Abstract
Escherichia coli tRNA contains four naturally occurring nucleosides modified with sulfur. Cysteine is the intracellular sulfur source for each of these modified bases. We previously found that the iscS gene, a member of the nifS cysteine desulfurase gene family, is required for 4-thiouridine biosynthesis in E. coli. Since IscS does not bind tRNA, its role is the mobilization and distribution of sulfur to enzymes that catalyze the sulfur insertion steps. In addition to iscS, E. coli contains two other nifS homologs, csdA and csdB, each of which has cysteine desulfurase activity and could potentially donate sulfur for thionucleoside biosynthesis. Double csdA csdB and iscS csdA mutants were prepared or obtained, and all mutants were analyzed for thionucleoside content. It was found that unfractionated tRNA isolated from the iscS mutant strain contained <5% of the level of sulfur found in the parent strain. High-pressure liquid chromatography analysis of tRNA nuclease digests from the mutant strain grown in the presence of [35S]cysteine showed that only a small fraction of 2-thiocytidine was present, while the other thionucleosides were absent when cells were isolated during log phase. As expected, digests from the iscS mutant strain contained 6-N-dimethylallyl adenosine (i6A) in place of 6-N-dimethylallyl-2-methylthioadenosine and 5-methylaminomethyl uridine (mnm5U) instead of 5-methylaminomethyl-2-thiouridine. Prolonged growth of the iscS and iscS csdA mutant strains revealed a gradual increase in levels of 2-thiocytidine and 6-N-dimethylallyl-2-methylthioadenosine with extended incubation (>24 h), while the thiouridines remained absent. This may be due to a residual level of Fe-S cluster biosynthesis in iscS deletion strains. An overall scheme for thionucleoside biosynthesis in E. coli is discussed.
Sulfur is found in the cell not only in essential cofactors, such as biotin and thiamine, but also in macromolecules, such as Fe-S proteins and tRNA (2, 22). The nearly universal source of sulfur for these molecules is l-cysteine. However, the biochemical reactions for the incorporation of the sulfur from cysteine are in many cases poorly understood. It was recently found that the cysteine desulfurase IscS is required for the biosynthesis of 4-thiouridine in tRNA both in vitro (14) and in vivo (17). In addition, iscS mutants are unable to synthesize thiazole and nicotinic acid and appear to be deficient in a number of other biosynthetic pathways. There now exists strong evidence from many laboratories that iscS is the major sulfur mobilization catalyst required for the biosynthesis of protein Fe-S clusters in both bacteria (32, 33, 40) and yeasts (19).
Base modification in tRNA is found in all organisms and represents the fine-tuning of the many functions of tRNA in protein translation (3, 4). Thionucleosides in particular have been found to influence tRNA aminoacylation (21, 37), codon-anticodon specificity (45), and reading frame maintenance (42) as well as the binding of tRNA to the ribosome (1). There have been several reports linking the biochemistry of tRNA modification to important pathways in primary or secondary metabolism. For example, Lipsett reported that Escherichia coli mutants lacking 4-thiouridine in their tRNA were also unable to synthesize thiamine (31). The thiI (27, 43) and iscS (17) genes, both required for 4-thiouridine and thiazole synthesis, were characterized as a result of these initial studies.
Some tRNA modifications are also thought to act as biochemical sensors for environmental stress. Interestingly, two of these involve sulfur modification. For example, 4-thiouridine has been proposed to act as a sensor for near-UV radiation (39), inducing intramolecular crosslinks into tRNA that result in poor aminoacylation (5) and the stringent response (16). The presence of 6-N-dimethylallyl-2-methylthioadenosine has been shown to require soluble iron and is absent in tRNA from bacteria isolated from lethally infected animals, in which iron is scarce (12). It has recently been shown that MiaB, which is implicated in sulfur insertion of 6-N-dimethylallyl-2-methylthioadenosine (8) is an Fe-S protein (30).
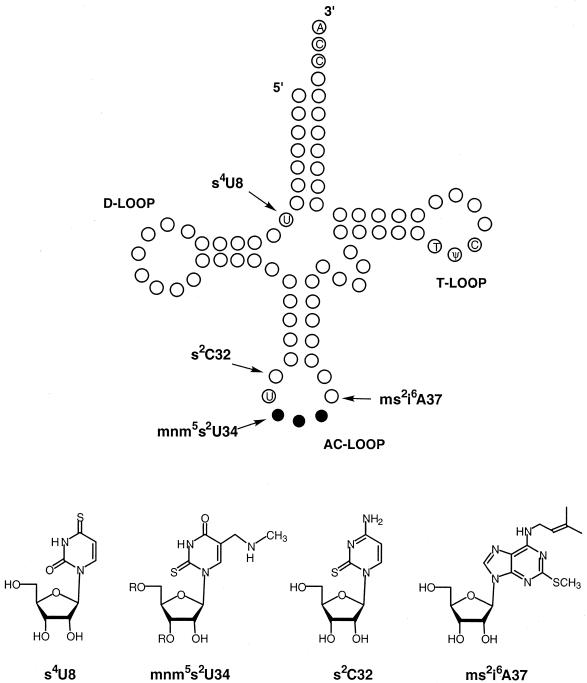
Four thionucleosides are naturally occurring in E. coli. Shown in Fig. 1, these are 4-thiouridine (s4U), 2-thiocytidine (s2C), 5-methylaminomethyl-2-thiouridine (mnm5s2U), and 6-N-dimethallyl-2-methylthioadenosine (ms2i6A). The biosynthesis of 4-thiouridine (Fig. 2A) has been shown to require both the thiamine pathway enzyme ThiI (27) and the cysteine desulfurase IscS (14). The mechanism of sulfur transfer (Fig. 2B) initially involves mobilization of sulfur from l-cysteine by IscS to form a persulfide at Cys328 in the active site (9). This persulfide, or sulfane sulfur, is then transferred to a cysteine on ThiI, which is in turn transferred to the tRNA (15) by a mechanism that likely involves oxidation of ThiI. Evidence for disulfide formation in ThiI during 4-thiouridine synthesis has recently been shown under single-turnover conditions (28). This type of mechanism may also be operating for the synthesis of 2-thiouridine by IscS and MnmA (R. Kambampati and C. T. Lauhon, submitted for publication).
FIG. 1.
Structures of the four naturally occurring thionucleosides and their locations in E. coli tRNA. Invariant nucleosides are shown in the tRNA secondary structure, and anticodon bases are represented by solid circles. s4U8, 4-thiouridine 8; mnm5s2U34, 5-methylaminomethyl-2-thiouridine 34; s2C32, 2-thiocytidine 32; ms2i6A37, 6-N-dimethylallyl-2-methylthioadenosine 37.
FIG. 2.
Biosynthesis of 4-thiouridine in E. coli. (A) Factors necessary for conversion of uridine (U8) to 4-thiouridine (s4U8) in E. coli. (B) Currently proposed mechanism for sulfur transfer from IscS to ThiI during 4-thiouridine biosynthesis. ThiI also utilizes MgATP for activation of the uridine O-4 and requires free thiol for reduction of a disulfide that is formed internally during turnover. IscS-SH, unmodified IscS protein; IscS-SSH, IscS persulfide; PLP, pyridoxal-l-phosphate.
E. coli contains three genes, iscS, csdA (24), and csdB (25), that code for cysteine desulfurases. Each gene product has been shown to catalyze cysteine desulfurase activity with varying efficiency in vitro (26). It seemed probable that one or more of these enzymes was required for biosynthesis of the other thionucleosides in E. coli. Therefore, deletion mutants of each of the genes were prepared, as well as a double mutants that lacked both csdA and csdB as well as iscS and csdA. High-pressure liquid chromatography (HPLC) traces of tRNA nuclease digests from these mutants were then compared. The results showed that the iscS gene in E. coli is required for the biosynthesis of each of the four thionucleosides and accounts for ≥95% of the sulfur content in E. coli tRNA.
MATERIALS AND METHODS
Materials.
4-Thiouridine, 2-thiocytidine, E. coli tRNALys and tRNAPhe, and bacterial alkaline phosphatase were obtained from Sigma. The deletion plasmid pKO3 was generously provided by George M. Church. Nuclease P1 was from Roche Biochemicals. Oligodeoxynucleotides were from Integrated DNA Technologies. The DE3 lysogenization kit used to prepare strain CL100(DE3) was from Novagen. Strains PK4331 (32) and PK5930 (ΔiscS ΔcsdA) were generous gifts of Patricia Kiley, University of Wisconsin.
General method of gene deletion in E. coli.
A combination of the methods of Link et al. (20) and Hamilton et al. (13) was used to perform the in-frame deletion of genes in E. coli. The method has previously been described in the deletion of E. coli iscS (17). The parent strain for all gene deletions was E. coli MC1061. Table 1 shows the primers used for the csdA and csdB genes.
TABLE 1.
Strains, plasmids, and primers used in this worka
| Strain, plasmid, or primer | Description or sequence | Source or reference |
|---|---|---|
| Strains | ||
| MC1061 | F−araD139 Δ(ara leu)7696 Δ(lacY74) galU galK hsdr hsdM+strA | BioRad |
| CL100 | ΔiscS | 17 |
| CL100(DE3) | DE3 lysogen of CL100 | 17 |
| CL201 | ΔcsdB | This work |
| CL102 | Δcsd ΔcsdB | This work |
| CL103 | ΔthiI | This work |
| CL250 | ΔmnmA | —b |
| CL260 | ΔmiaB | This work |
| PK4331 | MG1655 lacZΔ145 ΔiscS::Knr | 30 |
| PK5930 | MG1655 lacZΔ145 ΔiscS::Knr ΔcsdA::Cmr | —c |
| Plasmids | ||
| pCL010 | Wild-type E. coli iscS in pET21c | 14 |
| pCSD | Wild-type E. coli csd in pET21c | This work |
| pCSDB | Wild-type E. coli csdB in pET21c | This work |
| pKO3 | For deletion of genes in E. coli | 20 |
| Primers | ||
| CSD.N | 5′-ATC AAG CCG AGG AGT CAT ATG AAC GTT TTT AAT CCC GCG | |
| CSD.C | 5′-CGA ATT GCG GGT GAA TTC TTA ATC CAC CAA TAA TTC CAG | |
| CsdB.N | 5′-GCT GCC AGG AGG TGC CAT ATG ATT TTT TCC GTC GAC AAA | |
| CsdB.C | 5′-AGC CAT AGT GCC GGA TCC TTA TCC CAG CAA ACG GTG AAT | |
| CSD.No | 5′-AAG GAA AAA AGC GGC CGC TAC ATT TAC CCT GTC TGT CCA TAG TGAT T | |
| CSD.Ni | 5′-CAC GCA ATA ACC TTC ACA CTC CAA ATT TAT AAC CAT GGT ACT CCT CGG CTT G | |
| CSD.Co | 5′-CGC ACG CAT GTC GAC GTC TTA TCC GAC CCG GTT CT | |
| CSD.Ci | 5′-GTT ATA AAT TTG GAG TGT GAA GGT TAT TGC GTG TGA CCG CGC GCT GGA ATT A | |
| CsdB.No | 5′-AAG GAA AAA AGC GGC CGC ACG CAA CTC AAT GGC GAA AAC A | |
| CsdB.Ni | 5′-CAC GCA ATA ACC TTC ACA CTC CAA ATT TAT AAC AAT CAT CTT GCA CCT CCT GGC | |
| CsdB.Co | 5′-CGC ACG CAT GTC GAC CAT TGA CCA TCC GGC AAT GTG A | |
| CsdB.Ci | 5′-GTT ATA AAT TTG GAG TGT GAA GGT TAT TGC GTG CAC CGT TTG CTG GGA TAA CAG |
For primers, restriction sites are underlined, and the 33-bp gene replacement tag sequence is shown in bold (see text). Primers labeled No, Co, etc., were used to perform in-frame deletion by the method of Link et al. (20).
R. Kambampati and C. T. Lauhon, submitted.
P. J. Kiley, unpublished data.
Growth media and 35S labeling of tRNA.
Rich medium was Luria-Bertani (LB). For 35S labeling, 100-ml cultures containing LB supplemented with 1 mCi of l-[35S]cysteine were inoculated with 1 ml of an overnight culture of either the parent strain (MC1061), the iscS mutant [CL100(DE3)], orthe iscS mutant with a plasmid expressing wild-type E. coli iscS [CL100(DE3)/pCL010]. The cells were grown overnight, and tRNA was isolated and digested to nucleosides as outlined below.
Isolation of unfractionated tRNA.
Cells were recovered by centrifugation at 8,000 × g for 15 min at 4°C. The cell pellet was resuspended in 2 ml of 10 mM Tris (pH 7.5)-1 mM MgCl2. An equal volume of equilibrated buffered phenol was added, and the mixture was vortexed for 1 min. After separation of layers by centrifugation (13,000 × g, 20 min), the top aqueous layer was transferred to a polypropylene tube, and 0.1 volume of 3 M sodium acetate (pH 5.5) was added, followed by 2.5 volumes of cold ethanol. After storage at −20°C for at least 4 h (or overnight), the diffuse precipitate was isolated by centrifugation (13,000 × g, 4°C, 30 min), washed with 70% aqueous ethanol, dried, and resuspended in 0.5 ml of 10 mM Tris (pH 8) with vortexing. To this solution was added an equal volume of 8 M urea containing 0.01% (wt/vol) bromophenol blue. The resulting viscous solution was heated for 3 min at 85°C and loaded onto a 10% denaturing polyacrylamide gel.
The band corresponding to tRNA was excised and eluted by crushing and soaking in 0.5 M NaCl overnight. The tRNA was then precipitated with ethanol, resuspended in deionized water, and stored at −20°C. Quantitation was done by absorbance at 260 nm. The amounts of tRNA isolated from the parent and mutant strains were within the variation of the isolation procedure when cultures of equal cell density were compared. Amounts ranged from 100 to 200 μg for a 100-ml culture harvested at mid-log phase to double that amount for freshly saturated cultures. Simple “overnight” cultures will yield less tRNA for the iscS mutant due to the long time required to reach saturation (>24 h).
tRNA digestion and nucleoside analysis by HPLC.
A total of 100 μg of gel-purified unfractionated tRNA (concentration determined by A260) was treated with 3 U of nuclease P1 in a mixture of 30 mMsodium acetate (pH 5.3) and 20 mM zinc acetate in a total volume of 200 μl at 37°C. Reaction times could be as little as 4 h but were typically overnight. After reaction with nuclease P1, 20 μl of 1 M Tris (pH 7.5) was added, followed by 3 U of bacterial alkaline phosphatase, and the mixture was incubated at 37°C for at least 2 h.
For nucleoside analysis, the HPLC method of Gherke et al. (11) was used. Aliquots of the reaction mixture (typically corresponding to 25 μg of tRNA) were loaded directly onto a Supelco C18 column (catalog no. LC18-S). This column is specially designed for nucleoside separation. For elution of nucleosides that are more polar than adenosine, the following buffer system was chosen: buffer A contained 2.5% methanol in 10 mM ammonium phosphate, and buffer B contained 20% methanol in 10 mM ammonium phosphate, pH 5.3. The gradient was from 0 to 100% B over 45 min with the five-step program described by Gherke et al. (11) for high-resolution separation. For nucleosides less polar than adenosine (e.g., i6A and 6-N-dimethylallyl-2-methylthioadenosine), the following buffer system was used: buffer A contained 10% acetonitrile in 5 mM ammonium phosphate (pH 5.3), and buffer B contained 35% acetonitrile in 5 mM ammonium phosphate. A 30-min linear gradient from 0 to 100% B was used.
Nucleosides were detected by absorbance at 260 nm, with the additional use of 330 nm for detection of 4-thiouridine. HPLC standards were used for identifying 4-thiouridine, 2-thiocytidine, and mnm5U, whereas purified E. coli tRNAs could provide standards for 5-methylaminomethyl-2-thiouridine (tRNAGlu) and 6-N-dimethylallyl-2-methylthioadenosine (tRNAPhe). Quantitation (Table 2) was done by dividing the area of peaks from the thionucleosides by that of pseudouridine for 2-thiocytidine, mnm5U, 5-methylaminomethyl-2-thiouridine, and 4-thiouridine as previously described (6). For 6-N-dimethylallyl-2-methylthioadenosine and i6A, buffer A was replaced with 5% acetonitrile to allow separation of t6A and m2A from adenosine. The area of the 6-N-dimethylallyl-2-methylthioadenosine or i6A peak was divided by the area of the t6A peak, the latter of which was proportional to the amount of tRNA in my hands. These relative values served to minimize error due to loading variation.
TABLE 2.
Modified nucleoside levels for parent strain MC1061, iscS mutant CL100(DE3), and CL100(DE3) complemented with plasmid pCL010a
| Strain | Relative level
|
|||||
|---|---|---|---|---|---|---|
| s2C | mnm5U | mnm5s2U | s4U | i6A | ms2i6A | |
| MC1061 | 1.0 (0.2) | (0.015) | 1.0 (0.11) | 1.0 (1.2) | NDb | (0.52) |
| CL100(DE3) | 0.02 | (0.15) | ND | ND | (0.65) | ND |
| CL100(DE3)/pCL010 | 0.9 | (0.01) | 0.7 | 1.0 | ND | (0.47) |
Levels of 2-thiocytidine (s2C), 5-methylaminomethyl-2-thiouridine (mnm5s2U), and 4-thiouridine (s4U) were measured as the area ratio to pseudouridine (Ψ) and then normalized relative to the parent strain set at 1.0. Parenthetical values were not normalized. Levels of 6-N-dimethylalyl-2-methylthioadenosine (ms2i6A) and i6A are reported relative to t6A and were not normalized to the parent strain. All nucleosides were quantified at 260 nm except 4-thiouridine, which was quantitated at 330 mm.
ND, none detected.
The assigned thionucleoside peaks were found to be labeled with 35S when tRNA was analyzed from cells grown in medium supplemented with l-[35S]cysteine. In addition, the relative retention times for the nucleosides were very similar to those in the original work of Gherke et al. (11), since the same type of column and buffer system were used in both studies.
Growth of ΔiscS strain in medium supplemented with sulfide.
Cells of strain CL100(DE3) (ΔiscS) from 1 ml of overnight culture were added to 100 ml of LB supplemented with sodium sulfide (1 to 10 mM). Cultures were grown to saturation, and unfractionated bulk tRNA was isolated as described above.
RESULTS
Generation of mutants.
The strains used in this study are shown in Table 1. In addition to iscS, csdA, and csdB, mutations in the other known genes required for thionucleoside biosynthesis were also prepared. These included thiI, required for 4-thiouridine modification; mnmA, required for introduction of the sulfur at the 2 position of U34; and miaB, required for the introduction of sulfur at the 2 position of A37. The modified nucleoside profiles of these mutants were confirmed by HPLC analysis of nuclease digests of unfractionated tRNA isolated from the mutant strains (data not shown). thiI (27) and trmU (mnmA) (7, 36; A. T. Crescenzo, T. Hagerval, J. A. McCloskey, and D. Soll, unpublished data) mutants of E. coli and thiI (43) and miaB (8) mutants of Salmonella enterica serovar Typhimurium have been reported previously. The mutants prepared in this work are isogenic and provide control HPLC traces for the absence of specific thionucleosides. In each mutant, complementation with an expression plasmid containing the wild-type gene restored levels of the modified nucleoside to those found in the parent strain.
tRNA from ΔiscS strain lacks 95% of sulfur in tRNA
tRNA from the iscS mutant was analyzed for the presence of all thionucleosides. When the mutant and parent strains were grown in LB supplemented with l-[35S]cysteine and the tRNA was isolated, tRNA from the iscS mutant had only 2 to 5% of the sulfur level of tRNA of that of the parent strain. Figure 3 shows that expression of iscS from a multicopy plasmid in iscS mutant CL100(DE3) restored 87% of the sulfur in the tRNA. The reasons for less than complete complementation are not yet clear, but a similar observation has been made with Fe-S enzyme activity in another nonpolar iscS mutant (32).
FIG. 3.
Sulfur analysis of tRNA isolated from ΔiscS strain CL100(DE3). PhosphorImager scan of denaturing polyacrylamide gel of total tRNA isolated from parent and iscS mutant strain. The strains were grown to saturation in 100 ml of LB containing l-[35S]cysteine (1 mCi). Equal amounts of tRNA (10 μg) from each strain were loaded onto the gel. Left lane shows 35S-labeled tRNA from the parent strain E. coli MC1061. Middle lane is tRNA from strain CL100(DE3) (ΔiscS), and the right lane is tRNA from CL100(DE3) containing plasmid pCL010, which expresses wild-type iscS. The specific activities of tRNA from MC1061, CL100(DE3), and CL100(DE3)/pCL010 were 6,720, 330, and 5,789 cpm/μg, respectively.
The DE3 lysogen of strain CL100 that was used has significant basal expression of T7 polymerase in the absence of isopropylthiogalactopyranoside (IPTG). This results in overexpression of iscS from the T7 promoter-based plasmid pCL010, as measured by cysteine desulfurase activity of soluble cell extracts (not shown). Plasmid pCL010 completely complements both the slow growth rate and auxotrophic requirements of strain CL100(DE3). The presence of IPTG dramatically increased iscS expression but did not increase the level of thionucleosides by HPLC analysis (not shown).
Thionucleoside analysis of the ΔiscS strain.
tRNA digests from the iscS strain were examined for the presence of all thionucleosides. Figure 4 shows HPLC traces of the polar nucleoside region of tRNA digests from the parent strain MC1061, the iscS strain CL100(DE3), and CL100(DE3) complemented with a plasmid containing iscS (pCL010). The thiouridines 4-thiouridine and 5-methylaminomethyl-2-thiouridine were completely absent in the ΔiscS strain, and only a small amount of 2-thiocytidine was observed. A proportionate amount of mnm5U was found in the iscS mutant, confirming previous work suggesting that modification of the wobble uridine at the 5 position is independent of thiolation at the 2 position (7, 36; A. T. Crescenzo, T. Hagerval, J. A. McCloskey, and D. Soll, unpublished data).
FIG. 4.
HPLC profile of pyrimidine region of thionucleosides. (Top) Modified nucleoside HPLC profile of digested tRNA isolated from wild-type (wt) parent strain MC1061, with thionucleosides labeled. (Middle) Same profile from ΔiscS strain CL100. (Bottom) Profile from strain CL100 with plasmid pCL010 (wild-type iscS). The peak near 4-thiouridine in the middle panel is an unidentified compound that was present in all chromatograms but was revealed more clearly in the absence of 4-thiouridine. I independently verified at 330 nm that strain CL100 lacked 4-thiouridine (see Fig. 6) (16). s4U, 4-thiouridine; s2C, 2-thiocytidine; mnm5s2U, 5-methylaminomethyl-2-thiouridine.
Figure 5 shows that i6A was present in place of 6-N-dimethylallyl-2-methylthioadenosine in tRNA from strain CL100, a result that is identical to that with an E. coli miaB mutant prepared by me and previously described in S. enterica serovar Typhimurium by Esberg et al. (8). This strongly suggests that miaB activity is dependent on the presence of iscS in vivo. Such dependence is consistent with the observation, from both sequence analysis (35) and in vitro reconstitution (30), that MiaB is an Fe-S protein. The results are shown quantitatively in Table 2. Areas for 2-thiocytidine, 5-methylaminomethyl uridine (mnm5U), 5-methylaminomethyl-2-thiouridine, and 4-thiouridine are given as ratios with pseudouridine as an internal standard, whereas t6A was used as the standard for the hydrophobic nucleosides i6A and 6-N-dimethylallyl-2-methylthioadenosine. Since extinction coefficients vary, relative amounts of the thionucleosides are shown with that of the parent strain set to 1.0 when possible.
FIG. 5.
HPLC profile of modified adenosine region. (Top) Profile of digested tRNA isolated from parent strain MC1061. (Middle) Profile from ΔiscS strain CL100. (Bottom) Profile from strain CL100 with pCL010 (wild-type iscS). Approximately 25 μg of digested tRNA was loaded for each run. ms2i6A, 6-N-dimethlallyl-2-methylthioadenosine; i6A, 6-N-dimethylallyladenosine.
Increase in 2-thiocytidine and 6-N-dimethylallyl-2-methylthioadenosine levels with growth level.
tRNA was extracted from both the parent and strain CL100 at mid-log phase (A600 = 0.5), late log phase (A600 = 1.0), and saturation (A600 > 1.4) and analyzed. The time required for the iscS strain to reach saturation was >24 h. For the parent strain and the mutant strain CL100(DE3) complemented with wild-type iscS, final levels of all thionucleosides were reached by early log phase and remained essentially constant. As shown in Fig. 6 for the iscS mutant, only a very small fraction (ca. 2%) of 2-thiocytidine was present in the tRNA isolated from mid-log growth. A small increase in 2-thiocytidine levels to about 10% of the normal level was seen at late log phase, with no appearance in 6-N-dimethylallyl-2-methylthioadenosine. However, at saturation, 2-thiocytidine levels increased significantly to 70% of the wild-type level. Figure 7 shows that significant amounts of 6-N-dimethylallyl-2-methylthioadenosine were also seen at saturation but arose later relative to 2-thiocytidine, as none was detected at late log phase. Quantitation of these results is shown in Table 3.
FIG. 6.
Increase in 2-thiocytidine with growth (from upper left) of strain CL100 in LB medium. The A600 value is given next to each HPLC trace. The final trace, lower right, is of parent strain MC1061 and shows typical levels of 2-thiocytidine (s2C) and 5-methylaminomethyl-2-thiouridine (mnm5s2U) at all growth phases.
FIG. 7.
Increase in 6-N-dimethylallyl-2-methylthioadenosine in strain CL100 with extended growth. The top trace is for tRNA isolated at late log phase, while the bottom trace is after extended incubation. There was an additional relative decrease in the amount of 6-N-dimethylallyl adenosine (i6A), which is the precursor to 6-N-dimethylallyl-2-methylthioadenosine. Parent strain MC1061 or CL100(DE3) with pCL010 showed only 6-N-dimethylallyl-2-methylthioadenosine at all growth levels examined (see Fig. 5 for comparison and for abbreviations).
TABLE 3.
Levels of modified nucleosides in iscS mutant strains relative to the parent strain as a function of growtha
| Strain | Relative level
|
||||
|---|---|---|---|---|---|
| s2C | mnm5s2U | s4U | i6A | ms2i6A | |
| CL100, A600 = 0.5 | 0.01 | NDb | ND | 0.6 | ND |
| CL100, A600 = 1.0 | 0.4 | ND | ND | 0.5 | ND |
| CL100, t = 36 h | 0.7 | ND | ND | 0.2 | 0.2 |
| PK4331, A600 = 0.5 | 0.01 | ND | ND | 0.5 | ND |
| PK4331, t = 43 h | 0.9 | ND | ND | 0.1 | 0.1 |
| PK5930, A600 = 0.5 | 0.01 | ND | ND | 0.5 | ND |
| PK5930, t = 43 h | 0.7 | ND | ND | 0.1 | 0.1 |
2-Thiocytidine is reported as the area ratio to Ψ and normalized to the levels in MC1061, which was invariant with growth. Levels of 6-N-dimethylallyl adenosine (i6A) and 6-N-dimethylallyl-2-methylthioadenosine (ms2i6A) are reported directly as the area ratio to 6-N-threonylcarbamoyl adenosine (t6A). For other abbreviations, see Table 2, footnote a.
ND, none detected.
Possible supplementary role for iscS paralogs in E. coli.
Overexpression of the csdA and csdB genes in the iscS mutant did not complement any of the phenotypic characteristics of the iscS mutant. In addition, both the csdA and csdB mutants as well as a csdA csdB double mutant showed modified nucleoside profiles that were indistinguishable from that of the parent strain (data not shown). An E. coli iscS csdA double mutant (PK5930) was also obtained and analyzed. Thionucleoside levels were found to be identical to those in the isogenic iscS mutant PK4331 (32) in all respects, including the increase in 2-thiocytidine and 6-N-dimethylallyl-2-methylthioadenosine during stationary phase (Table 3). Thus, csdA is not involved in the residual 2-thiocytidine, 6-N-dimethylallyl-2-methylthioadenosine synthesis.
It is possible that spontaneous mutations may have caused the increase in these thionucleosides with extended incubation. Strains CL100 and PK4331 were both diluted after extended incubation (40 and 43 h, respectively) and regrown to mid log phase (A600 = 0.5). Thionucleoside analysis of tRNA showed no increase in 2-thiocytidine or 6-N-dimethylallyl-2-methylthioadenosine compared with the initially grown cultures harvested at the same growth level (Table 3). Thus, chromosomal mutations were not the cause of the increase in these thionucleoside levels.
Attempts to complement the iscS phenotype with exogenous sulfide.
Inorganic sulfide can replace IscS/cysteine in 4-thiouridine synthesis in vitro. The Km for sulfide is approximately 0.5 mM (C. T. Lauhon, unpublished data). To determine if intracellular sulfide could be the source for residual 2-thiocytidine and/or 6-N-dimethylallyl-2-methylthioadenosine synthesis, Strain CL100 was grown in rich medium supplemented with sulfide. Concentrations of 5 mM were tolerated by this strain; higher concentrations led to slow growth and eventual toxicity. HPLC analysis of digested tRNA isolated from strain CL100 grown in medium supplemented with sulfide (Fig. 8) showed a small amount of 4-thiouridine (1 to 2% of that in the parent strain). No increase in the other thionucleosides, including 2-thiocytidine, above those of the mutant grown in LB alone was observed. None of the previously documented slow-growth characteristics or auxotrophic requirements of the iscS mutant were complemented by exogenous sulfide up to 10 mM.
FIG. 8.
HPLC profiles of 4-thiouridine (s4U) region at 330 nm of strain CL100 (ΔiscS) grown in rich medium supplemented with sulfide. (Top) Profile from parent strain MC1061, which shows typical levels of 4-thiouridine. (Middle) tRNA nucleoside profile from strain CL100 grown in LB only. (Bottom) Profile from CL100 grown in LB supplemented with 5 mM sodium sulfide (HS−). Residual peaks from the large amounts of C, U, and G are visible for comparison between the three profiles. A similar amount of digested tRNA was loaded for each run shown.
DISCUSSION
The present results show that iscS is the major cysteine desulfurase required for thionucleoside synthesis in E. coli. The csdA csdB double mutant showed no variation in thionucleoside levels, and when overexpressed, neither allele can rescue the defects in thionucleoside levels caused by deletion of iscS. We (17) and others (32, 33, 40) have established that iscS mutants are defective in a wide range of metabolic pathways involving sulfur. These include the biosynthesis of vitamins, such as thiamine and nicotinic acid, as well as the amino acids isoleucine, valine, and methionine. There are undoubtedly other defects as well, since the growth rate of E. coli iscS mutants is half that of the parent strains in rich medium.
Many of the metabolic defects are the result of the key role that iscS plays in the formation and function of proteins of the isc gene cluster that is necessary for Fe-S cluster biosynthesis (46). A direct interaction and persulfide transfer (34, 41) have been shown between IscS and the isc cluster protein IscU. However, this is not the only function of iscS. IscS is also required for 4-thiouridine synthesis, and recently, Mihara et al. reported that iscS is required for both sulfur and seleniuminsertion into 5-methylaminomethyl-2-thio(seleno)uridine[mnm5s(se)2U] in E. coli (23). Xi et al. (44) showed evidence that IscS persulfide is involved in a novel reaction in the biosynthesis of thiazole.
Cysteine desulfurases are also required for sulfur transfer in the synthesis of the molybdopterin cofactor (18). These reactions do not involve Fe-S cluster chemistry. The purely “organic” sulfur transfer role of IscS (2) is distinct from that of iscS in isc cluster-dependent Fe-S biosynthesis and may be more prevalent than is currently known. Thus, IscS likely interacts with a number of other protein sulfur acceptors in addition to IscU and ThiI.
The similar growth dependence of 2-thiocytidine and 6-N-dimethylallyl-2-methylthioadenosine levels in the iscS mutant differentiates these thionucleosides from 4-thiouridine and 5-methylaminomethyl-2-thiouridine, which are almost certainly Fe-S cluster independent. The increase in levels of 6-N-dimethylallyl-2-methylthioadenosine with growth suggests that a residual level of Fe-S cluster biosynthesis exists in the iscS mutant. This is supported by the measurement of activities of Fe-S enzymes in E. coli iscS mutants (32, 33). A possible candidate for this auxiliary Fe-S synthesis is the suf operon. This operon contains the iscS paralog csdB, also termed sufS (29), as well as some homologs of other isc genes, and is arguably found in a wider range of organisms. It is clearly involved in Fe-S cluster synthesis, but the extent of its role is unclear.
Takahashi and Tokumoto (38) recently reported that iscS csdB (sufS) mutants of E. coli are not viable, and spontaneous mutations that increase expression of the suf operon (which contains csdB) can partially complement defects in isc operon mutants. To address the possibility of pseudorevertants, two independently isolated iscS mutant strains, CL100 and PK4331, with different genetic backgrounds were incubated for >36 h, diluted 200-fold into fresh medium, and harvested at mid-log phase for tRNA analysis. No differences in thionucleoside levels were observed from the original cultures harvested similarly. This argues against spontaneous mutations and in favor of an auxiliary Fe-S synthesis in the originally isolated iscS mutant strains. In addition, overexpression of csdB alone in strain CL100(DE3) did not affect thionucleoside levels; however, expression of the entire suf operon may be necessary to observe an increase in 2-thiocytidine and 6-N-dimethylallyl-2-methylthioadenosine levels.
Another possible source of sulfur for residual Fe-S synthesis in an iscS mutant is inorganic sulfide. Slow reconstitution of Fe-S enzymes in vitro has been demonstrated with soluble iron and sulfide under anaerobic conditions. Flint has reported the reconstitution of dihydroxy acid dehydratase as an assay for isolating the enzymes O-acetylserine sulfhydrolase and β-cystathionase in E. coli, which can mobilize sulfide from cysteine (10). Free sulfide is thought to be present at a low level in the cytoplasm of E. coli but may be sufficient to support very slow synthesis of Fe-S clusters for essential proteins. When strain CL100 was grown in rich medium supplemented with 5 mM sulfide, no difference in growth rate or auxotrophic requirements was observed. Thionucleoside analysis of tRNA showed that levels of 4-thiouridine were increased, which indicates that sulfide levels are likely to be elevated inside the cell. Since 4-thiouridine was observed under these conditions and no increase was observed in the rate of appearance or level of 2-thiocytidine or 6-N-dimethylallyl-2-methylthioadenosine, free sulfide is likely not the source for residual production of these thionucleosides.
A working hypothetical scheme for the role of iscS in thionucleoside biosynthesis may reflect the general role of iscS in sulfur metabolism. This scheme suggests a dual role for IscS as a sulfur source for both the isc Fe-S biosynthesis machinery and a more diverse non-Fe-S sulfurtransferase function. The non-isc sulfur transfer pathway has been shown definitively only with the 4-thiouridine and thiazole biosynthesis involving ThiI; however, it is likely that others will be found. MnmA, which has many conserved cysteines required for 2-thiouridine activity in vivo (C. T. Lauhon, unpublished data) is a likely IscS substrate for 2-thiouridine synthesis.
An unsolved question concerns the specificity of sulfur transfer. If a number of different protein substrates accept sulfane sulfur from IscS, there must be structural determinants for such specificity. Other putative protein substrates are currently being tested to provide a more complete explanation of the extensive role of IscS in sulfur metabolism.
Acknowledgments
I thank Alison Bednar for preparation of the csdB mutant. I also thank Glenn Bjork for sharing unpublished results and helpful discussion and Patricia Kiley for strains PK4331 and PK5930.
This work was supported by National Institutes of Health grant GM57002 and a grant from the University of Wisconsin Graduate School.
REFERENCES
- 1.Ashraf, S. S., E. Sochacka, R. Cain, R. Guenther, A. Malkiewicz, and P. F.Agris. 1999. Single atom modification (O->S) of tRNA confers ribosome binding. RNA 5:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beinert, H. 2000. A tribute to sulfur. Eur J. Biochem. 18:5657-5664. [DOI] [PubMed] [Google Scholar]
- 3.Bjork, G. R. 1995. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog. Nucleic Acid Res. Mol. Biol. 50:263-338. [DOI] [PubMed] [Google Scholar]
- 4.Bjork, G. R., J. U. Ericson, C. E. Gustafsson, T. G. Hagervall, Y. H. Jonsson, and P. M. Wikstrom. 1987. Transfer RNA modification. Annu. Rev. Biochem. 56:263-287. [DOI] [PubMed] [Google Scholar]
- 5.Blondel, M. O., and A. Favre. 1988. tRNAPhe and tRNAPro are the near-ultraviolet molecular targets triggering the growth delay effect. Biochem. Biophys. Res. Commun. 150:979-986. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M., M. Connick, and B. N. Ames. 1983. Complete analysis of tRNA-modified nucleosides by high-performance liquid chromatography: the 29 modified nucleosides of Salmonella typhimurium and Escherichia coli tRNA. Anal. Biochem. 129:1-13. [DOI] [PubMed] [Google Scholar]
- 7.Elseviers, D., L. A. Petrullo, and P. Gallaagher. 1984. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 12:3521-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esberg, B., H. C. E. Leung, H. C. T. Tsui, G. Bjork, and M. E. Winkler, 1999. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181:7256-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint, D. H. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068-16074. [PubMed] [Google Scholar]
- 10.Flint, D. H., J. F. Tuminello, and T. J. Miller. 1996. Studies on the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase in Escherichia coli crude extract. Isolation of O-acetylserine sulfhydrylases A and B and beta-cystathionase based on their ability to mobilize sulfur from cysteine and to participate in Fe-S cluster synthesis. J. Biol. Chem. 271:16053-16067. [DOI] [PubMed] [Google Scholar]
- 11.Gehrke, C. W., K. C. Kuo, R. A. McCune, and K. Gerhardt. 1982. Quantitative enzymatic hydrolysis of tRNAs. Reversed-phase high-perfirmance liquid chromatography of tRNA nucleosides. J. Chromatogr. 230:297-308. [PubMed] [Google Scholar]
- 12.Griffiths, E., J. Humphreys, A. Leach, and L. Scanlon. 1978. Alterations in the tRNAs of Escherichia coli recovered from lethally infected animals. Infect. Immun. 22:312-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kambampati, R., and C. T. Lauhon. 2000. IscS is a sulfurtransferase for the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. Biochemistry 38:16561-16568. [DOI] [PubMed] [Google Scholar]
- 15.Kambampati, R., and C. T. Lauhon. 2000. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J. Biol. Chem. 275:10727-10730. [DOI] [PubMed] [Google Scholar]
- 16.Kramer, G. F., J. C. Baker, and B. N. Ames. 1988. Near-UV stress in Salmonella typhimurium: 4-thiouridine in tRNA, ppGpp, and ApppGpp as components of an adaptive response. J. Bacteriol. 170:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauhon, C. T., and R. Kambampati. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamine, and NAD. J. Biol. Chem. 275:20096-21003. [DOI] [PubMed] [Google Scholar]
- 18.Leimkuhler, S., and K. V. Rajagopalan. 2001. A sulfurtransferase is required in the transfer of cysteine sulfur in the in vitro synthesis of molybdopterin from precursor Z in Escherichia coli. J. Biol. Chem. 276:22024-22031. [DOI] [PubMed] [Google Scholar]
- 19.Li, J., M. Kogan, S. A. B. Knight, D. Pain, and A. Dancis. 1999. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J. Biol. Chem. 274:33025-33034. [DOI] [PubMed] [Google Scholar]
- 20.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madore, E., C. Florentz, R. Giege, S. Sekine, S. Yokoyama, and J. Lapointe. 1999. Effect of modified nucleotides on Escherichia coli tRNAGlu structure and on its aminoacylation by glutamyl-tRNA synthetase. Predominant and distinct roles of the mnm5 and s2 modifications of U34. Eur. J. Biochem. 266:1128-1135. [DOI] [PubMed] [Google Scholar]
- 22.Marquet, A. 2001. Enzymology of carbon-sulfur bond formation. Curr. Opin. Chem. Biol. 5:541-549. [DOI] [PubMed] [Google Scholar]
- 23.Mihara, H., S. Kato, G. M. Lacourciere, T. C. Stadtman, R. A. Kennedy, T. Kurihara, U. Tokumoto, Y. Takahashi, and N. Esaki. 2002. The iscS gene is essential for the biosynthesis of 2-selenouridine in tRNA and the selenocysteine-containing formate dehydrogenase H. Proc. Natl. Acad. Sci. USA 99:6679-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihara, H., T. Kurihara, T. Yoshimura, K. Soda, and N. Esaki. 1997. Cysteine sulfinate desulfinase, an NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 272:22417-22424. [DOI] [PubMed] [Google Scholar]
- 25.Mihara, H., M. Maeda, T. Fujii, T. Kurihara, Y. Hata, and N. Esaki. 1999. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization, and preliminary x-ray crystallographic studies. J. Biol. Chem. 274:14768-14772. [DOI] [PubMed] [Google Scholar]
- 26.Mihara, H., T. Kurihara, T. Yoshimura, and N. Esaki. 2000. Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between l-cysteine desulfurase and l-selenocysteine lyase reactions. J. Biochem. (Tokyo) 127:559-567. [DOI] [PubMed] [Google Scholar]
- 27.Mueller, E. G., C. J. Buck, P. M. Palenchar, L. E. Barnhart, and J. L. Paulson. 1998. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res. 26:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller, E. G., P. M. Palenchar, and C. J. Buck. 2001. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem. 276:33588-33595. [DOI] [PubMed] [Google Scholar]
- 29.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierrel, F., G. R. Bjork, M. Fontecave, and M. Atta. 2002. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277:13367-13370. [DOI] [PubMed] [Google Scholar]
- 31.Ryals, J., R.-Y. Hsu, M. N. Lipsett, and H. Bremer. 1982. Isolation of single-Site Escherichia coli mutants deficient in thiamine and 4-thiouridine syntheses: identification of a nuvC mutant. J. Bacteriol. 151:899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase IscS has a major role in in vivo Fe-S cluster formation in Escherichia coli. J. Bacteriol. 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skovran, E., and D. M.Downs,. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, A. D., J. N. Agar, K. A. Johnson, J. Frazzon, I. J. Amster, D. R. Dean, and M. K. Johnson. 2001. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123:11103-11104. [DOI] [PubMed] [Google Scholar]
- 35.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization with new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, M. A., J. F. Cannon, F. H. Webb, and R. Bock. 1985. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J. Bacteriol. 161:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sylvers, L. A., K. C. Rogers, M. Shimizu, E. Ohtsuka, and D. Soll. 1993. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 32:3836-3841. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi, Y., and U. Tokumoto. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277:28380-28383. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, G., and A. Favre. 1975. 4-Thiouridine as the target for near-ultraviolet light induced growth delay in Escherichia coli. Biochem. Biophys. Res. Commun. 66:1454-1461. [DOI] [PubMed] [Google Scholar]
- 40.Tokumoto, U., and Y. Takahashi. 2001. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J. Biochem. (Tokyo) 130:63-71. [DOI] [PubMed] [Google Scholar]
- 41.Urbina, H. D., J. J. Silberg, K. G. Hoff, and L. Vickery. 2001. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276:44521-44526. [DOI] [PubMed] [Google Scholar]
- 42.Urbonavicius, J., Q. Qian, J. M. Durand, T. G. Hagervall, and G. R. Bjork. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb, E., K. Class, and D. M. Downs. 1997. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J. Bacteriol. 179:4399-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi, J., Y. Ge, C. Kinsland, F. W. McLafferty, and T. P. Begley. 2001. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein-protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc. Natl. Acad. Sci. USA 98:8513-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokoyama, S., T. Watanabe, K. Murao, H. Ishikura, Z. Yamaizumi, S. Nishimura, and T. Miyazawa. 1985. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. USA 82:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]
- 47.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Nat. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]