Abstract
Deficiency of a modified nucleoside in tRNA often mediates suppression of +1 frameshift mutations. In Salmonella enterica serovar Typhimurium strain TR970 (hisC3737), which requires histidine for growth, a potential +1 frameshifting site, CCC-CAA-UAA, exists within the frameshifting window created by insertion of a C in the hisC gene. This site may be suppressed by peptidyl-tRNAProcmo5UGG (cmo5U is uridine-5-oxyacetic acid), making a frameshift when decoding the near-cognate codon CCC, provided that a pause occurs by, e.g., a slow entry of the tRNAGlnmnm5s2UUG (mnm5s2U is 5-methylaminomethyl-2-thiouridine) to the CAA codon located in the A site. We selected mutants of strain TR970 that were able to grow without histidine, and one such mutant (iscS51) was shown to have an amino acid substitution in the l-cysteine desulfurase IscS. Moreover, the levels of all five thiolated nucleosides 2-thiocytidine, mnm5s2U, 5-carboxymethylaminomethyl-2-thiouridine, 4-thiouridine, and N-6-(4-hydroxyisopentenyl)-2-methylthioadenosine present in the tRNA of S. enterica were reduced in the iscS51 mutant. In logarithmically growing cells of Escherichia coli, a deletion of the iscS gene resulted in nondetectable levels of all thiolated nucleosides in tRNA except N-6-(4-hydroxyisopentenyl)-2-methylthioadenosine, which was present at only 1.6% of the wild-type level. After prolonged incubation of cells in stationary phase, a 20% level of 2-thiocytidine and a 2% level of N-6-(4-hydroxyisopentenyl)-2-methylthioadenosine was observed, whereas no 4-thiouridine, 5-carboxymethylaminomethyl-2-thiouridine, or mnm5s2U was found. We attribute the frameshifting ability mediated by the iscS51 mutation to a slow decoding of CAA by the tRNAGlnmnm5s2UUG due to mnm5s2U deficiency. Since the growth rate of the iscS deletion mutant in rich medium was similar to that of a mutant (mnmA) lacking only mnm5s2U, we suggest that the major cause for the reduced growth rate of the iscS deletion mutant is the lack of mnm5s2U and 5-carboxymethylaminomethyl-2-thiouridine and not the lack of any of the other three thiolated nucleosides that are also absent in the iscS deletion mutant.
tRNA from all organisms contains modified nucleosides, which are derivatives of the four normal nucleosides adenosine (A), guanosine (G), uridine (U), and cytidine (C). At present, more than 80 different modified nucleosides have been characterized (21). Thiolated nucleosides are present in tRNAs from organisms belonging to the domains Bacteria and Eucarya (2), and recently they have also been identified in tRNAs from organisms belonging to the domain Archaea (17). At present, 10 different thiolated nucleosides have been characterized in tRNAs from different organisms, and five, 2-thiocytidine (s2C), 4-thiouridine (s4U), 5-methylaminomethyl-2-thiouridine (mnm5s2U), 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U), and N-6-(4-hydroxyisopentenyl)-2-methylthioadenosine (ms2io6A), are present in tRNA from Salmonella enterica serovar Typhimurium. In Escherichia coli the same thiolated nucleosides are present, but instead of ms2io6A, tRNA from E. coli contains N-6-isopentyl-2-methylthioadenosine (ms2i6A) (7).
The sulfur source for the synthesis of the thiolated nucleosides is cysteine (1), but cysteine is also the sulfur source for a variety of cofactors, such as biotin, lipoic acid, and thiamine (4). Although the mechanism of incorporation into the various sulfur-containing molecules has been elusive, a major advance in our understanding of this process was the identification of the NifS protein from Azotobacter vinelandii as a cysteine desulfurase required for the maintenance of the metallosulfur cluster in nitrogenase (30). The NifS protein splits the cysteine into alanine and elemental sulfur, and the latter is transiently bound to a specific cysteine of NifS (29). E. coli has a similar enzyme, IscS, and its structural gene, iscS, is part of a gene cluster containing nine genes (25, 28).
The IscS protein is required for the synthesis of s4U in tRNA as well as the incorporation of sulfur into the thiazole ring of thiamine (15, 18). The sulfur of cysteine is transferred first to the IscS protein, thereby forming an IscS-SSH persulfide (SSH indicates a persulfide at a cysteine residue of IcsS), which in turn transfers the sulfur to the ThiI protein, forming a ThiI-SSH persulfide. In the presence of ATP-Mg, this modified protein transfers the sulfur to the tRNA, thus forming the s4U in position 8 of a subset of tRNAs. Alternatively, the IscS-SSH persulfide transfers the sulfur to the ThiS protein, which catalyzes the incorporation of sulfur into the thiazole ring of thiamine. The IscS protein is also involved in the synthesis of nicotinic acid, isoleucine, valine, and other Fe-S proteins (16).
Since the IscS protein is pivotal in the formation of Fe-S clusters in proteins and in the formation of s4U, the other four thiolated nucleosides present in tRNA from S. enterica may also require an Fe-S cluster protein. If so, thiolation of tRNA should be sensitive to the allelic state of the iscS gene. Indeed, the MiaB protein, which is required for the formation of the methylthio group of ms2io6A, is an Fe-S cluster protein (11, 12). Moreover, the formation of s2C, which is present in only four tRNA species in bacteria, requires an active stcA gene, the sequence of which reveals a highly conserved C-X1-X2-C motif common in the thioredoxin superfamily (G. Jäger, Q. Qian, and G R. Björk, unpublished results). However, the mnmA (asuE, trmU) gene encodes a protein required for the thiolation of mnm5s2U (24), and its sequence is similar to that of the ThiI protein but does not reveal any potential Fe-S cluster.
This article addresses the question of how and to what extent an active IscS protein is required for the synthesis of the five thiolated nucleosides present in tRNA from S. enterica.
MATERIALS AND METHODS
Bacteria and growth conditions.
The bacterial strains used were derivatives of either S. enterica serovar Typhimurium or E. coli K-12 (Table 1). As rich medium we used either Luria-Bertani (LB) (5) or NAA (Difco nutrient broth [0.8%]; Difco Laboratories, Detroit, Mich.) supplemented with the aromatic amino acids, aromatic vitamins, and adenine (8). The minimal medium was made from basal medium E (27) supplemented with 0.2% glucose and required amino acids or vitamins at the concentrations recommended earlier (8).
TABLE 1.
S. enterica and E. coli strains
| Strain | Genotype | Source or reference |
|---|---|---|
| S. enterica | ||
| TR970 | hisO1242 hisC3737 | J. Roth |
| GT1948 | his-644 (deletion of the his operon) serA790 lys-554 | J. Roth |
| GT6430 | hisO1242 hisC3737 zfh-2525::Tn10dTc | This work |
| GT6429 | hisO1242 hisC3737 zfh-2525::Tn10dTc iscS51 | This work |
| GT6408 | hisO1242 hisC3737 zcf-2524::Tn10dTc mnmA1 | This work |
| E. coli | ||
| MW100 | Wild type | Mikael Wikström |
| CL100 | ΔiscS | 16 |
| TH177 | mnmA+fadR::Tn10 | This work |
| TH178 | mnmA1 fadR::Tn10 | This work |
Genetic procedure.
Transduction with phage P22 HT105/1 (int-201) (22) and mutagenesis with nitrosoguanidine (50 μg/ml) were performed as described before (8).
DNA sequencing was performed on either chromosomal DNA or PCR products following the manual of the Applied Biosystems ABI Prism Big Dye cycle sequencing ready reaction kit.
Analysis of modified nucleosides in tRNA.
Bacterial strains were grown in LB medium at 37°C to about 4 × 108 to 6 × 108 cells/ml (100 to 150 Klett units). Cells were lysed, and total RNA was prepared (9), dissolved in buffer R200 (10 mM Tris-H3PO4 [pH 6.3], 15% ethanol, 200 mM KCl), and applied to a Nucleobond column equilibrated with the same buffer. tRNA was eluted with the same buffer except that the KCl concentration was raised to 650 mM. The tRNA was precipitated with 2.5 volumes of cold ethanol containing 1% potassium acetate, washed twice with 80% ethanol, and dried. The dried tRNA was dissolved in water, and a portion of it was degraded to nucleosides by nuclease P1, followed by treatment with bacterial alkaline phosphatase (14). The hydrolysate was analyzed by high-performance liquid chromatography (HPLC) (13).
RESULTS
Inactivation of iscS gene induces +1 frameshift errors.
We showed earlier how hypomodification of tRNA induces +1 frameshifts (26). Relevant for this study is how hypomodified tRNA may reduce the rate of A site selection and thus give the peptidyl-tRNA a longer time to make a +1 frameshift error. The hisC3737 mutation is a frameshift mutation which changes the hisC+ sequence CCC-AAU-AAU to the hisC3737 sequence CCC-CAA-UAA-U by insertion of a C (P. Chen and G. R. Björk, unpublished results; the boldface C is formally the inserted C in the mutant; the hyphens indicate the codons in the 0 frame). Thus, in the hisC3737 mutant, the ribosome will stop translating at the stop codon UAA, thereby creating a requirement for histidine. The near-cognate tRNAProcmo5UGG normally reads CCA/CCG and CCU codons due to the presence of the cmo5U modification in the wobble position. However, occasionally it may read the near-cognate codon CCC (19). Following translocation, such a peptidyl-tRNAProcmo5UGG is prone to shift frame when interacting with the near-cognate codon CCC, provided that a pause is induced by slow entry of the ternary complex at the A site codon (19). Accordingly, slow entry of a ternary complex containing a defective tRNAGlnmnm5s2UUG, which reads the A site codon CAA, may allow the error-prone peptidyl-tRNAProcmo5UGG to slip forward one nucleotide. This will reframe the ribosome into the 0 frame, and it will thereafter read codons as it does in the hisC+ strain. Thus, among His+ clones, mutations may be identified that affect the modification status of tRNAGlnmnm5s2UUG in such a way that a slow entry of this tRNA at the CAA codon would mediate a +1 frameshift by the peptidyl-tRNAProcmo5UGG.
In order to characterize mutations that suppress the +1 frameshift mutation hisC3737, we first isolated a pool of about 30,000 clones with randomly inserted Tn10dTc transposons in the chromosome of strain GT1948, which contains a deletion (his-644) of the his operon. Cells of this pool were then mutagenized with nitrosoguanidine. Phage P22 was grown on this culture, and the resulting phage stock was used to transduce various Tn10dTcs to the recipient strain TR970 (hisC3737). Among about 160,000 tetracycline-resistant (Tcr) transductants, several His+ transductants were obtained, of which one was denoted strain GT6429 (hisC3737 zfh-2525::Tn10dTc iscS51).
Phage P22 was grown on strain GT6429 and used to transduce the zfh-2525::Tn10dTc insertion to the parental strain TR970 (hisC3737). The results showed that the His+-inducing mutation (iscS51) was 97% linked to the Tn10dTc transposon. By using primers complementary to sequences within the Tn10dTc transposon, DNA sequences on both sides of the transposon were determined. Comparing these sequences with the DNA sequence of the S. enterica chromosome established that the transposon was inserted in the STM2545 gene, which is the gene immediately upstream of the isc operon. We also noticed that, compared to the parent strain, the mutant strain GT6429 (hisC3737 zfh-2525::Tn10dTc iscS51) grew well on rich medium plates but poorly on glucose minimal plates containing histidine. Growth on histidine plates was stimulated by the addition of thiamine, indicating a potential defect in the synthesis of thiamine in the mutant.
It was demonstrated earlier that inactivation of the iscS gene results in a requirement of thiamine for growth (16, 23). This fact and the close linkage between the mutation and the Tn10dTc insertion in the STM2545 gene, which is very close to iscS, prompted us to determine the sequence of the iscS gene in the mutant. Indeed, the iscS51 mutation caused a C-to-T base substitution, resulting in an amino acid change from Ala327 to Val327 in the IscS protein. Thus, this amino acid substitution in the IscS protein induced a deficiency of thiamine and suppression of the +1 frameshift mutation hisC3737.
iscS51 mutation reduces the level of all thiolated nucleosides in tRNA.
According to our model of how hypomodified tRNA induces frameshifting, we suspected that the cause of the +1 frameshift was a reduced entry rate of the ternary complex containing Gln-tRNAGlnmnm5s2UUG to the CAA codon. In the sequence CCC-CAA-UAA, the CAA codon would be in the A site when the tRNAProcmo5UGG is interacting with the CCC codon in the P site. Such slow entry of this ternary complex should stimulate the peptidyl-tRNAProcmo5UGG in the P site to frameshift (19). According to earlier results, lack of one modification of tRNAGlnmnm5s2UUG, ms2io6A, would lower the A site selection rate (26).
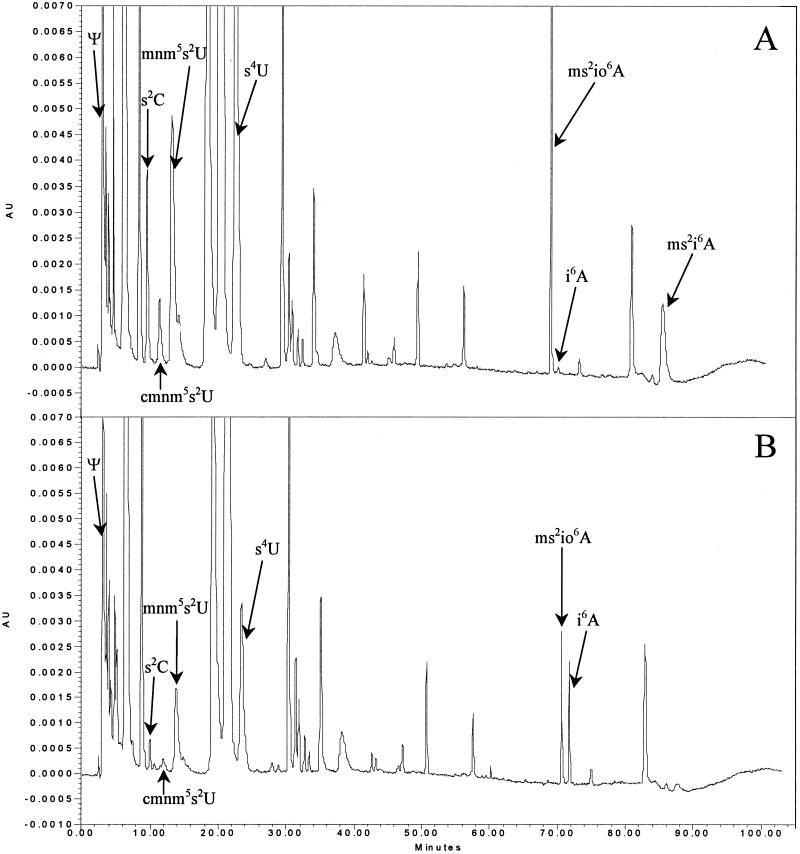
Therefore, we prepared tRNA from transductants differing only in the allelic state of the iscS gene and determined the tRNA modification pattern (Fig. 1). As expected, the iscS51 mutation decreased the level of mnm5s2U but also, surprisingly, the levels of s2C, s4U, and ms2io6A (Fig. 1; Table 1). The decreased level of mnm5s2U explains the ability to suppress the hisC3737 +1 frameshift mutation (26). The results also demonstrate that the IscS protein is required for synthesis of all the thiolated nucleosides present in the tRNA of S. enterica.
FIG. 1.
HPLC chromatograms of tRNA hydrolysates from the wild-type (A) and iscS51 mutant (B) strains. The nucleosides were monitored at 295 nm to maximize the detection of all thiolated nucleosides. mnm5s2U, cmnm5s2U, s2C, ms2io6A, s4U, ms2i6A, pseudouridine (Ψ), and N-6-isopentenyladenosine (i6A) were identified by comparing UV spectra with published spectra (13). For s2C, the molecular weight of the protonated form was determined by mass spectrometry (Jäger et al., unpublished data). AU, absorbance units.
iscS+ gene complements iscS51-mediated phenotypes.
Since the iscS51 mutation was induced following nitrosoguanidine treatment, the observed suppression of the +1 frameshift mutation hisC3737 might have been caused by a mutation other than the one found in the iscS gene. Therefore, we introduced plasmid piscS, which contains only the iscS+ gene, into strain GT6429 (hisC3737 iscS51). Indeed, this plasmid (but not the vector) complemented the +1 frameshifting phenotype and restored all the thiolated nucleosides to the wild-type level (Table 2). We conclude that the iscS51 mutation causes suppression of the +1 frameshift mutation hisC3737 and the reduced level of all thiolated nucleosides in tRNA.
TABLE 2.
Levels of thiolated nucleosides in tRNA from the iscS51 mutanta
| Strain | Relevant genotype | Plasmidb | Suppressionc of hisC3737 | Mean relative level ± SD (%)
|
||||
|---|---|---|---|---|---|---|---|---|
| s2C (247 nm) | cmnm5s2U (274 nm) | mnm5s2U (274 nm) | s4U (330 nm) | ms2i(o)6Ad (242 nm) | ||||
| GT6430 | iscS+hisC3737 | No | 0.135 ± 0.003 (100) | 0.017 ± 0.003 (100) | 0.111 ± 0.001 (100) | 1.11 ± 0.02 (100) | 0.230 ± 0.001 (100) | |
| GT6429 | iscS51 hisC3737 | Yes | 0.035 ± 0.002 (26) | 0.006 ± 0.001 (33) | 0.053 ± 0.001 (48) | 0.293 ± 0.015 (26) | 0.082 ± 0.004 (36) | |
| GT6453 | iscS51 hisC3737 | pSU19 | Yes | 0.040 ± 0.001 (29) | 0.005 ± 0.002 (31) | 0.051 ± 0.006 (46) | 0.272 ± 0.006 (24) | 0.079 ± 0.001 (34) |
| GT6452 | iscS51 hisC3737 | piscS | No | 0.133 ± 0.002 (99) | 0.015 ± 0.001 (85) | 0.133 ± 0.003 (102) | 1.12 ± 0.008 (101) | 0.228 ± 0.009 (99) |
The chromatograms were scanned at the indicated wavelengths to optimize the quantification of the five thiolated nucleosides. The levels of the various thiolated nucleosides at the indicated wavelengths are given relative to the level of ψ at 254 nm. The levels as a percentage of the wild-type level are shown in parentheses.
Plasmid pSU19 (3), which is a pACYC derivative, was the vector. piscS contains only the iscS+ gene, and its construction is described in reference 23.
Ability to suppress the hisC3737 mutation was scored as growth without histidine on agar plates containing medium E and 0.2% glucose.
The values for ms2i(o)6A are the combined values for ms2io6A and ms2i6A. A corresponding increase in i6A was noted when the level of ms2i(o)6A was decreased (see Fig. 1). 5-Methylaminomethyluridine was expected but not observed because it migrated in the crowded area between C and U.
Deletion of iscS gene abolishes synthesis of all thiolated nucleosides in tRNA.
To further verify the observation that a mutation in the iscS gene influenced the synthesis of all thiolated nucleosides in tRNA, we obtained an E. coli strain deleted for the iscS gene (16). This strain, CL100, was grown in LB medium at 37°C, and cells were harvested at a cell density of 4 × 108 cells/ml (100 Klett units; cells in logarithmic phase), after 24 h (cells at stationary phase), and after 32 h of incubation. tRNA was prepared, digested to nucleosides, and analyzed by HPLC. None of the thiolated nucleosides were detected in tRNA from logarithmically growing cells except a small amount of ms2i6A (1.6% of the wild-type level). However, tRNA from stationary-phase cells contained a small amount (20% of the wild-type level) of s2C and a small amount of ms2i6A (2% of the wild-type level; Table 3). No s4U or cmnm5s2U could be detected. Apparently, an inefficient IscS-independent pathway exists, at least for the formation of s2U and the methylthio group of ms2i6A. Clearly, a functional IscS protein is required for the efficient synthesis of all thiolated nucleosides in tRNA.
TABLE 3.
In-frame deletion of the iscS gene abolishes the synthesis of all four thiolated nucleosides in logarithmically growing cellsa
| Strain | Relevant genotype | Time of harvest | Relative level (% of ψ)
|
|||
|---|---|---|---|---|---|---|
| s2C (247 nm) | (c)mnm5s2U (274 nm) | s4U (330 nm) | ms2i(o)6A (242 nm) | |||
| E. coli MW100 | iscS+ | 100 Klett units | 100 (0.13) | 100 (0.10) | 100 (0.98) | 100 (0.19) |
| iscS+ | 24 h | 109 | 126 | 132 | 92 | |
| iscS+ | 32 h | 106 | 121 | 130 | 85 | |
| E. coli CL100 | ΔiscS | 100 Klett units | <9 | <0.4 | <0.04 | 1.6 |
| ΔiscS | 24 h | 22 | <0.4 | <0.04 | 2.0 | |
| ΔiscS | 32 h | 22 | <0.4 | <0.04 | 2.0 | |
Cells were grown in LB medium and harvested at 100 Klett units (4 × 108 cells) and after 24 or 32 h of incubation. tRNA was prepared, degraded to nucleosides, and analyzed by HPLC. Quantification of the various thiolated nucleosides is described in Table 2, footnote a, and they are expressed relative to the level in tRNA from the wild type (100%). (c)mnm5s2U represents the combined level of mnm5s2U and cmnm5s2U.
Slow growth induced by deletion of iscS is likely caused by the lack of mnm5s2U
The GT6429 (iscS51) mutant grows like the wild-type strain in rich medium (Table 4). This is in sharp contrast to the growth rate reduction observed for the iscS deletion strain of E. coli (Table 2), although the iscS deletion strain was also deficient in all thiolated nucleosides. Interestingly, the growth rate reduction caused by a mutation in the mnmA gene of both E. coli and S. enterica was similar to that induced by a deletion of the iscS gene (Table 4), and this growth reduction was correlated to the nondetectable level of (c)mnm5s2U. We suggest that the major cause of the growth rate reduction in the iscS deletion strains is the deficiency of (c)mnm5s2U and not the lack of any other thiolated nucleoside in tRNA.
TABLE 4.
Growth rate reduction is correlated to mnm5s2U deficiencya
| Strain | Relevant genotype | cmnm5s2U34 and mnm5s2U34 (% of wild type) | Growth rate k (h−1) in rich medium (% reduction in k) |
|---|---|---|---|
| S. enterica GT6409 | Wild type | 100 | 1.37 |
| S. enterica GT6429 | iscS51 | 26 | 1.23 (−10) |
| S. enterica GT6408 | mnmA1 | <0.4 | 0.79 (−42) |
| E. coli TH177 | Wild type | 100 | 1.32 |
| E. coli CL100 | ΔiscS | <0.4 | 0.62 (−53) |
| E. coli TH178 | mnmA1 | <0.4 | 0.80 (−41) |
Growth rates are expressed as the specific growth rate constant k, which is ln2/mass doubling time in hours. The reduction in k was calculated according to [(kGT6429 or GT6408 − kGT6409)/kGT6409] × 100 (S. enterica strains) or [(kCL100 or TH178 − kTH177)/kTH177] × 100 (E. coli strains).
DISCUSSION
We show here that an altered IscS protein influences reading frame maintenance and results in a reduced level of all thiolated nucleosides in tRNA. Our results also demonstrate that the l-cysteine desulfurase IscS is pivotal in the synthesis of all thiolated nucleosides in tRNA of S. enterica serovar Typhimurium and E. coli.
In tRNA from logarithmically growing strain CL100 (ΔiscS) cells, no (c)mnm5s2U or s4U was detected, whereas a low level of ms2i6A was apparent. Although we did not detect any s2C, it might be present at a similarly low level, since another compound migrated very close to s2C. Indeed, upon extended incubation of the cells in stationary phase, the presence of s2C was apparent (Table 3), but (c)mnm5s2U and s4U were still not detected. These results suggest that there is another, inefficient route to synthesizing s2C and ms2i6A that is not involved in the synthesis of s4U and (c)mnm5s2U.
In the synthesis of s4U, the sulfur is delivered from IscS to the ThiI protein, which in turn transfers sulfur to the tRNA and thereby forms s4U. The sequence of MnmA is similar to that of ThiI, suggesting that MnmA may donate sulfur to the tRNA, similar to ThiI. Thus, whereas the only route of sulfur transfer in the synthesis of (c)mnm5s2U and s4U is through the IscS pathway, an alternative path to transferring sulfur in the synthesis of s2C and ms2i6A exists. Alternatively, in the synthesis of these two thiolated nucleosides, a protein other than IscS may be the immediate donor of sulfur to the enzyme catalyzing the transfer of sulfur to the tRNA. In this alternative pathway, some of the other two desulfurases present in bacteria (CsdA and SufS [CsdB]) may be involved.
The iscS51 mutation was selected as a suppressor of the +1 frameshift mutation hisC3737. Moreover, this mutation also reduced the level of all thiolated nucleosides in tRNA, suggesting that the lack of all or one of those was the cause for the +1 frameshift suppressor phenotype. We suggest that the mnm5s2U deficiency in tRNAGlnmnm5s2UUG causes the suppressing phenotype for the following reasons. (i) An aroD mutation abolishes synthesis of cmo5U34 (6), including the one present in the tRNAProcmo5UGG. The frameshifting activity mediated by the iscS51 mutation is inhibited by the introduction of an aroD mutation (data not shown), demonstrating that a peptidyl-tRNAProcmo5UGG interacting with a CCC codon in the P site causes the frameshifting event (19).
There are two sites, CCC-GCG and CCC-CAA, within the 32-codon-long frameshifting window caused by the hisC3737 mutation where such a frameshifting event can occur (P. Chen and G. R. Björk, unpublished results). Although we cannot rule out that the frameshifting occurs at the CCC-GCG site caused by slow entry of the s4U-deficient tRNAcmo5UGCAla, we favor the CCC-CAA-UAA site, since we showed earlier that slow entry of the tRNAGlnmnm5s2UUG to the CAA codon caused by mnm5s2U deficiency in this tRNA results in frameshifting (26). (ii) The selection procedure used to isolate the iscS51 mutant also resulted in isolation of an mnmA mutant, which is deficient only in mnm5s2U (unpublished results). s2C and ms2io6A are not present in any of the tRNAs that would induce a frameshifting event at these two potential frameshifting sites.
The difference in growth rate between the point mutant iscS51 of S. enterica and the ΔiscS mutant of E. coli may be caused by a more severe reduction in sulfur metabolism in the ΔiscS strain than in the iscS51 strain. Alternatively, it may be caused by a growth reduction caused by a lower level of the thiolated nucleosides in the deletion strain. However, we know that complete lack of s2C does not induce any growth defects (G. Jäger, Q. Qian, and G. R. Björk, unpublished results), nor does lack of s4U or the methylthio group of ms2io6A (10, 20). Therefore, the severe growth reduction caused by a deletion of the iscS gene compared to the slight reduction of the growth rate mediated by the iscS51 mutation could be due to the difference in the level of mnm5s2U.
A mutation in either the S. enterica or the E. coli mnmA gene abolished the synthesis of mnm5s2U similarly to that in the ΔiscS mutant of E. coli (Table 2). Such a mutation caused an extensive reduction of the growth rate similar to that induced by the ΔiscS mutation. Since the growth reduction seems to be correlated to the level of mnm5s2U irrespective of whether this was caused by mutations in the iscS gene or in the mnmA gene, we suggest that the major cause for the severe reduction in growth rate in rich medium by a deletion of the iscS gene is the reduction of the level of mnm5s2U. Since the IscS protein is involved in the maintenance of several Fe-S proteins, the most pivotal role with respect to the growth rate in rich medium should be the sulfur transfer involved in the synthesis of mnm5s2U.
Acknowledgments
This work was supported by grants from the Swedish Cancer Foundation (Project 680) and Swedish Science Research council (Project BU-2930).
We are grateful for the generous gift of plasmid piscS from Diana Downs, University of Wisconsin, Madison, and strain CL100 (ΔiscS) from Charles Lauhon, University of Wisconsin, Madison. We thank Kerstin Jacobsson for skillful analysis of modified nucleosides by HPLC.
REFERENCES
- 1.Ajitkumar, P., and J. D. Cherayil. 1988. Thionucleosides in transfer ribonucleic acid: diversity, structure, biosynthesis, and function. Microbiol. Rev. 52:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auffinger, P., E. Westhof. 1998. Location and distribution of modified nucleotides in tRNA, p. 569-576. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 3.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 4.Begley, T. P., J. Xi, C. Kinsland, S. Taylor, and F. McLafferty. 1999. The enzymology of sulfur activation during thiamin and biotin biosynthesis. Curr. Opin. Chem. Biol. 3:623-629. [DOI] [PubMed] [Google Scholar]
- 5.Bertani, G. 1951. Studies on lysogenesis. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björk, G. R. 1980. A novel link between the biosynthesis of aromatic amino acids and transfer RNA modification in Escherichia coli. J. Mol. Biol. 140:391-410. [DOI] [PubMed] [Google Scholar]
- 7.Buck, M., J. A. McCloskey, B. Basile, and B. N. Ames. 1982. cis-2-Methylthio-ribosylzeatin (N-6-(4-hydroxyisopentenyl)-2-methylthioadenosine) is present in the transfer RNA of Salmonella typhimurium, but not Escherichia coli. Nucleic Acids Res. 10:5649-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Emilsson, V., and C. G. Kurland. 1990. Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J. 9:4359-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esberg, B., and G. R. Björk. 1995. The methylthio group (ms2) of N6-(4-hydroxyisopentenyl)-2-methylthioadenosine (ms2io6A) present next to the anticodon contributes to the decoding efficiency of the tRNA. J. Bacteriol. 177:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esberg, B., H. C. Leung, H. C. Tsui, G. R. Björk, and M. E. Winkler. 1999. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181:7256-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabien, P., G. R. Björk, M. Fontecave, and M. Atta. 2002. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277:13367-13370. [DOI] [PubMed] [Google Scholar]
- 13.Gehrke, C. W., and K. C. Kuo. 1990. Ribonucleoside analysis by reversed-phase high performance liquid chromatography, p. A3-A71. In C. W. Gehrke and K. C. T. Kuo (ed.), Chromatography and modification of nucleosides, part A: analytical methods for major and modified nucleosides. Elsevier, Amsterdam, The Netherlands.
- 14.Gehrke, C. W., K. C. Kuo, R. A. McCune, K. O. Gerhardt, and P. F. Agris. 1982. Quantitative enzymatic hydrolysis of tRNAs: reversed-phase high-performance liquid chromatography of tRNA nucleosides. J. Chromatogr. 230:297-308. [PubMed] [Google Scholar]
- 15.Kambampati, R., and C. T. Lauhon. 2000. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of s4U in Escherichia coli tRNA. J. Biol. Chem. 275:10727-10730. [DOI] [PubMed] [Google Scholar]
- 16.Lauhon, C. T., and R. Kambampati. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of s4U, thiamine, and NAD. J. Biol. Chem. 275:20096-20103. [DOI] [PubMed] [Google Scholar]
- 17.McCloskey, J. A., D. E. Graham, S. Zhou, P. F. Crain, M. Ibba, J. Konisky, D. Soll, and G. J. Olsen. 2001. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res. 29:4699-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palenchar, P. M., C. J. Buck, H. Cheng, T. J. Larson, and E. G. Mueller. 2000. Evidence that ThiI, an enzyme shared between thiamin and s4U biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J. Biol. Chem. 275:8283-8286. [DOI] [PubMed] [Google Scholar]
- 19.Qian, Q., J. N. Li, H. Zhao, T. G. Hagervall, P. J. Farabaugh, and G. R. Björk. 1998. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol. Cell 1:471-482. [DOI] [PubMed] [Google Scholar]
- 20.Ramabhadran, T. V., T. Fossum, and J. Jagger. 1976. Escherichia coli mutant lacking s4U in its transfer ribonucleic acid. J. Bacteriol. 128:671-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozenski, J., P. F. Crain, and J. A. McCloskey. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27:196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 23.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan, M. A., J. F. Cannon, F. H. Webb, and R. M. Bock. 1985. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J. Bacteriol. 161:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi, Y., and M. Nakamura. 1999. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. (Tokyo) 126:917-926. [DOI] [PubMed] [Google Scholar]
- 26.Urbonavicius, J., Q. Qian, J. M. Durand, T. G. Hagervall, and G. R. Björk. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 28.Zheng, L., V. L. Cash, D. H. Flint, and D. R. Dean. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273:13264-13272. [DOI] [PubMed] [Google Scholar]
- 29.Zheng, L., R. H. White, V. L. Cash, and D. R. Dean. 1994. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33:4714-4720. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NifS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]