Abstract
Bacteria of Shigella spp. use a virulence plasmid-encoded type III secretion (TTS) system to invade the colonic epithelium in humans. The activity of the TTS apparatus is tightly regulated in the wild-type strain and is induced upon contact of bacteria with epithelial cells, whereas it is deregulated, i.e., constitutively active, in some mutants. Under conditions of deregulated secretion, approximately 20 proteins are secreted, including VirA, OspB to OspG, and at least three members of the IpaH family, all of which are encoded by the virulence plasmid. Conditions inducing or deregulating the activity of secretion also induce the transcription of virA and four ipaH genes. The transcription of virA and ipaH9.8 requires both MxiE, a transcriptional activator of the AraC family, and IpgC, the chaperone of IpaB and IpaC, acting as a coactivator. Using reporter plasmids containing lacZ transcriptional fusions, we showed that the ipaH7.8. ipa4.5. ospC1, and ospF promoters are activated under conditions of deregulated secretion and that both MxiE and IpgC are necessary and sufficient for their activation in both Shigella flexneri and Escherichia coli. Promoter mapping and deletion analysis of the ipaH9.8. virA, and ospC1 promoters identified a 17-bp motif, the MxiE box, which overlaps the −35 region and is essential for the activation of these promoters. The presence of eight MxiE boxes on the virulence plasmid suggests that 11 genes encoding secreted proteins may be regulated by the activity of secretion. We also present evidence that at least one ipaH gene that is carried by the chromosome is controlled by MxiE and IpgC.
The type III secretion (TTS) pathway is used by numerous gram-negative pathogenic bacteria to deliver virulence proteins to the membrane or directly to the cytoplasm of host cells, where they interfere with cellular signaling pathways. The TTS system consists of (i) the secretion apparatus that spans the bacterial envelope, (ii) translocators that are inserted into the host cell membrane and effectors that are translocated into the host cell cytoplasm, (iii) specific cytoplasmic chaperones that associate with the translocators and some effectors within the bacterial cytoplasm, and (iv) transcriptional regulators required for the expression of components of the TTS apparatus and/or proteins secreted by this apparatus (21).
The TTS apparatus encoded by the virulence plasmids of Yersinia and Shigella spp. and the pathogenicity islands SPI-1 and SPI-2 of Salmonella spp. is not or is only weakly active during growth of bacteria in broth and is activated upon contact of bacteria with host cells (29, 34, 45). In vitro, increased secretion is obtained following exposure of bacteria to a medium (i) deprived of calcium for Yersinia spp. (31, 41), (ii) containing the dye Congo red for Shigella spp. (4), (iii) or at a low pH for Salmonella spp. (5). In addition, the inactivation of some genes, such as yopN. tyeA, or lcrG in Yersinia spp. and ipaB and ipaD in Shigella spp., results in deregulated secretion; i.e., the TTS apparatus of these mutants is active in the absence of inducers (8, 16, 30, 39). Conditions inducing or deregulating the activity of secretion also result in the increased transcription of some genes encoding secreted proteins, such as the yop genes in Yersinia spp. and the virA and ipaH genes in Shigella spp. (10, 13, 17, 42). In Yersinia spp., the mechanism by which the activity of secretion regulates the transcription of yop genes involves the secreted protein LcrQ and its cytoplasmic chaperone SycH but is not yet understood (18, 34, 36, 41, 44). In Shigella flexneri, the transcription of virA and ipaH genes requires both MxiE, a transcriptional activator of the AraC family, and its coactivator, IpgC, the chaperone for the proposed IpaB and IpaC translocators (28). Under conditions of nonsecretion, IpaB and IpaC act as anticoactivators by binding to and titrating IpgC, thereby rendering it unavailable to activate MxiE. This mechanism of regulation is reminiscent of that for the flagellin genes in Salmonella enterica serovar Typhimurium, in which the secretion of anti-sigma factor FlgM upon completion of the hook-basal body complex is required to liberate sigma factor σ28, which can then transcribe the late genes for flagellin (9, 22).
Bacteria of Shigella spp., the causative agents of bacillary dysentery in humans, use a TTS system to invade the colonic epithelium, resulting in tissue destruction and massive inflammation (25). Genes required for the entry of bacteria into epithelial cells are located on a 30-kb region, designated the entry region, of the 210-kb virulence plasmid (27, 38). This region contains genes for the Mxi-Spa TTS apparatus; the secreted IpaA to IpaD, IpgB1, and IpgD proteins; the chaperones IpgC, IpgE, and Spa15; and the transcriptional regulators VirB and MxiE (33). The transcription of genes of the entry region from the divergent icsB and ipgD promoters is under the control of both VirF and VirB. The expression of VirF, a member of the AraC family that is encoded by the virulence plasmid, is induced at 37°C (14) and activates transcription from the virB promoter (23). VirB, a member of the ParB family of partitioning proteins, is required for transcription of the icsB and ipgD promoters (1). Although the TTS apparatus is assembled and effector proteins are synthesized, wild-type S. flexneri secretes only a small proportion (∼5%) of IpaA to IpaD and IpgD upon growth at 37°C in laboratory medium (3). Secretion is induced by contact with cells (30), addition of the dye Congo red to the growth medium (4), or inactivation of ipaB or ipaD (29, 32). Under conditions of deregulated secretion, approximately 20 proteins are secreted, including VirA, OspB to OspG, and IpaH (6, 32). The virulence plasmid carries five ipaH genes, designated ipaH1.4. ipaH2.5. ipaH4.5. ipaH7.8, and ipaH9.8, according to the size of the HindIII fragment that carries each copy (20), and three ospC, three ospD, and two ospE genes (6).
It was previously shown that the transcription of the virA gene and four ipaH genes is regulated by the activity of the Mxi-Spa apparatus (13). The transcription of these genes is induced upon the entry of bacteria into epithelial cells, during the growth of bacteria in the presence of Congo red, and following the inactivation of ipaB or ipaD. Under conditions of active or deregulated secretion, activation of the virA and ipaH9.8 promoters is controlled by the MxiE transcriptional activator and the IpgC chaperone acting as a coactivator (28). However, neither the cis-acting element(s) involved in the activation of these promoters in response to secretion nor the repertoire of genes whose transcription is controlled by the activity of secretion has been characterized. In the present study, we used reporter plasmids containing the putative promoter regions of a number of genes encoding secreted proteins to investigate (i) which genes are regulated by the activity of secretion, (ii) the cis-acting region involved in this regulation, and (iii) whether or not these genes are under the control of MxiE and IpgC. Using promoter mapping and deletion analysis of the virA. ipaH9.8, and ospC1 promoters, we identified a motif, designated the MxiE box, which is essential for the activation of regulated promoters. Sequence analysis revealed the presence of eight MxiE boxes on the virulence plasmid, suggesting that 11 genes encoding secreted proteins may be regulated by the activity of secretion. We also present evidence that at least one ipaH gene carried by the chromosome is controlled by MxiE and IpgC.
MATERIALS AND METHODS
Bacterial strains and growth media.
S. flexneri strains are derivatives of wild-type strain M90T (serotype 5) (37); M90T-Sm (Smr), BS176 (a virulence plasmid-cured strain), SF622 (ipaD2), SF1076 (ipaB4), SF1070 (mxiE ipaB4) and SF1068 (ipgC ipaB4) have been described elsewhere (2, 28, 29). Escherichia coli strain DH5α was used for plasmids carrying the oriT origin of replication. Bacteria were grown in Luria-Bertani (LB) medium at 37°C. Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; chloramphenicol, 25 μg ml−1; and streptomycin, 100 μg ml−1.
Construction of reporter plasmids.
Briefly, a DNA fragment extending from the 3′ end of the nearest insertion sequence (IS) element or coding sequence located upstream from the gene of interest to approximately 100 bp downstream from the translation start site of that gene was amplified by PCR and cloned into the BamHI and HindIII sites of vector pQF50 (15), upstream from and in the same orientation as a promoterless lacZ gene. Coordinates, with respect to the sequence of pWR100 (6), of the 5′ and 3′ ends of the fragment cloned in each plasmid are indicated in Table 1. The construction of ipaH9.8-lacZ (pMM10) and virA-lacZ (pBD7) reporter plasmids, pMM71 (MxiE), and pKH128 (IpgC) has been described elsewhere (28).
TABLE 1.
Reporter plasmids used in this study
| Reporter plasmida | Transcriptional fusion | Cloned fragment
|
||
|---|---|---|---|---|
| Coordinateb
|
Size (bp) | |||
| 5′ End | 3′ End | |||
| pMM10 | ipaH9.8-lacZ | 173945 | 174493 | 547 |
| pMM33 | ipaH9.8-lacZ | 174183 | 174493 | 309 |
| pMM34 | ipaH9.8-lacZ | 174203 | 174493 | 289 |
| pMM11 | ipaH7.8-lacZ | 63585 | 64170 | 584 |
| pMM12 | ipaH4.5-lacZ | 65773 | 66399 | 625 |
| pMM13 | ipaH1.4-lacZ | 208426 | 208995 | 568 |
| pMM19 | ospC1-lacZ | 78233 | 78976 | 742 |
| pMM35 | ospC1-lacZ | 78233 | 78643 | 409 |
| pMM37 | ospC1-lacZ | 78233 | 78616 | 382 |
| pMM29 | ospD3-lacZ | 76410 | 76921 | 510 |
| pMM65 | ospF-lacZ | 12359 | 12488 | 128 |
| pMM20 | ospG-lacZ | 176488 | 176827 | 338 |
| pBD7 | virA-lacZ | 145165 | 146008 | 842 |
| pBD9 | virA-lacZ | 145165 | 145888 | 722 |
| pMM30 | icsB-lacZ | 111389 | 111804 | 414 |
The reporter plasmids were constructed with vector pQF50.
Coordinates of the 5′ end and the 3′ end of the cloned fragment are indicated with respect to the sequence of pWR100 (6). In pWR100, the direction of transcription of ipaH9.8. ipaH7.8. ipaH4.5, and ospG is clockwise, whereas that of ipaH1.4, ospC1. ospD3, and virA is counterclockwise.
Purification of RNA.
RNA preparations were performed essentially as described by Spohn et al. (40). S. flexneri strains M90T (wild type) and SF622 (ipaD) were grown in 25 ml of LB broth at 37°C to mid-log phase and harvested. Cells were lysed in 3.7 ml of a solution containing 100 mM Tris-HCl (pH 7.5), 2 mM disodium EDTA, and 1% sodium dodecyl sulfate (SDS) and incubated for 5 min at 95°C. Lysates were adjusted to 80 mM KCl and incubated for 10 min on ice. Cellular debris was removed by centrifugation at 8,000 rpm for 10 min in a JA20 rotor (Beckman). To 3.5 ml of supernatant, 4.56 g of CsCl was added, and the RNA was sedimented by centrifugation in an SW65 rotor (Beckman) for 15 to 20 h at 35,000 rpm. The RNA pellet was resuspended in 500 μl of Tris-EDTA (10 mM Tris-HCl [pH 8], 1 mM disodium EDTA), extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, resuspended in 200 μl of Tris-EDTA, reprecipitated, and stored at −20°C.
Primer extension analyses.
Oligonucleotides (10 pmol) were 5′ end labeled in the presence of [γ-32P]ATP (5,000 Ci mmol−1) and T4 polynucleotide kinase (New England Biolabs). Labeled oligonucleotides (0.4 to 1.0 pmol) were then coprecipitated with 15 μg of S. flexneri total RNA and resuspended in 5 μl of H2O-2 μl of each deoxynucleoside triphosphate (2 mM)- 2 μl of 5× reverse transcription buffer (cDNA synthesis kit; Boehringer Mannheim). The reaction mixture was incubated for 1 min at 95°C, 1 μl of reverse transcriptase (20 U ml−1; Boehringer Mannheim) was added, and reverse transcription was carried out at 45°C for 45 min. Samples were then incubated for 10 min at room temperature with 1 μl of RNase A (1 mg ml−1), extracted once with an equal volume of phenol-chloroform (1:1), ethanol precipitated, and resuspended in 5 μl of sequencing loading buffer. After denaturation at 95°C for 2 min, samples were subjected to urea-6% polyacrylamide gel electrophoresis (PAGE) together with the sequencing reaction products obtained by use of the same primers with plasmids containing fragments of the ospC1. virA, and ipaH9.8 promoters. Sequencing reactions were performed by the dideoxy chain termination method with [α-32P]dATP (Amersham) and a T7 sequencing kit (Pharmacia).
Enzyme assays.
The β-galactosidase activity present in bacteria growing in LB medium and harvested during the exponential phase of growth was assayed by using the substrate o-nitrophenyl-β-d-galactopyranoside as described previously (35)
Protein analysis.
To prepare whole-cell extracts, bacteria were grown in 3 ml of LB medium. One milliliter of bacteria was centrifuged and resuspended in 500 μl of Laemmli sample buffer (26). Protein samples were boiled for 3 min and analyzed by SDS-PAGE as described previously (26). After electrophoresis, proteins were transferred to a nitrocellulose membrane. Immunoblotting was carried out with a rabbit polyclonal anti-IpaH antibody (28). Horseradish peroxidase-labeled goat anti-rabbit antibodies were used as secondary antibodies and visualized by enhanced chemiluminescence.
RESULTS
Use of reporter plasmids to investigate the regulation of genes encoding secreted proteins.
To identify the cis-acting regions responsible for the regulation of transcription of the ipaH7.8. ipaH4.5, and ipaH1.4 genes in response to secretion (13), the putative promoter regions of these genes were amplified by PCR and cloned upstream from the promoterless lacZ gene of reporter plasmid pQF50 (Table 1). In each case, the cloned fragment extended from the 3′ end of the upstream IS element or coding sequence to 100 bp downstream from the translation start site of the gene of interest. As a control, we also cloned the icsB promoter, which is not regulated by the activity of secretion (13), into pQF50. The resulting reporter plasmids were each introduced into S. flexneri strain M90T (wild type), in which the activity of secretion is tightly controlled and promoters regulated by secretion are not active, and strain SF622 (ipaD), which displays a phenotype of deregulated secretion and in which promoters regulated by secretion are active (13, 28). β-Galactosidase activity was assayed during growth of the recombinant strains at 37°C (Table 2). As observed for reporter plasmids carrying the ipaH9.8 and virA promoters (28), the reporter plasmid containing a fusion with the ipaH7.8 putative regulatory region was activated more than 10-fold in the ipaD mutant compared to the wild-type strain, indicating that the regulatory region was present and functional on the reporter plasmid. In contrast, reporter plasmids containing fusions with ipaH4.5 and ipaH1.4 showed a moderate increase (threefold) and no increase, respectively, in β-galactosidase activity in the ipaD mutant. The very low level of β-galactosidase activity expressed from the ipaH1.4 reporter plasmid suggested that no promoters were present upstream from ipaH1.4, i.e., in the 389-bp ospE1-ipaH1.4 intergenic region. In contrast, the intermediate level of activation of the ipaH4.5-lacZ fusion suggested that the promoter and regulatory regions of ipaH4.5 were indeed present on the reporter plasmid, i.e., in the 429-bp ipaH7.8-ipaH4.5 intergenic region, and that this promoter was not as tightly regulated as that of the ipaH7.8 gene. As previously described for an icsB-lacZ transcriptional fusion carried by the virulence plasmid (13), the levels of expression of the icsB-lacZ fusion carried by the reporter plasmid were similar in the wild-type and ipaD strains.
TABLE 2.
Expression of lacZ transcriptional fusions from reporter plasmids in S. flexneri strains
| Transcriptional fusion | Reporter plasmida | Size of the cloned fragment (bp) | β-Galactosidase activity in derivatives of the following strainb:
|
Ratioc | |
|---|---|---|---|---|---|
| Wild type | ipaD | ||||
| ipaH7.8-lacZ | pMM11 | 584 | 30 | 1,700 | 55 |
| ipaH4.5-lacZ | pMM12 | 625 | 150 | 420 | 3 |
| ipaH1.4-lacZ | pMM13 | 568 | 45 | 40 | 1 |
| ospC1-lacZ | pMM19 | 740 | 30 | 440 | 15 |
| ospF-lacZ | pMM65 | 128 | 45 | 370 | 8 |
| ospD3-lacZ | pMM29 | 510 | 130 | 220 | 1.7 |
| ospG-lacZ | pMM20 | 338 | 220 | 250 | 1 |
| icsB-lacZ | pMM30 | 414 | 1,300 | 760 | 0.6 |
The reporter plasmids were constructed with vector pQF50 (Table 1).
β-Galactosidase activities assayed in derivatives of M90T (wild type) and SF622 (ipaD) are expressed in Miller units and are the means of at least four independent experiments. Standard deviations (not shown) were within 25% of the reported values.
Activity present in the ipaD mutant versus activity present in the wild-type strain.
To investigate the potential regulation of representatives of genes encoding proteins that were secreted under conditions of deregulated activity of the TTS apparatus (6), the putative promoter regions of ospC1. ospD3. ospF, and ospG were also cloned into pQF50, and the corresponding reporter plasmids were introduced into M90T (wild type) and SF622 (ipaD). As shown in Table 2, the levels of expression of both the ospC1-lacZ and the ospF-lacZ fusions were increased approximately 10-fold in the ipaD mutant compared to the wild-type strain, suggesting that the transcription of these two genes was also regulated by the activity of secretion. In contrast, the ospD3 and ospG reporter plasmids expressed similar levels of β-galactosidase activity in both backgrounds, suggesting that the transcription of these two genes was not regulated by the activity of secretion. In addition, similar levels of β-galactosidase activity were obtained in a S. flexneri strain cured of the virulence plasmid (data not shown), suggesting that the cloned promoters may not be controlled by regulators encoded by the virulence plasmid.
In conclusion, these and previous results (28) indicated that at least five promoters, including those for virA. ipaH9.8. ipaH7.8. ospC1, and ospF, and possibly that for ipaH4.5 were activated under conditions of deregulated secretion.
Primer extension analyses.
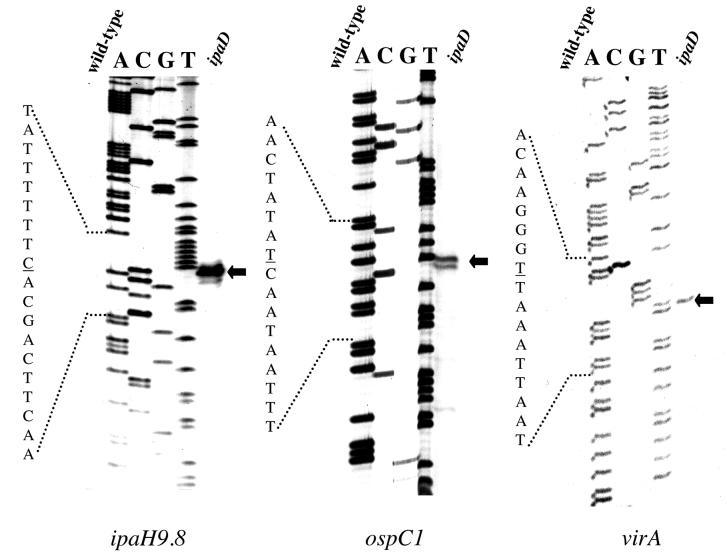
To determine the transcription start points for representative members of regulated promoters (ipaH9.8. ospC1, and virA), RNA was prepared from both wild-type and ipaD strains and used for primer extension analyses. No extension products were observed with RNA isolated from the wild-type strain, a result which was consistent with the lack of transcription of these genes under conditions of nonsecretion (Table 2) (28). The transcription start sites for the three genes were determined from the extension products obtained with the RNA samples from the ipaD mutant (Fig. 1). A strong similarity with the consensus sequence for the −10 region (TATAAT) was observed upstream from +1 for each of these promoters (Fig. 2). In contrast, no similarity or a very weak similarity was detected between the −35 regions of these promoters and the consensus −35 sequence (TTGACA).
FIG. 1.
Mapping of the ipaH9.8. ospC1, and virA promoters. Extension products obtained with RNA isolated from M90T (wild type) and SF622 (ipaD) by using primers specific for ipaH9.8. ospC1, and virA were analyzed by PAGE, together with sequencing reaction products obtained by using the same primers. The nucleotide sequence to the left of each panel corresponds to the antisense strand, and the underlined nucleotide corresponds to the major transcription start site. The arrows indicate the major primer extension products.
FIG. 2.
Comparison of the promoter regions of genes controlled by MxiE and IpgC. Sequences located upstream from the translation start site (ATG) of the various genes were aligned with respect to the transcription start site (+1). The nucleotides corresponding to the transcription start site determined for the ipaH9.8. ospC1, and virA promoters and proposed for the ospF. ipaH7.8. ospE1. ospE2, and ipaH4.5 promoters are indicated in bold and underlined characters, respectively. The number of nucleotides present between the transcription and the translation start sites is indicated. In each promoter region, nucleotides that are identical to those present in the consensus sequence of the MxiE box (last line) and the −10 region (TATAAT) are shown in uppercase bold characters. Coordinates with respect to the transcription start site are indicated at the top.
Identification of a putative regulatory region.
A comparison of the ipaH9.8. ospC1, and virA promoter sequences led to the identification of a 10-bp segment (5′-GTATCGTTTT-3′), starting at position −49 and extending to position −40, which was identical in the three promoters (Fig. 2). This 10-bp sequence was also detected upstream from the ipaH7.8 and ospF genes, both of which are also regulated by the activity of secretion (Table 2). Systematic analysis of the sequence of the virulence plasmid indicated that this conserved 10-bp sequence was also present upstream from the ospE1 and ospE2 genes, both of which encode proteins secreted by the TTS apparatus (6). A simultaneous comparison of all of these sequences indicated that the conserved region extended over 17 bp, from position −49 to position −33, and that, in each case, it was located 16 or 17 bp upstream from a sequence very similar to TATAAT (Fig. 2). The conservation of this 17-bp sequence, designated the MxiE box (for reasons detailed below), in regulated promoters and its location within these promoters suggested that it may be involved in their regulation. A sequence sharing similarities with the MxiE box was also detected upstream from the ipaH4.5 gene, the transcription of which was increased threefold in the ipaD mutant (Table 2). However, in this latter case, G and T bases, respectively, replaced the conserved T and C bases at positions −48 and −45.
Determination of the role of the MxiE box.
To investigate the potential role of the MxiE box in the regulation of the ipaH9.8 promoter, we constructed plasmids pMM33 and pMM34, in which the cloned regions started at nucleotides −63 and −43, respectively, in relation to the transcription start point and ended at nucleotide +259. In plasmid pMM33, the MxiE box was intact, whereas in plasmid pMM34, it was interrupted at position −43. The two plasmids were introduced into wild-type and ipaD strains, and β-galactosidase activity expressed in the recombinant strains was assayed (Table 3). The level of expression of the ipaH9.8-lacZ fusion from pMM33, in which the region located upstream from the consensus sequence had been deleted, was still higher in the ipaD mutant than in the wild-type strain. In contrast, similar low levels of β-galactosidase were expressed from pMM34, in which half of the MxiE box had been deleted, in both the ipaD mutant and the wild-type strain. This result indicated that the promoter carried by pMM34 was no longer regulated by the activity of secretion and suggested further that the MxiE box may represent the binding site for a transcriptional activator.
TABLE 3.
Deletion analyses of ipaH9.8. ospC1, and virA promoters
| Transcriptional fusion | Reporter plasmida | Coordinate of 5′ end of the cloned fragmentb | β-Galactosidase activity in derivatives of the following strainc:
|
Ratiod | |
|---|---|---|---|---|---|
| Wild type | ipaD | ||||
| ipaH9.8-lacZ | pMM10 | −312 | 400 | 4,000 | 10 |
| pMM33 | −63 | 480 | 1,400 | 3 | |
| pMM34 | −43 | 260 | 270 | 1 | |
| ospC1-lacZ | pMM19 | −398 | 40 | 570 | 15 |
| pMM35 | −63 | 110 | 1,100 | 10 | |
| pMM37 | −43 | 70 | 80 | 1 | |
| virA-lacZ | pBD7 | −163 | 70 | 530 | 8 |
| pBD9 | −43 | 135 | 55 | 0.4 | |
The reporter plasmids were constructed with vector pQF50 (Table 1).
Coordinates are indicated with respect to the transcription start site.
β-Galactosidase activities assayed in derivatives of M90T (wild type) and SF622 (ipaD) are expressed in Miller units and are the means of at least four independent experiments. Standard deviations (not shown) were within 25% of the reported values.
Activity present in the ipaD mutant versus activity present in the wild-type strain.
To confirm the involvement of the MxiE box in the regulation of the ospC1 and virA promoters, we constructed plasmids pMM35 and pMM37, in which the ospC1 promoter region started at nucleotides −63 and −43, respectively, and plasmid pBD9, in which the virA promoter region started at nucleotide −43. Each of these plasmids was introduced into wild-type and ipaD strains. As shown in Table 3, deletion of the region located upstream from the MxiE box in the ospC1 promoter had no effect on the regulation of this promoter. In contrast, deletion of part of the MxiE box abolished the activation of the ospC1 promoter in the ipaD mutant. Likewise, the level of expression of the virA-lacZ fusion from plasmid pBD9, in which part of the MxiE box of the virA promoter had been removed, was no longer increased in the ipaD mutant compared to the wild-type strain. Accordingly, for each of the ipaH9.8. ospC1, and virA promoters, a deletion within the MxiE box led to a loss of the activation of these promoters in the ipaD mutant.
Regulation of transcription by MxiE and the coactivator IpgC.
It was previously shown that both MxiE, a transcriptional activator of the AraC family, and IpgC, the chaperone of the IpaB and IpaC translocators, are required for the activation of transcription of the ipaH9.8 and virA promoters in response to secretion (28). To investigate whether the ospC1. ipaH7.8, and ospF promoters, which were activated in the ipaD mutant, were also under the control of MxiE and IpgC, reporter plasmids containing the ospC1. ospF, and ipaH7.8 promoters were introduced into S. flexneri strain SF1076 (ipaB4) which, like the ipaD mutant, displays a phenotype of constitutive secretion, and its derivatives, SF1068 (ipaB4 ipgC) and SF1070 (ipaB4 mxiE). As expected, each of these plasmids expressed much higher levels of β-galactosidase activity in the ipaB mutant than in the wild-type strain (Table 4). In addition, the absence of either MxiE (SF1070) or IpgC (SF1068) reduced the activity of the ospC1. ospF, and ipaH7.8 promoters to that observed in the wild-type background. These results indicated that both MxiE and IpgC were required for the transcription of these promoters in an ipaB background, i.e., under conditions of deregulated secretion.
TABLE 4.
Expression of lacZ transcriptional fusions from reporter plasmids in S. flexneri mxiE and ipgC strains
| Transcriptional fusion | Reporter plasmida | β-Galactosidase activity in derivatives of the following strainb:
|
|||
|---|---|---|---|---|---|
| Wild type | ipaB4 | ipaB4 ipgC | ipaB4 mxiE | ||
| ospC1-lacZ | pMM19 | 30 | 440 | 15 | 20 |
| ipaH7.8-lacZ | pMM11 | 30 | 1,000 | 10 | 25 |
| ospF-lacZ | pMM65 | 45 | 470 | 50 | 40 |
| ipaH4.5-lacZ | pMM12 | 150 | 500 | 140 | 130 |
| ospD3-lacZ | pMM29 | 130 | 160 | 190 | 140 |
The reporter plasmids were constructed with vector pQF50 (Table 1).
β-Galactosidase activities assayed in derivatives of M90T (wild type), SF1076 (ipaB), SF1068 (ipaB4 ipgC), and SF1070 (ipaB4 mxiE) are expressed in Miller units and are the means of at least four independent experiments. Standard deviations (not shown) were within 25% of the reported values. Data for the wild-type strain are from Table 2.
Likewise, to determine whether the ipaH4.5 promoter, which was activated only moderately in the ipaD mutant, and the ospD3 promoter, which was not activated in the ipaD mutant, were under the control of MxiE and IpgC, we introduced reporter plasmids carrying these promoters into SF1076, SF1068, and SF1070. As observed in the ipaD mutant, the level of expression of the ipaH4.5-lacZ fusion was increased threefold in the ipaB4 mutant compared to the wild-type strain (Table 4). Moreover, in the ipaB4 mxiE and ipaB4 ipgC mutants, the level of expression of this fusion was decreased to that observed in the wild-type strain. These results confirmed that the ipaH4.5 promoter was also regulated by the activity of secretion, although to a lesser extent than the ospC1. ospF, and ipaH7.8 promoters, and that this regulation was also dependent on MxiE and IpgC. In contrast, the level of expression of the ospD3-lacZ fusion was not increased in the ipaB4 mutant compared to the wild-type strain and was not affected by the inactivation of either mxiE or ipgC.
Activation of regulated promoters by MxiE and IpgC in E. coli.
As indicated above, both MxiE and IpgC were required for the activation of the ospC1. ospF, and ipaH7.8 promoters and, to a lesser extent, the ipaH4.5 promoter. To investigate whether MxiE and IpgC were the only virulence plasmid-encoded proteins directly involved in the activation of these promoters, the corresponding reporter plasmids were introduced into E. coli strains that expressed MxiE and IpgC from recombinant plasmids (28). Neither MxiE nor IpgC alone increased the levels of expression of the reporter fusions. In contrast, the levels of expression of the ospC1-lacZ. ospF-lacZ, and ipaH7.8-lacZ fusions were increased 4- to 20-fold in the E. coli strains producing both MxiE and IpgC (Table 5). These results indicated that MxiE and IpgC were sufficient and necessary for transcription from promoters possessing an MxiE box. The levels of expression of the ipaH4.5-lacZ fusion, which was regulated only moderately in S. flexneri, and the ospD3-lacZ fusion, which was not dependent on MxiE and IpgC in S. flexneri, were not increased in the presence of MxiE and IpgC. The lack of activation of transcription from the ipaH4.5 promoter may be due to its imperfect MxiE box, resulting in weaker recognition by MxiE or in weaker production of either MxiE or IpgC in the E. coli background than in the ipaD and ipaB mutants of S. flexneri.
TABLE 5.
Expression of lacZ transcriptional fusions from reporter plasmids in E. coli strains containing MxiE- and IpgC-producing plasmids
| Transcriptional fusion | Reporter plasmida | β-Galactosidase activity in derivatives of strains expressingb:
|
||
|---|---|---|---|---|
| MxiE | IpgC | MxiE + IpgC | ||
| ospC1-lacZ | pMM19 | 5 | 10 | 50 |
| ipaH7.8-lacZ | pMM11 | 190 | 280 | 5,500 |
| ospF-lacZ | pMM65 | 15 | 40 | 170 |
| ipaH4.5-lacZ | pMM12 | 50 | 90 | 45 |
| ospD3-lacZ | pMM29 | 20 | 60 | 50 |
The reporter plasmids were constructed with vector pQF50 (Table 1).
β-Galactosidase activities assayed in derivatives of E. coli strains harboring either pMM71 (MxiE), pKH128 (IpgC), or both (MxiE and IpgC) are expressed in Miller units and are the means of at least four independent experiments. Standard deviations (not shown) were within 25% of the reported values.
Regulation of expression of chromosomal ipaH genes by MxiE and IpgC.
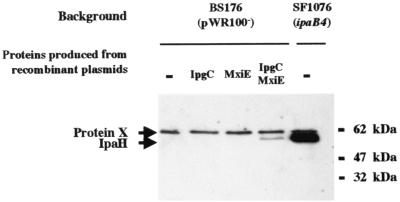
Southern hybridization with the constant portion of the ipaH genes as a probe indicated that five copies of ipaH also exist on the chromosome of wild-type S. flexneri strain M90T (7), although it is not known whether any of these genes is complete or expressed. To investigate whether the ipaH genes located on the chromosome were regulated in the same manner as those located on the virulence plasmid, we introduced into strain BS176, a virulence plasmid-cured derivative of M90T, plasmids expressing mxiE (pMM71) and ipgC (pKH128), either individually (MMCP536 and MMCP535) or together (MMCP537). Whole-cell extracts of recombinant strains were analyzed by SDS-PAGE and Western blotting with anti-IpaH serum (Fig. 3). No material cross-reacting with IpaH was detected in extracts of BS176 or its derivatives expressing either MxiE or IpgC alone. In contrast, one protein of approximately 60 kDa, i.e., the same size as IpaH proteins produced from the virulence plasmid, was detected in extracts of the derivative of BS176 expressing both MxiE and IpgC. These results indicated that at least one ipaH gene carried by the chromosome was complete and, more importantly, that it was under the control of both MxiE and IpgC.
FIG. 3.
Production of chromosomally encoded IpaH proteins is activated by MxiE and IpgC. Whole-cell extracts of BS176 (a virulence plasmid-cured derivative of wild-type S. flexneri strain M90T), its derivatives harboring plasmid pKH128 (which expresses IpgC), plasmid pMM71 (which expresses MxiE), or both plasmids, and SF1076 (an ipaB mutant) were analyzed by SDS-PAGE and probed with rabbit anti-IpaH serum. The positions of IpaH and protein X (a constitutively expressed, chromosomally encoded protein recognized by rabbit anti-IpaH serum) are indicated.
DISCUSSION
In the present study, we investigated further the repertoire of genes that are regulated by the activity of secretion and the cis-acting element involved in the activation of regulated promoters. We present evidence that six and potentially eight promoters carried by the virulence plasmid and at least one promoter carried by the chromosome are regulated by the activity of the TTS apparatus. The mechanism of regulation involves a conserved 17-bp cis-acting site overlapping the −35 region of each promoter and two coordinately acting regulators, MxiE and IpgC.
Reporter plasmids carrying the putative promoters and regulatory regions of a number of ipaH and osp genes were introduced into the wild-type strain, in which the TTS apparatus is not active during growth in broth, and into ipaD and ipaB mutants, in which the TTS apparatus is constitutively active. The DNA region located upstream from ipaH7.8. ospC1, and ospF contained promoters that were activated approximately 10-fold under conditions of secretion. As previously described for the ipaH9.8 and virA promoters (28), the activation of the ipaH7.8. ospC1, and ospF promoters under conditions of deregulated secretion required both MxiE and IpgC, as these promoters were no longer active in ipaB4 mxiE and ipaB4 ipgC mutants. Furthermore, each of these promoters was activated in an E. coli strain expressing both MxiE and IpgC. The lower β-galactosidase activity expressed from most reporter plasmids in the E. coli strain harboring pMM71 (MxiE) and pKH128 (IpgC) than in S. flexneri ipaD or ipaB mutants might have been due to a level of production of MxiE or IpgC from recombinant plasmids lower than from the virulence plasmid. Primer extension analyses were used to identify the transcription start site of the ospC1 promoter and those of the ipaH9.8 and virA promoters that are also regulated by the activity of secretion (28). A sequence comparison revealed a conserved motif, designated the MxiE box, overlapping the −35 region of each promoter, and deletion analyses indicated that this conserved sequence was essential for the activation of these promoters under conditions of secretion. An MxiE box was also detected upstream from the ipaH7.8 and ospF genes, both of which are regulated by the activity of secretion, at an appropriate distance from a putative −10 region and probably represents the regulatory region for each of these genes.
The characteristic features of regulated promoters include (i) a −10 region closely resembling the consensus sequence TATAAT, (ii) no similarities with a canonical −35 region, and (iii) the presence of an MxiE box between positions −49 and −33. The location of the MxiE box is consistent with previous observations indicating that target sequences of most members of the AraC family of transcriptional activators are located adjacent to or overlapping the −35 regions of regulated promoters (19). In S. enterica serovar Typhimurium, InvF, which is homologous to MxiE, and SicA, which is homologous to IpgC, are both required for activation of the sicA. sigD, and sopE promoters (12). The target sequence, designated the InvF box, is 11 bp long and is located between positions −51 and −41 with respect to the transcription start point (11, 12, 43). The InvF and MxiE boxes are located at the same positions in regulated promoters and exhibit some limited sequence similarities, including a stretch of six T's between nucleotides −43 and −38.
Using lacZ fusions carried by the virulence plasmid, Demers et al. previously showed that the transcription of ipaH1.4 was also regulated by the activity of secretion (13). However, the region located immediately upstream from ipaH1.4, i.e., the 389-bp ospE1-ipaH1.4 intergenic region, did not appear to contain any promoter, suggesting that the promoter and regulatory region involved in the expression of ipaH1.4 are located upstream from ospE1. Indeed, the region located immediately upstream from ospE1 has an MxiE box 16 nucleotides upstream from a putative −10 region, suggesting that ospE1 and ipaH1.4 belong to the same operon and are transcribed from a regulated promoter located upstream from ospE1. The region encompassing the ospE2 and ipaH2.5 genes is almost identical to the ospE1-ipaH1.4 region, except for the presence of IS elements inserted between ospE2 and ipaH2.5 (6). The presence of an MxiE box upstream from ospE2 suggests that the transcription of ospE2 is also regulated by the activity of secretion. Due to the presence of the IS elements inserted between ospE2 and ipaH2.5, it seems unlikely that ipaH2.5 is regulated or even transcribed.
The expression of an ipaH4.5-lacZ fusion carried by the virulence plasmid was increased 12-fold in an ipaD mutant compared to the wild-type strain (13), whereas the expression of the ipaH4.5-lacZ fusion carried by the reporter plasmid was increased only 3-fold in ipaD and ipaB mutants compared to the wild-type strain. As for other regulated fusions, no activation of the reporter plasmid-carried ipaH4.5-lacZ fusion was observed in the absence of either MxiE or IpgC, indicating that activation of the promoter upstream from ipaH4.5, although less efficient than that of other regulated promoters, was dependent on both MxiE and IpgC. Sequence analysis of the ipaH4.5 upstream region revealed the presence of a potential MxiE box that, however, differs from the consensus MxiE box by having a G instead of a T at position −48 and a T instead of a C at position −45. In contrast to other promoters that have an MxiE box, the ipaH4.5 promoter was not activated in the E. coli strain expressing both MxiE and IpgC. This result suggests that differences between the MxiE boxes of the ipaH4.5 promoter and other regulated genes may affect the binding affinity of MxiE which, as discussed above, may be produced in smaller amounts in E. coli than in S. flexneri. The ipaH4.5 gene is located 429 bp downstream from ipaH7.8, and the greater amplitude of regulation of the virulence plasmid-carried fusion than of the reporter plasmid-carried fusion suggests that, under conditions of secretion, the transcription of ipaH4.5 occurs mostly through activation of the upstream ipaH7.8 promoter.
While this study was under review, Kane et al. (24) reported an analysis of the expression of ospB. ospC1. ospE2. ospF. virA, and ipaH9.8 with recombinant plasmids and green fluorescent protein as a reporter system. These authors compared the amounts of the reporter protein present in intracellular bacteria following entry into epithelial cells and in extracellular bacteria growing in broth. Increased amounts of the reporter protein were present in intracellular bacteria harboring virA. ipaH9.8. ospB. ospC1. ospE2, and ospF fusions, and this increase was dependent on a functional mxiE gene. These authors concluded that these promoters were regulated by MxiE. These results are consistent with the previous demonstrations that the virA. ipaH9.8. ipaH7.8. ipaH4.5, and ipaH1.4 promoters are activated upon entry of bacteria into epithelial cells (13) and that the virA. ipaH9.8. ipaH7.8. ipaH4.5. ospC1, and ospF promoters are controlled by both MxiE and IpgC (28; this study) and with the above hypothesis that the target for the regulation of ipaH1.4 lies upstream from ospE1. No conserved MxiE boxes are present upstream from ospB, and the mechanism by which MxiE may control the ospB promoter remains to be investigated. Kane et al. (24) proposed that MxiE-regulated genes are activated in the intracellular compartment. However, using kinetic analysis, Demers et al. (13) showed that, although the transcription of virA was activated upon entry of bacteria into epithelial cells, it was subsequently repressed during growth of bacteria in the intracellular environment. As shown here and previously (28), the capacity of MxiE to act as a transcriptional activator is dependent on the activity of the TTS apparatus that is sensed by the presence of free IpgC, i.e., not associated with IpaB and IpaC.
It was previously shown that several copies of ipaH genes are carried by both the virulence plasmid and the chromosome (20). The 5′ portion of ipaH genes carried by the virulence plasmid exhibits a low GC content similar to those of mxi and spa genes, encoding the components of the TTS apparatus, and the ipa operon and osp genes, encoding secreted proteins, suggesting that ipaH genes were acquired from the same source as other genes of the virulence plasmid-encoded TTS system (6). Thus, it seems likely that chromosomal ipaH genes may result from the duplication of plasmid genes. Using recombinant plasmids expressing MxiE and IpgC in an S. flexneri strain cured of the virulence plasmid, we showed that the expression of at least one chromosomal ipaH gene is also under the control of MxiE and IpgC. The amount of IpaH proteins produced from the chromosome by the derivative of the virulence plasmid-cured strain expressing MxiE and IpgC was much lower than the amount of IpaH proteins produced by an ipaB mutant. This result suggests that chromosomally encoded IpaH proteins may represent only a small proportion of the IpaH proteins that are produced from the virulence plasmid. Alternatively, as in E. coli, the low amount of IpaH proteins produced in the virulence plasmid-cured strain containing pMM71 and pKH128 might have been due to a low level of production of MxiE or IpgC from the recombinant plasmids.
In conclusion, we have identified the cis-acting regulatory element, or MxiE box, which is involved in the activation of promoters that are regulated by the activity of the Mxi-Spa TTS apparatus. Analysis of the sequence of the entire virulence plasmid indicated that eight MxiE boxes are located upstream from ipaH9.8. ipaH7.8. ipaH4.5. ospC1. ospE1. ospE2. ospF, and virA. As is the case for ipaH1.4, which is located 389 bp downstream from ospE1 and is proposed to be regulated by the MxiE box located upstream from ospE1, the ospD2 and ospD3 genes, which are located 431 and 329 bp downstream from ospF and ospC1, respectively, may also be controlled by MxiE boxes located in front of the upstream genes. This hypothesis suggests that the transcription of 11 genes that are carried by the virulence plasmid and encode proteins secreted by the TTS apparatus is regulated by the activity of secretion. Determination of the complete sequence of the chromosomes of S. flexneri will help to determine which chromosomal ipaH genes exhibit an MxiE box and whether other chromosomal genes are potentially controlled by MxiE and IpgC.
Acknowledgments
We are indebted to and pleased to acknowledge Brigitte Demers for the construction of plasmids pBD7 and pBD9 and Julie Viala and the team of Enzo Scarlato at Chiron, Siena, Italy, for assistance with primer extension. We also thank Dana Philpott for critical reading of the manuscript.
This work was supported in part by grants from the European Community (contract ERBFMRXCT98-0164), GIP-Hoechst Marion Roussel (contract FRHMR1/9715-A2), and the Fondation pour la Recherche Medicale.
REFERENCES
- 1.Adler, B., C. Sasakawa, T. Tobe, S. Makino, K. Komatsu, and M. Yoshikawa. 1989. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol. Microbiol. 3:627-635. [DOI] [PubMed] [Google Scholar]
- 2.Allaoui, A., J. Mounier, M. C. Prevost, P. J. Sansonetti, and C. Parsot. 1992. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol. Microbiol. 6:1605-1616. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui, A., P. J. Sansonetti, and C. Parsot. 1993. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri lpa invasins. Mol. Microbiol. 7:59-68. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuzon, C. R., G. Banks, J. Deiwick, M. Hensel, and D. W. Holden. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33:806-816. [DOI] [PubMed] [Google Scholar]
- 6.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 7.Buysse, J. M., A. B. Hartman, N. Strockbine, and M. Venkatesan. 1995. Genetic polymorphism of the ipaH multicopy antigen gene in Shigella spps. and enteroinvasive Escherichia coli. Microb. Pathog. 19:335-349. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, L. W., and O. Schneewind. 2000. Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182:3183-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis, G., J. C. Vanootegem, and C. Sluiters. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb. Pathog. 2:367-379. [DOI] [PubMed] [Google Scholar]
- 11.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demers, B., P. J. Sansonetti, and C. Parsot. 1998. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 17:2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand, J. M., B. Dagberg, B. E. Uhlin, and G. R. Bjork. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924-935. [DOI] [PubMed] [Google Scholar]
- 15.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg, A., A. M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg, A., and H. Wolf-Watz. 1988. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-plB1-encoded trans-acting elements controlled by temperature and calcium. Mol. Microbiol. 2:121-133. [DOI] [PubMed] [Google Scholar]
- 18.Francis, M. S., S. A. Lloyd, and H. Wolf-Watz. 2001. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42:1075-1093. [DOI] [PubMed] [Google Scholar]
- 19.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman, A. B., M. Venkatesan, E. V. Oaks, and J. M. Buysse. 1990. Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of Shigella flexneri. J. Bacteriol. 172:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 23.Jost, B. H., and B. Adler. 1993. Site of transcriptional activation of virB on the large plasmid of Shigella flexneri 2a by VirF, a member of the AraC family of transcriptional activators. Microb. Pathog. 14:481-488. [DOI] [PubMed] [Google Scholar]
- 24.Kane, C. D., R. Schuch, W. A. Day, Jr., and A. T. Maurelli. 2002. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184:4409-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaBrec, E. H., H. Schneider, T. J. Magnani, and S. B. Formal. 1964. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J. Bacteriol. 88:1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavris, M., A.-L. Page, R.Tournebize, B. Demers, P. J. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 29.Ménard, R., P. Sansonetti, and C. Parsot. 1994. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michiels, T., P. Wattiau, R. Brasseur, J. M. Ruysschaert, and G. Cornelis. 1990. Secretion of Yop proteins by yersiniae. Infect. Immun. 58:2840-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsot, C., R. Ménard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 33.Parsot, C., and P. J. Sansonetti. 1999. The virulence plasmid of shigellae: an archipelago of pathogenicity islands? p. 151-165. In J. B. Kaper and J. Kacker (ed.), Pathogenicity islands and other mobile virulence elements. American Society for Microbiology, Washington, D.C.
- 34.Pettersson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf-Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231-1233. [DOI] [PubMed] [Google Scholar]
- 35.Platt, T., B. Müller-Hill, and J. H. Miller. 1972. Assay of beta-galactosidase, p. 352-355. In J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Rimpilainen, M., A. Forsberg, and H. Wolf-Watz. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J. Bacteriol. 174:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasakawa, C., K. Kamata, T. Sakai, S. Makino, M. Yamada, N. Okada, and M. Yoshikawa. 1988. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J. Bacteriol. 170:2480-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skryzpek, E., and S. C. Straley. 1993. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J. Bacteriol. 175:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spohn, G., I. Delany, R. Rappuoli, and V. Scarlato. 2002. Characterization of the HspR-mediated stress response in Helicobacter pylori. J. Bacteriol. 184:2925-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stainier, I., M. Iriarte, and G. R. Cornelis. 1997. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol. Microbiol. 26:833-843. [DOI] [PubMed] [Google Scholar]
- 42.Straley, S. C., G. V. Plano, E. Skrzypek, P. L. Haddix, and K. A. Fields. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol. Microbiol. 8:1005-1010. [DOI] [PubMed] [Google Scholar]
- 43.Tucker, S. C., and J. E. Galan. 2000. Complex function for SicA, a Salmonella enterica serovar Typhimurium type III secretion-associated chaperone. J. Bacteriol. 182:2262-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol. Microbiol. 43:411-423. [DOI] [PubMed] [Google Scholar]
- 45.Zierler, M. K., and J. E. Galan. 1995. Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect. Immun. 63:4024-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]