Abstract
We developed a transcript profiling methodology to elucidate expression patterns of the cyanobacterium Synechocystis sp. strain PCC 6803 and used the technology to investigate changes in gene expression caused by irradiation with either intermediate-wavelength UV light (UV-B) or high-intensity white light. Several families of transcripts were altered by UV-B treatment, including mRNAs specifying proteins involved in light harvesting, photosynthesis, photoprotection, and the heat shock response. In addition, UV-B light induced the stringent response in Synechocystis, as indicated by the repression of ribosomal protein transcripts and other mRNAs involved in translation. High-intensity white light- and UV-B-mediated expression profiles overlapped in the down-regulation of photosynthesis genes and induction of heat shock response but differed in several other transcriptional processes including those specifying carbon dioxide uptake and fixation, the stringent response, and the induction profile of the high-light-inducible proteins. These two profile comparisons not only corroborated known physiological changes but also suggested coordinated regulation of many pathways, including synchronized induction of D1 protein recycling and a coupling between decreased phycobilisome biosynthesis and increased phycobilisome degradation. Overall, the gene expression profile analysis generated new insights into the integrated network of genes that adapts rapidly to different wavelengths and intensities of light.
While sunlight provides the energy for life, intermediate-wavelength UV light (UV-B) and long-wavelength UV light (UV-A) injure many organisms. UV-B (280- to 320-nm-wavelength) light generates radicals that damage proteins, nucleic acids, lipids, and the photosynthetic apparatus. The latter damage appears in the form of impaired photosystems I and II, decreased oxygen evolution and CO2 fixation, inactivation of ATPase activity, reduction in chlorophyll content, and decreased biomass (33). Notably, the D1 protein of the light energy converting complex photosystem II is quite sensitive to UV-B light (10). Although less harmful than UV-B irradiation, UV-A irradiation (320- to 400-nm-wavelength light) also damages proteins, nucleic acids, lipids, and photosystems (11). UV-A irradiation leads to the intramolecular cross-linking of several tRNA species (2), which results in poor aminoacylation and triggering of the stringent response—a phenomenon that reduces the rate of stable RNA, ribosome, and translation factor synthesis, thus arresting growth (32). Higher fluxes of white light also cause a loss of photosynthetic productivity (photoinhibition) (30).
In cyanobacteria, more than 99% of UV-B is absorbed by chlorophyll-binding proteins and the light-harvesting complexes (phycobilisomes) (20). Carotenoids protect cells against photooxidative damage by absorbing triplet state energy from chlorophyll and quenching singlet state oxygen (15). In response to changes in light quantity or quality, cyanobacteria modulate the abundance of chlorophyll, phycobilisomes, and carotenoids and generate antioxidants and enzymes involved in radical scavenging (33). Upon irradiation with UV, chlorophyll levels decrease (33), while carotenoid content increases (8). Phycobilisomes adapt to different spectral qualities of light by adjusting their size and content (18). In addition, phycobilisomes are also used by cyanobacteria as nutrient reserves (4).
The photosystem II reaction center polypeptides D1 and D2 are involved in a degradation-resynthesis cycle. Photodamaged D1 and D2 polypeptides are degraded and replaced by newly synthesized polypeptides. Both psbA2 and psbA3 transcripts specifying the identical D1 polypeptide in the cyanobacterium Synechocystis sp. strain PCC 6803 are elevated significantly by UV-B irradiation (23). The recycling of damaged D1 polypeptide is likely to involve multiple enzymes, of which the FtsH protease has been identified (22).
Upon exposure to intense white light, cyanobacteria accumulate transcripts of several proteins that belong to the high-light-inducible protein (HLIP) family (13). HLIPs are present in cyanobacteria and plants and belong to the superfamily that also includes plant chlorophyll a or b-binding proteins and early light-induced proteins (7). HLIPs and early light-induced proteins have been proposed to protect photosynthetic pigments from photooxidative damage (24).
Although much is known about the physiological changes of photosynthetic organisms in response to UV-B and white light, little is known about the genetic regulatory networks that enable the organisms to respond to such environmental changes. Recently, DNA microarray technology has become a powerful tool for studying gene expression and regulation at the genomic level (5). Such comprehensive methods have been extended to prokaryotic organisms (27). The complete genomic sequence of Synechocystis sp. strain PCC 6803, a unicellular cyanobacterium, has been determined (16). It is composed of 3,168 open reading frames (ORFs), among which are genes involved in photosynthesis and other cellular processes highly homologous to those of photosynthetic algae and plants. We sought to characterize the global mRNA profiles of Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light.
MATERIALS AND METHODS
PCR amplification of Synechocystis sp. strain PCC 6803 ORFs and construction of a DNA microarray.
Synechocystis genomic DNA was a gift from Dexter Chisholm (DuPont Central Research and Development). ORF definitions and gene functional categorizations were according to the genomic database of Synechocystis sp. strain PCC 6803 (CyanoBase [http://www.kazusa.or.jp/cyano/cyano.html]). PCR primers were designed for the amplification of each Synechocystis ORF on the basis of a primer design program for bacterial genomes (Shiping Zhang, DuPont Central Research and Development, personal communication). The forward primer contains 5′-GGCGCGCCAAGGTCTCAC-3′, followed by the coding sequence starting at the first base of an ORF. The reverse primer starts with 5′-CTCTCGAGAAGGCGCGCC-3′, followed by the complementary sequence starting at the last base of the ORF or at 1,500 bp from the start codon if the ORF size is longer than 1.5 kb. The lengths of the primers are adjusted to give a melting temperature value of 68°C. Oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, Tex.). Each PCR mixture included 60 ng of Synechocystis genomic DNA as a template, 1.0 μM (each) PCR primer, 250 μM (each) of the four deoxynucleoside triphosphates (Amersham Pharmacia Biotech), and AmpliTaq DNA polymerase (1.0 U) (Perkin-Elmer, Boston, Mass.) in a 50-μl reaction mixture volume. PCR products were purified by using a QiaQuick 96 PCR purification kit (Qiagen, Valencia, Calif.) and analyzed by agarose gel electrophoresis (29), and the molecular weights of the DNA bands were estimated using the Eagle Eye Imaging system (Stratagene, La Jolla, Calif.) and compared to their expected sizes. Those PCR products that gave single bands and had estimated sizes within 10% of their expected lengths were included in the collection of ORFs to be arrayed. Failed PCRs were repeated using TaKaRa Taq DNA polymerase (Panvera, Madison, Wis.). Using the two polymerases, 3,117 (98%) ORFs were successfully amplified by PCR.
PCR samples dissolved in 5 M NaSCN (0.05 to 0.2 μg/μl) were spotted in duplicate onto microarray glass slides (Molecular Dynamics, Mountain View, Calif.) using a Molecular Dynamics GenIII DNA microarray spotter. Detailed methods for preparing glass slides have been described elsewhere (34).
Light source.
Synechocystis sp. strain PCC 6803 cultures were illuminated with white light from cool fluorescent light tubes. The UV-B light source was an 8-W UV lamp (UVM-28; Ultra-Violet Products, Upland, Calif.) which gives UV-B light at a wavelength of 290 to 330 nm. Residual UV-C light with a wavelength of <290 nm was effectively blocked by the plastic petri dish covers (VWR, Pittsburgh, Pa.) used in all experiments. The white and UV-B light penetrating through the petri dish cover was measured using a LI-250 light meter (Li-COR Inc., Lincoln, Nebr.) for white light and a UVX-31 radiometer (Ultra-Violet Products) for UV-B light.
Strains and methods.
Synechocystis sp. strain PCC 6803 cells were grown with illumination of 25 microeinsteins of white light m−2 s−1 in BG-11 medium (28) at 30°C, with shaking at 100 rpm. Fifty milliliters of a Synechocystis culture grown to mid-logarithmic phase (optical density at 730 nm, 0.8 to 1.0) was divided into two 25-ml portions, and each portion was transferred to a plastic petri dish (150 by 15 mm) with the lid on. One petri dish was shaken at 100 rpm with illumination of 25 microeinsteins of white light m−2 s−1 and UV-B light of the desired intensity (20 microeinsteins m−2 s−1 for low UV-B and 60 microeinsteins m−2 s−1 for high UV-B). The second petri dish containing the other portion was shaken at 100 rpm under 25 microeinsteins of white light m−2 s−1 without UV-B irradiation. White light illumination was performed at 200 microeinsteins m−2 s−1. The trace level of UV-B irradiation in 200 microeinsteins of white light m−2 s−1 was about 1.0 microeinstein m−2 s−1.
RNA isolation and fluorescence-labeled cDNA synthesis.
RNA isolation and fluorescence-labeled cDNA synthesis protocols were adapted from methods optimized for Escherichia coli (34). Immediately after the UV-B or high-intensity white light treatment, Synechocystis cells were cooled on ice and centrifuged at 3,200 × g for 5 min at 4°C. Total RNA samples were isolated using a Qiagen RNeasy mini-kit and digested with RNase-free DNase (Qiagen). Microarray analysis was performed using cDNA derived from total RNA isolations. Cyanine-3 (Cy3) and cyanine-5 (Cy5) fluorescent dye-labeled cDNA probes were prepared individually from 6-μg total RNA samples isolated from both control and treated cultures in reverse transcription reactions primed by random hexanucleotides. The purified probes were quantified by measuring the absorbance at 260 nm (RNA yield), 550 nm (Cy3 dye incorporation), and 650 nm (Cy5 dye incorporation). cDNA probes with equal amounts of incorporated labels (100 to 200 pmol) were dried under vacuum and redissolved in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 50% formamide, 0.1% sodium dodecyl sulfate, 0.03 mg of salmon sperm DNA per ml) (Gibco BRL, Gaithersburg, Md.).
Hybridization to microarray slides and data analysis.
For the RNA sample from each Synechocystis culture, two hybridization reactions were performed. The first reaction used equal amounts (typically 100 to 200 pmol of florescent dye) of Cy5-labeled cDNA from a treated sample and Cy3-labeled cDNA synthesized from a control sample; the second hybridization used reciprocally labeled cDNAs. It has been found previously (34) that a skew of signal intensities introduced by the differences in Cy3 and Cy5 florescent dyes can be eliminated by swapping the dyes in two separate hybridization reactions and averaging the signals from both data sets. Because each spotted slide contains a near-complete set of spotted ORFs of the Synechocystis genome, four independent measures of each mRNA in the RNA samples were performed in each two-slide hybridization experiment.
Two separate Synechocystis cultures were used in the UV-B treatment experiments with 20 microeinsteins of UV-B irradiation m−2 s−1 and an exposure period of 20 min, which resulted in two pairs of control and treated RNA samples and eight independent measurements. Similarly, eight independent measurements were obtained for 2 h of treatment with 20 microeinsteins of UV-B m−2 s−1. In both experiments, data analysis showed that the duplicate RNA samples gave essentially the same transcription profile. For the UV-B irradiation at 60 microeinsteins m−2 s−1 and intense white light treatment at 200 microeinsteins m−2 s−1, one pair of control and treated RNA samples was used in each experiment to generate four independent measurements of each transcript.
Methods for hybridization and scanning of microarray slides have been described previously (34). The hybridized and washed slides were scanned using a Molecular Dynamics laser scanner for imaging of Cy3- and Cy5-labeled cDNA probes, and the images were analyzed using Array Vision 4.0 software (Molecular Dynamics). The fluorescence signal of each spot above the local background level was calculated before being summed, and the sums were normalized, assuming that the total transcript level in the cell remains unchanged after irradiation. The relative expression level of each ORF was determined by dividing the fluorescence signal associated with each ORF by the fluorescence signals of the entire set of ORFs in the array.
The expression ratio of each transcript (relative expression level from the treated sample versus relative expression level of the control sample [treated/control ratio]) was calculated for each pair of RNA samples. The average expression ratios were calculated from the multiple measurements and listed in Tables 1 and 2. A treated versus control ratio of >1 indicates elevated mRNA abundance, and a ratio of <1 indicates reduced mRNA abundance. The standard deviations of the multiple measurements from single RNA samples or duplicate RNA samples were included in Tables 1 and 2. An average induction or repression ratio of 2 (treated/control ratio above 2 or below 0.5) was used as an arbitrary cutoff to assess whether a particular gene's expression level had changed. Genes with expression levels that did not change (expression ratio between 2 and 0.5) in all four experiments are not included in Tables 1 and 2. DNA microarrays cannot yet accurately measure very low abundance transcripts, and relative expression levels of less than 10% of average expression level were assumed to be 10% in order to reduce errors in expression ratios caused by inaccurate measurement of very low signals.
TABLE 1.
Transcripts induced in UV-B and intense white light irradiation experimentsa
| Category and function | Gene | ORF | Treated/control ratiob
|
|||
|---|---|---|---|---|---|---|
| UV low, 20 min | UV low, 2 h | UV high, 2 h | White light, 2 h | |||
| Amino acid biosynthesis | ||||||
| Acetolactate synthase | ilvB | sll1981 | 1.38 ± 0.22 | 1.35 ± 0.26 | 2.00 ± 0.34 | 1.93 ± 0.28 |
| Cofactor biosynthesis | ||||||
| Geranylgeranyl pyrophosphate synthase | crtE | slr0611 | 1.8 ± 0.55 | 1.82 ± 0.17 | 5.11 ± 1.08 | 1.57 ± 0.27 |
| Phytoene desaturase | crtP | slr1254 | 1.96 ± 0.95 | 2.68 ± 0.87 | 4.33 ± 2.52 | 4.67 ± 1.44 |
| ζ-Carotene desaturase | crtQ | slr0940 | 1.42 ± 0.51 | 1.82 ± 0.33 | 2.29 ± 0.76 | 2.51 ± 0.63 |
| Ferrochelatase | hemH | slr0839 | 1.27 ± 0.36 | 1.47 ± 0.29 | 2.2 ± 0.81 | 1.52 ± 0.18 |
| UbiH protein | ubiH | slr1300 | 1.63 ± 0.24 | 1.7 ± 0.18 | 3.45 ± 1.08 | 2.03 ± 0.46 |
| GTP cyclohydrolase II | ribA | sll1894 | 1.51 ± 0.26 | 1.69 ± 0.33 | 3.09 ± 1.49 | 1.36 ± 0.32 |
| Glutaredoxin 3 | grxC | ssr2061 | 1.1 ± 0.23 | 1.35 ± 0.13 | 1.98 ± 0.42 | 1.32 ± 0.26 |
| Thioredoxin M | trxM | slr0233 | 1.06 ± 0.21 | 1.14 ± 0.49 | 2.01 ± 0.52 | 1.23 ± 0.19 |
| Glutathione peroxidase | slr1171 | 1.48 ± 0.55 | 1.61 ± 0.53 | 3.69 ± 1.63 | 1.78 ± 0.77 | |
| Glutathione peroxidase | slr1992 | 2.55 ± 0.81 | 1.98 ± 0.35 | 3.03 ± 0.64 | 3.9 ± 1.17 | |
| Cell envelope | ||||||
| Rare lipoprotein A | rlpA | slr0423 | 0.9 ± 0.16 | 0.84 ± 0.29 | 0.72 ± 0.34 | 3.79 ± 0.92 |
| Penicillin-binding protein 4 | pbp | sll1167 | 2.16 ± 0.34 | 1.26 ± 0.39 | 2.08 ± 0.2 | 1.01 ± 0.27 |
| Cellular processes | ||||||
| Cell division protein FtsH | ftsH | slr1390 | 2.22 ± 0.54 | 1.75 ± 0.34 | 3.39 ± 1.29 | 1.89 ± 0.68 |
| Cell division protein FtsH | ftsH | slr0228 | 3.26 ± 0.97 | 2.52 ± 0.38 | 5.3 ± 2.69 | 1.84 ± 0.65 |
| Cell division protein FtsH | ftsH | slr1604 | 3.63 ± 1.12 | 2.74 ± 0.52 | 6.07 ± 3.03 | 2.17 ± 0.8 |
| Cell division protein FtsH | ftsH | sll1463 | 1.25 ± 0.10 | 1.33 ± 0.23 | 2.03 ± 0.4 | 1.45 ± 0.24 |
| MAF | maf | sll0905 | 1.34 ± 0.3 | 1.45 ± 0.43 | 2.09 ± 0.55 | 1.05 ± 0.05 |
| DnaJ | dnaJ | sll0897 | 1.72 ± 0.25 | 1.49 ± 0.23 | 3.22 ± 1.03 | 1.63 ± 0.15 |
| DnaJ | dnaJ | slr0093 | 2.13 ± 0.35 | 1.31 ± 0.4 | 2.35 ± 0.64 | 1.53 ± 0.22 |
| DnaK protein | dnaK | sll1932 | 2.08 ± 0.37 | 1.54 ± 0.25 | 2.19 ± 0.81 | 1.63 ± 0.31 |
| DnaK protein | dnaK | sll0170 | 3.48 ± 0.64 | 1.72 ± 0.34 | 3.89 ± 1.29 | 1.94 ± 0.52 |
| 60-kDa chaperonin 1 | groEL | slr2076 | 2.25 ± 0.87 | 1.6 ± 0.63 | 1.82 ± 1.09 | 4.33 ± 2.42 |
| 60-kDa chaperonin 2 | groEL-2 | sll0416 | 2.76 ± 1.01 | 1.87 ± 0.64 | 3.74 ± 2.08 | 3.2 ± 1.26 |
| 10-kDa chaperonin | groES | slr2075 | 2.27 ± 0.55 | 1.53 ± 0.47 | 3.23 ± 1.64 | 3.94 ± 1.8 |
| Heat shock protein | htpG | sll0430 | 3.73 ± 1.42 | 1.93 ± 0.74 | 6.23 ± 2.89 | 1.85 ± 0.8 |
| Small heat shock protein | hsp17 | sll1514 | 7.8 ± 4.18 | 3.52 ± 3.49 | 21.49 ± 11.89 | 2.89 ± 0.81 |
| Superoxide dismutase | sodB | slr1516 | 2.59 ± 0.52 | 1.31 ± 0.42 | 6.14 ± 3.38 | 1.81 ± 0.5 |
| Energy metabolism | ||||||
| P protein of glycine cleavage complex | gcvP | slr0293 | 1.51 ± 0.32 | 1.34 ± 0.12 | 2.2 ± 0.34 | 2.11 ± 0.25 |
| Glucose dehydrogenase B | gdhB | slr1608 | 1.66 ± 0.36 | 1.65 ± 0.11 | 2.89 ± 0.77 | 1.49 ± 0.27 |
| Mannose-1-phosphate guanyltransferase | sll1558 | 1.46 ± 0.29 | 1.72 ± 0.37 | 3.51 ± 1.03 | 1.42 ± 0.18 | |
| 3-Ketoacyl-acyl carrier protein reductase | fabG | sll0330 | 2.47 ± 1.1 | 1.31 ± 0.25 | 4.48 ± 1.84 | 5.15 ± 2.22 |
| Squalene-hopene-cyclase | shc | slr2089 | 1.72 ± 0.4 | 1.5 ± 0.24 | 1.95 ± 0.66 | 2.50 ± 0.88 |
| Photosynthesis and respiration | ||||||
| Ribulose bisphosphate carboxylase large subunit | rbcL | slr0009 | 0.69 ± 0.26 | 0.6 ± 0.27 | 0.26 ± 0.15 | 2.1 ± 1.22 |
| Ribulose bisphosphate carboxylase small subunit | rbcS | slr0012 | 0.65 ± 0.22 | 0.69 ± 0.34 | 0.2 ± 0.11 | 2 ± 0.93 |
| Chaperone | rbcX | slr0011 | 0.7 ± 0.31 | 0.79 ± 0.42 | 0.2 ± 0.13 | 2.12 ± 1.11 |
| Bicarbonate transporter | cmpA | slr0040 | 1.59 ± 0.70 | 1.31 ± 1.01 | 1.93 ± 1.48 | 7.74 ± 4.48 |
| Bicarbonate transporter | cmpB | slr0041 | 1.18 ± 0.45 | 0.95 ± 0.21 | 1.54 ± 1.14 | 4.79 ± 1.41 |
| Bicarbonate transporter | cmpC | slr0043 | 1.34 ± 0.28 | 1.34 ± 0.75 | 1.44 ± 0.97 | 2.88 ± 1.19 |
| CO2-concentrating mechanism protein CcmK | ccmK | sll1028 | 0.7 ± 0.25 | 0.74 ± 0.23 | 0.36 ± 0.1 | 2.47 ± 0.9 |
| CO2-concentrating mechanism protein CcmK | ccmK | sll1029 | 0.71 ± 0.2 | 0.71 ± 0.22 | 0.38 ± 0.05 | 2.52 ± 0.75 |
| CO2-concentrating mechanism protein CcmL | ccmL | sll1030 | 0.8 ± 0.21 | 0.71 ± 0.19 | 0.51 ± 0.24 | 2.56 ± 0.19 |
| CO2-concentrating mechanism protein CcmM | ccmM | sll1031 | 0.81 ± 0.2 | 0.73 ± 0.12 | 0.34 ± 0.02 | 2.67 ± 0.42 |
| NADH dehydrogenase | ndhB | slr1743 | 1.34 ± 0.24 | 1.41 ± 0.22 | 2.17 ± 0.54 | 2.13 ± 0.67 |
| NADH dehydrogenase subunit 4 | ndhD3 | sll1733 | 0.81 ± 0.32 | 0.68 ± 0.27 | 0.43 ± 0.07 | 5.88 ± 3.02 |
| NADH dehydrogenase subunit 4 | ndhD | slr2007 | 0.89 ± 0.16 | 1.00 ± 0.33 | 0.95 ± 0.48 | 3.49 ± 0.94 |
| NADH dehydrogenase subunit 4 | ndhD2 | slr1291 | 1.81 ± 0.47 | 1.62 ± 0.21 | 2.37 ± 1.00 | 6.9 ± 3.65 |
| NADH dehydrogenase subunit 5 | ndhF | sll1732 | 0.74 ± 0.29 | 0.66 ± 0.17 | 0.57 ± 0.12 | 4.89 ± 2.23 |
| Photosystem II D1 protein | psbA2 | slr1311 | 4.41 ± 1.54 | 4.04 ± 1.27 | 5.89 ± 3.82 | 2.57 ± 1.82 |
| Photosystem II D1 protein | psbA3 | sll1867 | 4.04 ± 0.67 | 3.85 ± 0.57 | 5.46 ± 3.02 | 2.26 ± 0.98 |
| Phycobilisome degradation protein NblA | nblA | ssl0452 | 1.97 ± 0.39 | 2.13 ± 1.11 | 4.2 ± 1.77 | 1.2 ± 0.51 |
| Phycobilisome degradation protein NblA | nblA | ssl0453 | 1.5 ± 0.24 | 1.36 ± 0.27 | 2.26 ± 1.18 | 1.14 ± 0.29 |
| Phycobilisome degradation protein NblB | nblB | slr1687 | 3.9 ± 0.61 | 2.73 ± 0.79 | 19.91 ± 6.33 | 1.72 ± 0.41 |
| Cytochrome CytM | cytM | sll1245 | 1.35 ± 0.21 | 1.36 ± 0.14 | 1.97 ± 0.68 | 1.21 ± 0.21 |
| Plastocyanin | petE | sll0199 | 0.95 ± 0.12 | 1.03 ± 0.18 | 1.97 ± 0.57 | 0.89 ± 0.1 |
| Photosystem II reaction center W protein | psbW | slr1739 | 1.29 ± 0.37 | 0.92 ± 0.87 | 2.2 ± 0.66 | 1.52 ± 0.36 |
| Regulatory functions | ||||||
| Transcription regulatory protein | lumQ | slr1213 | 1.18 ± 0.24 | 1.11 ± 0.37 | 0.9 ± 0.73 | 3.16 ± 0.6 |
| Transcription regulator Fur family | slr1738 | 1.63 ± 0.54 | 1.5 ± 0.26 | 2.92 ± 0.97 | 1.55 ± 0.32 | |
| Two-component sensor histidine kinase | slr1285 | 3.13 ± 0.65 | 1.38 ± 0.74 | 4.08 ± 1.69 | 1.25 ± 0.19/PICK> | |
| Transcription | ||||||
| RNase E | me | slr1129 | 2.18 ± 0.57 | 2.15 ± 0.57 | 2.16 ± 1.01 | 1.27 ± 0.44 |
| RNA polymerase sigma factor | rpoD | sll2012 | 2.45 ± 1.03 | 3.03 ± 0.66 | 10.68 ± 5.07 | 2.21 ± 1.06 |
| RNA polymerase sigma factor | rpoD | sll0184 | 1.36 ± 0.41 | 1.17 ± 0.22 | 2.49 ± 0.76 | 1.74 ± 0.25 |
| RNA polymerase sigma factor | rpoD | sll0306 | 4.21 ± 0.6 | 1.68 ± 1.08 | 5.4 ± 0.59 | 2.8 ± 0.32 |
| Translation | ||||||
| ClpB | clpB | slr1641 | 6.57 ± 2.15 | 1.79 ± 0.64 | 9.01 ± 2.76 | 1.56 ± 0.08 |
| ATP-dependent Clp protease regulatory subunit | clpC | sll0020 | 1.91 ± 0.24 | 1.7 ± 0.4 | 2.61 ± 0.71 | 2.3 ± 0.24 |
| Carboxyl-terminal processing protease | ctpA | slr0008 | 1.97 ± 0.54 | 1.6 ± 0.46 | 3.59 ± 1.40 | 2.03 ± 0.73 |
| Protease HhoA | hhoA | sll1679 | 1.28 ± 0.29 | 1.25 ± 0.51 | 2.18 ± 0.42 | 2.66 ± 0.3 |
| Protease HhoB | hhoB | sll1427 | 1.37 ± 0.25 | 1.29 ± 0.06 | 2.08 ± 0.40 | 2.18 ± 0.24 |
| Carboxyl-terminal protease | prc | slr1751 | 1.2 ± 0.31 | 1.21 ± 0.41 | 1.79 ± 0.43 | 2.5 ± 0.59 |
| Elongation factor EF-G | fus | slr1105 | 1.3 ± 0.37 | 1.55 ± 0.38 | 1.42 ± 0.48 | 2.31 ± 0.58 |
| Methionine aminopeptidase | map | slr0918 | 1.2 ± 0.34 | 1.47 ± 0.36 | 2.05 ± 0.82 | 1.05 ± 0.23 |
| Ribosomal-protein-alanine acetyltransferase | riml | slr0853 | 2.17 ± 1.07 | 1.26 ± 0.22 | 2.97 ± 0.38 | 1.48 ± 0.42 |
| Transport and binding proteins | ||||||
| Ferrichrome-iron receptor | fhuA | sll1409 | 0.93 ± 0.15 | 0.86 ± 0.13 | 0.82 ± 0.17 | 2.73 ± 0.51 |
| ABC transporter subunit | ycf24 | slr0074 | 0.85 ± 0.15 | 1.37 ± 0.38 | 3.27 ± 1.56 | 1.19 ± 0.42 |
| ABC transporter | sll0759 | 1.54 ± 0.5 | 2.23 ± 0.48 | 3.32 ± 1.53 | 2.57 ± 0.87 | |
| NaH-antiporter protein | sll0689 | 1.00 ± 0.13 | 0.9 ± 0.09 | 0.92 ± 0.15 | 2.79 ± 0.56 | |
| Periplasmic iron-binding protein | slr0513 | 0.91 ± 0.12 | 0.85 ± 0.14 | 0.83 ± 0.06 | 2.97 ± 1.09 | |
| ABC-transporter membrane fusion protein | sll1481 | 1.45 ± 0.6 | 1.37 ± 0.47 | 3.19 ± 0.95 | 2.29 ± 0.29 | |
| Other categories | ||||||
| High-light-inducible protein | hliA | ssl2542 | 4.07 ± 0.66 | 2.53 ± 1.89 | 4.54 ± 0.74 | 1.71 ± 0.11 |
| High-light-inducible protein | hliB | ssl2595 | 7.33 ± 0.81 | 2.94 ± 0.56 | 31.00 ± 6.00 | 1.88 ± 0.62 |
| High-light-inducible protein | hliC | ssl1633 | 3.43 ± 0.9 | 2.24 ± 1.00 | 8.23 ± 1.63 | 3.73 ± 0.83 |
| Acriflavin resistance protein | acrF | slr2131 | 1.69 ± 0.31 | 1.45 ± 0.13 | 2.02 ± 0.09 | 3.29 ± 0.54 |
| Quinolene resistance protein NorA | norA | sll1154 | 1.48 ± 0.18 | 1.53 ± 0.34 | 4.09 ± 0.59 | 1.91 ± 0.28 |
| Hydrogenase expression/formation protein | hypA | slr1675 | 3.77 ± 0.92 | 1.55 ± 0.44 | 8.28 ± 1.18 | 2.24 ± 1.01 |
| Probable ferredoxin | ssl3044 | 2.36 ± 0.99 | 1.71 ± 0.35 | 4.63 ± 2.98 | 1.79 ± 0.27 | |
| Secreted protein MPB70 | sll1735 | 0.98 ± 0.38 | 0.8 ± 0.19 | 0.81 ± 0.04 | 2.21 ± 0.68 | |
| Transforming growth factor-induced protein | sll1483 | 4.96 ± 1.5 | 4.13 ± 0.77 | 21.68 ± 2.35 | 21.97 ± 5.14 | |
| Esterase | slr1916 | 2.88 ± 0.48 | 1.31 ± 0.37 | 2.95 ± 0.54 | 0.96 ± 0.31 | |
| Peroxiredoxin family protein | sll1621 | 1.34 ± 0.28 | 1.47 ± 0.5 | 3.85 ± 2.10 | 5.32 ± 1.75 | |
| ABC1-like protein | sll1770 | 1.98 ± 0.27 | 2.23 ± 0.41 | 2.34 ± 0.87 | 2.29 ± 0.69 | |
| Zinc-containing alcohol dehydrogenase family | slr1192 | 1.44 ± 0.22 | 1.2 ± 0.3 | 2.59 ± 0.42 | 1.23 ± 0.22 | |
| Lignostilbene-α,β-dioxygenase | sll1541 | 2.29 ± 0.77 | 1.65 ± 0.65 | 6.32 ± 1.80 | 3.55 ± 1.04 | |
| CP12 polypeptide | ssl3364 | 1.82 ± 0.44 | 1.34 ± 0.31 | 2.13 ± 0.21 | 1.26 ± 0.11 | |
| Hypothetical proteins | ||||||
| Ycf21 | ycf21 | sll1797 | 1.55 ± 0.28 | 1.33 ± 0.33 | 2.76 ± 0.53 | 1.93 ± 0.26 |
| Ycf34 | ycf34 | ssr1425 | 1.56 ± 0.27 | 1.34 ± 0.65 | 1.97 ± 0.45 | 1.46 ± 0.53 |
| Ycf38 | ycf38 | sll0760 | 1.48 ± 0.53 | 1.32 ± 0.26 | 1.79 ± 0.81 | 2.16 ± 0.66 |
| Putative arsenical pump-driving ATPase | sll0086 | 1.72 ± 0.71 | 1.11 ± 0.55 | 2.8 ± 0.58 | 2.15 ± 1.06 | |
| sll0088 | 1.14 ± 0.27 | 1.32 ± 0.75 | 1.97 ± 0.47 | 1.03 ± 0.17 | ||
| sll0141 | 1.61 ± 0.45 | 1.63 ± 0.93 | 2.45 ± 1.48 | 3.2 ± 0.91 | ||
| sll0157 | 1.62 ± 0.4 | 1.8 ± 1.01 | 3.31 ± 0.81 | 1.82 ± 0.95 | ||
| sll0185 | 2.03 ± 0.47 | 1.81 ± 0.33 | 4.85 ± 2.81 | 3.19 ± 0.98 | ||
| Potential FMN protein | sll0217 | 1.08 ± 0.13 | 1.14 ± 0.24 | 3.13 ± 0.84 | 1.22 ± 0.23 | |
| sll0297 | 1.42 ± 0.23 | 1.59 ± 0.18 | 3.05 ± 0.26 | 1.64 ± 0.28 | ||
| sll0314 | 1.42 ± 1.16 | 1.14 ± 0.27 | 2.17 ± 0.67 | 2.42 ± 0.97 | ||
| sll0355 | 1.37 ± 0.48 | 1.42 ± 0.37 | 2.35 ± 0.96 | 1.32 ± 0.33 | ||
| sll0470 | 1.22 ± 0.14 | 1.06 ± 0.23 | 1.77 ± 0.72 | 2.61 ± 0.21 | ||
| sll0528 | 7.35 ± 2.87 | 3.27 ± 3.24 | 32.54 ± 7.82 | 2.23 ± 0.28 | ||
| sll0549 | 1.6 ± 0.47 | 1.5 ± 0.32 | 3.78 ± 1.24 | 1.78 ± 0.17 | ||
| sll0761 | 1.37 ± 0.44 | 1.26 ± 0.22 | 1.42 ± 0.6 | 2.13 ± 0.43 | ||
| sll0814 | 2.09 ± 0.45 | 1.52 ± 0.12 | 3.28 ± 1.34 | 2.71 ± 1.02 | ||
| sll0846 | 5.62 ± 1.01 | 1.82 ± 1.69 | 10.66 ± 3.29 | 1.43 ± 0.22 | ||
| sll0898 | 1.53 ± 0.43 | 1.53 ± 0.31 | 2.07 ± 0.66 | 1.62 ± 0.5 | ||
| sll0939 | 1.93 ± 0.5 | 1.3 ± 0.19 | 3.00 ± 1.23 | 1.65 ± 0.35 | ||
| sll1022 | 1.8 ± 0.37 | 1.29 ± 0.33 | 3.1 ± 0.87 | 1.03 ± 0.19 | ||
| sll1071 | 1.71 ± 0.51 | 2.2 ± 0.49 | 3.4 ± 1.3 | 1.43 ± 0.48 | ||
| sll1106 | 1.73 ± 0.27 | 0.89 ± 0.14 | 3.73 ± 1.19 | 1.34 ± 0.17 | ||
| sll1150 | 1.07 ± 0.12 | 1.11 ± 0.08 | 1.99 ± 0.72 | 1.6 ± 0.5 | ||
| sll1155 | 1.25 ± 0.21 | 1.26 ± 0.25 | 2.07 ± 0.51 | 1.52 ± 0.17 | ||
| sll1201 | 1.24 ± 0.2 | 1.68 ± 0.51 | 2.49 ± 0.78 | 1.2 ± 0.21 | ||
| sll1268 | 1.37 ± 0.24 | 0.95 ± 0.16 | 1.3 ± 0.3 | 2.1 ± 0.43 | ||
| sll1289 | 1.33 ± 0.52 | 1.46 ± 0.44 | 3.02 ± 1.64 | 1.7 ± 0.54 | ||
| sll1426 | 1.21 ± 0.14 | 1.12 ± 0.16 | 1.57 ± 0.13 | 2.4 ±0.35 | ||
| sll1515 | 1.28 ± 0.3 | 2.74 ± 1.29 | 2.72 ± 0.73 | 2.07 ± 0.58 | ||
| sll1620 | 1.13 ± 0.29 | 1.24 ± 0.24 | 2.1 ± 0.8 | 0.98 ± 0.15 | ||
| sll1722 | 1.02 ± 0.41 | 1.42 ± 0.81 | 2.19 ± 1.78 | 2.27 ± 0.65 | ||
| sll1769 | 1.64 ± 0.33 | 2.08 ± 0.47 | 3.3 ± 1.26 | 1.7 ± 0.42 | ||
| sll1774 | 1.38 ± 0.41 | 1.64 ± 0.53 | 2.47 ± 1.17 | 1.24 ± 0.3 | ||
| sll1884 | 1.84 ± 0.47 | 1.42 ± 0.3 | 3.84 ± 1.91 | 1.28 ± 0.32 | ||
| sll1891 | 1.07 ± 0.18 | 1.02 ± 0.16 | 1.7 ± 0.49 | 2.01 ± 0.62 | ||
| sll1911 | 2.08 ± 0.32 | 1.56 ± 0.37 | 1.9 ± 0.49 | 1.62 ± 0.11 | ||
| sll2013 | 1.5 ± 0.83 | 1.39 ± 0.18 | 2.09 ± 0.71 | 0.98 ± 0.24 | ||
| slr0006 | 0.96 ± 0.29 | 0.99 ± 0.39 | 1.42 ± 0.77 | 2.28 ± 0.74 | ||
| slr0112 | 1.18 ± 0.19 | 1.21 ± 0.13 | 2.76 ± 0.44 | 1.92 ± 0.39 | ||
| slr0270 | 1.42 ± 0.3 | 1.39 ± 0.5 | 2.67 ± 0.38 | 1.11 ± 0.11 | ||
| slr0292 | 1.44 ± 0.18 | 1.42 ± 0.49 | 2.45 ± 0.51 | 1.42 ± 0.31 | ||
| slr0320 | 2.62 ± 0.68 | 2.74 ± 0.67 | 6.55 ± 2.36 | 2.68 ± 0.86 | ||
| slr0364 | 1.64 ± 0.36 | 1.68 ± 0.35 | 3.18 ± 0.87 | 1.28 ± 0.18 | ||
| slr0551 | 2 ± 0.73 | 2.4 ± 0.88 | 2.32 ± 1.12 | 1.15 ± 0.41 | ||
| slr0582 | 1.28 ± 0.54 | 1.2 ± 0.43 | 2.47 ± 0.81 | 1.73 ± 0.32 | ||
| slr0594 | 1.18 ± 0.26 | 1.16 ± 0.17 | 2.48 ± 1.07 | 1.35 ± 0.37 | ||
| slr0642 | 1.62 ± 0.27 | 1.84 ± 0.64 | 3.66 ± 1.02 | 2.07 ± 0.36 | ||
| slr0852 | 2.23 ± 1.6 | 1.26 ± 0.38 | 3.67 ± 1.27 | 1.82 ± 0.77 | ||
| slr0863 | 1.53 ± 0.25 | 1.39 ± 0.49 | 2.00 ± 0.72 | 1.09 ± 0.14 | ||
| slr0959 | 3.1 ± 0.85 | 1.77 ± 0.73 | 4.29 ± 1.07 | 1.5 ± 0.12 | ||
| slr0967 | 1.94 ± 0.2 | 1.65 ± 0.34 | 6.34 ± 1.83 | 2.77 ± 0.5 | ||
| slr1074 | 0.99 ± 0.38 | 1.26 ± 0.21 | 0.46 ± 0.34 | 3.69 ± 0.79 | ||
| slr1119 | 1.82 ± 0.48 | 1.52 ± 0.83 | 3.27 ± 0.88 | 1.67 ± 0.58 | ||
| Similar to chlorobenzene dioxygenase | slr1205 | 1.03 ± 0.29 | 1.06 ± 0.28 | 0.97 ± 0.29 | 9.23 ± 3.29 | |
| slr1235 | 1.47 ± 0.27 | 1.5 ± 0.28 | 2.35 ± 0.13 | 1.41 ± 0.14 | ||
| slr1259 | 1.24 ± 0.21 | 1.57 ± 0.45 | 3.31 ± 1.59 | 0.97 ± 0.17 | ||
| Similar to to phytoene dehydrogenase | slr1293 | 1.37 ± 0.37 | 1.15 ± 0.37 | 2.03 ± 0.43 | 0.97 ± 0.23 | |
| Putative modulator of DNA gyrase | slr1322 | 1.31 ± 0.29 | 1.21 ± 0.18 | 1.98 ± 0.71 | 1.86 ± 0.35 | |
| slr1383 | 1.27 ± 0.56 | 1.48 ± 0.54 | 2.41 ± 0.69 | 1.57 ± 0.48 | ||
| slr1397 | 1.23 ± 0.35 | 1.24 ± 0.36 | 2.15 ± 1.02 | 1.21 ± 0.4 | ||
| slr1413 | 2.7 ± 0.55 | 1.53 ± 0.43 | 6.27 ± 2.44 | 1.43 ± 0.48 | ||
| slr1512 | 1.29 ± 0.35 | 1.1 ± 0.23 | 1.1 ± 0.86 | 11.63 ± 3.93 | ||
| slr1513 | 1.1 ± 0.17 | 1.05 ± 0.16 | 1.71 ± 1.51 | 3.68 ± 0.66 | ||
| slr1544 | 6.54 ± 2.57 | 3.3 ± 2.56 | 17.60 ± 5.10 | 2.73 ± 0.56 | ||
| slr1557 | 1.46 ± 0.32 | 1.44 ± 0.28 | 2.38 ± 1.12 | 1.45 ± 0.41 | ||
| slr1603 | 3.64 ± 0.66 | 1.79 ± 0.71 | 10.53 ± 3.77 | 1.68 ± 0.36 | ||
| slr1674 | 3.9 ± 1.11 | 1.91 ± 0.39 | 13.47 ± 2.89 | 2.71 ± 0.83 | ||
| slr1676 | 2.58 ± 0.49 | 1.74 ± 0.46 | 4.16 ± 2.36 | 2.53 ± 1.05 | ||
| slr1677 | 2.47 ± 0.48 | 1.87 ± 0.41 | 2.83 ± 1.2 | 2.13 ± 0.6 | ||
| slr1686 | 3.11 ± 1.29 | 1.84 ± 0.43 | 5.47 ± 3.03 | 1.59 ± 0.44 | ||
| Cell death suppressor protein Lls1 homolog | slr1747 | 2.1 ± 0.41 | 1.85 ± 0.91 | 3.32 ± 0.39 | 2.64 ± 0.56 | |
| Probable DNA-binding stress protein | slr1894 | 1.57 ± 0.24 | 1.42 ± 0.23 | 4.79 ± 1.92 | 2.44 ± 0.55 | |
| slr1915 | 2.18 ± 1.1 | 0.9 ± 0.16 | 1.61 ± 0.91 | 1.5 ± 1.32 | ||
| slr1917 | 1.55 ± 0.44 | 1.27 ± 0.48 | 2.28 ± 0.64 | 0.76 ± 0.15 | ||
| slr1926 | 1.51 ± 0.14 | 1.23 ± 0.24 | 2.24 ± 0.94 | 0.94 ± 0.33 | ||
| slr1930 | 1.09 ± 0.18 | 1.25 ± 0.42 | 2.3 ± 1.15 | 1.12 ± 0.31 | ||
| Water-soluble carotenoid protein | slr1963 | 2.59 ± 0.99 | 1.88 ± 0.72 | 6.36 ± 2.45 | 2.33 ± 0.85 | |
| slr1971 | 1.36 ± 0.41 | 1.05 ± 0.34 | 2.17 ± 0.64 | 2.00 ± 0.65 | ||
| slr1990 | 1.7 ± 0.27 | 1.61 ± 0.26 | 2.81 ± 0.5 | 0.87 ± 0.04 | ||
| slr2048 | 0.93 ± 0.18 | 1.18 ± 0.57 | 1.37 ± 0.65 | 3.57 ± 1.17 | ||
| slr2071 | 1.01 ± 0.28 | 1.01 ± 0.22 | 1.97 ± 0.49 | 1.29 ± 0.14 | ||
| ssl0242 | 1.5 ± 0.49 | 1.3 ± 0.45 | 2.05 ± 0.73 | 1.19 ± 0.39 | ||
| ssl2971 | 2.54 ± 0.37 | 1.24 ± 0.24 | 1.65 ± 0.87 | 1.38 ± 0.67 | ||
| ssl3446 | 1.71 ± 0.51 | 1.31 ± 0.36 | 3.93 ± 1.51 | 0.95 ± 0.07 | ||
| ssl3769 | 1.25 ± 0.13 | 1.32 ± 0.11 | 2.82 ± 1.45 | 1.23 ± 0.43 | ||
| ssr0349 | 1.11 ± 0.3 | 1.09 ± 0.2 | 2.72 ± 0.97 | 1.12 ± 0.2 | ||
| ssr1256 | 1.27 ± 0.26 | 1.18 ± 0.27 | 2.01 ± 0.31 | 1.36 ± 0.39 | ||
| ssr2016 | 1.81 ± 0.41 | 1.64 ± 1.2 | 2.77 ± 0.69 | 1.32 ± 0.39 | ||
| ssr3188 | 2.45 ± 0.52 | 1.24 ± 0.17 | 3.03 ± 1.17 | 1.15 ± 0.36 | ||
Synechocystis gene transcripts induced more than twofold by UV-B or high-intensity white light irradiation. Average expression ratios (treated/control ratio explained in Materials and Methods) and standard deviations from four experiments are included. The treatments were as follows: 20 microeinsteins of UV-B m−2 s−1 (UV low) for 20 min or 2 h; 60 microeinsteins of UV-B m−2 s−1 (UV high) for 2 h; and 200 microeinsteins of intense white light m−2 s−1 for 2 h. Standard deviations were calculated from eight independent measurements of duplicate RNA samples (20 microeinsteins of UV-B m−2 s−1 for 20 min or for 2 h) or four independent measurements of single RNA samples (60 microeinsteins of UV-B m−2 s−1 for 2 h and 200 microeinsteins of white light m−2 s−1 for 2 h). Genes with expression levels unchanged (expression ratio between 2 and 0.5 or difference between ratio and standard deviation is less than 1.0) in all four experiments are not included in the tables. Transcript levels that were induced more than twofold are shown in bold type.
TABLE 2.
Transcripts repressed in UV-B and intense white light irradiation experimentsa
| Category and function | Gene | ORF | Treated/control ratiob
|
|||
|---|---|---|---|---|---|---|
| UV low, 20 min | UV low, 2 h | UV high, 2 h | White light, 2 h | |||
| Amino acid biosynthesis | ||||||
| Anthranilate synthetase α subunit | trpE | slr0738 | 0.67 ± 0.11 | 0.63 ± 0.23 | 0.41 ± 0.2 | 0.53 ± 0.2 |
| Threonine synthase | thrC | sll1688 | 0.76 ± 0.18 | 0.42 ± 0.08 | 0.8 ± 0.19 | 0.91 ± 0.17 |
| Nitrate reductase | narB | sll1454 | 1.51 ± 0.2 | 1.08 ± 0.4 | 0.47 ± 0.22 | 0.63 ± 0.11 |
| Cell envelope | ||||||
| Lipoprotein NlpD | nlpD | slr0993 | 0.67 ± 0.34 | 0.64 ± 0.31 | 0.24 ± 0.12 | 1.31 ± 0.57 |
| UDP-MurNac-tripeptide synthetase | murE | slr0528 | 0.65 ± 0.12 | 0.69 ± 0.12 | 0.49 ± 0.11 | 0.77 ± 0.14 |
| Mannosyltransferase B | rfbU | slr1064 | 1.04 ± 0.26 | 0.89 ± 0.1 | 0.47 ± 0.06 | 0.88 ± 0.11 |
| Cellular processes | ||||||
| HlyD family of secretion proteins | hlyD | sll1181 | 0.85 ± 0.27 | 0.93 ± 0.47 | 0.49 ± 0.14 | 1.14 ± 0.33 |
| Hemolysin | sll1951 | 0.72 ± 0.3 | 0.82 ± 0.5 | 0.46 ± 0.29 | 1.09 ± 0.54 | |
| Chemotaxis protein CheA | cheA | sll1296 | 0.89 ± 0.19 | 0.9 ± 0.48 | 0.19 ± 0.08 | 1.09 ± 0.46 |
| General secretion pathway protein D | gspD | slr1277 | 0.77 ± 0.25 | 0.77 ± 0.24 | 0.32 ± 0.1 | 0.61 ± 0.2 |
| Preprotein translocase SecY subunit | secY | sll1814 | 0.83 ± 0.2 | 0.82 ± 0.29 | 0.41 ± 0.1 | 1.04 ± 0.4 |
| Central intermediary metabolism | ||||||
| Glycogen phosphorylase | glgP | sll1356 | 0.71 ± 0.15 | 0.58 ± 0.11 | 0.44 ± 0.11 | 0.69 ± 0.13 |
| Glycogen operon protein GlgX | glgX | slr1857 | 0.3 ± 0.09 | 0.37 ± 0.08 | 0.16 ± 0.03 | 0.38 ± 0.12 |
| Cofactor biosynthesis | ||||||
| Protoporphyrin IX magnesium chelatase subunit | bchH | slr1055 | 0.82 ± 0.16 | 1.02 ± 0.31 | 0.5 ± 0.19 | 1.4 ± 0.44 |
| Light-independent protochlorophyllide reductase iron protein | chlL | slr0749 | 0.27 ± 0.06 | 0.24 ± 0.06 | 0.11 ± 0.01 | 0.48 ± 0.53 |
| Geranylgeranyl reductase | chlP | sll1091 | 0.42 ± 0.13 | 0.46 ± 0.25 | 0.13 ± 0.05 | 0.39 ± 0.13 |
| Protochlorophyllide reductase ChlB subunit | chlB | slr0772 | 0.46 ± 0.09 | 0.38 ± 0.08 | 0.17 ± 0.02 | 0.34 ± 0.03 |
| Protochlorophillide reductase subunit ChlN | chlN | slr0750 | 0.45 ± 0.19 | 0.46 ± 0.1 | 0.41 ± 0.1 | 0.69 ± 0.54 |
| Transfer RNA-Gln reductase | hemA | slr1808 | 0.69 ± 0.2 | 0.71 ± 0.33 | 0.29 ± 0.15 | 0.37 ± 0.13 |
| Coproporphyrinogen III oxidase | hemF | sll1185 | 0.43 ± 0.15 | 0.46 ± 0.17 | 0.21 ± 0.02 | 0.23 ± 0.04 |
| Heme oxygenase | ho | sll1184 | 0.36 ± 0.16 | 0.46 ± 0.27 | 0.11 ± 0.04 | 0.23 ± 0.06 |
| Pyridine nucleotide transhydrogenase α subunit | pntA | slr1239 | 0.54 ± 0.05 | 0.55 ± 0.09 | 0.5 ± 0.17 | 0.67 ± 0.16 |
| Thioredoxin | trxA | slr0623 | 0.59 ± 0.19 | 0.7 ± 0.22 | 0.29 ± 0.1 | 0.85 ± 0.27 |
| DNA replication, modification, and repair | ||||||
| DNA ligase | lig | sll1583 | 0.69 ± 0.18 | 0.84 ± 0.31 | 0.42 ± 0.14 | 0.58 ± 0.08 |
| Formamidopyrimidine-DNA glycosylase | mutM | slr1689 | 0.69 ± 0.09 | 0.63 ± 0.09 | 0.43 ± 0.11 | 0.47 ± 0.05 |
| Energy metabolism | ||||||
| Agmatine ureohydrolase | speB | sll1077 | 0.7 ± 0.25 | 0.55 ± 0.08 | 0.53 ± 0.13 | 0.4 ± 0.1 |
| Phosphofructokinase | pfkA | sll1196 | 0.49 ± 0.05 | 0.5 ± 0.07 | 0.5 ± 0.13 | 0.49 ± 0.11 |
| Glucose-6-phosphate dehydrogenase | zwf | slr1643 | 0.65 ± 0.18 | 0.64 ± 0.15 | 0.5 ± 0.14 | 1.01 ± 0.3 |
| Fatty acid desaturase | desA | slr1350 | 0.76 ± 0.23 | 0.84 ± 0.37 | 0.45 ± 0.15 | 1.25 ± 0.36 |
| Photosynthesis and respiration | ||||||
| ATP synthase a subunit | atpA | sll1326 | 0.88 ± 0.29 | 0.96 ± 0.48 | 0.16 ± 0.02 | 0.94 ± 0.26 |
| ATP synthase b subunit | atpB | slr1329 | 0.87 ± 0.27 | 1.12 ± 0.52 | 0.48 ± 0.24 | 1.35 ± 0.56 |
| ATP synthase g subunit | atpC | sll1327 | 0.82 ± 0.2 | 0.87 ± 0.41 | 0.14 ± 0.02 | 1.03 ± 0.37 |
| ATP synthase d subunit | atpD | sll1325 | 0.99 ± 0.18 | 0.98 ± 0.35 | 0.35 ± 0.23 | 0.96 ± 0.28 |
| ATP synthase e subunit | atpE | slr1330 | 0.96 ± 0.11 | 1.02 ± 0.2 | 0.39 ± 0.06 | 1.07 ± 0.19 |
| ATP synthase subunit b | atpF | sll1324 | 1.01 ± 0.21 | 1.03 ± 0.43 | 0.33 ± 0.05 | 0.97 ± 0.26 |
| ATP synthase subunit b | atpG | sll1323 | 1.07 ± 0.23 | 1.01 ± 0.46 | 0.35 ± 0.03 | 0.91 ± 0.27 |
| ATP synthase subunit a | atpI | sll1322 | 0.94 ± 0.16 | 1.03 ± 0.49 | 0.47 ± 0.25 | 0.91 ± 0.39 |
| CO2-concentrating concentrating mechanism protein CcmK | ccmK | sll1028 | 0.7 ± 0.25 | 0.74 ± 0.23 | 0.36 ± 0.1 | 2.47 ± 0.9 |
| CO2-concentrating mechanism protein CcmK | ccmK | sll1029 | 0.71 ± 0.2 | 0.71 ± 0.22 | 0.38 ± 0.05 | 2.52 ± 0.75 |
| CO2-concentrating mechanism protein CcmM | ccmM | sll1031 | 0.81 ± 0.2 | 0.73 ± 0.12 | 0.34 ± 0.02 | 2.67 ± 0.42 |
| CO2-concentrating mechanism protein CcmN | ccmN | sll1032 | 0.92 ± 0.22 | 0.91 ± 0.21 | 0.41 ± 0.08 | 2.13 ± 0.85 |
| Ribulose bisphosphate carboxylase large subunit | rbcL | slr0009 | 0.69 ± 0.26 | 0.6 ± 0.27 | 0.26 ± 0.15 | 2.1 ± 1.22 |
| Ribulose bisphosphate carboxylase small subunit | rbcS | slr0012 | 0.65 ± 0.22 | 0.69 ± 0.34 | 0.2 ± 0.11 | 2.0 ± 0.93 |
| Chaperone | rbcX | slr0011 | 0.7 ± 0.31 | 0.79 ± 0.42 | 0.2 ± 0.13 | 2.12 ± 1.11 |
| NADH dehydrogenase subunit 4 | ndhD | sll1733 | 0.81 ± 0.32 | 0.68 ± 0.27 | 0.43 ± 0.07 | 5.88 ± 3.02 |
| P700 apoprotein subunit Ia | psaA | slr1834 | 0.3 ± 0.13 | 0.24 ± 0.06 | 0.05 ± 0.02 | 0.18 ± 0.07 |
| P700 apoprotein subunit Ib | psaB | slr1835 | 0.33 ± 0.14 | 0.25 ± 0.06 | 0.05 ± 0.02 | 0.21 ± 0.08 |
| Photosystem I subunit VII | psaC | ssl0563 | 0.44 ± 0.13 | 0.39 ± 0.12 | 0.1 ± 0.03 | 0.21 ± 0.07 |
| Photosystem I subunit II | psaD | slr0737 | 0.36 ± 0.11 | 0.49 ± 0.25 | 0.09 ± 0.04 | 0.26 ± 0.1 |
| Photosystem I subunit IV | psaE | ssr2831 | 0.4 ± 0.05 | 0.45 ± 0.16 | 0.22 ± 0.07 | 0.35 ± 0.12 |
| Photosystem I subunit III | psaF | sll0819 | 0.36 ± 0.18 | 0.53 ± 0.28 | 0.12 ± 0.08 | 0.23 ± 0.13 |
| Photosystem I subunit IX | psaJ | sml0008 | 0.43 ± 0.09 | 0.52 ± 0.15 | 0.23 ± 0.05 | 0.49 ± 0.08 |
| Photosystem I subunit X | psaK | sll0629 | 0.93 ± 0.46 | 0.58 ± 0.31 | 0.46 ± 0.23 | 0.36 ± 0.1 |
| Photosystem I subunit X | psaK | ssr0390 | 0.52 ± 0.06 | 0.55 ± 0.03 | 0.41 ± 0.02 | 0.67 ± 0.12 |
| Photosystem I subunit XI | psaL | slr1655 | 0.36 ± 0.07 | 0.28 ± 0.07 | 0.09 ± 0.01 | 0.18 ± 0.06 |
| Photosystem II P680 chlorophyll A apoprotein | psbB | slr0906 | 0.51 ± 0.18 | 0.85 ± 0.4 | 0.26 ± 0.17 | 0.62 ± 0.37/PICK> |
| Cytochrome b559 a subunit | psbE | ssr3451 | 0.79 ± 0.27 | 0.98 ± 0.42 | 0.47 ± 0.28 | 0.79 ± 0.34 |
| Photosystem II manganese-stabilizing polypeptide | psbO | sll0427 | 0.55 ± 0.12 | 0.72 ± 0.25 | 0.48 ± 0.24 | 0.39 ± 0.16 |
| Cytochrome c550 | psbV | sll0258 | 0.66 ± 0.13 | 0.74 ± 0.14 | 0.84 ± 0.68 | 0.42 ± 0.09 |
| Allophycocyanin a chain | apcA | slr2067 | 0.28 ± 0.08 | 0.48 ± 0.26 | 0.07 ± 0.04 | 0.2 ± 0.09 |
| Allophycocyanin b chain | apcB | slr1986 | 0.27 ± 0.08 | 0.52 ± 0.27 | 0.08 ± 0.03 | 0.16 ± 0.08 |
| Phycobilisome LC linker polypeptide | apcC | ssr3383 | 0.27 ± 0.05 | 0.41 ± 0.12 | 0.16 ± 0.03 | 0.2 ± 0.04 |
| Phycobilisome LCM core-membrane linker polypeptide | apcE | slr0335 | 0.35 ± 0.11 | 0.49 ± 0.29 | 0.1 ± 0.05 | 0.27 ± 0.13 |
| Phycobilisome core component | apcF | slr1459 | 0.47 ± 0.18 | 0.74 ± 0.38 | 0.19 ± 0.1 | 0.3 ± 0.13 |
| Phycocyanin a subunit | cpcA | sll1578 | 0.24 ± 0.03 | 0.24 ± 0.09 | 0.06 ± 0.03 | 0.07 ± 0.02 |
| Phycocyanin b subunit | cpcB | sll1577 | 0.23 ± 0.03 | 0.23 ± 0.09 | 0.08 ± 0.06 | 0.08 ± 0.03 |
| Phycocyanin-associated linker protein | cpcC | sll1579 | 0.19 ± 0.02 | 0.2 ± 0.06 | 0.05 ± 0.02 | 0.06 ± 0.01 |
| Phycocyanin-associated linker protein | cpcC | sll1580 | 0.23 ± 0.05 | 0.19 ± 0.05 | 0.06 ± 0.03 | 0.06 ± 0.00 |
| Phycocyanin-associated linker protein | cpcD | ssl3093 | 0.46 ± 0.15 | 0.38 ± 0.07 | 0.15 ± 0.01 | 0.23 ± 0.13 |
| Phycobilisome rod-core linker polypeptide CpcG | cpcG | sll1471 | 0.35 ± 0.27 | 0.14 ± 0.03 | 0.27 ± 0.07 | 0.31 ± 0.35 |
| Phycobilisome rod-core linker polypeptide CpcG | cpcG | slr2051 | 0.43 ± 0.16 | 0.55 ± 0.29 | 0.19 ± 0.13 | 0.27 ± 0.1 |
| Cytochrome c553 | petJ | sll1796 | 0.98 ± 0.13 | 0.79 ± 0.28 | 0.22 ± 0.07 | 0.75 ± 0.08 |
| Regulatory functions | ||||||
| Negative aliphatic amidase regulator | amiC | slr0447 | 0.72 ± 0.33 | 0.43 ± 0.15 | 0.2 ± 0.12 | 0.46 ± 0.22 |
| Regulatory components of sensory transduction system | copR | sll0789 | 0.92 ± 0.07 | 0.9 ± 0.14 | 1.02 ± 0.21 | 0.4 ± 0.05 |
| Regulatory components of sensory transduction system | lcfG | slr1860 | 0.54 ± 0.09 | 0.68 ± 0.16 | 0.44 ± 0.05 | 0.58 ± 0.07 |
| SOS function regulatory protein | lexA | sll1626 | 0.48 ± 0.1 | 0.53 ± 0.22 | 0.39 ± 0.15 | 0.53 ± 0.14 |
| Transcription | ||||||
| RNase III | mc | slr0346 | 0.66 ± 0.12 | 0.82 ± 0.22 | 0.59 ± 0.12 | 0.49 ± 0.18 |
| RNA polymerase alpha subunit | rpoA | sll1818 | 1.11 ± 0.13 | 1.16 ± 0.32 | 0.48 ± 0.15 | 0.99 ± 0.14 |
| Anti-sigma B factor antagonist | slr1856 | 0.37 ± 0.12 | 0.57 ± 0.07 | 0.32 ± 0.11 | 0.47 ± 0.11 | |
| Translation | ||||||
| Seryl-tRNA synthetase | serS | slr1703 | 0.76 ± 0.14 | 1.11 ± 0.29 | 0.33 ± 0.06 | 1.00 ± 0.43 |
| tRNA(m1G37)methyltransferase | trmD | sll1198 | 0.7 ± 0.3 | 0.62 ± 0.16 | 0.34 ± 0.08 | 0.46 ± 0.08 |
| Processing protease | ymxG | slr1331 | 0.99 ± 0.18 | 1.2 ± 0.38 | 0.44 ± 0.11 | 1.21 ± 0.24 |
| DNA-binding protein HU | sll1712 | 0.51 ± 0.19 | 0.58 ± 0.13 | 0.38 ± 0.13 | 0.58 ± 0.13 | |
| Elongation factor EF-G | fus | sll0830 | 0.56 ± 0.05 | 0.63 ± 0.08 | 0.49 ± 0.03 | 0.55 ± 0.05 |
| Protein synthesis elongation factor Tu | tufA | sll1099 | 0.81 ± 0.27 | 0.79 ± 0.37 | 0.17 ± 0.09 | 1.52 ± 0.74 |
| 50S ribosomal protein L1 | rpl1 | sll1744 | 0.87 ± 0.27 | 0.79 ± 0.32 | 0.27 ± 0.12 | 1.00 ± 0.41 |
| 50S ribosomal protein L2 | rpl2 | sll1802 | 0.92 ± 0.18 | 0.77 ± 0.29 | 0.19 ± 0.04 | 0.87 ± 0.21 |
| 50S ribosomal protein L4 | rpl4 | sll1800 | 0.95 ± 0.23 | 0.93 ± 0.32 | 0.28 ± 0.08 | 0.89 ± 0.22 |
| 50S ribosomal protein L5 | rpl5 | sll1808 | 0.77 ± 0.29 | 0.91 ± 0.48 | 0.08 ± 0.02 | 0.92 ± 0.39 |
| 50S ribosomal protein L6 | rpl6 | sll1810 | 0.76 ± 0.2 | 0.74 ± 0.32 | 0.13 ± 0.05 | 0.88 ± 0.29 |
| 50S ribosomal protein L10 | rpl10 | sll1745 | 0.72 ± 0.18 | 0.74 ± 0.3 | 0.15 ± 0.07 | 0.94 ± 0.46 |
| 50S ribosomal protein L11 | rpl11 | sll1743 | 0.97 ± 0.35 | 0.97 ± 0.41 | 0.45 ± 0.26 | 1.1 ± 0.58 |
| 50S ribosomal protein L12 | rpl12 | sll1746 | 0.87 ± 0.23 | 0.71 ± 0.3 | 0.18 ± 0.09 | 0.9 ± 0.4 |
| 50S ribosomal protein L13 | rpl13 | sll1821 | 0.99 ± 0.13 | 1.05 ± 0.38 | 0.43 ± 0.11 | 1.01 ± 0.32 |
| 50S ribosomal protein L14 | rpl14 | sll1806 | 0.79 ± 0.22 | 0.87 ± 0.33 | 0.14 ± 0.03 | 0.91 ± 0.35 |
| 50S ribosomal protein L15 | rpl15 | sll1813 | 0.75 ± 0.16 | 0.67 ± 0.12 | 0.18 ± 0.04 | 0.88 ± 0.29 |
| 50S ribosomal protein L16 | rpl16 | sll1805 | 0.84 ± 0.33 | 0.95 ± 0.48 | 0.1 ± 0.04 | 0.89 ± 0.37 |
| 50S ribosomal protein L17 | rpl17 | sll1819 | 1.18 ± 0.11 | 0.94 ± 0.17 | 0.47 ± 0.17 | 0.96 ± 0.08 |
| 50S ribosomal protein L18 | rpl18 | sll1811 | 0.72 ± 0.19 | 0.71 ± 0.2 | 0.19 ± 0.11 | 0.87 ± 0.24 |
| 50S ribosomal protein L22 | rpl22 | sll1803 | 0.85 ± 0.26 | 0.88 ± 0.44 | 0.15 ± 0.04 | 0.96 ± 0.39 |
| 50S ribosomal protein L23 | rpl23 | sll1801 | 0.92 ± 0.35 | 1.1 ± 0.57 | 0.22 ± 0.11 | 1.00 ± 0.45 |
| 50S ribosomal protein L24 | rpl24 | sll1807 | 0.8 ± 0.29 | 0.88 ± 0.37 | 0.12 ± 0.03 | 0.88 ± 0.35 |
| 50S ribosomal protein L29 | rpl29 | ssl3436 | 0.87 ± 0.27 | 1.04 ± 0.59 | 0.18 ± 0.05 | 0.94 ± 0.33 |
| 50S ribosomal protein L31 | rpl31 | ssl3445 | 1.11 ± 0.22 | 1.04 ± 0.43 | 0.46 ± 0.18 | 1.09 ± 0.22 |
| 30S ribosomal protein S3 | rps3 | sll1804 | 0.86 ± 0.27 | 0.8 ± 0.32 | 0.13 ± 0.04 | 0.89 ± 0.34 |
| 30S ribosomal protein S5 | rps5 | sll1812 | 0.77 ± 0.2 | 0.72 ± 0.28 | 0.19 ± 0.06 | 0.91 ± 0.28 |
| 30S ribosomal protein S8 | rps8 | sll1809 | 0.74 ± 0.23 | 0.8 ± 0.34 | 0.13 ± 0.02 | 0.95 ± 0.41 |
| 30S ribosomal protein S9 | rps9 | sll1822 | 1.04 ± 0.32 | 1.19 ± 0.59 | 0.29 ± 0.08 | 1.07 ± 0.44 |
| 30S ribosomal protein S10 | rps10 | sll1101 | 0.89 ± 0.23 | 0.89 ± 0.41 | 0.18 ± 0.07 | 0.98 ± 0.43 |
| 30S ribosomal protein S17 | rps17 | ssl3437 | 0.88 ± 0.21 | 0.93 ± 0.45 | 0.14 ± 0.04 | 1.00 ± 0.27 |
| 30S ribosomal protein S19 | rps19 | ssl3432 | 0.93 ± 0.12 | 0.76 ± 0.27 | 0.21 ± 0.03 | 0.98 ± 0.27 |
| Transport and binding proteins | ||||||
| Ammonium transporter | amt1 | sll1017 | 0.89 ± 0.15 | 0.74 ± 0.19 | 0.68 ± 0.14 | 0.5 ± 0.08 |
| NH4+ transporter | amt1 | sll0108 | 0.75 ± 0.11 | 0.72 ± 0.3 | 0.72 ± 0.11 | 0.33 ± 0.06 |
| Glucose transport protein | glcP | sll0771 | 0.65 ± 0.12 | 0.54 ± 0.11 | 0.35 ± 0.11 | 0.57 ± 0.2 |
| High-affinity branched-chain amino acid transport protein | livH | slr1200 | 0.72 ± 0.15 | 0.62 ± 0.1 | 0.74 ± 0.28 | 0.38 ± 0.15 |
| Nitrate transport protein NrtD | nrtD | sll1453 | 1.63 ± 0.21 | 1.2 ± 0.46 | 0.45 ± 0.05 | 0.75 ± 0.13 |
| Glutamine-binding periplasmic protein | sll1270 | 0.57 ± 0.18 | 0.59 ± 0.1 | 0.4 ± 0.17 | 0.49 ± 0.11 | |
| Binding protein of ABC transporter | sll1762 | 0.56 ± 0.16 | 0.55 ± 0.15 | 0.31 ± 0.08 | 0.53 ± 0.12 | |
| Probable sodium/calcium exchanger protein | slr0681 | 0.71 ± 0.11 | 0.79 ± 0.12 | 0.48 ± 0.05 | 0.65 ± 0.15 | |
| Permease protein of branched-chain amino acid ABC transporter | slr1201 | 0.79 ± 0.23 | 0.75 ± 0.09 | 0.56 ± 0.14 | 0.39 ± 0.11 | |
| Other categories | ||||||
| AT103 | sll1214 | 0.77 ± 0.32 | 0.94 ± 0.61 | 0.33 ± 0.2 | 0.61 ± 0.35 | |
| Light-repressed protein | lrtA | sll0947 | 0.63 ± 0.18 | 0.8 ± 0.23 | 0.46 ± 0.17 | 0.67 ± 0.11 |
| Phytochrome | cph1 | slr0473 | 0.51 ± 0.12 | 0.52 ± 0.18 | 0.26 ± 0.11 | 0.59 ± 0.14 |
| Dienelactone hydrolase | clcD | sll1298 | 1.47 ± 1.15 | 1.16 ± 0.61 | 0.07 ± 0.01 | 0.92 ± 0.41 |
| Iron-regulated protein | frpC | sll1009 | 0.5 ± 0.1 | 0.71 ± 0.1 | 0.68 ± 0.23 | 0.37 ± 0.07 |
| GlpX protein | glpX | slr2094 | 0.64 ± 0.22 | 0.7 ± 0.34 | 0.28 ± 0.16 | 0.99 ± 0.48 |
| Cytochrome b subunit of nitric oxide reductase | norB | sll0450 | 0.52 ± 0.13 | 0.59 ± 0.14 | 0.65 ± 0.31 | 0.34 ± 0.02 |
| Membrane protein | pilM | slr1274 | 0.87 ± 0.13 | 0.84 ± 0.26 | 0.49 ± 0.25 | 1.00 ± 0.21 |
| Twitching mobility protein | pilT | sll1533 | 0.6 ± 0.17 | 0.63 ± 0.15 | 0.28 ± 0.08 | 0.5 ± 0.08 |
| Phenoxybenzoate dioxygenase | pobA | sll1297 | 0.9 ± 0.17 | 0.7 ± 0.38 | 0.46 ± 0.09 | 1.07 ± 0.58 |
| Rehydrin | slr1198 | 0.67 ± 0.3 | 0.92 ± 0.49 | 0.46 ± 0.28 | 1.06 ± 0.55 | |
| GDP-fucose synthetase | sll1213 | 1.01 ± 0.24 | 1.08 ± 0.5 | 0.5 ± 0.19 | 1.19 ± 0.36 | |
| Carboxymuconolactone decarboxylase | slr1853 | 0.39 ± 0.06 | 0.45 ± 0.16 | 0.15 ± 0.04 | 0.43 ± 0.09 | |
| Hypothetical proteins | ||||||
| sll0172 | 1.14 ± 0.9 | 0.71 ± 0.35 | 0.89 ± 0.35 | 0.5 ± 0.23 | ||
| sll0173 | 1.06 ± 0.73 | 0.74 ± 0.12 | 0.78 ± 0.18 | 0.49 ± 0.1 | ||
| sll0513 | 0.84 ± 0.1 | 0.79 ± 0.16 | 0.49 ± 0.13 | 0.86 ± 0.14 | ||
| sll0543 | 0.46 ± 0.1 | 0.59 ± 0.14 | 0.28 ± 0.11 | 0.73 ± 0.29 | ||
| sll0630 | 0.13 ± 0.08 | 1.12 ± 1.55 | 1.51 ± 1.08 | 0.69 ± 0.28 | ||
| sll0735 | 0.75 ± 0.11 | 0.8 ± 0.21 | 0.45 ± 0.06 | 0.76 ± 0.3 | ||
| sll0788 | 0.8 ± 0.16 | 0.8 ± 0.22 | 1.22 ± 0.34 | 0.35 ± 0.1 | ||
| sll0822 | 0.87 ± 0.19 | 0.71 ± 0.22 | 0.47 ± 0.22 | 0.74 ± 0.27 | ||
| sll1358 | 0.42 ± 0.12 | 0.38 ± 0.1 | 0.22 ± 0.05 | 0.29 ± 0.05 | ||
| sll1359 | 0.56 ± 0.09 | 0.49 ± 0.05 | 0.47 ± 0.1 | 0.36 ± 0.05 | ||
| sll1531 | 0.98 ± 0.32 | 0.77 ± 0.18 | 0.37 ± 0.09 | 0.78 ± 0.15 | ||
| sll1586 | 0.66 ± 0.08 | 0.68 ± 0.07 | 0.77 ± 0.3 | 0.38 ± 0.03 | ||
| sll1665 | 0.73 ± 0.28 | 0.81 ± 0.38 | 0.5 ± 0.24 | 0.86 ± 0.29 | ||
| sll1783 | 0.38 ± 0.13 | 0.62 ± 0.12 | 0.23 ± 0.03 | 0.47 ± 0.07 | ||
| sll1784 | 0.37 ± 0.18 | 0.5 ± 0.32 | 0.13 ± 0.04 | 0.29 ± 0.1 | ||
| sll1830 | 0.44 ± 0.27 | 0.35 ± 0.09 | 0.38 ± 0.27 | 0.3 ± 0.05 | ||
| sll1926 | 0.52 ± 0.09 | 0.57 ± 0.11 | 0.33 ± 0.04 | 0.79 ± 0.13 | ||
| slr0144 | 0.68 ± 0.19 | 0.51 ± 0.17 | 0.28 ± 0.05 | 0.62 ± 0.38 | ||
| slr0145 | 0.86 ± 0.52 | 0.5 ± 0.13 | 0.4 ± 0.04 | 0.76 ± 0.75 | ||
| slr0146 | 0.66 ± 0.23 | 0.56 ± 0.12 | 0.31 ± 0.06 | 0.69 ± 0.45 | ||
| slr0147 | 0.66 ± 0.37 | 0.43 ± 0.15 | 0.31 ± 0.05 | 0.58 ± 0.51 | ||
| slr0148 | 0.7 ± 0.37 | 0.52 ± 0.06 | 0.46 ± 0.04 | 0.89 ± 0.63 | ||
| slr0149 | 0.8 ± 0.53 | 0.68 ± 0.26 | 0.44 ± 0.13 | 0.83 ± 0.26 | ||
| slr0151 | 0.76 ± 0.18 | 0.74 ± 0.19 | 0.44 ± 0.12 | 0.68 ± 0.24 | ||
| Similar to glutathione S-transferase | slr0236 | 1.16 ± 0.19 | 1.09 ± 0.24 | 0.45 ± 0.08 | 0.89 ± 0.17 | |
| slr0244 | 0.52 ± 0.06 | 0.66 ± 0.09 | 0.39 ± 0.07 | 0.6 ± 0.09 | ||
| slr0284 | 0.59 ± 0.2 | 0.5 ± 0.33 | 0.64 ± 0.16 | 1.1 ± 0.14 | ||
| slr0572 | 0.52 ± 0.1 | 0.5 ± 0.1 | 0.48 ± 0.25 | 0.62 ± 0.21 | ||
| slr0708 | 0.49 ± 0.1 | 0.48 ± 0.1 | 0.38 ± 0.13 | 0.49 ± 0.06 | ||
| slr0779 | 0.94 ± 0.33 | 0.95 ± 0.49 | 0.19 ± 0.1 | 1.14 ± 0.51 | ||
| slr0888 | 0.55 ± 0.1 | 0.61 ± 0.24 | 0.35 ± 0.16 | 0.58 ± 0.07 | ||
| slr0914 | 1.04 ± 0.43 | 0.96 ± 0.18 | 0.45 ± 0.21 | 0.99 ± 0.29 | ||
| slr1074 | 0.99 ± 0.38 | 1.26 ± 0.21 | 0.46 ± 0.34 | 3.69 ± 0.79 | ||
| slr1240 | 0.78 ± 0.33 | 0.65 ± 0.11 | 0.47 ± 0.06 | 0.72 ± 0.32 | ||
| slr1275 | 0.79 ± 0.11 | 0.75 ± 0.11 | 0.33 ± 0.08 | 0.99 ± 0.33 | ||
| slr1276 | 0.81 ± 0.06 | 0.73 ± 0.2 | 0.36 ± 0.09 | 0.97 ± 0.22 | ||
| slr1437 | 0.54 ± 0.15 | 0.81 ± 0.26 | 0.35 ± 0.13 | 0.76 ± 0.09 | ||
| slr1438 | 0.62 ± 0.08 | 0.81 ± 0.1 | 0.45 ± 0.17 | 0.6 ± 0.12 | ||
| slr1485 | 0.51 ± 0.12 | 0.69 ± 0.09 | 0.24 ± 0.06 | 0.72 ± 0.38 | ||
| slr1535 | 0.65 ± 0.11 | 0.76 ± 0.3 | 0.38 ± 0.17 | 0.29 ± 0.13 | ||
| slr1618 | 1.03 ± 0.1 | 0.96 ± 0.15 | 0.48 ± 0.19 | 0.88 ± 0.25 | ||
| slr1619 | 1.04 ± 0.15 | 0.96 ± 0.21 | 0.49 ± 0.11 | 0.93 ± 0.2 | ||
| slr1634 | 0.86 ± 0.11 | 0.7 ± 0.08 | 0.56 ± 0.11 | 0.45 ± 0.11 | ||
| slr1852 | 0.35 ± 0.07 | 0.37 ± 0.1 | 0.19 ± 0.03 | 0.36 ± 0.08 | ||
| slr1854 | 0.36 ± 0.05 | 0.47 ± 0.16 | 0.12 ± 0.03 | 0.39 ± 0.08 | ||
| slr1855 | 0.26 ± 0.05 | 0.38 ± 0.17 | 0.08 ± 0.05 | 0.37 ± 0.08 | ||
| slr1958 | 0.65 ± 0.2 | 0.66 ± 0.09 | 0.28 ± 0.06 | 0.52 ± 0.1 | ||
| slr2052 | 0.5 ± 0.11 | 0.4 ± 0.13 | 0.27 ± 0.09 | 0.34 ± 0.07 | ||
| Hypothetical protein YCF50 | slr2073 | 0.76 ± 0.25 | 0.73 ± 0.32 | 0.46 ± 0.28 | 0.84 ± 0.33 | |
| slr2077 | 0.46 ± 0.1 | 0.58 ± 0.18 | 0.23 ± 0.05 | 0.33 ± 0.05 | ||
| ssl0483 | 0.76 ± 0.19 | 0.9 ± 0.38 | 0.49 ± 0.27 | 0.53 ± 0.21 | ||
| ssl1046 | 0.51 ± 0.09 | 0.55 ± 0.05 | 0.44 ± 0.1 | 0.87 ± 0.13 | ||
| ssl1792 | 0.95 ± 0.34 | 0.99 ± 0.00 | 0.52 ± 0.00 | 1.13 ± 0.65 | ||
Synechocystis gene transcripts repressed more than twofold by UV-B or high intensity white light irradiation.
Average expression ratios (treated/control ratio explained in Materials and Methods) and standard deviations from four experiments are included. The treatments were as follows: 20 microeinsteins of UV-B m−2 s−1 (UV low) for 20 min or 2 h; 60 microeinsteins of UV-B m−2 s−1 (UV high) for 2 h; and 200 microeinsteins of intense white light m−2 s−1 for 2 h. Standard deviations were calculated from eight independent measurements of duplicated RNA samples (20 microeinsteins of UV-B m−2 s−1 for 20 min or for 2 h) or four independent measurements of single RNA samples (60 microeinsteins of UV-B m−2 s−1 for 2 h and 200 microeinsteins of white light m−2 s−1 for 2 h). Genes with expression levels unchanged (expression ratio between 2 and 0.5, or sum of ratio and standard deviation is more than 1.0) in all four experiments are not included in the tables. Transcript levels that were repressed more than twofold are shown in bold type.
Northern blotting experiment.
Total RNA samples isolated as described previously were fractionated using 1.5 or 2% agarose gels containing 6.7% formaldehyde, transferred onto a BrightStar-Plus nylon membrane (Ambion, Austin, Tex.), and then subjected to probe hybridization. 32P-labeled DNA probes were synthesized using the RadPrime DNA labeling system (Life Technologies, Gaithersburg, Md.). Hybridization signals were detected with a PhosphorImager 445SI (Molecular Dynamics) and quantitated using ImageQuant software (Amersham Biosciences, Sunnyvale, Calif.).
RESULTS
(i) Quality control of Synechocystis sp. strain PCC 6803 DNA microarray.
In order to assess the quality of spotted glass slides, Synechocystis sp. strain PCC 6803 genomic DNA was used as a template for fluorescent Cy3-dCTP or Cy5-dCTP-labeled DNA synthesis primed by random oligonucleotide hexamers and amplified with the Klenow fragment of DNA polymerase I (34). This labeled DNA served as a surrogate for an equimolar mix of all mRNAs. All of the spotted PCR products gave detectable signals with probes derived from genomic DNA, with an average intensity of about eightfold over the background level (data not shown).
(ii) Global gene expression profiling of Synechocystis sp. strain PCC 6803 cells in response to UV-B irradiation.
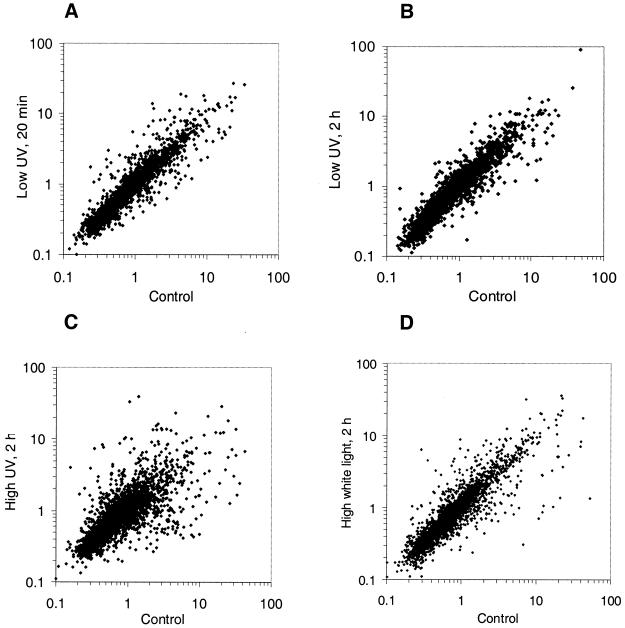
To elucidate the effects of different UV-B intensities and exposure times on the global gene transcription profile of exponential-phase Synechocystis, cells were treated with UV-B at a low intensity (20 microeinsteins m−2 s−1) for 20 or 120 min and at a high intensity (60 microeinsteins m−2 s−1) for 120 min. In Fig. 1, the average signal intensities of treated samples are plotted versus average signal intensities of control samples. With UV-B irradiation of 20 microeinsteins m−2 s−1, the number of induced or repressed mRNAs declined slightly with exposure time. Fifty-five genes were induced more than twofold and 44 genes were repressed more than twofold after 20 min of UV exposure, while 21 genes were induced more than twofold and 40 genes were repressed more than twofold after the 120-min exposure. This indicated a rapid transcriptional response to the UV-B insult, followed by a relaxation of the transcription levels towards an equilibrium state. Upon UV-B irradiation at 60 microeinsteins m−2 s−1 for 120 min, Synechocystis cells exhibited a more profound alteration in the transcription profile. Both the number of genes affected and the magnitude of changes increased, with 146 genes induced more than twofold and 159 genes repressed more than twofold. Treatment of exponential-phase Synechocystis cells with this intensity of UV-B for 2 h resulted in at most a small loss of viability (44% survival as measured by colony formation units).
FIG. 1.
Scatter plot of global gene expression profiles of Synechocystis in response to irradiation with UV-B (A to C) or white light (D). Synechocystis cultures were irradiated with 20 microeinsteins of UV-B m−2 s−1 for 20 min (A) or for 2 h (B) or with 60 microeinsteins of UV-B m−2s−1 for 2 h. (D) Synechocystis cultures were irradiated with 200 microeinsteins of white light m−2 s−1 for 2 h.
(a) Photosynthetic gene and accessory photosynthetic gene transcripts.
The D1 core protein of photosystem II is highly susceptible to damage by UV-B-generated radicals. Two genes, psbA2 and psbA3, encoding identical D1 polypeptides were induced four to sixfold by UV-B irradiation at 20 and 60 microeinsteins m−2 s−1, consistent with other results (23). Due to the sequence identity between the psbA2 and psbA3 ORFs, it is not possible to assign transcripts to either one of these two genes. The transcript of D1-processing protease (ctpA) was elevated by two- to threefold. Most other transcripts specifying components of photosystem II remained unchanged, except for the repression of psbB and psbO. In contrast, UV-B down-regulated the transcription of most genes encoding photosystem I components to 0.3- to 0.5-fold after 20 min of 20 microeinsteins of UV-B m−2 s−1 and 0.05- to 0.5-fold after 2 h of 60 microeinsteins of UV-B m−2 s−1 (Table 2). The repressed genes included psaA and psaB (the two major components of photosystem I) and photosystem I subunit-encoding genes psaC, psaD, psaE, psaF, psaJ, psaK, and psaL. Most significantly, the transcription of both psaA and psaB was repressed about 20-fold after 2 h of intense UV-B irradiation.
The FtsH protein of chloroplasts is an integral thylakoid membrane protease involved in degradation of D1 protein damaged by photooxidation (22). We showed that the expression of cyanobacterial ftsH was regulated by UV-B light. After exposure to UV-B irradiation, ftsH transcription increased two- to sixfold. Synechocystis has four ftsH genes, of which sll1463 is highly homologous to the Helicobacter pylori ftsH gene, while the others (slr1604, slr0228, and slr1390) are highly homologous to ftsH genes in other photosynthetic organisms. Since the cross hybridization between cDNA probes and different ftsH genes in DNA microarray cannot be ruled out, other methods such as nuclease protection assay are necessary for analyzing the relative expression profiles of the four ftsH genes.
In addition to photosystem I genes, there was a coordinate down-regulation of most genes encoding structural subunits of the phycobilisome light-harvesting antenna (Table 2). These genes included apcA, apcB, apcC, apcE, apcF, cpcA, cpcB, cpcC, cpcD, and cpcG. A putative operon formed by cpcB (sll1577), cpcA (sll1578), cpcC (sll1579 and sll1580) contained some of the most highly repressed genes, with expression levels of 0.05- to 0.08-fold. Biosyntheses of tetrapyrrole precursors leading to phycobilin and chlorophyll may have slowed, as indicated by the repression of hemA, hemF, and heme oxygenase genes. Phycocyanin-phycocyanobilin lyase, encoded by cpcE and cpcF, catalyzes covalent thioether bond formation between the light-absorbing tetrapyrrole molecule phycocyanobilin and apophycobiliproteins to give the functional phycocyanobilin protein (9). The transcription levels of cpcE and cpcF were not affected by UV-B. NblA and NblB are polypeptides involved in phycobilisome degradation. The transcription levels of nblA and nblB were up-regulated by three- to eightfold, contrary to the repression of phycobilisome structural genes. This suggested a coordinate slowdown of phycobilisome biosynthesis and activation of phycobilisome degradation in UV-B-treated Synechocystis. The net result is to minimize the cell's effective exposure to damage.
Chlorophylls and carotenoids are accessory pigments of the light-harvesting antenna in Synechocystis. Under UV-B exposure, chlorophyll biosynthesis-specifying RNAs were strongly repressed by 2- to 10-fold, including bchH, chlL, chlB, chlN, and chlP. In contrast to this repression, some carotenoid biosynthesis genes were induced (crtP [two- to fourfold], crtE [two- to fivefold], and crtQ [1.4- to 2-fold]). In addition to the singlet oxygen quenching of carotenoids, cellular detoxification of superoxide was suggested by the induction of the superoxide dismutase-specifying gene sodB (two- to sixfold).
(b) ATP generation.
Consistent with the down-regulation of photosystem I genes, transcripts allowing ATP synthesis by the Fo-F1 ATPase were repressed. A putative operon formed by atpG (sll1323), atpF (sll1324), atpD (sll1325), atpA (sll1326) and atpC (sll1327) had expression ratios of 0.1 to 0.5 (Table 2). This suggests a down-regulation of ATP generation, as a result of reduced photosynthetic activity.
(c) CO2 fixation.
The transcription of genes involved in carbon dioxide fixation is regulated by light (31). The transcription of ribulose bisphosphate carboxylase-oxygenase (Rubisco) genes (rbcL and rbcS) was synchronized with the level of photosynthetic activity: rbcL, rbcS, and rbcX were four- to fivefold down-regulated after 2 h of intense UV-B exposure. RbcX acts analogously to the GroEL/GroES chaperone proteins for Rubisco folding (21). An inorganic carbon-concentrating mechanism in cyanobacteria allows the cells to adapt to a wide range of CO2 concentrations in the environment via a light-dependent process (17). Under UV-B conditions, the operon composed of genes encoding carbon dioxide-concentrating mechanism proteins (ccmK [sll1028 and sll1029], ccmM [sll1031], and ccmN [sll1032]) was repressed (Table 2).
(d) HLIP family.
hliA (ssl2542), hliB (ssr2595) and hliC (ssl1633) encode for small 70-amino-acid HLIPs (7). Their transcript levels increase in high-intensity light and during nutrient starvation, and their presence has been proposed to be essential for acclimation to high-intensity white light (13). hliB of Synechocystis is one of the most highly induced genes (31-fold) after 2 h of 60 microeinsteins of UV-B m−2 s−1. hliA and hliC were induced by five- and eightfold, respectively. Therefore, it is likely that hliA, hliB, and hliC are important for UV-B acclimation in addition to high light acclimation.
(e) Heat shock response.
The heat shock response is triggered by unfolded cytoplasmic proteins (25). We have shown that UV-B led to induction of the heat shock response in Synechocystis. Heat shock regulon members whose mRNAs were elevated include dnaJ, dnaK, groEL, groES, clpB, hsp17, and htpG. Proteases HhoA, HhoB, Prc, and ClpC are involved in degradation of damaged or misfolded proteins (1). The transcripts specifying these proteins were induced moderately (two- to threefold) after 120 min of 60 microeinsteins of UV-B irradiation m−2 s−1 (Table 1).
(f) Stringent response.
The 2-h exposure of Synechocystis to intense UV-B led to a stringent response (3) in Synechocystis, a phenomenon not observed at 20 microeinsteins m− 2 s−1. There was an overall repression of the translation machinery, including genes encoding large and small ribosomal proteins (0.5-fold for most rbl and rbs genes) (Table 2). The transcript level of tufA, encoding protein synthesis elongation factor Tu, was down-regulated more than fivefold. In E. coli, the transcription of rpoD encoding the major sigma factor σ70 is positively regulated during the stringent response (3). The Synechocystis genome contains six copies of rpoD genes, three of which (sll2012, sll0184, and sll0306) were induced. Due to their sequence homology, the expression profiles of these three rpoD genes cannot be differentiated by DNA microarray analysis.
(g) Light-mediated signaling.
Plant phytochromes are photoreceptor proteins that regulate growth and development in response to changes in the ambient light environment (26). The cyanobacterial phytochrome Phy (slr0473) is responsible for sensing red or far red light and acts as a light-regulated histidine kinase in the initial step of the light-mediated signaling pathway (35). The transcription of slr0473 was repressed by a factor of 2 after 20 min of irradiation with 20 microeinsteins of UV-B m−2 s−1 and by a factor of 4 after 2 h of 60 microeinsteins of UV-B m−2 s−1. This suggests that Phy might be involved in the signaling pathways in response to UV-B and visible light. A phytochrome-regulated gene (sll1214), with homologous sequences in Porphyra purpurea and Arabidopsis thaliana, was down-regulated by UV-B, possibly as a result of phytochrome-mediated transcriptional repression.
(iii) Confirmation of DNA microarray data with Northern blot analysis.
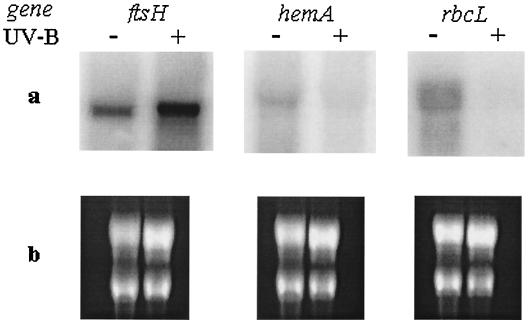
In order to validate the results from the microarray study, we performed Northern blot analysis for ftsH, hemA, and rbcL genes (Fig. 2). A comparison of Northern blot and DNA microarray results is listed in Table 3. For all three genes, expression ratios obtained by DNA microarray were consistent with ratios obtained by Northern blot analysis. This confirmed qualitatively the measurements made by microarray and suggested that nonspecific hybridization of rRNA to DNA probes is not significant.
FIG. 2.
Northern blot analysis of Synechocystis ftsH, rbcL, and hemA genes in response to 60 microeinsteins of UV-B irradiation m−2 s−1 for 2 h. (a) RNA blot images and (b) ethidium bromide-stained gel images indicating equal loading of RNA are shown. The RNA was from unlabeled control samples (−) and from UV-B irradiated samples (+).
TABLE 3.
Comparison of expression ratios from DNA microarray and Northern blot analysis
| Gene | ORF | Treated/control ratioa
|
|
|---|---|---|---|
| Microarray | Northern | ||
| ftsH | slr0228 | 5.3 ± 2.7 | |
| slr1390 | 3.4 ± 1.3 | 2.55 | |
| sll1463 | 2.0 ± 0.4 | ||
| hemA | slr1808 | 0.30 ± 0.15 | 0.26 |
| rbcL | slr0009 | 0.26 ± 0.15 | 0.13 |
For explanation of treated/control ratio, see Materials and Methods. For data derived from microarray analysis, the expression ratio (treated/control ratio) for each transcript is calculated from the average expression ratios based on multiple measurements. Data from Northern analysis are based on single hybridization experiments.
(iv) Gene expression profiling of Synechocystis sp. strain PCC 6803 in response to increased irradiation with white light.
We studied the change in the gene expression profile of Synechocystis in response to a shift from a low light growth condition (25 microeinsteins of white florescent light m−2 s−1) to a relatively high light fluence (200 microeinsteins m−2 s−1) for a period of 2 h. The overall gene expression pattern is different from that of the UV-B-irradiated Synechocystis cells, while similarities can be found in their effects on the photosystems and the heat shock modulon. Unlike UV-B, white light at 200 microeinsteins m−2 s−1 did not induce the stringent response in Synechocystis. This demonstrated the different mechanisms adopted by Synechocystis in response to variations in light quality (wavelength) as opposed to light intensity (fluence).
At an intense white light fluence, the most significantly induced gene families were involved in CO2 transport, concentration, and fixation. The transcripts of a bicarbonate transporter complex were elevated, including the genes encoding CmpA (slr0040, sevenfold), CmpB (slr0041, fivefold), and CmpC (slr0043, threefold). Genes encoding inorganic carbon-concentrating mechanism proteins, including CcmK (sll1028 and sll1029), CcmL (sll1030), CcmM (sll1031), and CcmN (sll1032) were induced by two- to threefold. Rubisco genes were also moderately induced: rbcL, rbcS, and rbcX were induced twofold each, indicating increased carbon fixation activity. In contrast, the ccm genes and rbc genes were repressed by the same fluence of UV-B light. NAD(P)H dehydrogenase type I complexes have been implicated in the CO2 uptake process (19). Consistent with this, intense white light-induced expression of NADH dehydrogenase subunits NdhD (slr2007), NdhD2 (slr1291), NdhD3 (sll1733), and NdhF (sll1732), with induction ratios of two- to sevenfold, was observed.
The effects of intense white light irradiation on the photosynthetic pathways were similar to UV-B irradiation of the same fluence. Increase of light intensity from 25 to 200 microeinsteins m−2 s−1 resulted in great repression of genes encoding phycobilisome core and linker proteins, including allophycocyanin subunit-specifying genes apcA, apcB, apcC, apcE, apcF, and phycocyanin subunit-specifying genes cpcA, cpcB, cpcC, cpcD, and cpcG. This suggests that phycobilisome size was reduced when more white light was available to the cells, minimizing the optical cross section of the antenna and hence the absorption of excessive photons. Photosystem I was down-regulated, as manifested by the repression of most psa genes (psaA, psaB, psaC, psaD, psaE, psaF, psaJ, psaK, and psaL) by two- to fivefold. Biosynthesis leading to phycobilin and chlorophyll was possibly reduced, as suggested by the repression of hemA, hemF, ho, chlB, chlL, and chlP.
Similar to the UV-B exposure, carotenoid biosynthesis genes crtP and crtQ were induced by intense white light, perhaps to increase singlet oxygen scavenging. Oxidative stress could also be countered by induction of glutathione peroxidase (slr1992) that detoxifies peroxides. slr1992 was up-regulated fourfold. Only one of the three hli transcripts elevated by UV-B, hliC, was induced fourfold by 200 microeinsteins of white light m−2 s−1. High-intensity white light also initiated a heat shock response in Synechocystis, as observed by the three- to fourfold induction of groEL/groES transcripts and two- to threefold induction of hhoA, hhoB, prc, and hsp17.
DISCUSSION
To accomplish photosynthetic electron transport and carbon fixation while avoiding damage from sunlight, cyanobacteria have evolved to adapt to constant fluctuations in solar irradiation, of which about 7% is in the UV-B range. We applied UV-B irradiation to Synechocystis cells at intensities that approximate the amount of UV-B in sunlight and studied the initial transcriptional response after a 20-min irradiation and the longer-term response to a 2-h exposure. After a 20-min UV-B exposure, changes in gene transcription profile were observed, consistent with previously described phenotypic responses of Synechocystis cells challenged by UV-B. The primary targets of UV-B irradiation are the photosystems and the accessory light-harvesting pigments. The heat shock regulon was induced, perhaps in response to the oxidative damage generated by UV-B treatment. At a higher UV-B intensity of 60 microeinsteins m−2 s−1, the triggering of the stringent response was observed, which could be due to inadequate concentrations of aminoacyl-tRNA molecules, perhaps caused by intramolecular tRNA cross-links. A further down-regulation of photosynthetic genes (except for psbA genes) suggested a decrease of photosynthetic activity as UV-B exposure intensified from 20 to 60 microeinsteins m−2 s−1. About half of Synechocystis cells survived the 60 microeinsteins of UV-B irradiation m−2 s−1 for 2 h, suggesting that the cells were able to recover from damage caused by UV-B.
By exposing Synechocystis to intense white light, we sought to understand the mechanisms by which Synechocystis adapted to changes in light intensity versus light quality. We chose to irradiate Synechocystis at 200 microeinsteins of white light m−2 s−1, which is comparable to the intensity of 60 microeinsteins of UV-B irradiation m−2 s−1. Although these two experiments cannot be directly compared for their light acclimation programs, it is reasonable to compare the transcriptional response to various light stresses and therefore differentiate their stress response mechanisms. As indicated in the profiles of certain stress response genes (e.g., those encoding glutathione peroxidase and heat shock proteins), Synechocystis cells experienced stress even when shifted from 25 microeinsteins of white light m−2 s−1 to the moderately high-intensity white light condition at 200 microeinsteins m−2 s−1. The intense white light- and UV-B-mediated gene expression profiles overlapped in the down-regulation of photosynthetic genes and induction of the heat shock response but differed in several other transcriptional processes, including those specifying carbon dioxide uptake and fixation, the stringent response, and the induction profile of the HLIPs.
Photosystem II D1 protein is a primary target of UV-B and white light damage, and transcription of the psbA genes encoding the D1 protein was shown previously to be induced by both UV-B and intense white light (23). Our results indicate that both D1 resynthesis and degradation respond similarly to UV-B and strong white light. We found that the transcript level of the Synechocystis FtsH protease is likely to be coregulated with the transcription of psbA. It is possible that one or more of the four copies of FtsH protease degrade D1 protein, although more-detailed studies are needed to identify the roles of all the FtsH proteins. The observed parallel responses of transcripts specifying D1, D1-processing protease CtpA, and D1-degrading FtsH protease suggested that the biosynthesis, processing, and degradation of D1 are coordinated.
A significant amount of the biosynthetic capacity of Synechocystis is committed to synthesis of proteins of the phycobilisome complex. For Synechocystis grown under 25 microeinsteins of white light m−2 s−1, phycobilisome genes are among the most highly expressed genes in the genome (data not shown). In nitrogen-depleted conditions, phycobilisomes are degraded to provide nitrogen and carbon for cellular biosynthesis (12). NblA activates phycobilisome degradation during nutrient limitation, whereas NblB encodes a polypeptide homologous to CpcE, a subunit of phycocyanin-phycocyanobilin lyase. While nblA is induced during nutrient limitation, the transcript level of nblB is constitutive (9). Our results indicated that under UV-B illumination, Synechocystis cells might inversely couple phycobilisome biosynthesis with the phycobilisome catabolism in the form of the production of phycobilisome degradation proteins NblA and NblB. The induction of nblB upon UV-B irradiation supports the suggestion that NblB plays a role in hydrolysis of the covalent bond between linear tetrapyrrole and apophycobiliproteins as part of the degradation process of phycobilisome complexes (6).
Although the heat shock response was evident in both UV-B- and high-light-irradiated cells, the numbers of genes induced were significantly different. Under 200 microeinsteins of white light m−2 s−1, only genes coding for the GroEL/GroES complex and Clp protease/chaperone family were induced, but upon irradiation with UV-B, additional genes including those coding for the DnaJ/DnaK and Hsp90 (HtpG) chaperone complexes were also induced, as well as the transcript which specifies the small heat shock protein Hsp17. The stronger heat shock responses could reflect greater damage caused by free radicals from UV-B illumination compared to white light and differential transcription of the heat shock regulon. Due to the increased radical levels and superoxide concentrations upon UV-B exposure, Synechocystis may also have elevated titers of protective molecules, including carotenoids, glutathione peroxidase, and superoxide dismutase, as suggested by the mRNA profiles.
HLIPs have been proposed to play roles in adaptation to both nutrient starvation and high-intensity white light (4). hliA (ssl2542), hliB (ssr2595), and hliC (ssl1633) were all highly induced by UV-B, but only hliC was induced by 200 microeinsteins of white light m−2 s−1. It is possible that the transcription of hliA, hliB, and hliC are regulated by oxidative damage caused by either UV-B or intense white light. Could hliC be induced by a relatively low level of oxidative damage in white light, which does not lead to elevated transcription of hliA and hliB? On the other hand, hliA and hliB have relatively higher induced levels under more-intense light stresses. At 500 microeinsteins of white light m−2 s−1, the concentration of free radicals may become comparable to radicals generated by 20 to 60 microeinsteins of UV-B m−2 s−1 and may have led to induction of hliA and hliB in the studies of He et al. (13). Although less likely, the minor UV-B component in white light irradiation (1 microeinstein of UV-B m−2 s−1 in 200 microeinsteins of white light m−2s −1) from the fluorescent light source might be another reason for the induction of hliC under intense white light irradiation.
In summary, we have performed a comprehensive analysis of global gene expression profiles in cyanobacterium Synechocystis in response to UV-B irradiation and high-intensity white light illumination, two common environmental factors for photosynthetic organisms. We showed that the DNA microarray approaches used for E. coli, Saccharomyces cerevisiae, and other organisms could be directly applied to Synechocystis. This work demonstrated an integrated global regulatory network in Synechocystis that suggests a dynamic orchestration of many pathways, including light harvesting and assimilation, photosynthesis, detoxification, heat shock response, and the stringent response. Both UV-B and high white light irradiation resulted in coordinate responses involving multiple strategies adopted by Synechocystis cells to avoid excessive light harvesting, reduce oxidative damage, and degrade and resynthesize damaged proteins. Since many Synechocystis genes have orthologs in algae and higher plants, the adaptation of cyanobacteria to UV-B and high-intensity white light will shed light as to how other photosynthetic organisms adapt to irradiation. Our results of gene expression profiles upon intense white light irradiation are consistent with the profiles described by Hihara et al. performed under similar conditions (14).
Acknowledgments
We thank Bruce Diner for thoughtful insights and critical reading of the manuscript. We also thank Zhixiong Xue, Tina Van Dyk, and Dexter Chisholm for helpful comments on the work. We are grateful to Steven Picataggio and Lee Heineman for developing a Microsoft Excel-based system for the analysis of transcriptional data. We thank Sean Siwen Huang and Dilip Rajagopalan for performing statistical analysis for the microarray data. Qun Zhu provided help with Northern blot analysis. We also thank Erik Gommeren for generating the figures and other computer support.
REFERENCES
- 1.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondel, M. O., and A. Favre. 1988. tRNAPhe and tRNAPro are the near-ultraviolet molecular targets triggering the growth delay effect. Biochem. Biophys. Res. Commun. 150:979-986. [DOI] [PubMed] [Google Scholar]
- 3.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 4.Collier, J. L., and A. R. Grossman. 1994. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13:1039-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 6.Dolganov, N., and A. R. Grossman. 1999. A polypeptide with similarity to phycocyanin alpha-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J. Bacteriol. 181:610-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolganov, N. A., D. Bhaya, and A. R. Grossman. 1995. Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: evolution and regulation. Proc. Natl. Acad. Sci. USA 92:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehling-Schulz, M., W. Bilger, and S. Scherer. 1997. UV-B-induced synthesis of photoprotective pigments and extracellular polysaccharides in the terrestrial cyanobacterium Nostoc commune. J. Bacteriol. 179:1940-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fairchild, C. D., and A. N. Glazer. 1994. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin alpha subunit phycocyanobilin lyase. J. Biol. Chem. 269:8686-8694. [PubMed] [Google Scholar]
- 10.Giacometti, G. M., R. Barbato, S. Chiaramonte, G. Friso, and F. Rigoni. 1996. Effects of ultraviolet-B radiation on photosystem II of the cyanobacterium Synechocystis sp. PCC 6083. Eur. J. Biochem. 242:799-806. [DOI] [PubMed] [Google Scholar]
- 11.Grether-Beck, S., G. Bonizzi, H. Schmitt-Brenden, I. Felsner, A. Timmer, H. Sies, J. P. Johnson, J. Piette, and J. Krutmann. 2000. Nonenzymatic triggering of the ceramide signalling cascade by solar UVA radiation. EMBO J. 19:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossman, A. R., M. R. Schaefer, G. G. Chiang, and J. L. Collier. 1993. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 57:725-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He, Q., N. Dolganov, O. Bjorkman, and A. R. Grossman. 2000. The Hli polypeptides in Synechocystis PCC6803: expression and function in high light. J. Biol. Chem. 276:306-314. [DOI] [PubMed] [Google Scholar]
- 14.Hihara, Y., A. Kamei, M. Kanehisa, A. Kaplan, and M. Ikeuchi. 2001. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschberg, J., and D. Chamovitz. 1994. Carotenoids in cyanobacteria, p. 559-579. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 16.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, A., R. Schwarz, J. Lieman-Hurwitz, M. Ronen-Tarazi, and L. Reinhold. 1994. Physiological and molecular studies on the response of cyanobacteria to changes in the ambient inorganic carbon concentration, p. 469-485. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Kehoe, D. M., and A. R. Grossman. 1994. Complementary chromatic adaptation: photoperception to gene regulation. Semin. Cell Biol. 5:303-313. [DOI] [PubMed] [Google Scholar]
- 19.Klughammer, B., D. Sultemeyer, M. R. Badger, and G. D. Price. 1999. The involvement of NAD(P)H dehydrogenase subunits, NdhD3 and NdhF3, in high-affinity CO2 uptake in Synechococcus sp. PCC7002 gives evidence for multiple NDH-1 complexes with specific roles in cyanobacteria. Mol. Microbiol. 32:1305-1315. [DOI] [PubMed] [Google Scholar]
- 20.Lao, K., and A. N. Glazer. 1996. Ultraviolet-B photodestruction of a light-harvesting complex. Proc. Natl. Acad. Sci. USA 93:5258-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L. A., and F. R. Tabita. 1997. Maximum activity of recombinant ribulose 1,5-bisphosphate carboxylase/oxygenase of Anabaena sp. strain CA requires the product of the rbcX gene. J. Bacteriol. 179:3793-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindahl, M., C. Spetea, T. Hundal, A. B. Oppenheim, Z. Adam, and B. Andersson. 2000. The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12:419-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mate, Z., L. Sass, M. Szekeres, I. Vass, and F. Nagy. 1998. UV-B-induced differential transcription of psbA genes encoding the D1 protein of photosystem II in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 273:17439-17444. [DOI] [PubMed] [Google Scholar]
- 24.Montane, M., and K. Kloppstech. 2000. The family of light-harvesting-related proteins (LHCs, ELIPs, HLIPs): was the harvesting of light their primary function? Gene 258:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Neidhardt, F. C., and R. A. VanBogelen. 1987. Heat shock response, p. 1334-1345. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 1st ed. American Society for Microbiology, Washington, D.C.
- 26.Quail, P. H., M. T. Boylan, B. M. Parks, T. W. Short, Y. Xu, and D. Wagner. 1995. Phytochromes: photosensory perception and signal transduction. Science 268:675-680. [DOI] [PubMed] [Google Scholar]
- 27.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Satoh, K. 1998. Photooxidative stresses, p. 3-111. In K. Satoh and N. Murata (ed.), Stress responses of photosynthetic organisms. Elsevier, Amsterdam, The Netherlands.
- 31.Tabita, F. R. 1994. The biochemistry and molecular regulation of carbon dioxide metabolism in cyanobacteria, p. 437-467. In D. A. Bryant (ed.), The molecular biology of cyanobacteria, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Thomas, G., and A. Favre. 1975. 4-Thiouridine as the target for near-ultraviolet light induced growth delay in Escherichia coli. Biochem. Biophys. Res. Commun. 66:1454-1461. [DOI] [PubMed] [Google Scholar]
- 33.Vass, I. 1996. Adverse effects of UV-B light on the structure and function of the photosynthetic apparatus, p. 931-949. In M. Pessarakli (ed.), Handbook of photosynthesis. Marcel Dekker, New York, N.Y.
- 34.Wei, Y., J.-M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh, K. C., S. H. Wu, J. T. Murphy, and J. C. Lagarias. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277:1505-1508. [DOI] [PubMed] [Google Scholar]