Abstract
Mycoplasma genomes contain compact gene sets that approach the minimal complement necessary for life and reflect multiple evolutionary instances of genomic reduction. Lateral gene transfer may play a critical role in shaping the mobile gene pool in these organisms, yet complex mobile elements have not been reported within this genus. We describe here a large (∼23-kb) genetic element with unique features that is present in four copies in the Mycoplasma fermentans PG18 chromosome, accounting for approximately 8% of the genome. These novel elements, designated ICEF (integrative conjugal elements of M. fermentans), resemble conjugative, self-transmissible integrating elements (constins) in that circular, nonreplicative extrachromosomal forms occur in which the left and right termini of the integrated element are juxtaposed and separated by a coupling sequence derived from direct repeats flanking chromosomal copies of ICEF as a result of target site duplication. ICEF contain multiple similarly oriented open reading frames (ORFs), of which some have homology to products of known conjugation genes but others have no known counterparts. Surprisingly, unlike other constins, ICEF lack homologs of known integrases, transposases, or recombinases, suggesting that a novel enzyme may be employed for integration-excision. Skewed distribution and varied sites of chromosomal integration among M. fermentans isolates suggest a role for ICEF in promoting genomic and phenotypic variation in this species. Identification of homologs of terminal ICEF ORFs in two additional mycoplasma species indicates that ICEF is the prototype member of a family of ICE-related elements that may be widespread among pathogenic mycoplasmas infecting diverse vertebrate hosts.
It is becoming increasingly appreciated that lateral gene transfer (LGT) has played a major role in bacterial evolution, disseminating key traits both within and among bacterial species. In contrast to evolution by random point mutation and subsequent selection for variants with improved fitness, acquisition of a new phenotype by LGT is considered a “quantum leap” in evolution (23), as the genes responsible for the phenotype are typically transferred laterally en bloc. Such a transfer also enables the trait to be more widely disseminated rather than relying on different bacterial clones to independently create de novo the desired mutation(s) that will confer increased fitness (41, 42).
The extent to which LGT has shaped bacterial genomes has been revealed by assessment of proliferating genome sequence data. Not only the diverse forms of mobilizable gene pools but also the mechanisms of transfer and the types of element involved are becoming better defined (reviewed in references 15 and 47). Among naturally competent bacteria, transformation of naked DNA is likely to mediate genetic exchange within and between bacterial genera. In the context of large pathogenicity islands, the presence of bacteriophage-related sequences and their insertion into known prophage attachment sites implicate transduction as one mechanism of gene transfer (25). Other genomic islands contain genes with homology to tra genes present on self-transmissible plasmids, indicating that conjugation is responsible for the transfer of certain islands (24). Indeed, although the spread of antibiotic resistance genes by conjugative plasmids and conjugative transposons has been known for some time, a number of additional, diverse phenotypes that have been shown to be disseminated by the conjugation of chromosomal regions have recently been reported (29, 44, 53). These include sucrose metabolism in Salmonella enterica serovar Seftenberg via cTnscr94, the symbiosis traits associated with the 500-kb symbiosis island in Mesorhizobium loti, and the degradation of chlorocatechol mediated by gene products resident on the 105-kb clc element of Pseudomonas putida. The term constin has recently been coined (31) to describe this diverse group of (conjugative, self-transmissible, integrating) elements to which previously reported conjugative transposons such as Tn916 (49), the Bacteroides tetracycline resistance elements (46), and the large SXT unit of Vibrio cholerae (31) also belong.
Knowledge of the distribution of constins, their ability to mediate lateral exchange of genetic information, and their full potential for altering the phenotype of an organism is still relatively incomplete. Among the better characterized of these diverse elements, some have been shown to be inserted at specific sites in the chromosome. In the case of the >60-kb element SXT, an antibiotic resistance-conferring element, a member of the λ integrase protein family catalyzes site-specific integration into a single site within the N-terminal coding sequence of the prfC gene (31). Integration of the M. loti symbiosis island is site specific also, with insertion into a Phe-tRNA gene mediated by a phage P4-related integrase (53). Two distinct Gly-tRNA genes are the known insertion sites for the P. putida clc element, a conjugative unit that also employs a P4-related integrase (44). In contrast, other elements, such as Tn916, exhibit a more promiscuous range of integration sites and can be transferred between a remarkably broad range of hosts, illustrating the importance of certain constins in the dissemination of genetic information across diverse genus boundaries (49). In addition to self-transfer, some constins are able to mobilize other genetic loci, such as mobilizable plasmids (both in cis and in trans), discrete integrative portions of a chromosome (in the case of transfer of the nonreplicating Bacteroides units in Bacteroides spp.) (51), or chromosomal loci that are linked to the constin integration site (as described for the Hfr-like transfer of chromosomal markers by the SXT element) (30).
Although conjugative elements have been described for members of many bacterial genera, indigenous self-transmissible molecules have not been reported in the genus Mycoplasma. This diverse group of more than 100 species is characterized by their small genome sizes, which range from 580 to 1,300 kb (61). Accompanying their reductive evolution from the low G+C content group of gram-positive eubacteria (20, 33, 38) has been the loss of several functions that are widespread among other bacterial genera. These include the absence of genes for cell wall biogenesis and many biosynthetic pathways, including those for amino acids, fatty acids, and de novo nucleotide synthesis. Understandably, known mycoplasmas typically are obligate parasites of their vertebrate hosts. Mycoplasmas can occupy host niches for long periods during chronic infections, and many cause important diseases in a broad range of animal hosts, including humans (34, 52). Mycoplasmas generally adhere to the surfaces of host cells and tissues. The absence of a cell wall, their single limiting membrane, and the chronic nature and communal environment of most infections are predicted to be conducive to genetic exchange between mycoplasmas. Scrutiny of completed genome sequences (11, 19, 21, 28) from four mycoplasmas, however, has not revealed the presence of any conjugative elements and has raised the prospect that LGT by such elements may be minimal compared to that observed in bacterial genomes of greater size (41). As these first four published genomes were at the lower end of the range of mycoplasma genome sizes (61), it is predicted that they may reflect features of “minimal genomes,” with a relative preponderance of essential genes encoding housekeeping functions (33, 38). In contrast, Mycoplasma species with larger genomes or those showing intraspecies differences in genome size may be predicted to have larger “mobile” gene pools (24).
With the exception of several simple insertion sequences (ISs) (9, 37, 57), three cryptic plasmids (16, 59), and two mycoplasma bacteriophages (56, 60), no other mobile genetic elements had been identified in the genus Mycoplasma and no indigenous conjugative elements have been described. In the present report, a novel constin-like element is described in the genome of Mycoplasma fermentans, a human infectious agent that is under investigation as a primary pathogen or copathogen in a number of clinical diseases (34, 35). This genetic element, designated ICEF (integrative conjugal element of M. fermentans), is approximately 23 kb long, is flanked by 8-bp direct repeats that are associated with element insertion, and is variable in its distribution among isolates and strains of the species. In the type strain, PG18, ICEF is present as four dispersed copies at distinct integration sites in the chromosome, accounting for 8% of the 1,200-kb genome. Furthermore, an extrachromosomal form can be detected by Southern hybridization analysis and by inverse PCR methods. ICEF contains multiple open reading frames (ORFs), including some with homology to signature genes encoded by other conjugal elements, but lacks a recognizable gene encoding an integrase function. Taken together, these data support the identification of ICEF as a novel mobile DNA in the genus Mycoplasma and a novel member of the constin group of mobile elements.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
M. fermentans strains PG18 (clone 39) and II-29/1 were grown in Hayflick medium and GBF-3 medium, respectively, as described elsewhere (8). Escherichia coli DH10B (Invitrogen, Carlsbad, Calif.) was used as a cloning host for recombinant plasmids derived from vectors pZerO-1, pZerO-2 (from Invitrogen), and pLITMUS38 (New England Biolabs, Beverly, Mass.). E. coli was grown at 37°C in Luria-Bertani medium supplemented with either 10 μg of Zeocin per ml, 50 μg of kanamycin per ml, or 100 μg of ampicillin per ml, as appropriate.
DNA preparation and cloning of ICEF-related restriction fragments.
Genomic DNAs from M. fermentans PG18 and II-29/1 were prepared as described previously (8). A standard, hybridization-driven chromosome walking approach was taken to obtain a series of overlapping restriction fragments that encompassed the ICEF. All restriction fragments were cloned in E. coli plasmid vectors by using routine ligation and transformation protocols. The following restriction fragments (as designated also in Fig. 2) were introduced into the vectors indicated and detected by using the oligonucleotides shown in parentheses. BamHI fragments IA-2 and N1 were inserted into the BamHI site of pZerO-1 and identified as two of the five different restriction fragments that were recovered following shotgun cloning of small BamHI fragments; BamHI-BglII fragments IA-1 and IIC-1 (primer 1; 5′ GAA AGA AGT GTG TTG AAC ACT T) and N3 (primer 3; 5′ CGA GTA TAT GCC TAA AAT CAA TGT) and BglII fragments N5 (primer 5; 5′ AGC CTC AAA AAA CGG CTC TAA ATG) and IIA-2 (primer 7; 5′ CTA TTC AAG TAG CGC GTG TG) were ligated into the BamHI site of pZerO-2. BssHII fragments IA-3 and N2 (the only small BssHII fragments in the genome of M. fermentans PG18) plus MluI-BssHII fragments IIA-1 (primer 2; 5′ GTA TCC TTG CTT TTC AAG CTT TTT), IA-5 and N4 (primer 4; 5′ TGG AGC ATA TGG TGT ATT GCC), and IIC-2 and IA-6 (primer 6; 5′ GAA CAA CTT CGT TTT GTA TAT CC) were cloned into the MluI site of pLITMUS38. NheI fragment IIB-1 (primer 1) and XbaI fragments IA-7, IIA-3, IIB-2, and IIC-3 (all primer 7) were inserted into the XbaI site of plasmid pZerO-2. A pdhA-containing BglII fragment from M. fermentans strain II-29/1 was cloned into the BamHI site of pZerO-2 and identified by colony hybridization with primer 8 (5′ GCA TAC GTC CTT GAC GTT GCA). The cloning and sequencing of a Sau3AI fragment encompassing the gene for a previously described putative lipoprotein (14) (fragment IA-4 in Fig. 2) were performed by Tonghua Lu (36).
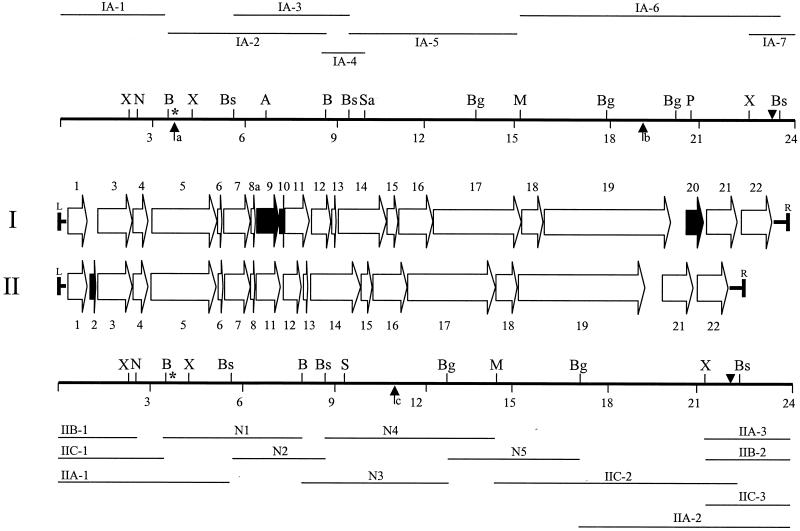
FIG. 2.
Restriction map and ORF complement of the two ICEF configurations in M. fermentans PG18. The locations and directions of ORFs within ICEF units are shown by arrows, and the ORFs are numbered consecutively. Each of the two ICEF configurations (designated I and II) has a slightly different ORF complement; ORFs that are unique to one type of element are shown as black horizontal arrows. The left and right boundaries of the ICEF units are indicated by the letters L and R, respectively. Restriction fragments that have been cloned and sequenced are shown as narrow horizontal lines above (for ICEF-IA) or below (for ICEF-IIA, -IIB, and -IIC) the ORFs. Each fragment is assigned an identifier with a prefix (IA-, IIA-, IIB-, or IIC-) to indicate the ICEF copy from which it was derived or given an N designation if it is an internal fragment that has not been assigned to a specific copy of the type II configuration. Restriction fragments IA-1, IIA-1, IIB-1, IIC-1, IA-7, IIA-2, IIA-3, IIB-2, and IIC-3 contain genomic regions flanking ICE units in addition to the regions of ICE shown. Restriction maps (thick horizontal lines) are shown representing the single type I configuration and the three variants of the type II configuration, with 3-kb size intervals marked below. Abbreviations for restriction sites: A, AflII; B, BamHI; Bg, BglII; Bs, BssHII; M, MluI; N, NheI; P, PstI; S, ScaI; Sa, Sau3AI; X, XbaI. Only the restriction sites used in fragment cloning or hybridization analysis or those that represent characteristic polymorphisms between ICEF configurations I and II are shown. The locations of Southern hybridization probes used in the analyses shown in Fig. 1B (primer 2; asterisks) and C (primer 7; triangles) are shown above the restriction maps. Locations at which comparative sequence analysis revealed the presence of an in-frame stop codon in ORF5 of ICEF-IA (a), a frameshift mutation in ICEF-IA ORF19 (b), and a frameshift mutation in ORF16 (c) within the internal N4 restriction fragment are highlighted by vertical arrows.
Hybridization of genomic restriction fragments.
Standard Southern blot and colony hybridization protocols were used to identify suitable DNA fragments for cloning and for identification of recombinant clones. High-stringency hybridization and washing conditions were used as described by the supplier of the nylon hybridization membranes (Roche Molecular Biochemicals, Indianapolis, Ind.). In all cases, hybridization was carried out by using oligonucleotide probes labeled at the 3′ end with digoxigenin (DIG), and hybridization bands or colonies were detected by nonradioactive detection methods as detailed in the manual provided by the manufacturer (Roche). Fragments separated by pulsed-field gel electrophoresis were prepared from fresh cells and separated with a CHEF (contour-clamped homogeneous electric field) Mapper XA system (Bio-Rad, Hercules, Calif.) under parameters controlled by the autoalgorithm function.
PCR amplification.
Oligonucleotide primers 1 (see above) and 9 (5′ TTA TAG TAG CTT TCA CCG AGT A) were used to amplify an approximately 420-bp portion of ICEF from the genomic DNAs of various M. fermentans strains. These included DNA preparations from strains K7, MT-2, M39A, M70B, SK5, SK6, and Incognitus (all kindly provided by S.-C. Lo, Armed Forces Institute of Pathology, Washington, D.C.) plus genomic DNA from M. fermentans strain II-29/1 (8). Template DNA (10 ng) was used in a standard PCR consisting of 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. The resulting amplicons were analyzed following electrophoresis through ethidium bromide-containing agarose gels and by direct sequencing following purification through QIAquick spin columns (Qiagen Inc., Valencia, Calif.). As a control, primers 10 (5′ TTG AGA TAT TTA AGC AAA ATA TCT A) and 11 (5′ ATT TTC CAG CAT TTT TTT GAT TAA) were used to amplify the previously described malp gene, which had been shown to be present in each of these isolates (8). To amplify the regions of the M. fermentans II-29/1 genome that correspond to the insertion sites for ICEF-IA and ICEF-IIC (in PG18), a long-range PCR with the Expand Long Template PCR system (Roche) was performed as described previously (9). Primers 12 (5′ AAC TTC ATG CAG CTG ATG TTT CAA) and 13 (5′ TTT GTA GAC ATA TTT CCT CCT TA) were used to amplify an approximately 1.5-kb region occupied by ICEF-IA in strain PG18. Primers 14 (5′ GGA GGA TTA ATT TTA GTT GAA TCA) and 15 (5′ GGC AAG TAA AGG TTG AGA AGT TCT) were used to amplify an approximately 2.9-kb region of the strain II-29/1 genome corresponding to the ICEF-IIC integration site in strain PG18.
Inverse PCR methods.
To identify an extrachromosomal form of ICEF, a PCR assay was employed with primers 7 and 9 (see above), which were outwardly oriented toward the left and right junctions, respectively, of ICEF in the chromosomally integrated form of the element. A standard PCR was employed, with approximately 50 ng of M. fermentans PG18 genomic DNA as the template and 30 amplification cycles with the following profile: 94°C for 1 min, 50°C for 30 s, and 72°C for 1 min. To selectively amplify the extrachromosomal form of ICEF-IA, a similar PCR assay was employed, with primer 7 paired with primer 16 (5′ ACA GGT CGA ACG GGT AGT AA). The latter primer amplifies ICEF-IA specifically, as the sequence corresponds to a region of microheterogeneity between ICEF-IA and three type II elements.
Oligonucleotide synthesis and DNA sequencing.
All of the oligonucleotides used in this study were synthesized on a model 3948 Nucleic Acid Synthesis and Purification System (Applied Biosystems, Inc., Foster City, Calif.), and all DNA sequencing was performed with Taq dye terminators and either a Prism 373 or a Prism 377 automated DNA sequencer (Applied Biosystems, Inc.). Both of these services were carried out at the University of Missouri Molecular Biology Program DNA Core Facility. DNA and deduced peptide sequences were manipulated and analyzed by using the Genetics Computer Group (Madison, Wis.) software package through the Pittsburgh Supercomputing Center (http://www.psc.edu/biomed) and the PSORT program (http://psort.nibb.ac.jp/) and by using the BLAST program (and linked databases) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession numbers.
The DNA sequences reported here have been deposited in the GenBank database and assigned the following accession numbers: ICEF-IA, AY168953; a composite type II ICEF, AY168954; fragment IIA-1, AY168955; fragments IIA-2 and IIA-3, AY168956; fragment IIB-1, AY168957; fragment IIB-2, AY168958; fragment IC-1, AY168960; fragments IIC-2 and IIC-3, AY168959; fragment N1, AY168961; fragment N2, AY168962; fragment N3, AY168963; fragment N4, AY168964; fragment N5, AY168965.
RESULTS AND DISCUSSION
Identification of a novel repeated sequence in the genome of M. fermentans PG18.
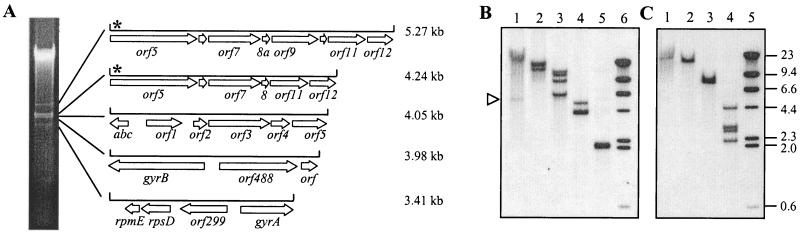
During an ongoing analysis of genes encoding surface components of M. fermentans, a repeated sequence was serendipitously identified within genomic DNA from a clonal isolate of the type strain, PG18. This sequence was revealed during agarose gel electrophoresis of genomic DNA restriction fragments generated with BamHI, recognizing the GGATCC site, which is predicted to be rare in the A+T-rich (28% G+C) M. fermentans genome (27). Unexpectedly, the BamHI restriction profile of M. fermentans PG18 DNA contained a distinctive cluster of several fragments ranging in size from 3.4 kb to approximately 5 kb, with a predominant band at approximately 4 kb (Fig. 1A). Because such patterns can reflect a tandemly repeated sequence, a dispersed repetitive sequence, or a hitherto unrecognized replicon, the BamHI fragments were cloned in E. coli to determine whether any are related. Restriction analysis of the recombinant plasmids indicated that five different BamHI fragments had been cloned. The nucleotide sequence of a representative of each fragment was determined and analyzed (Fig. 1A; Tables 1 and 2).
FIG. 1.
Identification of a repetitive sequence in the genome of M. fermentans PG18. (A) Genomic DNA from M. fermentans PG18 (clone 39) was digested with BamHI, and the resulting fragments were separated by standard agarose electrophoresis. Five BamHI fragments, corresponding to visible ethidium bromide-stained bands in the 3- to 5-kb size range, were cloned, and the nucleotide sequences were determined. The size of each fragment (calculated from the nucleotide sequence) is indicated in kilobases. The locations and directions of ORFs within each fragment are shown by open arrows, and the ORFs are labeled with the standard gene names for ORFs that are housekeeping genes with known functions. ORF1 to ORF12 correspond to ORF1 to ORF12 in Fig. 2. ORF488 and ORF299 are designated by length (in amino acid residues) and have significant homology (BLAST P < 1E-16) to conserved hypothetical ORFs MG443 (from M. genitalium) and Mypu4350 (from M. pulmonis), respectively. The asterisk represents the location of the hybridization probe (primer 2; see Materials and Methods) used in the Southern analysis in panel B. Some terminal ORFs indicated by arrows (for example, orf5) are incomplete by virtue of the BamHI restriction site by which their fragments were cloned. (B) Southern hybridization analysis of multiple copies of ICEF in M. fermentans PG18. Genomic DNA was digested with BssHII (lane 1), BglII (lane 2), BglII and BssHII (lane 3), BamHI (lane 4), or XbaI (lane 5) and probed with DIG-labeled oligonucleotide primer 2 (see Materials and Methods). Lane 6 contained DIG-labeled λ HindIII markers (Roche), the sizes of which are indicated in kilobase pairs at the right of panel C. The open triangle highlights a reproducible but weakly hybridizing BssHII restriction fragment of approximately 5.7 kb that was present in lane 1. (C) Southern analysis of multiple copies of the right terminal portion of ICEF. Genomic DNA from M. fermentans PG18 was digested with MluI (lane 1), BssHII (lane 2), MluI-BssHII (lane 3), or XbaI (lane 4) and probed with DIG-labeled primer 7 (see Materials and Methods and Fig. 2). Lane 5 contained DIG-labeled markers as in panel B.
TABLE 1.
Selected features of ICEF-encoded deduced gene productsa
| ORF | No. of amino acid residues | Mass (Da) | pI | Cellular locationb | Feature(s)c |
|---|---|---|---|---|---|
| ORF1 | 239 | 28,164 | 10.86 | C | Weak homology to short M. capricolum contig MC293 |
| ORF2 | 63 | 7,235 | 10.59 | C | Absent from ICEF-IA |
| ORF3 | 400 | 47,249 | 8.17 | C | Contains Tral relaxase motif III (43) |
| ORF4 | 155 | 17,610 | 9.34 | M | |
| ORF5 | 739 | 84,981 | 10.04 | M | TraG family (pfam02534; P = 3E-10); NTP-binding site |
| ORF6 | 62 | 7,549 | 10.36 | C | Weak homology to putative exported protein (accession no. YP01989) of Y. pestis |
| ORF7 | 315 | 35,628 | 6.63 | M | |
| ORF8 | 79 | 9,481 | 10.42 | C | Identical to ORF8a in first 52 residues |
| ORF8a | 62 | 7,209 | 10.14 | C | Present in ICEF-IA only |
| ORF9 | 288 | 34,455 | 9.77 | C | Present in ICEF-IA only |
| ORF10 | 54 | 6,541 | 10.54 | C | Present in ICEF-IA only |
| ORF11 | 238 | 27,904 | 4.27 | C | |
| ORF12 | 183 | 21,063 | 6.58 | C | Single-stranded DNA-binding protein family (pfam00436; P = 4E-14) |
| ORF13 | 84 | 10,160 | 10.34 | M | |
| ORF14 | 522 | 61,800 | 5.82 | LP | P57 lipoprotein; signal sequence |
| ORF15 | 93 | 9,987 | 9.22 | M | |
| ORF16 | 396 | 43,672 | 10.51 | M | Frameshift detected in one type II element |
| ORF17 | 937 | 108,137 | 8.05 | M | Homology to TraE/TrsE conjugation proteins; NTP-binding site |
| ORF18 | 227 | 26,633 | 6.94 | M | |
| ORF19 | 1,409 | 162,788 | 8.66 | M | Frameshift in ICEF-IA; signal sequence |
| ORF20 | 159 | 19,023 | 9.59 | C | Present in ICEF-IA only |
| ORF21 | 308 | 36,026 | 9.73 | C | |
| ORF22 | 388 | 45,786 | 10.24 | C | Homology to M. pulmonis P16 (31% identity over 137 amino acid residues) |
The relative position and orientation of each orf are shown in Fig. 2.
As predicted by the PSORT program (C, cytoplasm; M, contains transmembrane domain; LP, putative lipoprotein).
P represents the BLAST probability score for homology to consensus for protein families (pfam).
TABLE 2.
Deduced gene productsa encoded in chromosomal regions flanking ICEF copies in M. fermentans PG18
| Gene product or IS | ICEF copy | Sizeb | Closest homolog | % Identity (length)c | Pd | Comment |
|---|---|---|---|---|---|---|
| LepA | IA | 599 | LepA, M. pulmonis | 66 (598) | 0 | Conserved GTPase |
| LicA | IA | 242 | LicA, M. pulmonis | 48 (237) | 2E-60 | Putative choline kinase |
| ORF228 | IIA | 228 | CysS, M. pulmonis | 42 (183) | 2E-36 | Apparent fusion of conserved hypothetical ORF and cysS in M. pulmonis |
| ISMf1 | IIA, IIB | 1,570 | IS285 transposase, Yersinia pestis | 41 (143) | IE-51 | Multiple frameshifts in Tnp reading frame |
| ORF700 | IIA | 700 | ND | Signal sequence, strain variable among M. fermentans isolates (Fig. 3) | ||
| ORF251 | IIA | 251 | OrfD1, M. fermentans | 38 (239) | 4E-37 | ABC transporter, ATP-binding protein; ICEF-IIA insertion at residue 47 |
| Tnp | IIA | 136 | UU372 integrase, U. urealyticum | 68 (119) | 6E-41 | Fragment of IS3 family transposase gene |
| ORF349 | IIA | 349 | RC0529, Riskettsia conorii | 25 (247) | 7E-07 | Hypothetical ORF |
| PdhA | IIA | >101 | PdhA, M. pulmonis | 67 (101) | 1E-32 | Pyruvate dehydrogenase E1 α subunit |
| ORF | IIB | >469 | NDe | |||
| ORF406 | IIB | 406 | ND | |||
| ORF472 | IIC | 472 | CQ002, S. aureus | 21 (507) | 5E-15 | Strain-variable ORF of S. aureus |
| ORF242 | IIC | 242 | HI1038, Haemophilus influenzae | 32 (127) | 1E-22 | orf242 contains multiple frameshift mutations |
| ORF375 | IIC | 375 | RC0529 R. conorii | 28 (187) | 2E-09 | Hypothetical ORF |
Three of these BamHI fragments contained distinct sets of genes with strong homology (BLAST P < 1E-14) to orthologous genes of known function that are typically located on bacterial chromosomes. Among these were genes encoding GyrA together with ribosomal proteins L31 and S4 (from the 3.41-kb fragment) and GyrB (from the 3.98-kb fragment). Parenthetically, this demonstrated that the genes encoding the DNA gyrase subunits of M. fermentans are not tandemly arranged, as is the case in many bacterial chromosomes, including those of some mycoplasma species (19, 21, 28). One BamHI fragment of 4.05 kb had, at one end, an ORF (abc) encoding an ATP-binding cassette of an ABC transporter. This fragment will be further discussed below in the context of a junction region. Analysis of two other BamHI fragments formally identified a novel repetitive sequence in the M. fermentans PG18 chromosome. The sequences of the 5.27- and 4.24-kb BamHI fragments (Fig. 1A) were greater than 99.9% identical, except for an additional 1-kb sequence (encoding ORF9 and ORF10) that was present within the larger DNA fragment. Within these nearly identical fragments were several similarly oriented ORFs without homologs in the current databases, but at each end were ORFs with significant homology to proteins encoded by mobile genetic elements. Thus, one end of each fragment encoded the C-terminal portion of an ORF strongly resembling the conserved TraG family of proteins (ORF5, Table 1). At the other end, the BamHI site was within an ORF with significant homology to the family of single-stranded DNA-binding proteins (SSBs) (ORF12, Table 1). These comparisons suggested that, within the M. fermentans PG18 genome, there are at least two copies of a repeated unit with sequence characteristics of a conjugative mobile genetic element. This was of considerable interest because of the paucity of known complex elements in mycoplasmas and prompted a more complete characterization of the repeated sequence.
To determine the copy number of the repeated sequence, M. fermentans genomic DNA was restricted and subjected to Southern analysis. With an oligonucleotide probe corresponding to a sequence within ORF5 encoding the TraG family homolog, different hybridization profiles were observed, depending on the restriction enzyme employed. As expected, the probe hybridized to both a 5.3-kb and a 4.2-kb BamHI fragment, but the hybridization signal for the latter was reproducibly greater, even though the target sequence selected in each fragment was the same. These data suggested a greater copy number for the 4.2-kb BamHI fragment. Such disproportionate hybridization was also obtained with Southern blots of EcoRI or ClaI fragments (data not shown). Some restriction digests yielded a single hybridizing band (for example, XbaI [Fig. 1B, lane 5]). Through systematic analysis of restriction fragments, Southern analysis of DNA doubly digested with BglII (a moderately frequent cutter) and BssHII (a rare cutter, but for which a restriction site exists within each of the two related BamHI fragments) ultimately revealed four hybridizing bands of similar intensities (Fig. 1B, lane 3), indicating that there are four genomic copies of the repeated sequence in M. fermentans PG18. This was confirmed by the data in Fig. 1C (discussed below). A band of fainter intensity could also be detected following hybridization of certain restriction fragments (BssHII [lane 1] and BssHII-BglII [lane 3]; indicated by the open triangle in Fig. 1B) and is discussed below. Southern hybridization of large (>20-kb) PstI, SacI, and KpnI restriction fragments that had been separated by pulsed-field gel electrophoresis also indicated that there are four copies of the repeated sequence (data not shown). Importantly, the approximately equivalent hybridization of the probe to four large and distinct DNA fragments strongly suggested that the repeated sequences reside within the chromosome of M. fermentans. This was subsequently confirmed following the isolation and sequence analysis of junction fragments linking the repeated sequence to flanking genes in the chromosome (discussed below).
The repeated sequence is a 22- to 23-kb genetic element.
To determine the length and nature of the repeated sequence, overlapping clones were identified by traditional hybridization-driven walking techniques (Fig. 2). The combined data from extensive hybridization studies and sequence analyses of >20 cloned restriction fragments (the most pertinent of which are shown in Fig. 2) indicated that the four repeated elements were approximately 23 kb in length. Although very closely related in sequence, one of the four copies had a configuration (designated type I) distinct from that of the other three copies (designated type II), on the basis of ORF composition and originally revealed by multiple characteristic restriction fragment length polymorphisms (for example, as in the two largest fragments in Fig. 1A). These polymorphisms and the single-copy number facilitated the identification and assembly of sequences from the type I element (Fig. 2). However, no distinctive restriction site polymorphisms have been identified among the three copies of the type II element, and so, for many internal restriction fragments, it was not possible to determine from which copy these cloned fragments were derived. Nevertheless, comparison of approximately 4-kb homologous sequences from distinct copies of the type II elements (assigned by linkage to unique junction fragments) revealed only four nucleotide differences. Taken together, these data support the consensus model shown in Fig. 2, representing the organization and features common to all of the type II elements.
Analysis of the entire set of sequences revealed that if the termini of the four elements are defined as the first position at which the sequences of the four junction fragments diverge, the single type I element is 23,773 bp in length and the three type II elements are 22,331 bp in length. As the M. fermentans PG18 chromosome has been reported to be 1,245 kb (48), the approximately 91 kb of ICEF sequence accounts for approximately 8% of this small genome. Although each element contains several G+C-rich restriction sites, overall, the A+T content of each element is 71%, which is within the range of mycoplasmas (27) and is similar to the 73% A+T composition reported for one 15-kb genomic region of M. fermentans (8). This feature and the predominant use of the UGA codon to encode Trp within coding sequences indicated that these repeated elements represent bona fide components of the M. fermentans genome rather than a relatively recent LGT from a bacterium with the standard genetic code.
Overall, configurations I and II of the element (Fig. 2) have very high sequence similarity. Two regions, of 167 and 291 bp, are absent from the type I element, which is nevertheless larger than type II elements because of the presence of unique 1,040- and 865-bp sequences in the type I configuration. Within the approximately 21 kb of sequence that occurs in both configurations, the nucleotide sequences show a similarity of greater than 99.5% (95 nucleotide differences overall). The similarity among copies of type II elements is even more striking. Comparison of the approximately 2.8-kb DNA sequences that have been assigned to the copies of type II elements (fragments IIA-1, IIB-1, and IIC-1 in Fig. 2) revealed differences at only two nucleotides. A similar analysis of the approximately 1.1 kb that is present in each of fragments IIA-3, IIB-2, and IIC-3 identified only one nucleotide difference. These data, together with the conservation of multiple restriction sites among the type II elements (evident during cloning of overlapping fragments and Southern analysis [Fig. 1B and C, lanes 2 and 3]), implied that only minor differences occur among type II elements.
Consistent with these nucleotide similarities, type I and II elements contain similar but not identical complements of ORFs (Fig. 2). With 50 amino acids as a minimum, 22 different ORFs were identified, ranging in length from 54 to 1,409 amino acid residues. The type I configuration differs from the type II configuration by the presence of three type I-specific ORFs and the absence of small ORF2, which is present in all copies of the type II configuration. Overall, the coding density is high (92%), with many ORFs predicted to reside within operons (i.e., with short intergenic regions or overlapping start-stop codons indicative of translational coupling and likely reflecting a related function requiring coregulation). The extreme bias in gene orientation and compact organization are features that resemble the genomes of mobile DNAs, including bacteriophages (58), conjugative transposons (46), and tra regions of self-transmissible plasmids (5, 17).
ICEF contains multiple genes associated with DNA mobility and conjugation-related functions.
As expected from the near nucleotide sequence identity, the deduced polypeptide sequences of ORFs that are conserved between the type I and II configurations are greater than 98.5% identical (some paralogs have complete amino acid sequence identity). Pairwise comparison of corresponding ORFs from the two configurations also revealed the presence of one in-frame stop codon and two frameshift mutations (Fig. 2 and Table 1).
Considering the recent proliferation of gene sequences represented in current databases, it was somewhat surprising that BLAST analysis identified only three ORFs with significant homology (P < 1E-06 [2]) to proteins in the GenBank database. As mentioned, two of these, ORF5 and ORF12, were similar to gene products having DNA mobility-related function. Thus, TraG-related ORF5 has the greatest homology to the PCP51 protein of Clostridium perfringens plasmid pCP13 (P = 8E-13) and TrsK of Enterococcus faecalis transposon Tn1549 (P = 4E-11). The prototype of this family, TraG of plasmid RP4, is essential for conjugation (54) and has been proposed to couple the relaxosome to the conjugal mating pore (7). Members of the TraG family have conserved nucleotide-binding sites and, where studied, have been shown to localize to the cytoplasmic membrane (22). The latter is consistent with the presence of transmembrane domains in the N-terminal portion of the TraG polypeptide. The presence in ORF5 (Table 1) of conserved nucleoside triphosphate (NTP)-binding motifs and two putative membrane-spanning domains indicates that this gene product may also function as a membrane-linked NTPase.
ORF12 encodes a small basic polypeptide with significant homology to SSBs. The deduced ORF12 sequence exhibits the greatest similarity to three SSBs encoded within the Clostridium acetobutylicum genome (CAC3723, CAC1919, and CAC0945; P = 1E-08) and those encoded by Staphylococcus aureus prophages phiPV83 (P= 1E-09) and phiPVL (P = 2E-09). As with comparisons among other SSBs, the greatest similarity is localized to the first 100 residues from the N terminus (45), a region that has been shown for some family members to be sufficient for DNA binding (12). The four published mycoplasma genome sequences each contain a chromosomal housekeeping gene encoding SSB, a feature that is shared by most (if not all) bacteria. Many mobile elements, however, encode an additional ssb gene. In the context of conjugative elements, SSBs may protect from nucleases or stabilize the single-stranded DNA intermediate that is formed during the DNA-processing phase of element transfer.
The only other ORF within the element with significant overall similarity to proteins in the databases was ORF17, which had homology to conjugation proteins of the TraE/TrsE family, with the greatest similarity to the chromosomally encoded TraE protein of Mycoplasma pulmonis (P = 9E-53) and TraE of Lactococcus lactis plasmid pMRC01(P = 9E-20). TraC of the E. coli F plasmid is also a member of this family. Although the function of TraE proteins is unknown, family members are membrane associated and possess a conserved NTP-binding site (18, 39), both features that are predicted to be shared by the ORF17 polypeptide.
Our finding that these large repeated elements contain multiple homologs of conjugation-related proteins strongly suggests that they represent previously unrecognized conjugative elements of M. fermentans. In light of the potential for conjugation and the chromosomal residency of the element, the designation ICEF was adopted to describe the element. The four copies of ICEF in M. fermentans PG18 are identified as ICEF-IA (a single copy of the type I configuration) and ICEF-IIA, -IIB, and -IIC (three copies of the element with the type II configuration). The possibility of additional characteristic motifs associated with this class of element was examined. The deduced ORF3 polypeptide contained a sequence, HGNTDNPHIH, that closely resembles the sequence HHDTDNLHIH of the TraI protein of RP64, which forms motif III, the most conserved of the three motifs characteristic of relaxases (43). Appropriately spaced regions with limited sequence similarity to relaxase motifs I and II can also be identified in ORF3, strengthening the notion that this polypeptide serves as the relaxase for ICEF. With the absence of other homologs in the protein database, each ORF sequence of ICEF was queried against six-frame translations of the current databases. In this way, two additional homologs were identified. ORF1 had weak homology to a short sequence contig (MC293) from a genome sequence survey for M. capricolum (6). Although the homology score was low, this was likely due to frameshifts in the 684-bp M. capricolum sequence. The second homology (31% over 137 amino acid residues) occurred between terminal ORF22 and the reading frame encoding the P16 ORF of M. pulmonis strain KD 735-15 (50). The function of P16 is unknown, but it is of interest because within the complete genome sequence of M. pulmonis strain CT (11) is a previously unrecognized p16 gene sequence that is present in a different chromosomal location, consistent with a previous association with mobile DNA. The possibility that p16-containing elements exist in other M. pulmonis isolates is under investigation.
ORFs within ICEF were analyzed for short sequence motifs (with the “motifs” function of the Genetics Computer Group package) and sequence characteristics indicative of the subcellular location of each putative gene product (Table 1). Ten of the 22 ORFs are predicted to contain transmembrane-spanning domains. The largest predicted product, that of ORF19, contains a signal sequence at the N terminus. As little is known about mycoplasmal signal peptidases and their cognate cleavage specificities, it is not possible to predict whether the product of ORF19 is cleaved (and possibly released from the cell surface) or whether the hydrophobic signal peptide serves as a membrane-spanning anchor that may tether the ORF19 product to the single limiting membrane of M. fermentans.
A single lipoprotein is encoded by each copy of the genetic element. ORF14 encodes a 57-kDa polypeptide predicted to be the mature lipoprotein following removal of a signal peptide of 25 amino acid residues. This signal sequence was originally identified through a systematic screen for export functions of M. fermentans in E. coli, and the predicted lipid-modified amphipathic P57 product was confirmed by Western analysis with an antibody to a corresponding synthetic peptide (14). Subsequent work in this laboratory led to the cloning of two distinct alleles of the gene encoding P57, designated p57 and p57′ (36). The latter represents the allele found in the sequence of the single-copy type I configuration, ICEF-IA, on the basis of restriction fragment length polymorphism analysis for the restriction enzymes AflII and FspI (data not shown). In contrast to other ORFs in ICEF, the two alleles are comparatively divergent between ICEF configurations. The two p57 alleles contain 23 nucleotide differences, of which 13 are nonsynonymous, with amino acid substitutions dispersed throughout the P57 sequence. No differences in p57 sequences were identified in multiple clones from type II elements.
The P57 lipoprotein lacks homology to any other database entries, although as previously reported, a proline-rich region close to the N terminus is similar to an analogous region of the pMGA lipoproteins of M. gallisepticum (14). Lipoproteins are encoded by other mobile elements. TraV of the E. coli F plasmid is proposed to be an outer membrane anchor for a transenvelope transfer structure (26). The function of TraH of Staphylococcus aureus plasmid pSK41 is unknown (18), but as with certain plasmid-encoded lipoproteins from E. faecalis, the cleaved signal peptides are precursors for signaling pheromones (3).
ICEF integrates at multiple distinct sites in the M. fermentans chromosome.
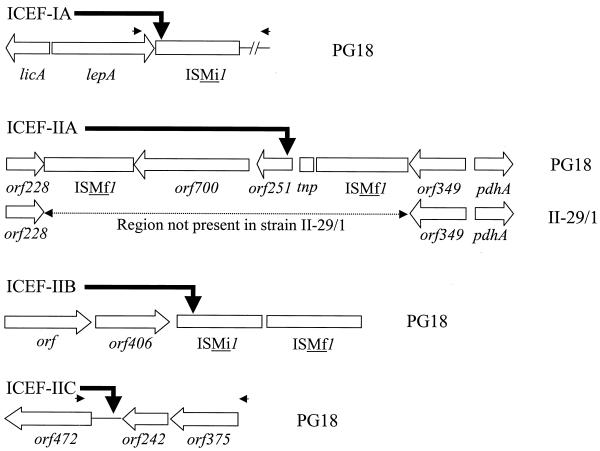
Initial hybridization analyses indicated that ICEF units were chromosomal in location. To confirm their chromosomal residency and to better understand the mechanism by which these elements integrated into the genome, junction fragments were sought that would contain both the ICEF termini and adjacent flanking sequences. Hybridization with oligonucleotide probes that anneal to sequences close to the ICEF termini enabled four genomic junction fragments of chromosomal DNA to be identified and cloned. These contained the left (IA-1, IIA-1, IIB-1, and IIC-1) or right (IA-7, IIA-2, IIB-2, and IIC-3) termini of the element (Fig. 2). Sequence analysis of these junction fragments, together with a comparison to the equivalent regions from an M. fermentans isolate, II-29/1 (8), that lacks ICEF, enabled pairs of left and right junctions to be assigned to a specific copy of ICEF (Fig. 3), thereby identifying individual insertion sites for each ICEF copy. Two ICEF units were inserted into different sites in distinct copies of ISMi1, a previously described multicopy IS of M. fermentans (32, 37). ICEF-IA was inserted into the left inverted repeat of an ISMi1 copy that is adjacent to the M. fermentans lepA and licA genes (Fig. 3 and Table 2). ICEF-IIB was embedded within the transposase coding sequence of ISMi1, disrupting synthesis of the enzyme from this IS copy. Interestingly, this copy of ISMi1 is adjacent to a previously unrecognized IS of M. fermentans that belongs to the IS256 family of elements (37). Two additional copies of this IS, herein designated ISMf1, were also identified in the vicinity of the ICEF-IIA insertion site. Upstream of ISMi1-containing ICEF-IIB were two ORFs that lack significant homology to entries in the current databases.
FIG. 3.
Genomic locations of ICEF insertion sites in M. fermentans. The locations of ICEF insertion sites (solid arrows) in M. fermentans PG18 are shown, together with the equivalent region (for ICEF-IIA) of the strain II-29/1 chromosome. The locations and directions of ORFs (open arrows) and individual copies of multicopy ISs ISMi1 and ISMf1 (open rectangles) that flank each of the four ICEF units in M. fermentans PG18 are shown. ORFs for coding sequences that either lack homology to known proteins or encode homologs of conserved hypothetical proteins are designated by ORF length (in amino acid residues, where this is known). ORFs with significant homology to housekeeping genes are labeled with standard gene abbreviations. A truncated transposase coding sequence is indicated (tnp) by an open square. For ICEF-IA and ICEF-IIC, horizontal arrows indicate the relative positions and orientations of the primers used for PCR amplification of the corresponding sites in the chromosome of M. fermentans II-29/1 (which lacks ICEF units). The chromosomal region of strain II-29/1 that is equivalent to that occupied by ICEF-IIA in strain PG18 was analyzed by sequencing a cloned genomic fragment (see Materials and Methods). ICEF-IIB is inserted into one copy of ISMi1 that is flanked by ISMf1 and two hypothetical genes.
Analysis of the ICEF-IIA junction fragments revealed that this ICEF copy was inserted in the vicinity of the housekeeping gene pdhA, encoding the E1α subunit of pyruvate dehydrogenase. Coincidentally, the 4.05-kb BamHI fragment (one of the five initially identified; Fig. 1A) was also a junction fragment for ICEF-IIA. In this case, ICEF had not disrupted an IS but was inserted into the 5′ region of a gene encoding an ATP-binding cassette of an ABC transporter (ORF251; abc in Fig. 1A). Interestingly, this region of the M. fermentans chromosome is strain variable. In attempting to confirm the linkage of the pdhA region to orf251 (ABC transporter gene containing ICEF-IIA in strain PG18), the pdhA region of strain II-29/1 was cloned and sequenced. As can be seen in Fig. 3, the pdhA clone from II-29/1 also contained the genes encoding the conserved hypothetical proteins encoded by ORF228 and ORF349 but lacked the two copies of ISMf1, a truncated transposase gene, and the intervening ORFs that occurred in PG18. Whether this observed genomic variation was due to the insertion en bloc of a composite structure that was flanked by ISMf1 elements or whether each of the integrative elements was acquired independently could not be determined.
The fourth copy, ICEF-IIC, was inserted into an intergenic region between conserved ORFs of unknown function (Table 2). To assign the left and right junction fragments to ICEF-IIC, a PCR was used to confirm the ordered linkage of hypothetical genes orf472-orf242-orf375 in the chromosome of strain II-29/1.
ICEF insertion generates direct repeats by target site duplication.
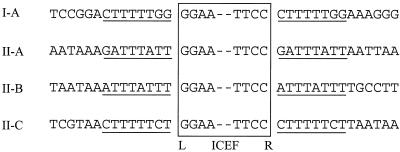
The observation that three of the four ICEF copies in strain PG18 are located within other genes or genetic units suggested that ICEF units are integrative DNAs. A hallmark of such elements is the presence of flanking direct repeats that are generated during DNA integration. The direct repeats result from asymmetric cleavage of the two DNA strands by the integration machinery and subsequent repair once the element has been inserted (37). Target site duplications vary in length but typically range from 2 to 13 bp. Generally, however, the length of the duplication is fixed for any given integrative DNA and is a characteristic feature of that element. Inspection of the sequences immediately abutting the termini of ICEF revealed that each copy is flanked by an 8-bp sequence that is present as a direct repeat (Fig. 4). Although this pattern is consistent with target duplication by an integrative element, it is also possible formally that ICEF is inserted between direct repeats. However, since two ICEF molecules had been inserted into copies of a known IS element and a third had been inserted into a gene for which the nondisrupted sequence was subsequently determined, it was possible to ascertain that for three of the four known ICEF insertion sites, the two 8-bp sequences flanking the respective ICEF are present only as a single copy in the “empty” site.
FIG. 4.
ICEF insertion generates 8-bp direct repeats by target site duplication. The nucleotide sequences flanking each copy of ICEF in strain PG18 are shown, together with the terminal nucleotides of ICEF (boxed). Underlining highlights the 8-bp sequence that is present in single copy in an unoccupied context and as a flanking direct repeat upon ICEF insertion.
These data indicate that ICEF are integrating DNAs. Clearly, multiple copies of the element can be comaintained within a single genome (since the organisms used in this study were isolated from low passage of a clonal isolate). The occupancy of multiple distinct insertion sites indicates that ICEF integration is not site specific, but a larger number of target sites need to be identified before the possibility of insertion site preference can be ascertained.
The available data indicate that ICEF units are novel genetic elements of M. fermentans that most closely resemble conjugative transposons and other members of the constin family of self-transmissible integrative DNAs. In contrast to other members of the family, however, ICEF units lack genes that encode homologs of known integrases, transposases, or recombinases. This raises the interesting possibility that these unusual elements have novel genes and encoded enzymes that carry out the critical integration-excision functions. As the genes encoding mobility functions of elements are typically located close to the sites of action (the ICEF termini in the present context) and would be predicted to encode basic cytoplasmic proteins, the terminal ORFs, ORF1 and ORF22, are candidates for this requisite function.
ICEF occurs in extrachromosomal forms.
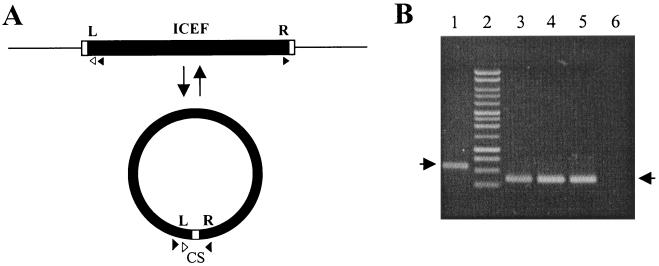
In the absence of systems for the genetic manipulation of M. fermentans, it has not been possible to demonstrate the mobility of ICEF directly. However, consideration of a faint, approximately 5.5-kb BssHII fragment that was reproducibly detected by Southern hybridization (Fig. 1B, open triangle) suggested that the dynamics of ICEF might be monitored by alternative approaches. The faint band (identified with probe 2; location indicated by an asterisk in Fig. 1A and 2) was intriguing since there was no evidence of the presence of a BssHII fragment of that size in the M. fermentans chromosome. However, cleavage of a circular extrachromosomal form of ICEF would yield such a fragment. The presence and identity of this fragment were confirmed by reprobing the same blot with a probe corresponding to the ICEF right terminal region (data not shown). These data suggested a model (Fig. 5A) similar to that proposed for other constins in which, at low frequency, ICEF is excised from the chromosome, circularized as a nonreplicative intermediate, and transferred by conjugation into a suitable recipient. Once in the latter host, the element is integrated into the chromosome. For several constins, including Tn916 (10), the SXT element (31), and CTnDOT (13), the intermediate can be detected by PCR. A similar assay was adopted to confirm that an extrachromosomal form of ICEF was indeed present in genomic DNA preparations from M. fermentans. Accordingly, outwardly facing PCR primers were employed under standard PCR conditions that would only yield an amplicon if permuted ICEF molecules exist in which the termini are juxtaposed (tandemly arranged ICEF units would also provide a suitable amplification template, but contour-clamped homogeneous electric field mapping studies had shown that the two closest ICEF units are separated by >30 kb). With this assay, the expected 570-bp amplicon was obtained (Fig. 5B), the sequence of which indicated that the termini of ICEF are indeed juxtaposed and separated by a 6-bp region that is heterogeneous in sequence. This heterogeneity suggested that multiple elements contribute to a “pool” of extrachromosomal molecules. To specifically amplify the extrachromosomal form of ICEF-IA, advantage was taken of a region of sequence microheterogeneity between type I and II elements. Substitution of a primer corresponding to the single copy of this type I sequence resulted in the expected 330-bp product (Fig. 5B). Within the sequence of this amplicon, the sequence of the 6-bp spacer or coupling sequence between the ICEF termini could be discerned, allowing the origin of this spacer to be traced to the 8-bp direct repeats that flank ICEF-IA.
FIG. 5.
Detection of an extrachromosomal form of ICEF. (A) Model for integration-excision of ICEF units. The direct repeats flanking ICEF are shown as open rectangles abutting the left (L) and right (R) termini of the integrated form of ICEF (solid bar). In the extrachromosomal form, the termini are juxtaposed and separated by a 6-bp coupling sequence (CS). Orientations of primers (triangles) used to amplify the unique configuration of the extrachromosomal form are indicated. The solid triangles represent primers 7 and 9 (see Materials and Methods) that together amplify terminal regions of extrachromosomal forms of all known ICEF units. PCR with primer 16 (open triangle) in combination with primer 7 (right-facing solid triangle in integrated form) selectively amplified a product from an extrachromosomal form of ICEF-IA. (B) Amplicons derived from genomic DNA containing extrachromosomal ICEF following agarose gel electrophoresis. Lanes: 1, PCR amplicon obtained with primers 7 and 9 with M. fermentans PG18 DNA as the template; 3 to 6, amplicons generated with primers 7 and 16 by using as the template three M. fermentans PG18 DNA preparations purified from different cultures (lanes 3 to 5) or DNA from M. fermentans II-29/I, which lacks ICEF units (lane 6); 2, 1-kb ladder (Promega Corporation, Madison, Wis.). The positions of the anticipated ∼570-bp (primers 7 and 9) and ∼330-bp (primers 7 and 16) amplicons are indicated by arrows at left and right, respectively.
The ICEF terminal regions contain distinctive motifs.
Several features of the untranslated region between the terminal ORFs in the extrachromosomal form of the element warrant further mention. The untranslated regions at the left and right termini and the juxtaposed termini in the extrachromosomal form are evidently the sites of action for DNA mobility functions. The region was analyzed for distinctive features that might be candidate DNA-binding sites for such DNA-binding proteins. One striking finding was the presence of four SphI restriction sites within a 250-bp section of ICEF close to the right terminus. The recognition sequence for this endonuclease (GCATGC) is relatively rare in the A+T-rich M. fermentans genome and is not present elsewhere in ICEF. Further inspection of this untranslated region revealed that the restriction site is within a repeated motif with a consensus of AAAGTGCATGC. Of the five similarly oriented copies of this repeat, three conform perfectly to the consensus whereas two have single-nucleotide deviations. Also of interest is the finding that the region immediately downstream of p16 in M. pulmonis (the homolog of terminal ORF22 in ICEF) also contains an untranslated region with distinctive features that are shared with the ICEF terminal region (50). Within the M. pulmonis region, there are also four SphI sites within an ∼120-bp region. Furthermore, each copy of ICEF and the p16-linked locus has a rare BssHII site within the sequence GTGCGCGCA, located 200 to 250 bp downstream of the stop codon for either ORF22 or P16. The repetitive nature, location, and distinctiveness suggest that these short sequence motifs are important components of ICE units, possibly functioning as cognate binding sites for the hypothesized DNA-binding proteins that mediate ICE mobility.
ICEF is variably distributed among M. fermentans isolates.
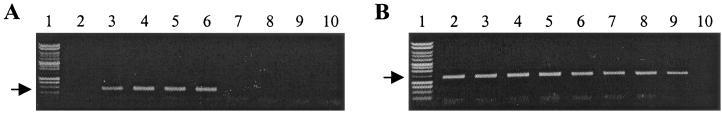
To determine the distribution of ICEF among M. fermentans isolates, PCR analysis was performed with primer pairs corresponding to a short intergenic region 5′ to ORF1 that is conserved between the type I and II configurations of ICEF. As a positive control for the functional integrity of the genomic template DNAs from various strains, the previously described primers for amplification of the malp gene of M. fermentans were used (8). As shown in Fig. 6, the malp gene was amplified from each of the eight isolates tested, as expected on the basis of the previously reported strain distribution of this gene. In addition, ICEF sequences could be amplified from strains SK5, SK6, M39B, and M70B with the ICEF-specific primers but not from the four remaining isolates. Further PCR analysis indicated that p57 sequences could also be amplified from the isolates that yielded ICEF amplicons (data not shown). Sequence analysis of the population of p57 amplicons revealed that strains M39B and M70B contained a mixture of the p57 and p57′ alleles, whereas SK5 and SK6 contained only the p57′ allele similar to that in ICEF-IA of strain PG18. Further PCR analysis demonstrated that only the type I element can be detected in SK5 and SK6 (on the basis of the absence of ORF2 between ORF1 and ORF3) but that there is no insertion within the lepA region of the genome (on the basis of amplification with the primers shown in Fig. 3). Taken together, these data demonstrate variable distribution and sites of integration of ICEF among M. fermentans isolates, with some strains having both type I and type II elements, others having only type I units, and still others lacking ICEF. Interestingly, of the limited number of strains tested, all of the ICEF-negative isolates have been either identified as tissue culture isolates or are suspected tissue culture contaminants, raising the intriguing possibility that ICEF units are dispensable during multiple passages in culture but that they may contribute to host adaptation during M. fermentans infection of humans.
FIG. 6.
Strain-variable distribution of ICEF among M. fermentans isolates. (A) Agarose gel electrophoresis of amplicons generated with ICEF-specific primers 1 and 9 (see Materials and Methods) using genomic DNA templates from M. fermentans isolates SK6 (lane 3), SK5 (lane 4), M70 (lane 5), M39 (lane 6), Incognitus (lane 7), MT-2 (lane 8), K7 (lane 9), and II-29/1 (lane 10). Lane 1 contained a 1-kb ladder; lane 2 contained a negative control (water as the template). The location of the anticipated 420-bp amplicon is indicated by an arrow. (B) Amplicons generated with malp-specific primers 10 and 11 (see Materials and Methods) using the same M. fermentans DNA templates as in panel A. Lanes 1 and 10 contained a molecular size ladder and a negative control reaction mixture, respectively. Lanes 2 to 9 correspond to lanes 3 to 10 in panel A. The anticipated 1.4-kb amplicon is indicated by an arrow.
ICEF contributes to genomic plasticity and phenotypic diversity.
One striking feature of this element (and the one that contributed to its identification) is that ICEF constitutes approximately 8% of the M. fermentans genome. Previous studies based on genomic analyses of M. genitalium and M. pneumoniae suggested that the “flexible” or “mobile” gene pool might be less than 1% (41). The data presented here clearly show that whereas some mycoplasma species may indeed contain minimal genomes in the context of the “core” gene pool, the flexible gene pool may show greater diversity (size variation) between species. Within a species (as in other genera), genome size can vary significantly between isolates (40). In the case of M. fermentans, isolates have been identified that contain genomes that are estimated to be 115 kb (strain K7) or 210 kb (strain Incognitus) smaller than that of type strain PG18 (48). As both of these strains are known to lack ICEF units, the presence or absence of ICEF repeats must contribute significantly to the genome size variation between M. fermentans isolates.
In other species, differences in chromosomal gene complement and organization have been identified and termed plasticity zones (1). Mobile DNAs and selfish DNAs, including those encoding restriction-modification systems, are associated with the boundaries of these zones. Repeated sequences may play a role in determining chromosome structure, as the presence of large repeats provides recombination substrates that may promote gene deletion, gene duplication, and DNA inversion. It will be of interest to determine whether ICEF has been involved in such large-scale rearrangements. From comparative analysis of mycoplasma genomes, one general emerging feature is the lack of conservation of gene order, even within regions encoding housekeeping genes. It is tempting to speculate that insertion of ICE-related elements into genomes that are undergoing reductive evolution can reduce the recombinational linkage of gene clusters by increasing the distance between genes.
In addition to contributing to genetic diversity, ICEF may contribute to surface diversity between M. fermentans isolates. ICEF encodes multiple proteins that are predicted to be surface localized on the basis of the presence of putative transmembrane domains and signal peptides. Among these is a single lipoprotein that has previously been shown to be expressed and membrane associated. Since the function of this lipoprotein is unknown, it is not clear whether it plays a role in the proposed mobility of the element or whether it has a direct role in interactions between M. fermentans and the human host. The same pertains to other surface proteins encoded by genes resident within ICEF. Inadvertently, ICEF and other mobile DNAs may also bring about mutation by direct inactivation of genes encoding surface proteins. We note that ICEF-IIA is inserted into the coding sequence for a putative ATP-binding cassette of an ABC transporter. Although this gene product is not predicted to be expressed on the surface, the downstream reading frame, ORF700, contains a putative signal peptide and is probably transcriptionally linked to the upstream transporter gene.
ICE-like elements in LGT in mycoplasmas?
Understanding the potential for inter- or intraspecies exchange is clearly important in the context of bacteria like mycoplasmas that co-occupy host niches for extended periods during chronic infection. The lack of a rigid cell wall might facilitate close interaction between mycoplasmas and promote genetic exchange. The presence of short regions of homology between ICEF ORF1 and the short sequence contigs from a genome sequence survey of M. capricolum has led to the identification of an analogous ICE unit in this caprine host that, although incompletely characterized, has features in common with the ICEF prototype (M. J. Calcutt and K. S. Wise, 13th Int. Congr. Int. Org. Mycoplasmol., abstr. 41, 2000), strengthening the likelihood that related elements are more widespread in nature than is presently appreciated. Data derived from an ongoing genome sequencing project supports this notion, as tra genes related to those in ICEF have been identified in Spiroplasma citri (F. Laigret, P. Carle, N. Carrère, M. Garnier, and J.-M. Bové, 13th Int. Congr. Int. Org. Mycoplasmol., abstr. 48, 2000). As the complete sequences of this and other mycoplasma genomes become available, it will be of interest to ascertain whether these loci are associated with ICE-like units. The finding of Tra homologs in S. citri is intriguing, as the transfer of chromosomal resistance markers via a DNase I-insensitive mechanism in this species has been reported (4). The pathway and/or vector involved in this genetic exchange are not known, although conjugation or direct fusion was proposed. Whether ICE-related sequences play a role in marker exchange, perhaps by encoding components of a specific conjugal apparatus or by promoting transfer of nearby chromosomal markers, is important to determine.
tra genes are not ubiquitous among mycoplasmas, as the complete genome sequences of M. genitalium (19), M. pneumoniae (28), and Ureaplasma urealyticum (21) lack identifiable tra-like genes. The complete data set of one M. pulmonis isolate contains a traE homolog and p16-related sequences, although neither was present within a larger genetic unit. The relationship of these genes to each other and to the recently reported demonstration (55) of gene transfer among certain M. pulmonis isolates is not known.
In the absence of a transformation system for M. fermentans, it has not been possible to directly demonstrate ICEF integration or to show transfer of ICEF between isolates. Furthermore, the codon usage of mycoplasmas precludes the facile use of E. coli as a surrogate recipient for studies of integration functions. Our finding that ICE-related sequences exist in other mycoplasma species, including some that are genetically tractable, is therefore important not only in surveying the distribution and organization of ICE-related sequences but also in determining the possible contribution of ICE units to LGT, as well as to host adaptation and pathogenesis of the mycoplasmas in which they reside.
Acknowledgments
We thank Shyh-Ching Lo for providing DNA preparations from M. fermentans isolates and Tonghua Lu for providing p57-related sequences and hybridization probes.
This work was supported by U.S. Public Health Service grants AI32219 (to K.S.W.) and AI47937 (to M.J.C.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alm, R. A., L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Ura-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barroso, G., and J. Labarère. 1988. Chromosomal gene transfer in Spiroplasma citri. Science 241:959-961. [DOI] [PubMed] [Google Scholar]
- 5.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bork, P., C. Ouzounis, G. Casari, R. Schneider, C. Sander, M. Dolan, W. Gilbert, and P. M. Gillevet. 1995. Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol. Microbiol. 16:955-967. [DOI] [PubMed] [Google Scholar]
- 7.Cabezon, E., J. I. Sastre, and F. De La Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 8.Calcutt, M. J., M. F. Kim, A. B. Karpas, P. F. Mühlradt, and K. S. Wise. 1999. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of Mycoplasma fermentans. Infect. Immun. 67:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calcutt, M. J., J. L. Lavrrar, and K. S. Wise. 1999. IS1630 of Mycoplasma fermentans, a novel IS30-type insertion element that targets and duplicates inverted repeats of variable length and sequence during insertion. J. Bacteriol. 181:7597-7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caparon, M. G., and J. R. Scott. 1989. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 59:1027-1034. [DOI] [PubMed] [Google Scholar]
- 11.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewsky, A. Viari, E. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chase, J. W., and K. R. Williams. 1986. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 55:103-136. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cleavinger, C. M., M. F. Kim, J. H. Im, and K. S. Wise. 1995. Identification of mycoplasma membrane proteins by systematic TnphoA mutagenesis of a recombinant library. Mol. Microbiol. 18:283-293. [DOI] [PubMed] [Google Scholar]
- 15.De La Cruz, F., and J. Davies. 2000. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 8:128-133. [DOI] [PubMed] [Google Scholar]
- 16.Djordjevic, S. P., W. A. Forbes, J. Forbes-Faulkner, P. Kuhnert, S. Hum, M. A. Hornitzky, E.-M. Vilei, and J. Frey. 2001. Genetic diversity among Mycoplasma species bovine group 7: clonal isolates from an outbreak of polyarthritis, mastitis, and abortion in dairy cattle. Electrophoresis 22:3551-3561. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 18.Firth, N., K. P. Ridgway, M. E. Byrne, P. D. Fink, L. Johnson, I. T. Paulsen, and R. A. Skurray. 1993. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene 136:13-25. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 20.Freundt, E. A., and D. G. Edward. 1979. Classification and taxonomy, p. 1-41. In M. F. Barile and S. Razin (ed.), The mycoplasmas. Academic Press, Inc., New York, N.Y.
- 21.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, and E. Y. C. G. H. Chen. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 22.Grahn, A. M., J. Haase, D. H. Bamford, and E. Lanka. 2000. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 182:1564-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 24.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 26.Harris, R. L., V. Hombs, and P. M. Silverman. 2001. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42:757-766. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann, R. 1992. Genome structure and organization, p. 157-168. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 28.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochhut, B., K. Jahreis, J. W. Lengeler, and K. Schmid. 1997. CTnscr94, a conjugative transposon found in enterobacteria. J. Bacteriol. 179:2097-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochhut, B., J. Marrero, and M. K. Waldor. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J. Bacteriol. 182:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochhut, B., and M. K. Waldor. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 32.Hu, W. S., R. Y. Wang, R. S. Liou, J. W. Shih, and S. C. Lo. 1990. Identification of an insertion-sequence-like genetic element in the newly recognized human pathogen Mycoplasma incognitus. Gene 93:67-72. [DOI] [PubMed] [Google Scholar]
- 33.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 1:99-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause, D. C., and D. Taylor-Robinson. 1992. Mycoplasmas which infect humans, p. 417-444. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 35.Lo, S. C. 1992. Mycoplasmas and AIDS, p. 525-545. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC.
- 36.Lu, T. 1998. Characterization and strain distribution of multicopy allelic variants of the M. fermentans membrane lipoprotein gene, p57. Ph.D. thesis. University of Missouri—Columbia.
- 37.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 39.Morton, T. M., D. M. Eaton, J. L. Johnston, and G. L. Archer. 1993. DNA sequence and units of transcription of the conjugative transfer gene complex (trs) of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 175:4436-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niemark, H. C., and C. S. Lange. 1990. Pulse-field electrophoresis indicates full-length mycoplasma chromosomes vary widely in size. Nucleic Acids Res. 18:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 42.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1098. [DOI] [PubMed] [Google Scholar]
- 43.Pansegrau, W., W. Schröder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 44.Ravatn, R., S. Studer, D. Springael, A. J. B. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruvolo, P. P., K. M. Keating, K. R. Williams, and J. W. Chase. 1991. Single-stranded DNA binding proteins (SSBs) from prokaryotic transmissible plasmids. Proteins 9:120-134. [DOI] [PubMed] [Google Scholar]
- 46.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saunders, N. J., D. W. Hood, and E. R. Moxon. 1999. Bacterial evolution: bacteria play pass the gene. Curr. Biol. 9:R180-R183. [DOI] [PubMed] [Google Scholar]
- 48.Schaeverbeke, T., M. Cleric, L. Lequen, A. Charron, C. Bébéar, B. De Barbeyrac, B. Bannwarth, and J. Dehais. 1998. Genotypic characterization of seven strains of Mycoplasma fermentans isolated from synovial fluids of patients with arthritis. J. Clin. Microbiol. 36:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, J. R. 1993. Conjugative transposons, p. 597-614. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
- 50.Shen, X., J. Gumulak, H. Yu, C. T. French, N. Zou, and K. Dybvig. 2000. Gene rearrangements in the vsa locus of Mycoplasma pulmonis. J. Bacteriol. 182:2900-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoemaker, N. B., G.-R. Wang, and A. A. Salyers. 1996. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J. Bacteriol. 178:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simecka, J. W., J. K. Davis, M. K. Davidson, S. E. Ross, C. T. K. H. Städtlander, and G. H. Cassell. 1992. Mycoplasma diseases of animals, p. 391-416. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 53.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500 kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szpirer, C. Y., M. Faelen, and M. Couturier. 2000. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 37:1283-1292. [DOI] [PubMed] [Google Scholar]
- 55.Teachman, A. M., C. T. French, H. Yu, W. L. Simmons, and K. Dybvig. 2002. Gene transfer in Mycoplasma pulmonis. J. Bacteriol. 184:947-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tu, A. H., L. L. Voelker, X. Shen, and K. Dybvig. 2001. Complete nucleotide sequence of the mycoplasma virus P1 genome. Plasmid 45:122-126. [DOI] [PubMed] [Google Scholar]
- 57.Vilei, E. M., J. Nicolet, and J. Frey. 1999. IS1634, a novel insertion element creating long, variable-length direct repeats which is specific for Mycoplasma mycoides subsp. mycoides small-colony type. J. Bacteriol. 181:1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voelker, L. L., and K. Dybvig. 1998. Characterization of the lysogenic bacteriophage MAV1 from Mycoplasma arthritidis. J. Bacteriol. 180:5928-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voelker, L. L., and K. Dybvig. 1998. Demonstration of extrachromosomal elements. Methods Mol. Biol. 104:239-246. [DOI] [PubMed] [Google Scholar]
- 60.Voelker, L. L., and K. Dybvig. 1999. Sequence analysis of the Mycoplasma arthritidis bacteriophage MAV1 genome identifies the putative virulence factor. Gene 233:101-107. [DOI] [PubMed] [Google Scholar]
- 61.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]