5 | ushAe | − | − | − | − | nadN | + | + | + |
| Pyridine nucleoside transporter | pnuC | + | + | + | − | + | + | + | + | |
| Tripartite NadR protein: | ||||||||||
| Ribosylnicotinamide kinase; domain 3 | 2.7.1.22 | nadR | + | + | + | + | + | + | + | + |
| NMN adenylyltransferase; domain 2 | 2.7.7.1 | nadR | + | + | − | + | + | + | + | + |
| Transcription regulator; domain 1 | nadR | − | − | − | − | − | + | + | + | |
| Universal pathways: NaMN to NAD | Yes | Yes | Yes | Yes | Yes | No | No | No | No | |
| NaMN adenylyltransferase | 2.7.7.18 | nadDd | + | + | + | + | − | − | − | − |
| NAD synthetase | 6.3.5.1 | nadE | + | + | + | + | − | − | − | − |
| Universal pathways: NAD to NADP | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| NAD kinase | 2.7.1.23 | nadFd | + | + | + | + | + | + | + | + |
Representative genomes containing NadR homologs are shown. The presence or absence of an orthologous gene in a specific genome is marked by a + or −, respectively. The deduced presence or absence of corresponding pathways is shown by “Yes” or “No”, respectively. A ? means “unknown.”
E. coli and S. enterica serovar Typhimurium have identical genes and pathway patterns.
In spite of the presence of all required orthologs, M. tuberculosis is believed to be unable to salvage exogenous NAD precursors (19).
ydjB, ybeN, and yfgB are the previous gene names for pncA, nadD, and nadF, respectively, in E. coli.
Bacterial strains and DNA.
E. coli strains DH5α (Life Technologies, Rockville, Md.), BL21, and BL21 (DE3) (Stratagene, La Jolla, Calif.) were used for cloning and protein overexpression. H. influenzae Rd (ATCC 51907) was used for genetic studies. For PCR amplification of the target genes, we used total genomic DNA purchased from the American Type Culture Collection (Manassas, Va.): H. influenzae Rd (KW20) (ATCC 51907D) and Salmonella enterica subsp. enterica (Smith) Weldin serotype Typhimurium (S. enterica serovar Typhimurium) LT2 (ATCC 700720D). Genomic DNA of P. aeruginosa PAO1 was a kind gift from O. Zaborina (University of Illinois, Chicago). In cloning for sequencing we used pGEM 3Z(+) (Promega, Madison, Wis.). A pET-derived plasmid vector containing a T7 promoter, N-terminal His6 tag, and TEV protease cleavage site described in reference 37 or a similar vector, pProEX HTa (Life Technologies) with a Trc promoter, was used for protein expression. Enzymes for DNA manipulations were from New England Biolabs (Beverly, Mass.) and Fermentas (Vilnius, Lithuania). Plasmid purification kits and Ni-nitrilotriacetic acid (NTA) resin were purchased from QIAGEN (Valencia, Calif.). Reagents for DNA sequencing were from PE Biosystems (Foster City, Calif.). Oligonucleotides for PCR and sequencing were ordered from Sigma-Genosys (Woodlands, Tex.) and MWG Biotech Inc. (High Point, N.C.).
Reagents.
Buffer components, EDTA, dithiothreitol, and all reagents for enzymatic assays were from Sigma (St. Louis, Mo.). Tiazofurin was a kind gift from V. Marquez (National Cancer Institute, Frederick, Md.). Recombinant NAD synthetase from Corynebacterium glutamicum was a kind gift from K. Shatalin (Integrated Genomics, Inc.).
PCR amplification, cloning, and sequencing.
Pairs of 5′ and 3′ primers used for PCR amplification of target genes and gene fragments are listed below. Mutations and added nucleotides are shown in lowercase letters, and engineered restriction sites are shown in bold.
(i) H. influenzae primers.
The primers for the full-size coding region of hiNadRwere gggccATGgCAAAAACAAAAGAGAAAAAAGTCGGTGTCATTTTC (NcoI) and ggggtcgacTCATTGAGATGTCCCTTTTATAGGAAAGGTTGTG (SalI). The primers for the coding fragment for the NMNAT domain of hiNadRwere gggccATGgCAAAAACAAAAGAGAAAAAAGTCGGTGTCATTTTC(NcoI) and ggggtcgactcaAAAGAAAGGACGAGCTTCTTTCGGAATAAACTTC (SalI). Those for the pnuC locus were ggggaaTTCTACACCTTGACATTACCAATTCACAATGC (EcoRI) and ggggaattcATCATTTAACGGGGATTTTTTATAAGCTATTTACTG (EcoRI).
(ii) S. enterica serovar Typhimurium primers.
The primers for the full-size coding region of stNadR were gggtcaTGaCATCGTTCGACTATCTCAAAACCGCG (BspHI) and ggggtcgacTTAactcgagCCCTGCTCGCCCATCATCTCTTTC (SalI, XhoI). For site-directed mutagenesis of the ATP-binding site in the stNadR protein (mutations H77A and H80A), the primers were CCATTGgcTACCGGAgcCATCTACTTGATCC (sense) and GGATCAAGTAGATGgcTCCGGTAgcCAATGG (antisense).
(iii) P. aeruginosa primers.
For PnuC-NadR of P. aeruginosa (paPnuC-NadR), the primers were gggGAATTCTTGCCAGCCGGCGTCTG (EcoRI) and ggggaattcGCGCCGAGACAGCTTTCCGC (EcoRI).
PCR amplification used genomic DNA as the template and Pfu DNA polymerase (Stratagene). PCR fragments were purified, digested with restriction enzymes, and cloned into appropriately digested vectors. All clones were verified by DNA sequencing by standard procedures. Additional internal primers were synthesized when needed to complete DNA sequencing. No differences from the originally reported genomic sequences were found, except for the correction of a presumed sequencing error in the H. influenzae pnuC locus. Two insertions were found: a single G at 70 bases upstream of the start codon and a single C at position 394 of ORF HI1077.1 (17). The revised H. influenzae pnuC locus can be translated into a 226-residue contiguous ORF with high end-to-end sequence similarity with E. coli PnuC protein (gi|1651338).
Site-directed mutagenesis.
Amino acid substitutions (H77A and H80A) in the ATP-binding site (HTGH) of stNadR were produced using the sequential PCR steps technique (3). Briefly, mutant primers (sense and antisense) containing the separate base substitutions were synthesized. In the first step, two separate PCRs were performed with two sets of primers, 5′-end-flanking and antisense mutant primers and 3′-end-flanking and sense mutant primers, and 10 ng of the expression plasmid encoding full-sized stNadR. Both amplified mutated fragments were purified, mixed in a 1:1 molar ratio, and used as the template for a second step of amplification with flanking primers under standard conditions to produce the full-length mutated fragment.
Protein overexpression and purification.
All proteins were expressed in BL21 or BL21/DE3 strains of E. coli as N-terminal His6 tag fusion proteins with a TEV protease cleavage site. The cultures were grown in Luria-Bertani medium with 100 μg of ampicillin per ml at 37°C in shaking flasks (200 rpm) to an optical density at 600 nm of ∼0.8. The temperature was adjusted to 20°C, and expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.8 mM. Cells were harvested after ∼12 h of shaking at 20°C.
A rapid partial purification of recombinant proteins used Ni-NTA agarose minicolumns (38). Briefly, cells harvested from a 50-ml culture were resuspended and lysed in 10 mM HEPES-NaOH (pH 7.2) containing 100 mM NaCl, 2 mM β-mercaptoethanol, 0.03% Brij 35, 2 mM phenylmethylsulfonyl fluoride, and 1 mg of lysozyme per ml. After lysis by freezing-thawing and sonication, the lysate was centrifuged and pellets and supernatants were processed separately. Supernatants (adjusted to a pH of 8 with 50 mM Tris-HCl) were loaded onto 200-μl Ni-NTA agarose minicolumns that had been equilibrated with the same buffer. After being washed with starting buffer containing 1 M NaCl and 0.3% Brij 35, bound proteins were eluted with 300 μl of starting buffer containing 250 mM imidazole. Original pellets were resuspended in the same buffer containing 7 M urea before they were processed by the same minicolumn procedure with 7 M urea included in all buffers. The yield and distribution between soluble and insoluble fractions were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. For further enzymatic analysis, proteins were purified from 6-liter cultures by chromatography on a Ni-NTA agarose column followed by gel filtration on a HiLoad Superdex 200 16/60 column (Pharmacia) with an AKTA fast protein liquid chromatography system, as described previously (14, 37).
Substrates for RNK activity assay.
N-Ribosylnicotinamide was produced by alkaline phosphatase (Fermentas) treatment of 1 mM solutions of NMN (Sigma) at 37°C for ∼12 h in 200 mM Tris-HCl (pH 8.0) and 10 mM MgCl2 (described in reference 23). The reaction was monitored by high-performance liquid chromatography (HPLC) (as described below). N-Ribosylnicotinate was synthesized from NaMN by the same procedure.
Enzymatic characterization of NadR proteins. (i) Direct (HPLC-based) assay.
An HPLC-based protocol was used to directly verify the NMNAT and RNK activities of NadR proteins. Assays were carried out with 100 mM HEPES-KOH (pH 7.5) with 10 mM MgCl2. The enzymes (0.04 mg of hiNadR per ml and 0.02 or 0.2 mg of stNadR per ml) were incubated with 2 mM concentrations of the substrates and 2 mM ATP at 37°C overnight. After the reaction, protein was removed by microultrafiltration with Microcon YM-10 centrifugal filters (Amicon, Bedford, Mass.) and the filtrates were analyzed on 50- by 4.6-mm C18 columns (Supelco, Bellerofonte, Pa.) after appropriate dilution. Ion pair separation was carried out isocratically in 50 mM sodium phosphate (pH 5.5) containing 8 mM tetrabutylammonium bromide and 8% methanol at 1 ml/min. Samples were monitored at 254 nm with a Gilson (Middleton, Wis.) HPLC system. The same procedure was used to monitor the synthesis of N-ribosylnicotinamide and N-ribosylnicotinate.
(ii) NMNAT- and NaMNAT-coupled assay.
The continuous NMNAT assay coupling NAD formation to the alcohol dehydrogenase-catalyzed conversion of NAD to NADH was adapted from reference 4. The assay was performed with UV-transparent plastic cuvettes in the six-cuvette autosampler of a model DU-640 spectrophotometer (Beckman, Fullerton, Calif.) at 37°C. The 500-μl reaction mixture contained 100 mM HEPES-KOH (pH 7.5), 115 mM ethanol, 40 mM semicarbazide, 2 mM ATP, 3 U of alcohol dehydrogenase (Sigma), and 0.1 to 10 μg of NadR (depending on the source and specific activity). The reaction was started by adding 10 μl of 50 mM NMN and monitored at a wavelength of 340 nm over a 20-min period. The NADH extinction coefficient of 6.22 mM−1 cm−1 was used for rate calculations. One unit of enzyme was defined as the amount capable of producing 1 μmol of NADH per min. To measure NaMNAT activity, the procedure was modified by introducing an additional enzymatic step: conversion of β-nicotinic acid adenine dinucleotide (NaAD) to NAD by NAD synthetase (EC 6.3.5.1). NAD synthetase (0.3 U) from C. glutamicum and 10 mM NH4Cl were added to the same reaction mixture, and the reaction was initiated by adding 10 μl of 50 mM NaMN. The pH optimum for hiNadR and stNadR NMNAT activity was determined to be in the range 7.0 to 9.0 by the same assay.
NMNAT steady-state kinetic parameters were determined by the same protocol in a reduced volume (250 μl) in a 96-well plate format with an automated pipetting station (Quadra-96; Tomtec, Hamden, Conn.) in combination with a microplate reader from Spectrafluor Plus (Tecan-US, Durham, N.C.) with a 340-nm-pore-size filter at 37°C. NMN and ATP concentrations were varied between 20 and 20,000 μM and 200 and 2,000 μM, respectively. Data was acquired, and initial rate calculations were performed with Magellan software (version 2.22; Tecan-US). Further analysis was performed with the software Sigma-Plot 2000 (SPSS Science, Chicago, Ill.). A standard Michaelis-Menten model was used to determine apparent values of Km (Km,app) and kcat (kcat,app) at various NMN concentrations and several fixed ATP concentrations.
(iii) RNK-coupled assay.
An original continuous spectrophotometric assay was developed coupling RNK activity to NADH formation via two additional enzymatic steps: the conversion of NMN to NAD with an excess of recombinant human NMNAT (O. V. Kurnasov and A. L. Osterman, unpublished data) and the alcohol dehydrogenase-catalyzed conversion of NAD to NADH. The assay was performed as described for the NMNAT assay, except that the preincubation mixture contained 0.8 mM N-ribosylnicotinamide instead of NMN and 0.15 U of human NMNAT and did not contain ATP. The reaction was initiated by adding 50 μl of 50 mM ATP. Data were collected and analyzed as described for the NMNAT assay.
Analysis of gene essentiality. The essentiality of the nadR and pnuC genes in H. influenzae was determined by the GAMBIT (genomic analysis and mapping by in vitro transposition) technique (1) with some modifications. A 5.3-kb DNA fragment containing nadR was PCR amplified with the primers TGGCTTTGCGGGGAGTGGTTCAT (FP0) and ACGACGGTTCACCCATCATCACAAG (RP1) and genomic DNA from H. influenzae. An 11.6-kb DNA fragment containing pnuC was amplified with primers GAAGGCATCACAAATGGGAACATTGA and TTGCGCTGTTCAATGGTTATGATT. The PCR products were mutagenized by in vitro transposition with the EZ::TN<KAN-2> transposon and the hyperactive Tn5 transposase (Epicentre Technologies, Madison, Wis.) as recommended by the manufacturer. The 20-μl reaction mixture containing 20 ng of target DNA per μl, 15 ng of transposon DNA per μl, 0.1 U of transposase per μl, 50 mM Tris-acetate (pH 7.5), 150 mM potassium glutamate, 10 mM magnesium acetate, and 40 mM spermidine was incubated at 37°C for 2 h. The reaction was stopped by the addition of 0.1% SDS, and transposase was removed by phenol extraction. Gaps in transposition products were repaired with T4 DNA polymerase and 0.1 mM concentrations of each deoxynucleoside triphosphate, followed by ligation with T4 DNA ligase. Repaired transposition products were transformed into H. influenzae (5). Transformants were selected on brain heart infusion (BHI) plates supplemented with 10-μg/ml concentrations (each) of hemin, NAD, and ribostamycin (50). Colonies were pooled after growth for 24 h, and total genomic DNA was extracted and used to generate genetic footprints of the region by PCR with outwardly directed transposon-specific primers and primers specific for each of the two nadR and pnuC chromosomal loci. The PCR products were separated on 0.65% agarose gel. The images were captured and analyzed with 1D Image Analysis Software (Eastman Kodak Company, Rochester, N.Y.). Chromosomal mapping of inserts was performed as described previously (22).
Nucleotide sequence accession number.
The revised H. influenzae pnuC locus, translated into a 226-residue contiguous ORF, was deposited in GenBank under accession no. AF503632.
RESULTS
Reconstruction of NAD metabolism from genomic data and RNK prediction.
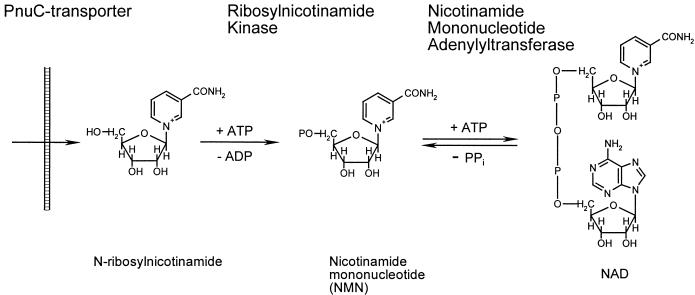
A simplified diagram of NAD biosynthesis based on biochemical and genetic data, largely from E. coli and S. enterica serovar Typhimurium, is shown in Fig. 1. Most of the E. coli genes involved with the biochemical pathways shown in this diagram (except for NMN deamidase and NMN glycohydrolase) are currently known. When we began this study, a gene for RNK had not been identified in any species, although the corresponding enzymatic activity had been detected in E. coli (41) and H. influenzae (13). In E. coli, RNK is presumably involved with NMN recycling and does not play a key role in the biosynthetic machinery of NAD. The two major sources of NAD are de novo biosynthesis and salvage of exogenous niacin (Fig. 1). Based on genomic evidence, neither of these pathways is expected to occur in H. influenzae (Table 1). This expectation is consistent with the requirement of H. influenzae for the presence of the so-called V factors in the growth medium (13). The minimal V factor that can support the growth of H. influenzae is N-ribosylnicotinamide. Other acceptable V factors include NADP, NAD, and NMN. Before uptake, the other V factors must be degraded to N-ribosylnicotinamide by recently characterized extracellular and periplasmic phosphohydrolases (25, 45, 48). In contrast to most bacteria, H. influenzae is incapable of attaching the pyridine ring to a ribose and cannot amidate the nicotinate moiety (13). H. influenzae lacks the corresponding genes (nadC, pncB, and nadV as well as nadE orthologs) (Table 1). The only pathway for NAD biosynthesis from exogenous N-ribosylnicotinamide anticipated in H. influenzae consists of (i) transport of N-ribosylnicotinamide across the periplasmic membrane, (ii) phosphorylation to NMN, and (iii) formation of NAD by NMN adenylation, as illustrated in Fig. 2.
FIG. 2.
Schematic of the N-ribosylnicotinamide salvage pathway of NAD biosynthesis.
We used a comparative genomics approach to search for a missing RNK gene candidate in the genomes of H. influenzae and related bacteria, based on the fact that this activity is expected to be indispensable for NAD metabolism in these organisms. None of the genes involved in the N-ribosylnicotinamide salvage pathway was previously identified in H. influenzae. Among all of the genes involved in NAD metabolism in other organisms (Table 1), only two, nadR and pnuC, have reliable homologs in the H. influenzae genome. Relatively low NMNAT activity (0.05 U/mg) and an approximately 170-fold preference for NMN over NaMN were previously reported for recombinant ecNadR (44). These properties markedly contrast with NadD, the major adenylyltransferase involved with NAD biosynthesis in E. coli, which displays very high NaMNAT activity (120 U/mg) and a >500-fold preference for NaMN over NMN (O. V. Kurnasov and A. L. Osterman, unpublished data). A universally conserved NadD-NadE pathway (Table 1 and Fig. 1) represents the major NAD biosynthetic route in E. coli and many other bacteria. Neither NadD nor NadE orthologs are present in H. influenzae or any other sequenced members of the Pasteurellaceae (Table 1), suggesting that in these organisms NadR is the major (and likely the only) enzyme responsible for the adenylyl transfer step in NAD biosynthesis. Based on this hypothesis, we expected hiNadR to differ significantly from ecNadR in at least two aspects: (i) higher catalytic efficiency and (ii) essentiality for growth and survival.
In addition to the role of stNadR in the transcriptional regulation of early NAD biosynthetic genes, this protein was also implicated in PnuC transporter-mediated NMN salvage by an unknown mechanism (20, 58). PnuC (in the TC 9.A.4.11 family according to the existing classification [http://tcdb.ucsd.edu/tcdb]) was characterized as an integral membrane protein involved with the active transport of exogenous NMN (56). The pnuC gene was not properly identified in the original version of the H. influenzae genome (17) due to sequencing errors. We have cloned and sequenced the corresponding 800-bp fragment of the H. influenzae chromosome and shown that it contains a full-size homolog of the canonical pnuC gene. Based on analogy with E. coli and S. enterica serovar Typhimurium, we suggest that the H. influenzae PnuC transporter homolog is the most likely candidate for an N-ribosylnicotinamide transporter in the V factor salvage pathway of NAD biosynthesis (25). A key prediction of this hypothesis is that the pnuC gene is expected to be essential for the growth and survival of H. influenzae, although it is not essential in S. enterica serovar Typhimurium (56).
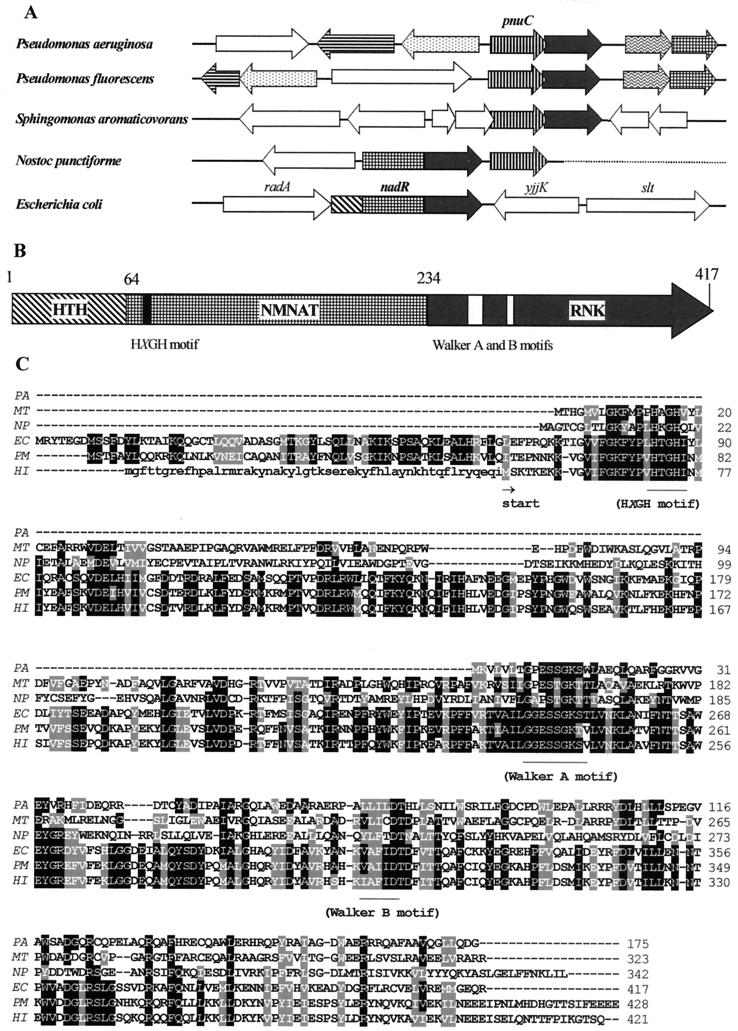
A putative identification of two components of H. influenzae NAD biosynthesis prompted us to use genome comparative analysis techniques, such as chromosomal clustering (39) and protein fusion events (29), to search for the last missing component of this pathway, the gene encoding RNK. Orthologs of pnuC and nadR are not clustered in the Enterobacteriaceae or the Pasteurellaceae, but they occur next to each other in some other genomes, as illustrated in Fig. 3. The pnuC orthologs in P. aeruginosa (PA1958) and some related species form a tight operon (Fig. 3A) with an uncharacterized protein (PA1957) corresponding to the “stand-alone” C-terminal domain of the NadR protein (residues 238 to 416 of ecNadR). Based on a multiple alignment of NadR homologs from various sources, this C-terminal domain represents a putative P-loop kinase (Fig. 3) with the predicted characteristic fold and conserved Walker-type ATP-binding motifs A and B (11, 26). The NadR protein was previously predicted to possess ATPase activity (35), but no specific functional role for this activity was proposed. Therefore, we considered the C-terminal domain of NadR protein to be a likely candidate for the missing RNK enzyme. Some representatives of the nucleoside and nucleotide kinase superfamily, uridine kinase (EC 2.7.1.48) and thymidylate kinase (EC 2.7.4.9), are distantly related to the NadR C-terminal domain, indirectly supporting this prediction.
FIG.3.
Gene clustering, domain organization, and amino acid sequence conservation in NadR homologs. (A) Homologous ORFs that clustered in the vicinity of pnuC genes on the chromosomes of several species are outlined by matching patterns. For nadR homologs, the presence of the corresponding domains (HTH, NMNAT, and RNK) is shown. (B) Domain organization and approximate domain boundaries in ecNadR protein. (C) Alignment of multiple amino acid sequences of representative NadR homologs from P. aeruginosa (PA; gi|15597153), M. tuberculosis (MT; gi|15607353), N. punctiforme (NP; http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?taxid = 63737), E. coli (EC; gi|730107), P. multocida (PM; gi|15603252), and H. influenzae (HI; gi|1171638). Conserved amino acids are outlined by black (for identity) and gray (for similarity) backgrounds based on a larger alignment of 15 NadR homologs produced by using ClustalW (http://dot.imgen.bcm.tmc.edu:9331/multi-align/Options/clustalw.html) and Boxshade 3.21 (http://www.isrec.isb-sib.ch:8080/software/BOX_form.html). Conserved sequence motifs characteristic of nucleotidyltransferases (HXGH motif) and P-loop kinases (Walker A and B motifs) are indicated. The N-terminal extension in hiNadR preceding the alternative translation start (Met-53) is shown in lowercase letters.
Enzymatic characterization of NadR protein.
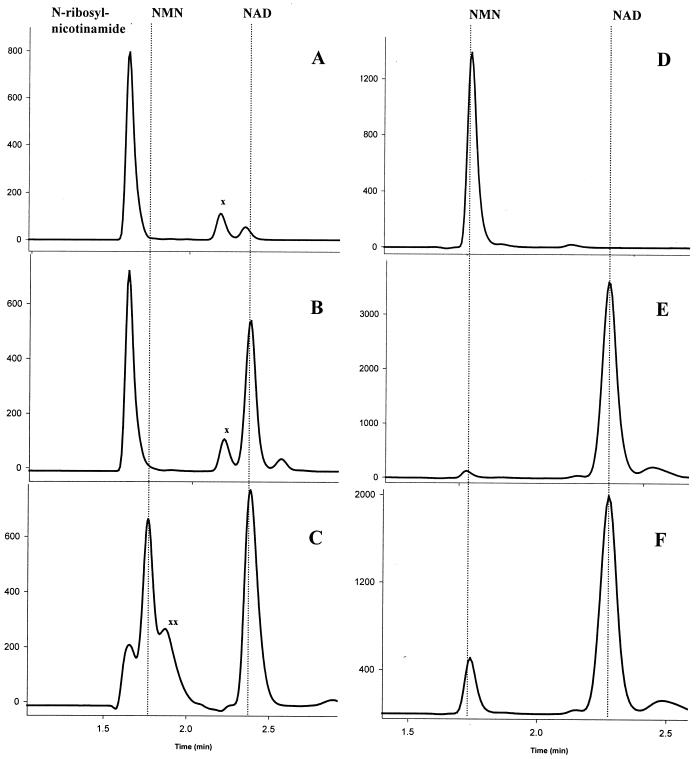
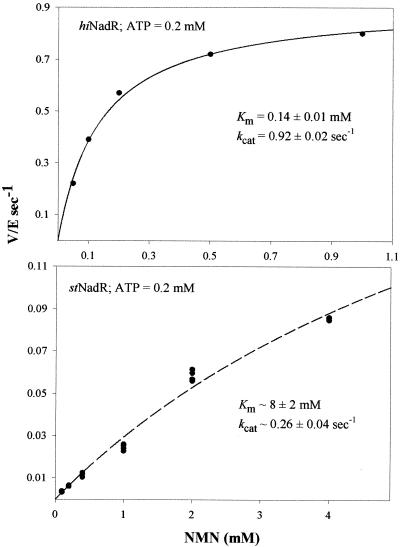
We cloned, expressed, purified, and characterized stNadR and hiNadR. An expected, NMNAT activity was experimentally confirmed for both proteins by HPLC analysis (Fig. 4). Steady-state kinetic data were generated by using a continuous coupled assay (Fig. 5). Due to the very poor affinity of stNadR for its substrate, only rough estimates of NMNAT kinetic parameters could be obtained (Km,app, ∼8 ± 2 mM, and kcat,app, ∼0.26 ± 0.04 s−1 at a saturating concentration of 0.2 mM ATP). The NMNAT enzymatic efficiency of hiNadR is ∼200-fold higher than that of stNadR, based on the kcat/Km ratio, in agreement with the presumed biological significance of NMNAT for H. influenzae NAD biosynthesis. This difference is manifested mostly at the substrate affinity level (Km,app = 0.14 ± 0.01 mM and kcat,app = 0.92 ± 0.02 s−1 at 0.2 mM ATP). Both the stNadR and hiNadR proteins reveal equally strong (∼250-fold) preferences for NMN-to-NaMN substrates, based on their respective specific activities at a 1 mM concentration of either substrate (Table 2). This substrate preference is consistent with the biological role of the NadR protein in NMN salvaging and recycling in each organism (Fig. 1).
FIG. 4.
Direct verification of NMNAT and RNK activities of hiNadR and stNadR. Reaction products were analyzed by ion-pair HPLC. Shown are the HPLC profiles of the N-ribosylnicotinamide substrate before (A) and after incubation with hiNadR (B) and stNadR (C) and of the NMN substrate before (D) and after incubation with hiNadR (E) and stNadR (F). Substrates and product retention times are shown with dotted vertical lines. Unidentified peaks are marked by “x” and “xx.” ATP and ADP were eluted at higher retention times (data not shown).
FIG. 5.
NMNAT steady-state kinetics of hiNadR and stNadR. Apparent kinetic parameters derived at ATP concentrations close to saturation (0.2 mM) are shown for each enzyme. Only an estimate of kinetic parameters could be obtained for stNadR, due to its poor affinity for NMN. V/E, ratio of initial reaction rate (V, in micromoles per second) to enzyme concentration (E, micromolar).
TABLE 2.
NadR substrate preferencesa
| Protein, size | Sp act of NMNAT (U/mg) | NMNAT/ NaMNAT ratio | Sp act of RNK (U/mg) | RNK/ RNaK ratio | RNK/ NMNAT ratio |
|---|---|---|---|---|---|
| stNadR, full size | 0.04 | ∼250 | 1 | ≥10,000 | 25 |
| hiNadR, full size | 0.90 | ∼250 | 0.04 | ≥10,000 | 0.04 |
| hiNadR, NMNAT domain | 3 | None |
Specific activities of stNadR and hiNadR were determined at 1 mM NaMN, NMN, and N-ribosylnicotinamide in the presence of 1 mM ATP by coupled assays. No N-ribosylnicotinate kinase (RNaK) activity could be detected by HPLC after overnight incubation with the corresponding substrate and 1 mM ATP. In the case of the NMNAT domain of hiNadR, no RNK activity could be detected; NaMNAT and N-ribosylnicotinate kinase activities were not measured.
In addition to NMNAT activity, both proteins possess the predicted RNK activity, directly confirmed by HPLC analysis (Fig. 4). In the case of stNadR, formation of an NMN intermediate could be observed, while in the case of hiNadR, the two-step reaction proceeded to NAD without detectable NMN accumulation. These results are also consistent with a difference in the relative efficiencies of the NMNAT and RNK half-reactions observed between stNadR and hiNadR (Table 2), when they were measured independently by the coupled assay. In this test, stNadR displayed significantly higher RNK activity than hiNadR (Table 2).
In order to refine the functional assignment for the NadR RNK domain, we analyzed its substrate specificity towards structurally similar nucleosides. For this purpose, we used stNadR, which displays significantly higher RNK activity than hiNadR. Based on HPLC analysis, no nucleoside phosphorylation was detected after overnight incubation of 0.2 mM concentrations of ribosyl derivatives of nicotinic acid, uracil, and cytosine in the presence of 5 mM ATP with 0.02 mg of stNadR per ml. The same stringent preference towards the natural substrate N-ribosylnicotinamide is also characteristic of hiNadR, evidenced by its inability to phosphorylate N-ribosylnicotinate (Table 2). This result is in contrast to the substrate specificity of human RNK purified from placenta, which efficiently phosphorylates a broad spectrum of nucleoside analogs, including the antitumor agent tiazofurin (47). No tiazofurin phosphorylation could be detected after extensive incubation with hiNadR or stNadR protein.
Domain organization of NadR protein.
A multiple alignment of NadR proteins from various sources suggests the existence of three distinct functional domains (Fig. 3). (i) The N-terminal helix-turn-helix (HTH) domain (corresponding to amino acids [aa] 1 to 63 in ecNadR) is characteristic of the DNA-binding proteins involved in the transcriptional regulation of the early NAD biosynthetic genes nadB, nadA, and pncB (42). This assertion is in agreement with the localization of mutations affecting the regulatory function of NadR protein from S. enterica serovar Typhimurium (21). (ii) The central NMNAT domain (aa 64 to 233 in ecNadR) has a recognizable Rossman fold with a conserved ATP-binding site in the N-terminal part of the sequence, characteristic of the nucleotidyltransferase superfamily (9). (iii) The C-terminal RNK domain (aa 234 to 417 in ecNadR) has a Rossman-like fold and recognizable Walker A and B motifs typical of members of the P-loop kinase family (11, 26).
The HTH domain is not preserved in hiNadR, and the reliable sequence similarity between hiNadR and other NadR homologs starts only within the boundaries of the NMNAT domain (Fig. 3). In the absence of direct experimental data, this observation may be explained by imprecise identification of the translational start site, a known limitation of the existing ORF-prediction algorithms. Among the two possible alternative hiNadR start positions at Met-15 and Met-52 (by numeration of the corresponding protein [gi|1573771] in GenBank), the latter is more consistent with the predicted start sites of N. punctiforme and M. tuberculosis NadR homologs (Fig. 3).
Overexpression of the extended version of hiNadR (N-terminal His6 tag fusion at Met-15) generated two products (revealed by SDS-polyacrylamide gel electrophoresis [data not shown]), consistent with an alternative translation initiation point at Met-52. A similar construct with a fusion at Met-52 produced a single protein of the predicted size, supporting its role as a possible hiNadR translation start site. Both short and extended forms of hiNadR have the same NMNAT and RNK specific activities (data not shown).
To verify the distribution of NMNAT and RNK activities within the domain boundaries predicted from the multiple alignment, we have attempted overexpressing the corresponding hiNadR domains individually. The NMNAT domain (aa 52 to 226) has the same specific NMNAT activity as the full-size protein, but no RNK activity could be detected (Table 2). All attempts to overexpress the C-terminal RNK domain failed to produce any soluble and active protein, regardless of the source (hiNadR or stNadR), type of fusion (His6 or thioredoxin tag), or expression protocol (different temperatures). The NMNAT domain may be required for the proper folding of the RNK domain or to prevent its self-aggregation or sequestration within the insoluble membrane fraction. Earlier experimental data implicated the C-terminal part of stNadR in PnuC-mediated association with membranes (21). As mentioned before, a stand-alone RNK gene in P. aeruginosa (encoding a paNadR protein corresponding to the ecNadR C-terminal domain) occurs in an operon with the pnuC gene. All paNadR protein was found in an insoluble form when it was overexpressed in E. coli, either as a single ORF or as a part of the pnuC-nadR operon.
Full-size NadR contains two putative ATP-binding sites in the NMNAT and the RNK domains (Fig. 3). Enzymatic characterization of C-terminally truncated hiNadR protein confirmed that only the first ATP-binding site is required for NMNAT activity (Table 2). Directed mutagenesis of this site (H77A and H80A of the HXGH motif) in stNadR produced the full-size protein completely lacking NMNAT activity. These mutations had no effect on RNK activity (data not shown), confirming that the first ATP-binding site is not involved with the enzymatic activity of the RNK domain. Crystallographic studies of hiNadR (51) show that the NMNAT and the NRK domains are well separated in the tertiary structure, consistent with their independent action.
Essentiality of pnuC and nadR genes in H. influenzae.
Mutations of both pnuC and nadR genes were previously described for S. enterica serovar Typhimurium as having no effect on growth in rich medium (20, 55, 56). Both of these genes are also dispensable in E. coli in rich medium, as shown by a genetic footprinting technique (22). We used a similar technique to address the essentiality of pnuC and nadR homologs in H. influenzae. As illustrated in Fig. 6, no transposon insertions were detected within nadR after aerobic growth in BHI medium, while multiple inserts were present in ORFs HI0762, HI0765, and HI0766. This result suggests that transposon insertions in nadR were lethal under the experimental conditions but that inserts in HI0762, HI0765, and HI0766 could be tolerated. Likewise, we detected no insertion within the pnuC gene (data not shown), demonstrating that this gene is also required for H. influenzae growth in BHI media. These results are in agreement with recently published data on genome-scale gene essentiality in H. influenzae (2).
FIG. 6.
Genetic footprinting of the H. influenzae chromosomal region containing nadR. (A) Random insertions were introduced into the nadR locus via transposition in vitro and the transformation of H. influenzae. (B) Transformants capable of forming colonies on BHI plates were pooled, and their genomic DNA was isolated and used for insert detection. Detection of insertions was performed by PCR as illustrated. In each PCR, one chromosome-specific primer (FP0, FP1, or RP1) and one transposon (Tn10 or Tn12)-specific primer were used (marked above each lane of the gel image; only three out of six used primer combinations are shown). Each band on the gel originates from a specific transposon insertion, which can thus be mapped in the H. influenzae chromosome (C). Note that areas on the gel that correspond to nadR do not contain any PCR products. This indicates that all insertions in this gene were lethal and were not represented in the analyzed pool of viable mutants. Lane M, molecular size markers. (C) ORFs of the H. influenzae chromosome containing transposon insertions (nonessential) are shown by white arrows, and those without insertions (essential) are shown by black arrows. PCR primers used for the fragment amplification and for insert detection are shown with arrows under the map. Detected transposon insertions are shown with vertical lines above the map.
The difference between Enterobacteriaceae and V factor-dependent Pasteurellaceae with respect to the essentiality of the pnuC and nadR genes is consistent with their expected differences in levels of NAD biosynthesis (Fig. 1). Since the hiNadR protein contains two functional domains involved in the same pathway, our transposon mutagenesis results do not allow us to assert whether only one (NMNAT domain) or both are essential. However, the absence of transposon insertions in the C-terminal part of the gene (Fig. 6) may be an indication of the essentiality of RNK activity in H. influenzae.
DISCUSSION
Using a comparative genomics approach, we established that RNK, involved with the N-ribosylnicotinamide salvage pathway of NAD biosynthesis (Fig. 1), is encoded in the previously uncharacterized C-terminal domain of NadR protein. This pathway was hypothesized to be of critical importance for V factor-dependent Pasteurellaceae, such as H. influenzae, due to its role in the only biosynthetic route of NAD and NADP biosynthesis in these organisms (13). At least one of the four known V factors, NADP, NAD, NMN, and N-ribosylnicotinamide, must be present in the medium to support the growth of H. influenzae. However, only a nonphosphorylated V factor, N-ribosylnicotinamide, can be transported across the inner membrane and converted to NAD or NADP in the cytoplasm (13). Extracellular and periplasmic enzymes degrading phosphorylated V factors to N-ribosylnicotinamide prior to uptake were recently identified (25, 45, 48), while genes encoding other components of NAD biosynthesis remained unknown. Based on genomic reconstruction of the NAD biosynthetic machinery in various species, including H. influenzae, we propose that the pnuC and nadR genes encode all the necessary components of the N-ribosylnicotinamide-NAD salvage pathway (Fig. 1, Fig. 2, and Table 1).
After correction of a sequencing error, the reconstructed H. influenzae PnuC homolog was identified as the most likely candidate for a putative N-ribosylnicotinamide transporter. This integral membrane protein, previously implicated in the NMN salvage of S. enterica serovar Typhimurium, is conserved in all Enterobacteriaceae and Pasteurellaceae and in a limited number of other gram-negative bacteria (Table 1). Local saturating transposon mutagenesis studies confirmed that the pnuC gene is essential for the growth and survival of H. influenzae, consistent with its proposed role in V factor utilization. In E. coli (22) and S. enterica serovar Typhimurium (56) the pnuC gene is not essential, due to the existence of alternative pathways such as de novo biosynthesis from aspartate and niacin salvage (Table 1 and Fig. 1).
Our model, implicating PnuC protein in N-ribosylnicotinamide transport, seems to contradict the earlier results for S. enterica serovar Typhimurium (27), which suggested a PnuC-dependent transport of intact NMN. This apparent contradiction may be the result of (i) fundamental functional differences between H. influenzae and S. enterica serovar Typhimurium PnuC transporters, (ii) the presence of an NMN transport mechanism unrelated to PnuC in S. enterica serovar Typhimurium, or (iii) the existence of a PnuC-unrelated mechanism of N-ribosylnicotinamide transport in H. influenzae. However, all of these alternative explanations are inconsistent with other observations and considerations. For example, the sequence similarity between PnuC homologs in H. influenzae and S. enterica serovar Typhimurium appears to be too high to accommodate such a dramatic difference in transporter specificities. Earlier genetic data for S. enterica serovar Typhimurium consistently identify PnuC as the only transporter involved with NMN salvage (20, 56). In addition, the identification of pnuC as an essential gene in H. influenzae is consistent with its unique role in the V factor salvage pathway.
Additional experiments are required to reconcile these contradictory observations. By a combination of arguments, we assume that the model of PnuC-mediated N-ribosylnicotinamide transport can be generalized and projected to E. coli and other related bacteria. Notably, NMN phosphohydrolase homologs (nadN gene of H. influenzae) are present in E. coli and S. enterica serovar Typhimurium (Table 1), supporting the idea that they are able to convert exogenous NMN to N-ribosylnicotinamide prior to transport.
The next step of the pathway, phosphorylation of N-ribosylnicotinamide to NMN, is catalyzed by RNK. We predicted the C-terminal domain of NadR to be a candidate for this functional role based on cumulative genomic evidence, such as (i) clustering on the chromosome with the pnuC gene (Fig. 3), (ii) translational fusion with the NMNAT domain, (iii) cooccurrence of pnuC and nadR in all Pasteurellaceae and Enterobacteriaceae, and (iv) a recognizable Rossman-like fold with Walker A and B sequence motifs, characteristic of the P-loop kinase superfamily. The predicted functional activity was verified and quantitatively characterized for two representatives of the NadR family from Pasteurellaceae (hiNadR) and Enterobacteriaceae (stNadR).
Sequence similarity searches revealed that this RNK family is present in a limited set of bacteria (Table 1) and not present in human or any other eukaryotic genome. Human RNK was previously partially purified and characterized (47). This enzyme, together with human NMNAT (15, 54), presumably form a similar N-ribosylnicotinamide salvage pathway, implicated in the activation of the anticancer prodrug tiazofurin. Thus, the human RNK, by phosphorylating the nucleoside analog tiazofurin, displays relatively broad substrate specificity (47) in contrast to the very stringent preference of bacterial RNK for its natural substrate N-ribosylnicotinamide (Table 2). This observation is consistent with the assertion that the unidentified human RNK gene is structurally unrelated or very divergent from its bacterial counterpart.
Early genetic studies of S. enterica serovar Typhimurium implicated the C-terminal segment of NadR in the salvage of exogenous NMN mediated by the PnuC transporter (21). This finding indicates that RNK activity may be directly involved with the PnuC transporter mode of action. Although the mechanism is unknown, one may hypothesize that ATP-dependent phosphorylation of N-ribosylnicotinamide by RNK occurs as a concerted process with PnuC-mediated transport, “trapping” a transported molecule within the cell. Therefore, RNK activity may play a dual role in assisting vectorial transport by preventing the efflux of negatively charged NMN through the phospholipid bilayer and providing an intermediate substrate for the next step of NAD biosynthesis.
Based on metabolic reconstruction from genomic data, we suggested that the last step of the N-ribosylnicotinamide salvage pathway in H. influenzae is performed by the NMNAT domain of hiNadR. We have shown that the NMNAT specific activity of hiNadR is at least 200-fold higher than the corresponding activities of ecNadR (44) and stNadR (Table 2) in correlation with our previous data, suggesting that the NMNAT activity of NadR is not sufficient to support the growth of an E. coli nadD mutant on the media supplemented with NMN (22). These observations are consistent with the unique role of the NadR-dependent NAD biosynthetic pathway in H. influenzae (Fig. 1). Another remarkable difference between hiNadR and stNadR is the relative catalytic efficiencies of the two consecutive enzymatic steps, RNK and NMNAT (Table 2). In the case of H. influenzae, where the NadR-dependent pathway is the only route for NAD formation, the higher catalytic efficiency of NMNAT compared to that of RNK is critically important for avoiding the potentially harmful accumulation of NMN (36).
In E. coli and S. enterica serovar Typhimurium, the major flux to NAD involves a two-step transformation of NaMN by NaMNAT (the nadD gene product) and NAD synthetase (the nadE gene product). In these bacteria, NMN utilization occurs mostly via NMN deamidase (18) (Fig. 1), which alleviates the risk of NMN accumulation due to the higher catalytic efficiency of RNK than that of NMNAT as observed with stNadR (Table 2). A gene encoding NMN deamidase has not been identified, and based on the absence of the NadD-NadE pathway, it is not expected to occur in H. influenzae. In P. aeruginosa and a few other bacteria containing RNK without NMNAT (Table 1), the NMN deamidase-dependent pathway appears to be the only possible route of N-ribosylnicotinamide salvage.
Comparative analysis and reconstruction of the NAD biosynthetic machinery suggest that the PnuC-NadR salvage pathway is the only route for NAD formation in H. influenzae. This pathway is expected to be present in Enterobacteriaceae, presumably with a minor flux, unlike the de novo biosynthesis and niacin salvage pathways (Fig. 1). In agreement with this hypothesis, both pnuC and nadR are essential genes in H. influenzae (our data and that in reference 2) while neither is essential in E. coli (22) or S. enterica serovar Typhimurium (20, 55, 56).
H. influenzae is the only representative of the V factor-dependent Pasteurellaceae with a publicly available genome sequence, although other related V factor-dependent species were previously described (53). Recently, the nadV gene encoding a nicotinamide phosphoribosyltransferase (EC 2.4.2.12), a novel enzyme of the niacin salvage pathway, was identified in V factor-independent H. ducreyi (30). This enzyme provides V factor-independent Pasteurellaceae with the ability to grow in the presence of nicotinamide via an alternative route to NAD (Fig. 1). This is likely to make the PnuC transporter and RNK activity dispensable in these organisms. In contrast, the NMNAT component of NadR is expected to remain essential in the V factor-independent Pasteurellaceae, since this activity is involved with the salvage of either precursor, nicotinamide or N-ribosylnicotinamide. NadV homologs are conserved in a number of diverse species (30), including humans (pre-B-cell colony-enhancing factor protein [32]), where they probably have a similar functional role (A. Rongvaux, R. J. Shea, M. H. Mulks, D. Gigot, J. Urbain, O. Leo, and F. Andris, Abstracts of the Meeting of the Belgian Society of Biochemistry and Molecular Biology, abstr. 10, 2002; [online]http://www.biochemistry.be/22feb2002/list_of_abstracts.htm).
In contrast to hiNadR, all of the sequenced NadR homologs from V factor-independent Pasteurelaceae contain a recognizable HTH domain (Table 1). However, none of these organisms contain the nadA, nadB, or pncB gene, known to be regulated by NadR in Enterobacteriaceae. It is tempting to propose that the nadV gene may provide a point of regulation by NadR in H. ducreyi and similar organisms. However, insufficient sequence data on V factor-dependent Pasteurellacae, as well as the absence of recognizable NAD boxes upstream of nadV (or elsewhere in Pasteurellacae genomes), make this suggestion highly speculative.
In summary, a comparative genome analysis and reconstruction of NAD metabolism from genomic data allowed us to predict and experimentally confirm that the C-terminal domain of the multifunctional NadR protein constitutes the previously uncharacterized RNK. Based on the characterization of RNK and NMNAT enzymatic activities of recombinant NadR proteins and on genetic analysis, we propose a model for the complete PnuC-NadR salvage pathway from exogenous N-ribosylnicotinamide to NAD. The essentiality of the pnuC and nadR genes in H. influenzae, together with their absence in the human genome, identifies the corresponding proteins as potential narrow-spectrum drug targets.
Acknowledgments
We are indebted to Ross Overbeek and all of the ERGO development team at Integrated Genomics for providing the technical support and inspiration which made this study possible. We are grateful to Tadhg Begley (Cornell University, Ithaca, N.Y.), Nick Grishin, and Hong Zhang (University of Texas, Southwestern Medical Center) for valuable discussions, help, and support throughout this project. We thank Robert Haselkorn (University of Chicago), John Campbell, and Matt Daugherty (Integrated Genomics) for discussing and helping with the manuscript. We also thank Shamim Chowdhury and Stephen Dobkowski for technical assistance.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. 1999. Short protocols in molecular biology: a compendium of methods from Current protocols in molecular biology, 4th ed. Wiley, New York, N.Y.
- 4.Balducci, E., M. Emanuelli, N. Raffaelli, S. Ruggieri, A. Amici, G. Magni, G. Orsomando, V. Polzonetti, and P. Natalini. 1995. Assay methods for nicotinamide mononucleotide adenylyltransferase of wide applicability. Anal. Biochem. 228:64-68. [DOI] [PubMed] [Google Scholar]
- 5.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 6.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-119. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. ColladoVides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 8.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bork, P., L. Holm, E. V. Koonin, and C. Sander. 1995. The cytidylyltransferase superfamily: identification of the nucleotide-binding site and fold prediction. Proteins 22:259-266. [DOI] [PubMed] [Google Scholar]
- 10.Burns, D. M., and I. R. Beacham. 1986. Nucleotide sequence and transcriptional analysis of the E. coli ushA gene, encoding periplasmic UDP-sugar hydrolase (5′-nucleotidase): regulation of the ushA gene, and the signal sequence of its encoded protein product. Nucleic Acids Res. 14:4325-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheek, S., H. Zhang, and N. V. Grishin. 2002. Sequence and structure classification of kinases. J. Mol. Biol. 320:855-881. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. (Erratum, 396: 190.) [DOI] [PubMed] [Google Scholar]
- 13.Cynamon, M. H., T. B. Sorg, and A. Patapow. 1988. Utilization and metabolism of NAD by Haemophilus parainfluenzae. J. Gen. Microbiol. 134:2789-2799. [DOI] [PubMed] [Google Scholar]
- 14.Daugherty, M., V. Vonstein, R. Overbeek, and A. Osterman. 2001. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 183:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emanuelli, M., F. Carnevali, F. Saccucci, F. Pierella, A. Amici, N. Raffaelli, and G. Magni. 2000. Human NMN adenylyltransferase: molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties. J. Biol. Chem. 276:406-412. [DOI] [PubMed] [Google Scholar]
- 16.Enright, A. J., I. Iliopoulos, N. C. Kyrpides, and C. A. Ouzounis. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402:86-90. [DOI] [PubMed] [Google Scholar]
- 17.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 18.Foster, J. W., D. M. Kinney, and A. G. Moat. 1979. Pyridine nucleotide cycle of Salmonella typhimurium: regulation of nicotinic acid phosphoribosyltransferase and nicotinamide deamidase. J. Bacteriol. 138:957-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster, J. W., and A. G. Moat. 1980. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol. Rev. 44:83-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, J. W., Y. K. Park, T. Penfound, T. Fenger, and M. P. Spector. 1990. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J. Bacteriol. 172:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, J. W., and T. Penfound. 1993. The bifunctional NadR regulator of Salmonella typhimurium: location of regions involved with DNA binding, nucleotide transport and intramolecular communication. FEMS Microbiol. Lett. 112:179-183. [DOI] [PubMed] [Google Scholar]
- 22.Gerdes, S. Y., M. D. Scholle, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. K. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godek, C. P., and M. H. Cynamon. 1990. In vitro evaluation of nicotinamide riboside analogs against Haemophilus influenzae. Antimicrob. Agents Chemother. 34:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai, S., S. Mori, T. Mukai, S. Suzuki, T. Yamada, W. Hashimoto, and K. Murata. 2000. Inorganic polyphosphate/ATP-NAD kinase of Micrococcus flavus and Mycobacterium tuberculosis H37Rv. Biochem. Biophys. Res. Commun. 276:57-63. [DOI] [PubMed] [Google Scholar]
- 25.Kemmer, G., T. J. Reilly, J. Schmidt-Brauns, G. W. Zlotnik, B. A. Green, M. J. Fiske, M. Herbert, A. Kraiss, S. Schlör, A. Smith, and J. Reidl. 2001. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183:3974-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165-1174. [DOI] [PubMed] [Google Scholar]
- 27.Liu, G., J. Foster, P. Manlapaz-Ramos, and B. M. Olivera. 1982. Nucleoside salvage pathway for NAD biosynthesis in Salmonella typhimurium. J. Bacteriol. 152:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magni, G., A. Amici, M. Emanuelli, N. Raffaelli, and S. Ruggieri. 1999. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 73:135-182. [DOI] [PubMed] [Google Scholar]
- 29.Marcotte, E. M., M. Pellegrini, M. J. Thompson, T. O. Yeates, and D. Eisenberg. 1999. A combined algorithm for genome-wide prediction of protein function. Nature 402:83-86. [DOI] [PubMed] [Google Scholar]
- 30.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNiece, I. K., R. A. Briddell, X. Q. Yan, C. A. Hartley, A. Gringeri, M. A. Foote, and R. G. Andrews. 1994. The role of stem cell factor in mobilization of peripheral blood progenitor cells. Leuk. Lymphoma 15:405-409. [DOI] [PubMed] [Google Scholar]
- 33.Mehl, R. A., C. Kinsland, and T. P. Begley. 2000. Identification of the Escherichia coli nicotinic acid mononucleotide adenylyltransferase gene. J. Bacteriol. 182:4372-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moat, A. G., and J. W. Foster. 1987. Biosynthesis and salvage pathways of pyridine nucleotides, p. 1-20. In D. Dolphin and O. Avramovich (ed.), Pyridine nucleotide coenzymes. Part B. John Wiley & Sons, New York, N.Y.P. R.
- 35.Mushegian, A. 1999. The purloined letter: bacterial orthologs of archaeal NMN adenylyltransferase are domains within multifunctional transcription regulator NadR. J. Mol. Microbiol. Biotechnol. 1:127-128. [PubMed] [Google Scholar]
- 36.Olivera, B. M., and F. Bonhoeff. 1972. Discontinuous DNA replication in vitro. I. Two distinct size classes of intermediates. Nat. New Biol. 240:233-235. [DOI] [PubMed] [Google Scholar]
- 37.Osterman, A., N. V. Grishin, L. N. Kinch, and M. A. Phillips. 1994. Formation of functional cross-species heterodimers of ornithine decarboxylase. Biochemistry 33:13662-13667. [DOI] [PubMed] [Google Scholar]
- 38.Osterman, A. L., D. V. Lueder, M. Quick, D. Myers, B. J. Canagarajah, and M. A. Phillips. 1995. Domain organization and a protease-sensitive loop in eukaryotic ornithine decarboxylase. Biochemistry 34:13431-13436. [DOI] [PubMed] [Google Scholar]
- 39.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 96:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrini, M., E. M. Marcotte, M. J. Thompson, D. Eisenberg, and T. O. Yeates. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 96:4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-730. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella. ASM Press, Washington, D.C.
- 42.Penfound, T., and J. W. Foster. 1999. NAD-dependent DNA-binding activity of the bifunctional NadR regulator of Salmonella typhimurium. J. Bacteriol. 181:648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raffaelli, N., F. M. Pisani, T. Lorenzi, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1997. Characterization of nicotinamide mononucleotide adenylyltransferase from thermophilic archaea. J. Bacteriol. 179:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reidl, J., S. Schlor, A. Kraiss, J. Schmidt-Brauns, G. Kemmer, and E. Soleva. 2000. NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 35:1573-1581. [DOI] [PubMed] [Google Scholar]
- 46.Sasiak, K., and P. P. Saunders. 1996. Purification and properties of a human nicotinamide ribonucleoside kinase. Arch. Biochem. Biophys. 333:414-418. [DOI] [PubMed] [Google Scholar]
- 47.Saunders, P. P., M. T. Tan, C. D. Spindler, and R. K. Robins. 1989. Phosphorylation of 3-deazaguanosine by nicotinamide riboside kinase in Chinese hamster ovary cells. Cancer Res. 49:6593-6599. [PubMed] [Google Scholar]
- 48.Schmidt-Brauns, J., M. Herbert, G. Kemmer, A. Kraiss, S. Schlor, and J. Reidl. 2001. Is a NAD pyrophosphatase activity necessary for Haemophilus influenzae type b multiplication in the blood stream? Int. J. Med. Microbiol. 291:219-225. [DOI] [PubMed] [Google Scholar]
- 49.Selkov, E., N. Maltsev, G. J. Olsen, R. Overbeek, and W. B. Whitman. 1997. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene 197:GC11-GC26. [DOI] [PubMed] [Google Scholar]
- 50.Sharetzsky, C., T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1991. A novel approach to insertional mutagenesis of Haemophilus influenzae. J. Bacteriol. 173:1561-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh, K., O. V. Kurnasov, B. Chen, H. Robinson, N. V. Grishin, A. L. Osterman, and H. Zhang. 2002. Crystal structure of Haemophilus influenzae NadR protein: a bifunctional enzyme endowed with NMN adenylyltransferase and ribosylnicotinamide kinase activities. J. Biol. Chem. 277:33291-33299. [DOI] [PubMed] [Google Scholar]
- 52.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 53.Windsor, H. M., R. C. Gromkova, and H. J. Koornhof. 1991. Plasmid-mediated NAD independence in Haemophilus parainfluenzae. J. Gen. Microbiol. 137:2415-2421. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, H., T. Zhou, O. Kurnasov, S. Cheek, N. V. Grishin, and A. Osterman. 2002. Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD. Structure (Cambridge) 10:69-79. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, N., B. M. Olivera, and J. R. Roth. 1988. Identification of a repressor gene involved in the regulation of NAD de novo biosynthesis in Salmonella typhimurium. J. Bacteriol. 170:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, N., B. M. Olivera, and J. R. Roth. 1989. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J. Bacteriol. 171:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, N., B. M. Olivera, and J. R. Roth. 1991. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J. Bacteriol. 173:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, N., and J. R. Roth. 1991. The nadI region of Salmonella typhimurium encodes a bifunctional regulatory protein. J. Bacteriol. 173:1302-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]