Abstract
Obligately aerobic tubercle bacilli are capable of adapting to survive hypoxia by developing into a nonreplicating or dormant form. Dormant bacilli maintain viability for extended periods. Furthermore, they are resistant to antimycobacterials, and hence, dormancy might play a role in the persistence of tuberculosis infection despite prolonged chemotherapy. Previously, we have grown dormant Mycobacterium bovis BCG in an oxygen-limited Wayne culture system and subjected the bacilli to proteome analysis. This work revealed the upregulation of the response regulator Rv3133c and three other polypeptides (α-crystallin and two “conserved hypothetical” proteins) upon entry into dormancy. Here, we replaced the coding sequence of the response regulator with a kanamycin resistance cassette and demonstrated that the loss-of-function mutant died after oxygen starvation-induced termination of growth. Thus, the disruption of this dormancy-induced transcription factor resulted in loss of the ability of BCG to adapt to survival of hypoxia. Two-dimensional gel electrophoresis of protein extracts from the gene-disrupted strain showed that the genetic loss of the response regulator caused loss of the induction of the other three dormancy proteins. Thus, the upregulation of these dormancy proteins requires the response regulator. Based on these two functions, dormancy survival and regulation, we named the Rv3133c gene dosR for dormancy survival regulator. Our results provide conclusive evidence that DosR is a key regulator in the oxygen starvation-induced mycobacterial dormancy response.
Mycobacteria are obligate aerobes, i.e., they require oxygen for growth. However, ample evidence from animal models and human studies suggests that tubercle bacilli encounter hypoxic environments in active disease as well as in latent infection (30, 32, 33, 38, 41, 42). L. G. Wayne established a link between starvation for oxygen and drug resistance. He demonstrated that upon gradual depletion of oxygen from a culture (oxygen-limited Wayne culture system), the bacillus exits the cell cycle and develops into a defined dormant form that is adapted to hypoxia and maintains viability for extended periods of time. Importantly, the dormant form of the bacterium is resistant to antimycobacterials. This phenotypic resistance could be due to the fact that antimycobacterials target mainly growth-related functions such as cell wall synthesis (15, 34-40). Hence, an emerging working model suggests that phenotypically drug-resistant dormant bacilli, similar to the ones observed in oxygen-starved cultures, exist in hypoxic microenvironments in the host and may cause persistent infection despite prolonged chemotherapy; currently, 6 to 9 months of treatment is required to cure the patient of tuberculosis (4, 8, 9, 18, 21, 24).
Obviously, this monocausal model of persistence oversimplifies the situation in vivo. However, if it does reflect some aspects of the life of this parasite in its host, the development of drugs that target hypoxic dormant bacilli could have a profound impact on tuberculosis therapy. A combination therapy consisting of conventional antimycobacterials (targeting growing bacilli) and antidormancy drugs (hitting the dormant subpopulation) might considerably shorten the time necessary for treatment (6, 41).
Mycobacterium tuberculosis is an aerosol-transmitted biosafety level 3 pathogen. To overcome the associated experimental limitations, we established the attenuated BCG strain of M. bovis as a model organism to study the hypoxic life of the tubercle bacillus. We demonstrated that BCG shows a dormancy response that is strikingly similar to the behavior shown by M. tuberculosis when grown in the oxygen-limited Wayne culture system (11, 12, 16, 20, 22, 29). To identify dormancy-induced proteins, we previously subjected BCG grown in the Wayne system to a proteome analysis (1). In this work, the response regulator Rv3133c, a putative phosphorylation-dependent transcription factor identified in the M. tuberculosis H37Rv genome project (2), was found to be upregulated immediately upon entry into dormancy. In contrast, this protein is not detectable in either aerated growing or aerated stationary-phase cultures. The temporal expression profile in the Wayne system and the apparent dormancy specificity, together with the probable biochemical function of the protein as a transcription factor, suggest that the molecule could play a key regulatory role in the adaptation of bacilli to survival under hypoxic conditions (1).
Together with the response regulator, three other proteins, α-crystallin (5, 14, 17, 26, 31, 36, 43, 44) and two “conserved hypothetical” proteins, universal stress protein domain-containing Rv2623 (7, 14, 19, 26, 31) and cystathionine β-synthase domain-containing Rv2626c (14, 26, 31) are upregulated upon entry into dormancy (1). The observed temporal coinduction of these three proteins along with the response regulator upon entry into dormancy could indicate that they are under the control of this transcription factor. To determine whether Rv3133c plays a role in adaptation to hypoxic survival and in the upregulation of the three coinduced proteins, we deleted its coding sequence from the BCG genome. We report the phenotypic consequences of this loss-of-function mutation on growth and viability and the proteome of the bacillus grown in the Wayne dormancy culture system.
MATERIALS AND METHODS
Strain, media, cultivation, and monitoring of growth and survival.
All experiments were conducted with Mycobacterium bovis BCG Pasteur strain ATCC 35734 at 37°C. Liquid medium was Dubos Tween-albumin broth; solid medium was Dubos oleic-albumin agar (Difco). Kanamycin, gentamicin, and hygromycin were from Sigma. Standard (i.e., oxygen-unlimited) cultures (100 ml) were grown in plastic roller bottles (10 by 14 cm) at 1 rpm and with a starting A600 of 0.05. The containers were opened daily to allow exchange of air (1). Oxygen-limited Wayne cultures were grown in screw-cap test tubes (20 mm by 125 mm), with a total volume of 25.5 ml. Precultures were diluted to an A600 of 0.005 in a final volume of 17 ml. Magnetic stirrers were added; the cultures were sealed by tightly screwing down solid caps with latex liners and stirred gently at 170 rpm on magnetic stirring platforms (1, 16, 39).
Oxygen depletion was monitored via decolorization of the redox indicator methylene blue as described previously (16). Growth and survival were determined by turbidity measurements in a Bacharach Coleman model 35 photometer (Thomas Scientific) and by determination of CFU on agar after plating of appropriate dilutions as described previously (16). In contrast to the hypoxic stationary-phase cells grown in the Wayne system, clumping of bacilli occurred throughout aerated stationary-phase growth in roller bottles. To reduce clumping for CFU determinations, the roller bottle cultures were sonicated five times for 15 s each with 1-min ice breaks at level 2 of an Ultrasonic processor XC sonicator (Misonix) before plating.
DNA manipulations.
DNA manipulations were done according to standard protocols (28). DNA purifications were carried out with Qiagen kits as recommended by the supplier. DNA was introduced into BCG by electroporation as described before (29) with minor modifications. A lambda ZAPII library of genomic DNA from BCG strain ATTC 35734 was constructed with Stratagene reagents (B. Murugasu-Oei and T. Dick, unpublished data). A 429-bp probe specific to dosR was generated by PCR with the primers TCGTAGGTGAGGCGGGTTC and CGGCGATCTGCTTGTTGGT, designed according to the sequence of the gene in M. tuberculosis H37Rv (Rv3133c [2]). The library was screened, and positive clones were excised as pBluescriptSK plasmids with helper phages according to the protocol provided by the supplier.
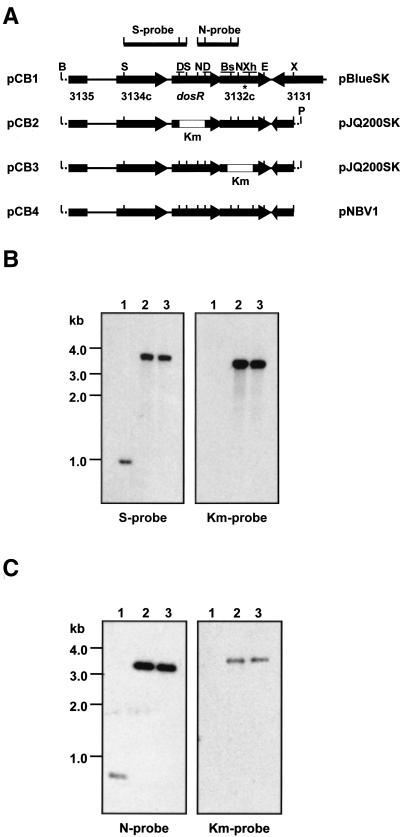
One clone (pCB1, Fig. 1A) carrying a 5.9-kb insert containing the dosR gene plus flanking regions was subjected to DNA sequencing on a Perkin-Elmer ABI Prism 377 sequencer. To facilitate subsequent manipulations of the dosR operon, a 4.7-kb BamHI-XbaI fragment (BamHI site in the polylinker of pBluescriptSK, XbaI site downstream of Rv3132c within the genomic fragment) was excised from pCB1 and ligated with BamHI- and XbaI-digested pNEB193 (New England Biolabs). The resulting plasmid was digested with DraIII, resulting in a 607-bp deletion within the 654-bp dosR coding sequence. The product was blunt ended with T4 DNA polymerase and ligated with the blunt-ended, 1.2-kb PstI fragment carrying the kanamycin resistance gene from pUCK4 (Pharmacia). The 5.4-kb genomic DNA fragment carrying the kanamycin resistance cassette replacing the dosR coding sequence was released via BamHI-PstI digestion (PstI site in the polylinker of pNEB193) and ligated into BamHI- and PstI-digested pJQ200SK (25), resulting in the gene replacement plasmid pCB2 (Fig. 1A).
FIG. 1.
dosR locus, gene replacement constructs, rescue plasmid, and Southern blot analyses of gene replacement mutants. (A) Schematic diagrams (not to scale) of the dosR locus in BCG(pCB1) and derivatives thereof (pCB2 to -4) are shown. pCB2, dosR gene replacement plasmid; pCB3, Rv3132c gene replacement plasmid; pCB4, rescue plasmid. Horizontal arrows show open reading frames, and the numbers below the arrows in pCB1 indicate the corresponding Rv numbers of open reading frames as annotated for the M. tuberculosis H37Rv genome (2). Empty boxes labeled Km represent the kanamycin resistance cassette. Only relevant restriction sites are indicated: B, BamHI; S, SphI; D, DraIII; N, NotI; Bs, BstXI; Xh, XhoI; E, EcoRI; X, XbaI; P, PstI. Underlined restriction sites were used for deletion of coding sequences. S-probe and N-probe indicate SphI and NotI fragments used as probes for the Southern blots shown in B and C, respectively. Solid line, genomic DNA; dotted line, polylinker sequence. Vector backbones of the plasmids are indicated on the rightside. pBlueSK, pBluescriptSK (Stratagene); pJQ200SK, suicide vector (25); pNBV1, E. coli-mycobacterium shuttle vector (10). The asterisk indicates the site of a single-base-pair polymorphism that was detected between BCG and M. tuberculosis H37Rv. Codon 283 of Rv3132c in H37Rv (ATT) is ACT in BCG, resulting in an Ile-to-Thr amino acid replacement in the sensor domain of the kinase. (B) Southern blot analyses of ΔdosR::km gene replacement mutants. S-probe, DNA from the wild type (lane 1) and two independently isolated dosR gene replacement mutants (lanes 2 and 3) was digested with SphI and EcoRI, transferred to a membrane, and probed with the 1-kb SphI fragment (S-probe) shown in A. BCG wild-type genomic DNA showed the expected hybridization band, corresponding to the size of the SphI fragment used as the probe. In contrast, the ΔdosR::km mutants, which have lost an SphI site due to deletion of the dosR coding sequence, showed a band of 3.7 kb, corresponding to the expected size of the genomic SphI-EcoRI fragment containing the disrupted ΔdosR::km allele (compare pCB1 with pCB2). Km-probe, probing of the blot with a 1.2-kb PstI fragment carrying the kanamycin resistance cassette detected the same hybridization band in the ΔdosR::km strains that was detected by using the 1-kb SphI fragment as the probe, confirming that the 3.7-kb SphI-EcoRI band observed for these strains indeed contained the kanamycin resistance cassette and, hence, the disrupted dosR gene. (C) Southern blot analyses of ΔRv3132c::km gene replacement mutants. N-probe, DNA from the wild type (lane 1) and two independently isolated Rv3132c gene replacement mutants (lanes 2 and 3) was digested with NotI and XbaI, transferred to a membrane, and probed with the 0.8-kb NotI fragment (N-probe) shown in A. Wild-type genomic DNA showed the expected hybridization band, corresponding to the size of the NotI fragment used as the probe. In contrast, the ΔRv3132c::km mutants, which have lost a NotI site due to deletion of the Rv3132c coding sequence, showed a band of 3.2 kb corresponding to the expected size of the genomic NotI-XbaI fragment containing the disrupted ΔRv3132c::km allele (compare pCB1 and pCB3). Km-probe, probing of the blot with the 1.2-kb PstI fragment carrying the kanamycin resistance cassette (Km-probe) detected the same hybridization band in the ΔRv3132c::km strains that was detected with the 0.8-kb NotI fragment as a probe, confirming that the 3.2-kb NotI-XbaI band in BCG ΔRv3132c::km genomic DNA contained the kanamycin resistance cassette and hence the disrupted gene. DNA from both the ΔdosR::km and ΔRv3132c::km mutants showed no hybridization signal with the pJK200SK vector as the probe, confirming that the strains had lost all vector sequences (data not shown). Size markers are from the 1-kb ladder; 1 to 2 μg of genomic DNAs was loaded.
pNEB193 carrying the 4.7-kb BamHI-XbaI fragment was also used to generate the replacement construct for the Rv3132c coding sequence. The plasmid was digested with BstXI and XhoI, resulting in an 895-bp deletion within the 1,737-bp Rv3132c coding sequence. The product was blunt ended and ligated with the kanamycin resistance cassette as before. The 5.1-kb genomic DNA fragment carrying the kanamycin resistance cassette replacing the Rv3132c coding sequence was transferred to pJQ200SK as before, resulting in the Rv3132c gene replacement plasmid pCB3 (Fig. 1A). Sequencing the deletion sites showed that the kanamycin resistance cassette was inserted in the same transcriptional orientation as dosR and Rv3132c. Gene replacement was carried out according to the two-step protocol, taking advantage of the properties of the suicide vector pJQ200SK to confer sensitivity to sucrose and resistance to gentamicin (23).
Briefly, BCG was transformed with 1 μg of the gene replacement constructs and selected on agar containing kanamycin (20 μg ml−1). Replica plating of kanamycin-resistant colonies on agar containing sucrose (2%) and gentamicin (5 μg ml−1) identified strains that were sucrose sensitive and gentamicin resistant, indicating genomic recombination of the whole-gene replacement constructs. These recombinants were grown in broth to allow a second, intrachromosomal recombination step to occur. For selection of second-step recombinants that had lost the pJQ200SK sequence and the flanking copy of the native, wild-type genes, cultures were plated on agar containing sucrose and kanamycin. Sucrose- and kanamycin-resistant colonies were replica plated on agar containing gentamicin to confirm gentamicin sensitivity and thus loss of the pJQ200SK sequence. For rescue of the phenotypes caused by disruption of dosR, the 4.7-kb BamHI-XbaI genomic fragment containing the dosR operon was released from pNEB193 (see above) via BamHI and XbaI digestion and ligated into BamHI- and XbaI-digested pNBV1, which confers resistance to hygromycin (50 μg ml−1) (10), resulting in pCB4 (Fig. 1A).
Two-dimensional gel electrophoretic analysis.
Bacilli were broken up by disrupting the cells with glass beads in a Mini Beadbeater cell disruptor as described previously (1). Then 100 μg of total protein was subjected to isoelectric focusing with pH 4 to 7 Immobiline dry strips and an IPGphor isoelectric focusing unit (Amersham Pharmacia) and separated in the second dimension by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (Protean IIxi system; Bio-Rad) as described before (1).
Nucleotide sequence accession number.
The sequence of the dosR locus as shown in pCB1 (Fig. 1A) has been deposited in GenBank under accession number AF522461.
RESULTS
Generation of BCG ΔdosR::km.
The dosR (Rv3133c) locus was isolated from a BCG genomic library and sequenced. Figure 1A (pCB1) shows the structural organization of the dosR region in BCG, which was found to be identical to that of M. tuberculosis H37Rv (2). At 30 bp upstream of the dosR coding sequence is the 3′ end of the Rv3134c open reading frame (conserved hypothetical protein). Overlapping the TGA stop codon of dosR is the ATG start codon (italic) of Rv3132c (CCA TGA) encoding a sensor histidine kinase (27). This genomic organization suggests that the three genes form an operon. In vivo promoter probing confirmed the presence of dormancy-inducible promoter activity in the intergenic region upstream of Rv3134c and the absence of such activity upstream of dosR and Rv3132c (B. H. Tan, C. Boon, and T. Dick, unpublished observations). This proposed operon structure is consistent with recent transcriptional studies of the locus (3, 31).
To determine the function of DosR, its coding sequence was replaced with a kanamycin resistance cassette (Fig. 1A, pCB2; see Materials and Methods for details). The replacement of the dosR gene by its inactivated allele (ΔdosR::km) was confirmed by Southern blotting, as shown in Fig. 1B.
Hypoxia-induced death of BCG ΔdosR::km.
To determine the role of the dosR gene in the dormancy response, BCG ΔdosR::km was grown in the oxygen-limited Wayne culture system. This system is based on growth of the bacilli in sealed tubes with stirring (16, 39). Initially the cultures grow exponentially and consume oxygen. A temporal oxygen gradient is self-generated, and upon encountering a microaerobic threshold level after about 4 to 5 days, the wild type is deflected from exponential growth and enters a transition phase in which the turbidity of the culture increases slowly up to day 10, after which the A600 stays constant (Fig. 2A) (1). Wild-type bacilli in the hypoxic stationary phase are in a state of dormancy, i.e., they are nonreplicating and maintain viability (16, 39).
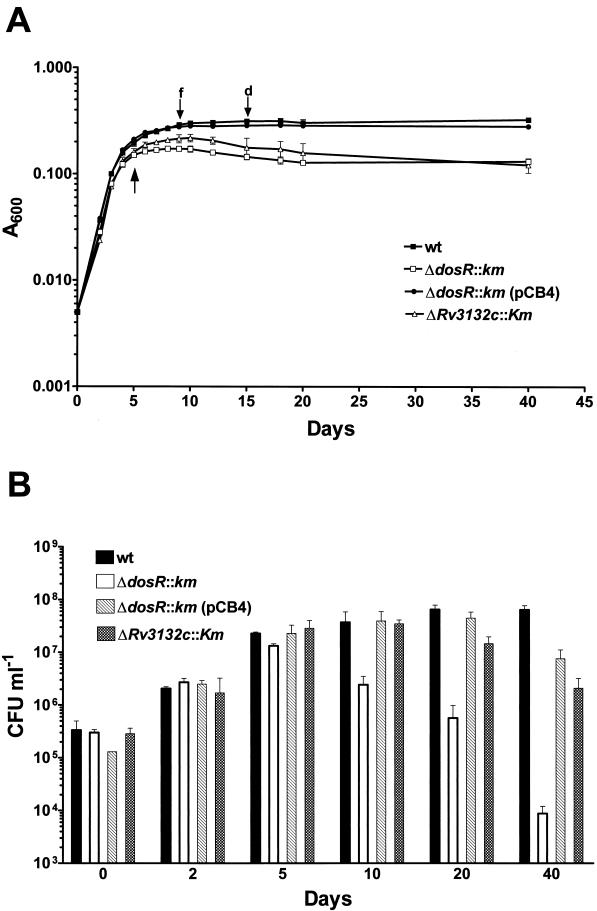
FIG. 2.
Growth and survival of wild-type BCG and ΔdosR::km, ΔdosR::km(pCB4), and ΔRv3132c::km strains in the Wayne dormancy culture system. Exponentially growing cultures were diluted and grown in sealed tubes with stirring. (A) Growth monitored by turbidity measurement. (B) Growth monitored by colony count determination. wt, wild type; ΔdosR::km, dosR gene replacement mutant; ΔdosR::km(pCB4), ΔdosR::km mutant transformed with the dosR rescue plasmid pCB4; ΔRv3132c::km, Rv3132c gene replacement mutant (see Fig. 1A). Growth and survival of ΔdosR::km transformed with pNBV1, i.e., the vector backbone used to carry the dosR genomic fragment in the pCB4 rescue construct (Fig. 1A), was indistinguishable from the behavior of the untransformed ΔdosR::km strain (data not shown). The experiments were carried out three times with duplicate cultures. Standard deviations are shown. The arrow pointing upwards indicates the time at which samples were taken for the protein analyses shown in Fig. 3. f and d, fading and complete decolorization of the oxygen indicator methylene blue, respectively.
In the initial aerobic exponential growth phase observed in this system, BCG ΔdosR::km was indistinguishable from the wild type (Fig. 2A). This shows that the loss of dosR did not affect the growth of the bacillus. BCG ΔdosR::km terminated aerobic exponential growth at the same time (i.e., after 4 to 5 days) and at the same A600 as the wild type. However, in contrast to the wild type, BCG ΔdosR::km underwent the transition from exponential growth to constant turbidity by day 6, i.e., the characteristic extended transition phase of the wild type has been lost in the ΔdosR::km strain. This result could indicate that dosR is involved in the transition (adaptation) from aerobic exponential growth to hypoxic nonreplicating survival.
To determine the effect of the dosR loss-of-function mutation on the actual viability of the hypoxic nongrowing culture, CFU were determined. Figure 2B shows that the CFU of BCG ΔdosR::km and the wild type were identical up to day 5 in the Wayne system, when both strains terminated aerobic exponential growth. Consistent with the slight increase in A600 over the next 5 days during the extended transition phase, a moderate increase in CFU was observed for the wild type at day 10. After that, the CFU of the wild type stayed constant for 40 days of total cultivation time, after which the experiments were terminated. In contrast to the survival of hypoxia shown by the wild type, Fig. 2B shows that the CFU of BCG ΔdosR::km started to decline at day 10. After 40 days of total incubation time, the viable-cell count of the BCG ΔdosR::km culture was 1,500-fold lower than that at day 5, when exponential growth terminated (Fig. 2B).
The growth and survival analyses were carried out with an independently isolated ΔdosR::km strain (Fig. 1B, lane 3), yielding identical results (data not shown). Figure 2 also shows that both phenotypes exhibited by BCG ΔdosR::km, loss of the extended transition phase and hypoxic death, were rescued by a wild-type copy of the dosR operon carried by plasmid pCB4 (Fig. 1A). Taken together, these results show that the dosR disruption affected the transition phase and caused massive death of the oxygen-starved culture. Hence, dosR is essential for the adaptation of BCG to survival of hypoxia.
Loss of dormancy proteins in BCG ΔdosR::km.
Previously we demonstrated that DosR is upregulated immediately upon oxygen starvation-induced termination of growth and that α-crystallin and the two conserved hypothetical proteins Rv2623 and Rv2626c are temporally coinduced with the response regulator (Fig. 3A) (1). To reveal a possible regulatory role of the dosR gene in the induction of these dormancy proteins, BCG ΔdosR::km was grown in the Wayne culture system, and a proteome analysis was carried out. Protein extracts were prepared from cultures that were terminating growth (Fig. 2A, arrow) and were subjected to two-dimensional gel electrophoresis. BCG ΔdosR::Km had lost the DosR protein, as expected (compare protein spot numbered 1 in Fig. 3A with empty circle numbered 1 in Fig. 3B).
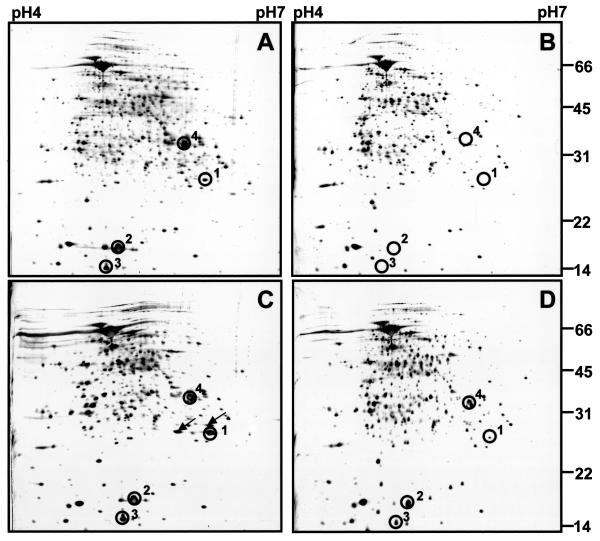
FIG. 3.
Two-dimensional gel electrophoretic analyses of protein extracts from wild-type BCG and ΔdosR::km, ΔdosR::km(pCB4), and ΔRv3132c::km strains grown in the Wayne dormancy culture system. Protein extracts were prepared at the time when cultures grown in the Wayne system terminated growth at day 5, as indicated by the arrow in Fig. 2A. Then 100-μg samples of total protein were subjected to two-dimensional gel electrophoresis. Silver-stained gels are shown. (A) Wild-type BCG. (B) BCG ΔdosR::km. (C) BCG ΔdosR::km(pCB4). Two protein spots that were detected in the pCB4-transformed ΔdosR::km strain upon termination of growth are marked by arrows; their identity remains to be determined. (D) BCG ΔRv3132c::km. Circles labeled 1 to 4 indicate dormancy-induced protein spots. In B, where the dormancy proteins were not detectable, the circles indicate their expected migration positions. 1, DosR; 2, α-crystallin; 3, conserved hypothetical protein Rv2626c; 4, conserved hypothetical protein Rv2623. Sizes are indicated in kilodaltons. The experiments were repeated twice with independently prepared cultures, yielding the same results. The experiments were also carried out for all four strains with protein extracts from 20-day-old Wayne cultures, yielding the same results that were obtained for 5-day-old cultures (data not shown).
Strikingly, Fig. 3B also shows the loss of induction of all three dormancy proteins that were coinduced with DosR in wild-type bacilli. The proteome analysis was carried out with the independently isolated ΔdosR::km strain (Fig. 1B, lane 3), yielding identical results (data not shown). Figure 3C shows that the regulatory phenotype shown by BCG ΔdosR::km could be rescued by a wild-type copy of the dosR operon carried by plasmid pCB4 (Fig. 1A). These results demonstrate that the induction of α-crystallin, Rv2623, and Rv2626c by dormancy depends on DosR and thus that DosR plays a regulatory role in the dormancy response.
Minor phenotypic consequences in BCG ΔRv3132c::km.
The apparent cotranscription of the histidine kinase Rv3132c (27) with the dosR response regulator (see Fig. 1A, pCB1) could indicate that the two proteins form a two-component system in which the kinase is involved in phosphorylation of DosR. Thus, Rv3132c could play the role of a dormancy sensor kinase, regulating DosR activity. To determine whether the kinase is indeed essential for the dormancy response, the gene was replaced with a kanamycin resistance cassette (Fig. 1A, pCB3; Fig. 1C), and the resulting ΔRv3132c::km strain was grown in the Wayne culture system.
Figure 2A shows that the ΔRv3132c::km strain appeared to maintain a wild-type-like extended transition phase, although the turbidity of the culture at day 10 was somewhat lower. Viability of the hypoxic, nongrowing BCG ΔRv3132c::km culture showed a moderate 15-fold reduction after 40 days of growth in the Wayne system, compared to the 1,500-fold hypoxic kill observed for the ΔdosR::km mutant under the same conditions. Consistent with the moderate effect of the loss of kinase function on growth and survival in the Wayne culture system, Fig. 3D shows that BCG ΔRv3132c::km maintained inducible expression of the dormancy proteins; although the induced level of the dormancy proteins appeared to be somewhat reduced compared to the induction of the proteins in the wild type. The growth, survival, and proteome analyses were carried out with an independently isolated ΔRv3132c::km strain (Fig. 1C, lane 3), yielding identical results (data not shown).
BCG ΔRv3132c::km thus showed a moderate phenotype compared to the loss of extended transition phase, loss of hypoxic viability, and loss of expression of dormancy proteins observed for BCG ΔdosR::Km when grown in the Wayne culture system. These results indicate that, in comparison to the key role observed for dosR, the Rv3132c kinase plays a minor role in the dormancy response. Furthermore, these results confirm that the phenotypic effects of the dosR inactivation were not due to polar effects on the downstream-located Rv3132c kinase gene.
Wild-type-like survival of BCG ΔdosR::km in aerated stationary phase.
Previously we demonstrated that upregulation of DosR is specific to oxygen starvation-induced termination of growth, i.e., to the dormancy response. Termination of growth by nutrient starvation under non-oxygen-limited conditions in aerated roller bottles is not accompanied by an increase in the steady-state level of DosR (1). Based on the lack of DosR induction in aerated stationary-phase cultures, one would predict that the protein does not play a role in the survival of aerated nongrowing culture. To test the dormancy specificity of dosR function, BCG ΔdosR::km was grown in aerated roller bottles, and growth and survival were determined. Both strains terminated exponential growth after 5 days of cultivation, at an A600 of 0.8. They showed an identical transition phase and the same viable-cell counts in the aerated stationary phase after a total cultivation time of 20 days (8 × 107 and 6 × 107 CFU ml−1 for the wild-type and BCG ΔdosR::km strains, respectively) and 40 days (3 × 106 and 2 × 106 CFU ml−1 for the wild-type and BCG ΔdosR::km strains, respectively). These results show that growth and survival of BCG ΔdosR::km under non-oxygen-limited conditions in roller bottles were indistinguishable from those of the wild type. Hence, the function of dosR in maintaining the viability of nongrowing cultures is specific for hypoxic dormancy, and dormancy-specific induction of DosR correlates with the dormancy specificity of its function.
DISCUSSION
Oxygen starvation triggers an adaptive dormancy response in the obligately aerobic tubercle bacillus. Survival of hypoxia and upregulation of the response regulator DosR (Rv3133c), α-crystallin, and the conserved hypothetical proteins Rv2623 and Rv2626c are hallmarks of the dormant organism (1). We show here that DosR is a key regulator of this response. Disruption of the dosR gene resulted in a more than 1,000-fold loss of viability in oxygen-starved nongrowing culture. This shows that dosR function is essential for adaptation to hypoxic survival. At the proteome level, we demonstrated that the genetic loss of dosR resulted in loss of induction of the other dormancy-induced proteins, indicating that expression of these proteins is under the control of DosR. Whether the dependence of expression of the dormancy proteins on DosR is direct (via activation of the promoters of the dormancy proteins) or indirect remains to be elucidated.
The deletion of the coding sequence of the histidine kinase Rv3132c located downstream of dosR had moderate consequences on hypoxic survival and induction of dormancy proteins. This suggests that the Rv3132c kinase plays only a minor role in the dormancy response and that other histidine kinases could be involved in dormancy signaling. The situation in the signal transduction pathway in Mycobacterium dormancy could thus be similar to sporulation signaling in Bacillus subtilis, in which multiple histidine kinases control (via a complex phosphorelay system) the phosphorylation status of the master regulator of sporulation, the response regulator Spo0A (13).
Our finding that α-crystallin induction depends on dosR but not on the Rv3132c histidine kinase is consistent with data from Sherman et al. (31). Those authors were unable to generate a dosR gene replacement mutant (as a “result of technical difficulties?” [31]). Hence, they interfered with the expression of dosR by interrupting the conserved hypothetical protein gene Rv3134c, which is located upstream of dosR (see Fig. 1A). By employing an anti-α-crystallin antibody, it was shown that this mutation resulted in loss of induction of α-crystallin under low-oxygen culture conditions. Complementation experiments suggested that this was due to reduced dosR expression and not to the inactivation of Rv3134c (31). In contrast, an Rv3132c histidine kinase-disrupted strain maintained inducible α-crystallin expression. Consistent with our data, the Rv3134c-disrupted strain did not show significant loss of viability in stationary-phase cultures grown in aerated roller bottles. Growth and survival of the Rv3134c and Rv3132c mutant strains in an oxygen-limited, Wayne-type culture system were not characterized by the authors (31).
The identification of DosR as an essential dormancy response regulator in our present study promises rapid progress in understanding the biochemistry underlying the quiescent hypoxic life of these obligate aerobes. The ΔdosR::km strain allows definition of the complete regulon (and thus the dormancy-essential enzymatic functions) under control of this transcription factor by comparative DNA microarray analyses. Even more important, perhaps, BCG ΔdosR::km represents the first Mycobacterium mutant that shows not only a strong but also an apparently specific survival phenotype in hypoxic dormant bacilli generated in the Wayne culture system. We demonstrated previously that DosR is not detectable in aerated stationary-phase culture (1), and we show here that, consistent with that observation, survival of the dosR loss-of-function mutant was indistinguishable from that of the wild type under these conditions. Thus, the ΔdosR::km mutant provides us with a probe for testing the relevance of hypoxic dormant bacilli for the life of the parasite in vivo.
If hypoxic dormant bacilli do play a role in the persistence of the bacilli during drug treatment, one would predict that chemotherapy of active disease and/or latency caused by the ΔdosR::km strain is more effective than the treatment of wild-type infection. DosR loss-of-function bacilli cannot weather the onslaught of conventional drugs by “hiding” in their dormant form, and hence, the time required to eradicate infection should be reduced. Furthermore, hypoxic dormancy of the bacillus might play a role in the progression of active disease and/or in the establishment and maintenance of latency of infection. If that is the case, the ΔdosR::km mutant is expected to show attenuated virulence. Animal models are in place to test these predictions (8). Thus, DosR and its dormancy regulon could provide a rich source of targets for a fundamentally new level of chemotherapeutic intervention against tuberculosis. Not hitting growth-related targets in proliferating bacilli but knocking out dormancy-related targets in sleepers may turn out to be the way to go in the development of future, more effective antimycobacterials.
Acknowledgments
We thank Alice Tay, Genome Analysis Laboratory, IMCB, for help with DNA sequencing and Indrajit Sinha and Michael Box for comments on the manuscript. We thank Bill Bishai for pNBV1 and M. F. Hynes for pJQ200SK.
T.D. is an adjunct staff member of the Department of Microbiology, National University of Singapore. This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore.
REFERENCES
- 1.Boon, C., R. Li, R. Qi, and T. Dick. 2001. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J. Bacteriol. 183:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 3.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterisation of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tubercle Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 4.DeMaio, J., Y. Zhang, C. Ko, D. B. Young, and W. R. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DesJardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the α-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick, T. 2001. Dormant tubercle bacilli: the key to more effective TB chemotherapy? J. Antimicrob. Chemother. 47:117-118. [DOI] [PubMed] [Google Scholar]
- 7.Florczyk, M. A., L. A. McCue, R. F. Stack, C. R. Hauer, and K. A. McDonough. 2001. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Global Alliance for TB Drug Development. 2001. Scientific blueprint for TB drug development. Tuberculosis 81(Suppl. 1):1-52. [DOI] [PubMed] [Google Scholar]
- 9.Höner zu Bentrup, K., and D. G. Russell. 2001. Mycobacterial persistence: adaptation to changing environment. Trends Microbiol. 9:597-605. [DOI] [PubMed] [Google Scholar]
- 10.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 11.Hutter, B., and T. Dick. 1999. Upregulation of narX, encoding a putative “fused nitrate reductase” in anaerobic dormant Mycobacterium bovis BCG. FEMS Microbiol. Lett. 178:63-69. [DOI] [PubMed] [Google Scholar]
- 12.Hutter, B., and T. Dick. 2000. Analysis of the dormancy-inducible narK2 promoter in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 188:141-146. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, M., W. Shao, M. Prego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 14.Jungblut, P. R., U. E. Schaible, H. J. Mollenkopf, U. Zimny-Arndt, B. Raupach, J. Mattow, P. Halada, S. Lamer, K. Hagens, and S. H. E. Kaufman. 1999. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol. Microbiol. 33:1103-1117. [DOI] [PubMed] [Google Scholar]
- 15.Kaprelyants, A. S., J. C. Gottschal, and D. B. Kell. 1993. Dormancy in non-sporulating bacteria. FEMS Microbiol. Rev. 104:271-286. [DOI] [PubMed] [Google Scholar]
- 16.Lim, A., M. Eleuterio, B. Hutter, B. Murugasu-Oei, and T. Dick. 1999. Oxygen depletion induced dormancy in Mycobacterium bovis BCG. J. Bacteriol. 181:2252-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manabe, Y. C., J. M. Chen, C. G. Ko, P. Chen, and W. R. Bishai. 1999. Conditional sigma factor expression, with the inducible acetamidase promoter, reveals that the Mycobacterium tuberculosis sigF gene modulates expression of the 16-kilodalton α-crystallin homologue. J. Bacteriol. 181:7629-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manabe, Y. C., and W. R. Bishai. 2000. Latent Mycobacterium tuberculosis—persistence, patience, and winning by waiting. Nat. Med. 6:1327-1329. [DOI] [PubMed] [Google Scholar]
- 19.Monahan, I. M., J. Betts, D. K. Banerjee, and P. D. Butcher. 2001. Differential expression of mycobacterial proteins following phagocytosis by macrophages. Microbiology 147:459-471. [DOI] [PubMed] [Google Scholar]
- 20.Murugasu-Oei, B., and T. Dick. 2000. Bactericidal activity of nitrofurans against growing and dormant Mycobacterium bovis BCG. J. Antimicrob. Chemother. 46:917-919. [DOI] [PubMed] [Google Scholar]
- 21.Parrish, N. M., J. D. Dick, and W. R. Bishai. 1998. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107-112. [DOI] [PubMed] [Google Scholar]
- 22.Peh, H. L., A. Toh, B. Murugasu-Oei, and T. Dick. 2001. In vitro activity of mitomycin C against growing and hypoxic dormant tubercle bacilli. Antimicrob. Agents Chemother. 45:2403-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicic, V., J. M. Reyrat, and B. Giquel. 1996. Generation of unmarked directed mutations in mycobacteria, with sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 24.Primm, T. P., S. J. Andersen, V. Mizrahi, D. Avarbock, H. Rubin, and C. E. Barry III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 26.Rosenkrands, I., R. A. Slayden, J. Crawford, C. Aagaard, C. E. Barry III, and P. Andersen. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol. 184:3485-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini, D. K., N. Pant, T. K. Das, and J. S. Tyagi. 2002. Cloning, overexpression, purification, and matrix-assisted refolding of DevS (Rv3132c) histidine protein kinase of Mycobacterium tuberculosis. Protein Expr. Purif. 25:203-208. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sander, P., K. G. Papavinasasundaram, T. Dick, E. Stavropoulos, K. Ellrott, B. Springer, M. J. Colston, and E. C. Böttger. 2001. Mycobacterium bovis BCG recA deletion mutant shows increased susceptibility to DNA-damaging agents but wild-type survival in a mouse infection model. Infect. Immun. 69:3562-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal, W. 1984. Growth dynamics of in vivo- and in vitro-grown mycobacterial pathogens, p. 547-573. In G. P. Kubica and L. G. Wayne (ed.), The mycobacteria: a source book. Marcel Dekker, New York, N.Y.
- 31.Sherman. D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wayne, L. G., and D. Salkin. 1956. The bacteriology of resected tuberculous pulmonary lesions. I. The effect of interval between reversal of infectiousness and subsequent surgery. Am. Rev. Respir. Dis. 74:376-387. [DOI] [PubMed] [Google Scholar]
- 33.Wayne, L. G. 1960. The bacteriology of resected tuberculous pulmonary lesions. II. Observations on bacilli which are stainable but which cannot be cultured. Am. Rev. Respir. Dis. 82:370-377. [DOI] [PubMed] [Google Scholar]
- 34.Wayne, L. G. 1976. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am. Rev. Respir. Dis. 114:807-811. [DOI] [PubMed] [Google Scholar]
- 35.Wayne, L. G. 1977. Synchronized replication of Mycobacterium tuberculosis. Infect. Immun. 17:528-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wayne, L. G., and H. A. Sramek. 1979. Antigenic differences between extracts of actively replicating and synchronized resting cells of Mycobacterium tuberculosis. Infect. Immun. 24:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wayne, L. G., and K. Y. Lin. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wayne, L. G. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur. J. Clin. Microbiol. Infect. Dis. 13:908-914. [DOI] [PubMed] [Google Scholar]
- 39.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne, L. G., and L. G. Hayes. 1998. Nitrate reduction as a marker for hypoxic shift down of Mycobacterium tuberculosis. Tubercle Lung Dis. 79:127-132. [DOI] [PubMed] [Google Scholar]
- 41.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]
- 42.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 43.Yuan, Y., D. D. Crane, and C. E. Barry III. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J. Bacteriol. 178:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry III. 1998. The 16-kDa α-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. USA 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]