Abstract
Nitrogen limitation induces the nitrogen-regulated (Ntr) response, which includes proteins that assimilate ammonia and scavenge nitrogen. Nitrogen limitation also induces catabolic pathways that degrade four metabolically related compounds: putrescine, arginine, ornithine, and γ-aminobutyrate (GABA). We analyzed the structure, function, and regulation of the gab operon, whose products degrade GABA, a proposed intermediate in putrescine catabolism. We showed that the gabDTPC gene cluster constitutes an operon based partially on coregulation of GabT and GabD activities and the polarity of an insertion in gabT on gabC. A ΔgabDT mutant grew normally on all of the nitrogen sources tested except GABA. The unexpected growth with putrescine resulted from specific induction of gab-independent enzymes. Nac was required for gab transcription in vivo and in vitro. Ntr induction did not require GABA, but various nitrogen sources did not induce enzyme activity equally. A gabC (formerly ygaE) mutant grew faster with GABA and had elevated levels of gab operon products, which suggests that GabC is a repressor. GabC is proposed to reduce nitrogen source-specific modulation of expression. Unlike a wild-type strain, a gabC mutant utilized GABA as a carbon source and such growth required σS. Previous studies showing σS-dependent gab expression in stationary phase involved gabC mutants, which suggests that such expression does not occur in wild-type strains. The seemingly narrow catabolic function of the gab operon is contrasted with the nonspecific (nitrogen source-independent) induction. We propose that the gab operon and the Ntr response itself contribute to putrescine and polyamine homeostasis.
Escherichia coli can use ammonia and a limited number of organic compounds as sole nitrogen sources (30). Ammonia supports the fastest growth rate, and a product of ammonia assimilation, glutamine, signals nitrogen sufficiency and prevents activation of the nitrogen-regulated (Ntr) response. Growth with organic nitrogen sources is slower, results in low glutamine (a signal of nitrogen limitation), and activates the Ntr response. Expression of Ntr genes often requires the alternate sigma factor σ54 and the response regulator nitrogen regulator I (NRI; also called NtrC) (30, 31). Sometimes Ntr gene expression also involves a σ70-dependent transcriptional activator, Nac, itself the product of an Ntr gene.
Microarray and computer analyses of E. coli genes have provided a catalog of Ntr genes and genes controlled by σ54 (29, 50). About half of the products of σ54-dependent genes are involved in nitrogen metabolism (29). Ntr proteins assimilate ammonia and transport nitrogen-containing compounds, such as amino acids and ammonia. Because of the number of transport systems induced by nitrogen limitation, nitrogen scavenging has been proposed as a major function of the Ntr response (50).
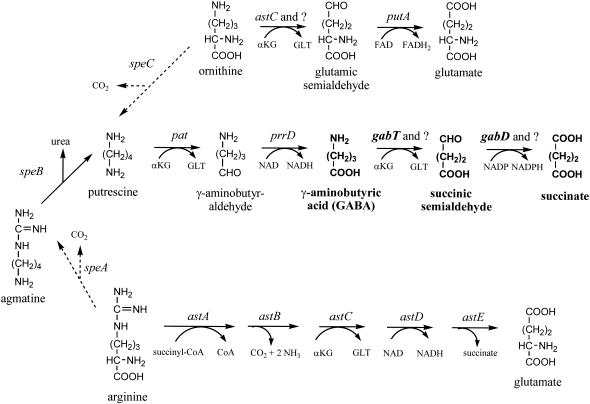
The proposed assimilatory and scavenging functions of the Ntr response do not account for the induction of a few catabolic pathways. It is remarkable that the majority of catabolic pathways induced by nitrogen limitation catabolize a group of metabolically related compounds: arginine, ornithine, putrescine, and γ-aminobutyrate (GABA) (Fig. 1). Arginine and ornithine are substrates for putrescine synthesis, while GABA is an intermediate in putrescine catabolism. This pattern of gene expression suggests that Ntr genes may contribute to putrescine and polyamine homeostasis during nitrogen-limited growth.
FIG. 1.
GABA, putrescine, agmatine, arginine, and ornithine metabolism. The solid arrows indicate catabolic reactions, while the dashed arrows are anabolic reactions. The genes that specify the enzymes involved are shown above the reaction arrows. The question mark indicates that more than one enzyme is involved and that the gene for the second enzyme has not been identified. Details of the arginine pathway are given elsewhere (38). Abbreviations: αKG, α-ketoglutarate; GLT, glutamate; FAD, flavin adenine dinucleotide.
To explore this possibility, we initially examined the gab operon and its relationship with GABA catabolism. GABA catabolism involves a two-step pathway that produces succinate (Fig. 1, middle line) (7, 8, 23). gabT codes for a GABA transaminase that generates succinic semialdehyde. gabD specifies an NADP-dependent succinic semialdehyde dehydrogenase, which oxidizes succinic semialdehyde to succinate (2, 7, 26). gabP encodes a GABA-specific permease (15, 26). Strains with missense mutations in these genes grow poorly with GABA as the nitrogen source (7, 41). In contrast to these mutations, a mutation in gabC stimulates GABA catabolism, which suggests that GabC is a repressor (8). The first three genes are clustered and coexpressed (50), which suggests that they form a gabDTP operon. Little is known about gabC, since previous reports on its location are not consistent (7, 23), and nothing is known about its expression.
GABA has been proposed to be an intermediate in the catabolism of arginine, agmatine, ornithine, and putrescine (Fig. 1) (41). However, recent results have shown that GABA is not an intermediate in arginine and ornithine catabolism (41). Instead, enzymes of the arginine succinyltransferase pathway (Fig. 1, bottom line), products of the astCADBE operon, are required for arginine catabolism (Fig. 1) (38). One product of the ast operon, AstC, has been implicated in ornithine catabolism, which suggests that glutamic semialdehyde, not GABA, is an intermediate in ornithine catabolism (Fig. 1) (38). Nonetheless, GABA catabolism could conceivably contribute to putrescine and agmatine metabolism.
In this paper, we describe an analysis of the structure, function, and regulation of the gab operon. We show that the gab operon has an exceptionally narrow catabolic function, considering the compounds that activate its expression and the number of pathways in which GABA might be an intermediate. We show that gabC is part of the gab operon and that its product does not participate in GABA-specific regulation. These properties are consistent with a broader physiological function than nitrogen source catabolism, such as modulation of putrescine and polyamine content during nitrogen-limited growth.
MATERIALS AND METHODS
Strains and plasmids.
The strains used in this study were derivatives of E. coli K-12 strain W3110 (Table 1).
TABLE 1.
Strains, plasmids, and phage used in this study
| Strain, phage, or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| AK3 | rpoS::tet in W3110 | Laboratory strain |
| BLS1 | ΔgabC (ygaE)::cat Δnac-28 | P1 lysate from SR5 (Kanr) into SR1 |
| BLS2 | ΔgabC (ygaE)::cat rpoS::tet | P1 lysate from SR1 (Camr) into AK3 |
| CP6 | ΔgabT | This work |
| EB3364 | Δnac-28::kan | 25 |
| HK1 | gabT::(mini-Tn5 lacZ-tet/1) in W3110 | P1 lysate from MT114 (Tetr) into W3110 |
| HK9 | Δnac::cat | This work |
| HK10 | Δnac::cat trp::(kan-gabD′-′lacZ) | P1 lysate from HK9 (Camr) into SR6 |
| K4633 | recD1903::Tn10 | D. Friedman (University of Michigan) |
| MT114 | gabT::(mini-Tn5 lacZ-tet/1) | 1 |
| SR1 | ΔgabC (ygaE)::cat in W3110 | This work |
| SR2 | ΔygaF::cat in W3110 | This work |
| SR4 | ΔgabDT::cat in W3110 | This work |
| SR5 | nac-28::Tn5 in W3110 | P1 lysate from EB3364 (Kanr) into W3110 |
| SR6 | trp::(kan-gabD′-′lacZ) in W3110 | This work |
| SR7 | trp::(kan-gabP′-′lacZ) in W3110 | This work |
| SR8 | trp::(kan-gabD′-′lacZ) ΔgabC (ygaE)::cat in W3110 | P1 lysate from SR1 (Camr) into SR6 |
| TE2680 | Δ(lac)X74 recD1903::Tn10 trpDC700::putPA1303::(cat-′lac) | 9 |
| W3110 | lacL8 lacIq | Laboratory strain |
| Kohara 443 | ygaF-gabDTP-ygaE (gabC) | F. Blattner (University of Wisconsin) |
| pACYC184 | cat | 3 |
| pGabp | Transcription template for gabD promoter | This work |
| pRS551 | kan-′lacZ | 42 |
| pWM529 | Cloning vector | 22 |
In strains SR1, SR2, and SR4, a gene coding for chloramphenicol resistance replaces part of a gene of interest. Their construction involved variations of the same procedure. For each, DNA was isolated from Kohara phage 443 (34), which F. Blattner (University of Wisconsin—Madison) generously supplied. The DNA of interest was cloned into the multicloning site of pWM529 (22), and part of the insert was replaced with a 1.2-kb chloramphenicol resistance cassette from pACYC184 (3). A linear fragment of the disrupted allele was electroporated into K4633 (recD1903::Tn10), and chloramphenicol resistance from the resulting strain was transduced into W3110. All constructs were confirmed by both Southern hybridization (35) and linkage to genetic markers in the vicinity (43). In the descriptions that follow, the numbers in parentheses after restriction enzymes refer to the coordinates of the sequence with GenBank accession number AE000351.
In SR1, the chloramphenicol cassette replaces 139 bp of ygaE (gabC), which deletes codons 122 to 168 of a 226-codon gene. The 3.2-kb BamHI (position 5788)-SphI (position 9025) fragment was isolated from Kohara phage 443 and cloned into pWM529. The resulting plasmid was digested with ClaI (position 7878) and StuI (position 7740), and the large fragment was isolated and ligated with the chloramphenicol resistance cassette.
SR2 has a 508-bp deletion of ygaF. The 4-kb Eco47III fragment (positions 319 to 4288) from phage 443 was cloned into the SmaI site of pWM529. The 508-bp region between the SalI (position 1746) and SnaBI (position 2252) sites was replaced with the 1.2-kb chloramphenicol resistance cassette. This deleted residues 38 to 206 of a putative 444-residue protein.
SR4 was constructed after ligation of the 4.8-kb PvuII (position 939)-BamHI (position 5788) fragment from phage 443 into pWM529. The DNA between two BstEII sites (positions 3619 to 5355) was replaced with the chloramphenicol resistance gene. This deletes codons 109 to 482 of the 482-codon gabD gene and codons 1 to 303 of the 426-codon gabT gene.
SR6 and SR7 contain an operon fusion to lacZ, which was transferred to the chromosome as previously described (9). This method involves ligation of promoter-containing DNA into a cloning site just upstream of a promoterless lacZ gene on pRS551 (42). Linearization of the plasmid, transformation into strain TE2680, and isolation of kanamycin-resistant cells selected for strains with the lacZ fusion integrated into the chromosome. Finally, the gene for kanamycin resistance, which is adjacent to lacZ, was transduced from the TE2680 derivative into W3110.
SR6 contains chromosomal lacZ under the control of the gabD promoter. The promoter fragment was isolated from a plasmid containing gab DNA between two Eco47III sites (positions 319 to 4288). This plasmid was digested with BsaAI (positions 2252 to 2933), and the resulting 681-bp fragment (extending from −640 to +41 with respect to the transcription start site) was ligated into the multicloning site of pWM529. This permitted removal of the promoter-containing DNA as an EcoRI-BamHI fragment, which was ligated into the same sites of pRS551, which are just upstream of a promoterless lacZ gene.
SR7 contains chromosomal lacZ fused to DNA upstream from gabP. The plasmid used to construct SR1 was digested with BamHI (position 5788) and ApaI (position 6104). This DNA, which extends from −187 to +131 relative to the 5′ end of the gabP structural gene, was ligated into the multicloning site of pWM529 and then ligated into pRS551 as described for SR6.
HK9 (nac::cat) was constructed by the method of Datsenko and Wanner (4). The P1 primer was TTAGCTCACCAATTGCCACTGCCTTTTTTCCATCACTGGAGAACGTGTAGGCTGGAGCTGCTTCG, and the P2 primer was CTCCTTTTATAGGGCAGGGGAACGCGACAGCTGATTAAAGGCATATGAATATCCTCCTTAG. This deleted all of nac from 144 bases upstream of the structural gene to the stop codon.
CP6 (ΔgabT) contains a nonpolar deletion that was constructed as previously described (4). The P1 primer was GTGGGACGTTGAAGGCCGTGAGTATCTTGATTTCGCGTGTAGGCTGGAGCTGCTTCG. The P2 primer was CGGTACAAGGATGCGCAGCACGTTGTAATACGGGCCCATATGAATATCCTCCTTAGTTCC. The antibiotic resistance cassette was removed, which left an in-frame deletion of residues 47 to 391 of a 426-residue protein.
Plasmid pGabp was used as a template for transcription in vitro. It was constructed by cloning a PCR product from chromosomal DNA with primers GCTCCGAATTCACCATCTACGCTCAGGACTGG (called gabD-L) and GGTCGAAGCTTGGCGTGTAAGGCATCCACAC (called gabD-R) into the EcoRI/HindIII site of plasmid pTE103. We verified the DNA insert by sequencing. The insert contained the gabD promoter region from −280 to +64 relative to the transcription start site.
Cell growth.
The minimal medium for growth was W salts (adjusted to pH 7.0) supplemented with a carbon source at 0.4% and a nitrogen source at 0.2% (33). Medium was supplemented with 100 μg of ampicillin per ml, 50 μg of kanamycin per ml, 25 μg of tetracycline per ml, or 10 μg of chloramphenicol per ml when appropriate.
Enzyme assays.
Cells were harvested late in the exponential growth phase, when the A600 of the culture reached either 0.6 or 100 Klett units (no. 42 filter). For the succinic semialdehyde dehydrogenase and GABA transaminase assays, cell pellets were frozen, resuspended in 0.1 M KPO4 buffer (pH 7.5)-1 mM dithiothreitol-1 mM phenylmethylsulfonyl fluoride-9% glycerol just before assay, and disrupted with three 5-s bursts of sonication. The crude extracts were ultracentrifuged for 90 min at 120,000 × g to remove NADH-oxidizing activity. NADP-dependent succinic semialdehyde dehydrogenase activity was assayed in 0.1 M KPO4 buffer (pH 7.8)-0.28 mM NADP at 30°C. The reaction was started with the addition of 0.6 mM succinic semialdehyde, and the A340 was monitored. The assay used to measure GABA transaminase activity was identical to that described previously (49), except that 100 mM HEPES buffer at (pH 7.0), 0.15 mM NADH, and 0.4 mM succinic semialdehyde were used. β-Galactosidase was assayed as previously described (24). Specific activity is reported as nanomoles of product formed per minute per milligram of protein.
In vitro transcription.
Single-round in vitro transcription reaction mixtures contained, in a final volume of 25 μl, transcription buffer (50 mM Tris · HCl [pH 7.5], 100 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA), 10 nM pGabp (template), 4 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.1 mM UTP, 1.25 μl of [α-32P]UTP (800 Ci/mmol, 10 mCi/ml; ICN), 150 μg of heparin, and 100 nM core RNA polymerase (Epicenter Technologies). When indicated, the reaction mixture also contained 300 nM σS, 100 nM Eσ70 (Epicenter Technologies), 200 nM Nac dimer, or 250 μM cyclic AMP (cAMP) and 100 nM cAMP receptor protein (CRP) dimer.
A mixture containing polymerase, cAMP, and 3 U of RNAGuard (Pharmacia) in transcription buffer was incubated for 5 min at 37°C. Nac and CRP were added together, and the mixture was incubated for another 10 min. All components were in transcription buffer, and the final volume after these additions was 20.5 μl. Transcription was initiated by addition of 4.5 μl of a solution containing GTP, CTP, labeled and unlabeled UTP, and heparin in transcription buffer and allowed to proceed for 15 min at 37°C, after which the reactions were stopped by addition of 25 μl of 50 mM EDTA-100 μg of yeast tRNA per ml and the mixtures were stored on ice. The reaction mixtures were extracted with 50 μl of acidic phenol (pH 4.2; Sigma), and 40 μl of the upper phase was added to 10 μl of sequencing loading dye. The tubes were heated to 90°C for 3 min and centrifuged briefly before being loaded onto a 5% acrylamide-8 M urea-0.5× Tris-borate-EDTA gel.
Primer extension.
Primers for primer extension and sequencing reactions were GGTCGAAGCTTGGCGTGTAAGGCATCCACAC for the gabD region and GCCCCATCCTGAATCCTCTCGAAA for the gabP region. Primer extension was based on previously described methods (17, 35), with the following modifications. For determination of the transcription start site in vitro, the transcription reaction was run without labeled UTP. The nucleic acids were precipitated, resuspended in water, mixed with labeled primer, dried, and resuspended in 30 μl of an annealing buffer consisting of 40 mM PIPES (pH 6.4), 1 mM EDTA, 400 mM NaCl, and 80% formamide. The mixture was heated for 2 min at 85°C, cooled slowly to 50°C, incubated for 2.5 h, and ethanol precipitated twice. The pellet was resuspended in 15 μl of a mixture containing the buffer supplied by the manufacturer for murine leukemia virus reverse transcriptase (NEB), 100 μg of bovine serum albumin per ml, 2 mM MgCl2, 1 mM each deoxynucleoside triphosphate, 7 U of RNAGuard, and 100 U of murine leukemia virus reverse transcriptase. The reaction was performed for 90 min at 40°C and stopped by addition of 15 μl of sequencing loading dye. The mixture was heated for 3 min at 95°C and loaded into a sequencing gel.
Other methods.
Routine recombinant DNA techniques were carried out as previously described (35). We carried out DNA sequencing by using chain-terminating dideoxy nucleotides with T7 DNA polymerase (Sequenase) on double-stranded templates (36). Sequencing reagents were from United States Biochemicals, and [γ-32P]ATP (7,000 Ci/mmol) was from ICN.
RESULTS
Phenotypes of gabT, gabDT, ygaE (gabC), and ygaF mutants.
Wild-type E. coli strains (W3110 or SR6) grew with a doubling time of 6 to 7 h with GABA as the sole nitrogen source (Table 2). CP6 (ΔgabT) and SR4 (ΔgabDT) could not utilize GABA as a nitrogen source. These strains grew normally with arginine, ornithine, putrescine, and agmatine, which suggests that the gab operon does not obviously contribute to their catabolism as nitrogen sources (Table 2). The growth with arginine and ornithine was expected (Fig. 1, top line) (38), but the growth with agmatine or putrescine was not, since GABA is a proposed intermediate in their catabolism (41).
TABLE 2.
Generation times for wild-type and mutant strains on various nitrogen sources
| N source | Avg generation time (min) ± SEa
|
||||||
|---|---|---|---|---|---|---|---|
| W3110 (wild type) | SR6 (wild type) | CP6 (ΔgabT) | SR4 (ΔgabDT) | SR1 (ΔgabC) | SR8 (ΔgabC) | SR5 (Δnac) | |
| Ammonia | NDb | 84 ± 11 | 83 ± 2 | ND | ND | 78 ± 6 | 122 ± 3 |
| Serine | ND | 184 ± 8 | ND | ND | ND | 171 ± 8 | 201 ± 3 |
| Alanine | ND | 115 ± 4 | ND | ND | ND | 116 ± 4 | 180 ± 10 |
| Proline | ND | 215 ± 11 | ND | ND | ND | 222 ± 14 | 235 ± 12 |
| Aspartate | ND | 227 ± 14 | ND | ND | ND | 217 ± 12 | 191 ± 11 |
| Glutamine | 103 ± 5 | 98 ± 1 | ND | ND | 103 ± 5 | 92 ± 4 | 137 ± 1 |
| GABA | 398 | 374 ± 12 | NGc | NG | 155 | 183 ± 24 | NG |
| GABA + aspartate | 134 | 164 ± 13 | 131 ± 5 | ND | 139 | 127 ± 17 | 162 ± 5 |
| Arginine | 440 ± 12 | 405 ± 19 | 513 ± 31 | 372 ± 1 | 444 ± 4 | 381 ± 7 | 252 ± 4 |
| Putrescine | 172 ± 2 | 169 ± 8 | 152 | 168 ± 1 | 184 ± 7 | 169 ± 9 | 149 ± 4 |
| Agmatine | 144 ± 6 | ND | ND | 144 ± 6 | 138 ± 12 | ND | 168 ± 10 |
| Ornithine | 228 ± 12 | ND | ND | 276 ± 1 | 240 ± 2 | ND | 318 ± 10 |
All values are for at least three separate cultures, except when no standard error is given, and these are for only one culture.
ND, not determined.
NG, no growth.
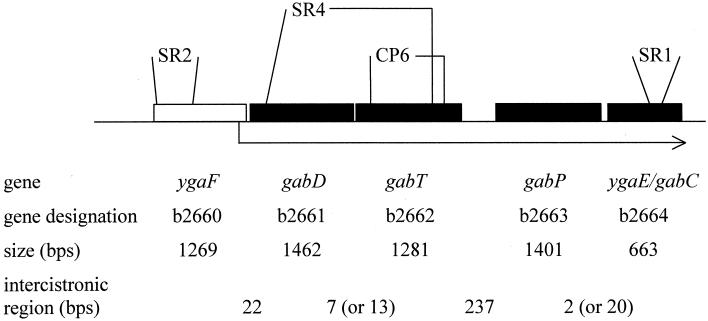
Nitrogen limitation induces gabD, gabT, and gabP expression coordinately (50). This, together with their proximity, suggests that they constitute an operon. The distance and potential products of the genes adjacent to gabD and gabP (Fig. 2) suggest that a putative gab operon might contain more than three genes—perhaps under certain conditions that were not assessed by the microarray analysis. Only 22 bases separate gabD from the preceding ygaF gene. BLAST analysis suggested that YgaF is a putative dehydrogenase, which could conceivably oxidize either γ-aminobutyraldehyde or succinic semialdehyde (Fig. 1). However, SR2 (ΔygaF) had the same doubling times (<15% difference) as W3110 (wild type) with GABA, agmatine, arginine, and ornithine as nitrogen sources (S. Ruback and L. Reitzer, unpublished data). Therefore, YgaF is not obviously involved in GABA catabolism.
FIG. 2.
The gab operon. The extents of the deletions and the insertion sites for SR1, SR2, SR4, and CP6 are indicated above the genes. The line below the genes indicates the proposed transcript. There is uncertainty concerning the start codons of three genes. The genes and coordinates (from the sequence with GenBank accession no. AE000351) of the first nucleotide of the start codons are as follows: ygaF, 1639 (GTG) or 1705 (ATG); gabT, 4452 or 4458 (both ATG); gabC/ygaE, 7379 or 7397 (both ATG). There are three repetitive extragenic palindromic elements between gabD and gabP.
BLAST analysis indicates that the gene downstream from gabP, ygaE, codes for a putative transcriptional repressor. It is the only potential repressor gene in the immediate vicinity. SR1 and SR8 (both ΔygaE mutants) grew twice as fast as W3110 and SR6 (isogenic wild-type strains) with GABA as the sole nitrogen source but otherwise grew normally with all of the other nitrogen sources tested (Table 2). The phenotype of these mutants, together with enzyme assays from these strains (results presented below), shows that ygaE codes for the gab repressor. Therefore, ygaE is the previously described gabC gene.
GabT transaminase and GabD dehydrogenase activities in nitrogen-limited wild-type and gabT and gabD mutant strains.
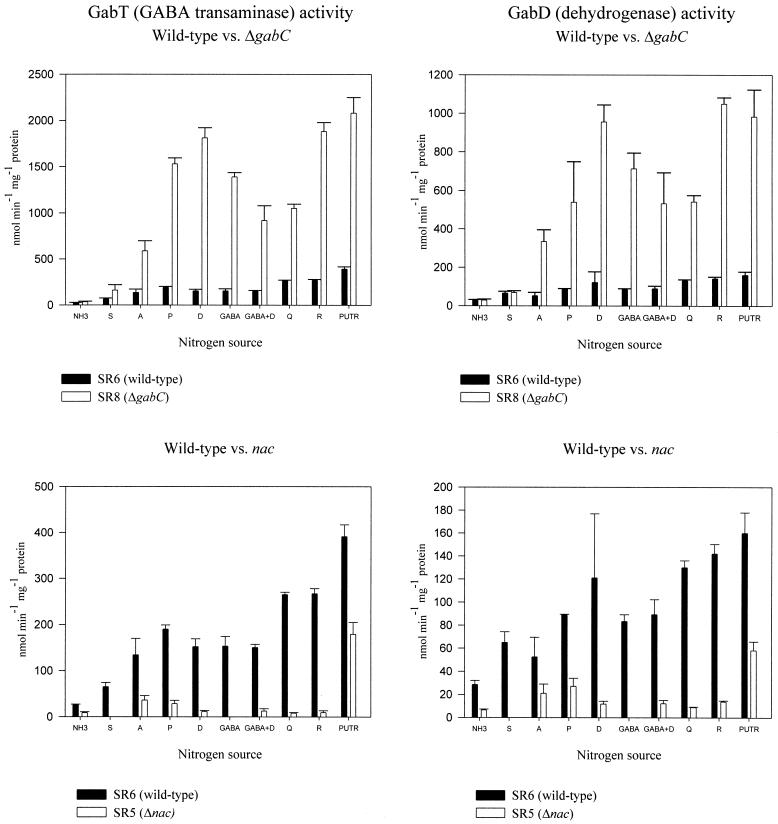
To examine gab operon expression, we assayed the gabT product, GABA transaminase (referred to as the transaminase), and the gabD product, NADP-dependent succinic semialdehyde dehydrogenase (referred to as the dehydrogenase), from extracts of exponentially growing cells. Transaminase activity was low (26 U) for cells grown in ammonia-containing nitrogen-rich media and higher in all nitrogen-limited media (65 to 267 U) (Table 3; Fig. 3, lower left panel). Dehydrogenase activity paralleled transaminase activity: it was low with ammonia (28 U) and higher for all cells grown in nitrogen-limited media (52 to 142 U) (Table 4; Fig. 3, lower right panel).
TABLE 3.
GabT (GABA-succinic semialdehyde transaminase) activity
| N sourcea | Avg activity (nmol min−1 mg of protein−1) ± SE (no. of determinations)
|
|||
|---|---|---|---|---|
| SR6 (wild type) | CP6 (ΔgabT) | HK1 (gabT::lacZ) | SR4 (ΔgabDT) | |
| Ammonia | 25.7 ± 1.4 (3) | 1.71 ± 0.84 (3) | 2.4 ± 0.6 (2) | NDb |
| Alanine | 134 ± 36 (5) | ND | 4.42 ± 0.24 (2) | 3.7 ± 1.3 (3) |
| Aspartate | 152 ± 17 (5) | 2.9 ± 1.5 (2) | ND | ND |
| GABA + aspartate | 150 ± 7.0 (4) | 25.4 ± 0.3 (3) | 3.3 ± 2.9 (3) | 7.88 ± 1.0 (4) |
| Glutamine | 265 ± 5.0 (3) | 3.72 ± 2.8 (3) | ND | ND |
| Arginine | 267 ± 11 (3) | 0.77 ± 0.39 (3) | ND | ND |
| Putrescine | 391 ± 26 (5) | 111 ± 8.2 (3) | 98.0 ± 2.9 (3) | 102 ± 2.1 (3) |
All nitrogen sources were at 0.2%.
ND, not determined.
FIG. 3.
GabT and GabD activities in wild-type, ΔgabC, and Δnac strains. Cells were grown with each of the indicated nitrogen sources at 0.2%. The errors bars are the standard errors of three to five determinations. Abbreviations: NH3, ammonia; S, serine; A, alanine; P, proline; D, aspartate; GABA+D, GABA plus aspartate; Q, glutamine; R, arginine; PUTR, putrescine.
TABLE 4.
GabD (succinic semialdehyde dehydrogenase) activitya
| N source | Avg activity (nmol min−1 mg of protein−1) ± SE (no. of determinations)
|
|||
|---|---|---|---|---|
| SR6 (wild type) | CP6 (ΔgabT) | HK1 (gabT::lacZ) | SR4 (ΔgabDT) | |
| Ammonia | 28.5 ± 3.5 (3) | 13.0 ± 4.2 (3) | 37.4 ± 11 (2) | ND |
| Alanine | 52.5 ± 17 (5) | ND | 396 ± 130 (2) | 2.1 ± 0.6 (3) |
| Aspartate | 121 ± 56 (5) | 58.8 ± 2.5 (2) | ND | ND |
| GABA + aspartate | 89.3 ± 13 (4) | 53.3 ± 6.6 (3) | 398 ± 51 (3) | 6.36 ± 1.1 (4) |
| Glutamine | 130 ± 6 (3) | 109 ± 8.6 (3) | ND | ND |
| Arginine | 142 ± 8.3 (4) | 97.5 ± 5.0 (3) | ND | ND |
| Putrescine | 160 ± 18 (5) | 139 ± 11 (3) | 704 ± 70 (3) | 65.0 ± 3.2 (3) |
See the footnotes to Table 3.
CP6 (ΔgabT), SR4 (ΔgabDT), and HK1 (gabT::lacZ) had low transaminase activity (2 to 8 U) after growth with ammonia, alanine, aspartate, arginine, and glutamine as nitrogen sources (Table 3). SR4 (ΔgabDT) had low dehydrogenase activity (2 to 6 U) with alanine or GABA plus aspartate as the nitrogen sources (Table 4). In contrast, all of the gabT mutants grown with putrescine had 100 U of transaminase activity (Table 3) and the ΔgabDT mutant had 65 U of dehydrogenase activity (Table 4), which are about 25 and 40% of the fully induced wild-type activities, respectively. These results suggest that putrescine, but not other nitrogen sources, induces a gab-independent transaminase and dehydrogenase. These gab-independent enzymes account for the normal growth of gab operon mutants with putrescine and agmatine as nitrogen sources. In summary, gabT and gabD specify essentially all of the transaminase and dehydrogenase activities, respectively, except when putrescine is the nitrogen source.
GabC-dependent regulation.
SR8 (ΔgabC) had higher levels of activities of both enzymes than those from SR6 (wild type), but only in nitrogen-limited cells (Fig. 3, top panels). The higher enzyme activities account for the faster growth with GABA as a nitrogen source and suggest that GabC is a gab operon-specific repressor.
Nac-dependent regulation.
Most Ntr genes have either a σ54-dependent promoter, which requires nitrogen regulator I (NRI; also called NtrC), or a σ70-dependent promoter, which requires Nac. nac is itself a σ54-dependent Ntr gene. The regulatory region of the gab operon does not have the distinctive sequence of σ54-dependent promoters, which is easily identified in all known σ54-dependent promoters (29). Instead, our results suggest Nac-dependent control.
SR5 (Δnac) could not grow with GABA as a nitrogen source, which suggests that gab operon expression requires Nac (Table 2). SR5 grew normally (<20% difference) with agmatine, aspartate, proline, or putrescine; at least 40% slower with ammonia, alanine, glutamine, or ornithine; and 43% faster with arginine (Table 2). The latter result had been previously observed (25). Transaminase and dehydrogenase activities in SR5 were dramatically lower than in the wild-type strain (Fig. 3, bottom panels). The only exception was for putrescine-grown cells, which have putrescine-inducible gab-independent isozymes.
Nitrogen source-specific regulation and the function of GabC.
Transaminase and dehydrogenase activities in the wild-type strain varied with the nitrogen source (most easily seen in the bottom panels of Fig. 3). Serine and alanine resulted in lower activities than did the other nitrogen sources. GABA induced enzyme activity as well as did several other nitrogen sources, including those that are not metabolized to GABA (e.g., aspartate, proline, and glutamine), which is not consistent with GABA-specific induction.
The absence of GABA-specific induction suggests that GABA does not bind GabC. To account for the lack of GABA-specific induction, we considered the possibility that GabC from E. coli has lost its responsiveness to GABA and that GabC from other organisms has retained GABA responsiveness. Therefore, we cloned the Klebsiella aerogenes gabC gene into SR8. If GABA inactivated the K. aerogenes GabC protein, then a strain with the K. aerogenes gabC gene would grow faster with GABA as a nitrogen source than a similar strain with the E. coli gabC gene. The K. aerogenes and E. coli gabC genes on a plasmid slowed the growth of SR8 with GABA as the nitrogen source to the same extent (B. Schneider and L. Reitzer, unpublished data). We conclude that GABA or a product of GABA catabolism does not inactivate K. aerogenes GabC. Therefore, GabC from at least two organisms fails to respond to GABA or compounds derived from GABA.
The range of the nitrogen source-specific regulation in nitrogen-limited media was much greater in SR8 (ΔgabC) than in SR6 (wild type) (Fig. 3). Transaminase activity from SR8 varied 12-fold (159 to 1,880 U), but that from SR6 varied only 4-fold (65 to 267 U). Dehydrogenase activity from SR8 varied 15-fold (68 to 1,050 U), but that from SR6 varied only 2.2-fold (65 to 142 U). (The values for putrescine-grown cells were excluded because of the complication of gab-independent isozymes.) These results suggest that the function of GabC may be to reduce nitrogen source-dependent variation.
A strain with a polar insertion in gabT is phenotypically GabC−.
GabD dehydrogenase was unexpectedly high in HK1, which contains an insertion in gabT (Table 4). Dehydrogenase activity in HK1 was four to eight times higher than that in SR6 (wild type) for cells grown with alanine, GABA (with aspartate), and putrescine and 70 to 100% of that from SR8 (ΔgabC). In other words, HK1 is phenotypically GabC−. In contrast, CP6, which contains an in-frame deletion of gabT, invariably had less dehydrogenase activity than the wild-type strain (Table 4). The simplest explanation for the difference in dehydrogenase activity between HK1 (gabT::lacZ) and CP6 (ΔgabT) is the polarity of the insertion in HK1. This result suggests that gabC is part of the gab operon.
Expression from a gabD-lacZ fusion.
We examined expression from a gab-lacZ fusion in order to confirm the results suggested by direct enzyme assays and to eliminate possible complications due to redundant enzymes. SR6 contains a chromosomal fusion of the gabD promoter (which was subsequently shown [see below] to contain DNA from −640 to +41 relative to the transcription start site) to lacZ. The gabD-lacZ fusion is inserted within the trp operon, which leaves the gab operon intact. Nitrogen limitation activated expression that varied with the nitrogen source (Table 5), which agrees with the results of direct assays (Fig. 3). β-Galactosidase activity was at least 10-fold lower in isogenic nac mutant cells grown in three different nitrogen-limited media (Table 5), which further confirms control by Nac.
TABLE 5.
β-Galactosidase activity from reporter strains with the gabD-lacZ operon fusiona
| N source | Avg activity (nmol min−1 mg of protein−1) ± SE (no. of determinations)
|
||
|---|---|---|---|
| SR6 (wild type) | SR8 (ΔgabC) | HK10 (Δnac) | |
| Ammonia | 63.6 ± 5.1 | 79.0 ± 15 | 83 ± 6 |
| Alanine | 236 ± 61 | 218 ± 33 | ND |
| Serine | 382 ± 86 | 322 ± 81 | ND |
| GABA | 1,530 ± 650 | 708 ± 72 | ND |
| GABA + aspartate | 1,280 ± 158 | ND | 120 ± 2 |
| Arginine | 2,070 ± 220 | 2,350 ± 420 | ND |
| Aspartate | 1,790 ± 71 | 1,650 ± 650 | 139 ± 25 |
| Glutamine | 1,660 ± 450 | 1,770 ± 410 | ND |
| Putrescine | 3,350 ± 140 | 3,220 ± 240 | 147 ± 11 |
See the footnotes to Table 3. All assays were performed at least three times.
Loss of GabC had no effect on β-galactosidase activity from cells grown with a variety of nitrogen sources and actually decreased β-galactosidase activity twofold when GABA was the nitrogen source (Table 5). This result is not consistent with the faster growth of gabC mutants (Table 2) or assays of transaminase and dehydrogenase activities (Fig. 3). One explanation for the discrepancy is that the promoter fragment fused to lacZ does not contain all of the GabC operator, even though the fusion contains DNA from −640 to +41 with respect to the gabD transcription start site. Attempts to clone a larger promoter fragment were unsuccessful. An alternate explanation is that the fusion's location in the trp operon affects the topology of the promoter and prevents GabC binding. In any case, the gabD-lacZ reporter recapitulated all aspects of regulation, except the control by GabC, which was lost.
GabT transaminase and GabD dehydrogenase activities in carbon-limited cells.
W3110 (wild type) could not utilize GABA as a carbon source, but SR1 (gabC) could (Table 6). Such growth is not consistent with Nac-dependent control. Therefore, we examined transaminase and dehydrogenase activities from carbon-limited cells. Carbon limitation induced both enzymes in W3110 about twofold with aspartate versus glucose as the carbon source (Table 6). In contrast, SR1 had low activities with glucose, three- to fourfold higher activities with glycerol, and 20- to 30-fold higher activities with GABA and aspartate as carbon sources. These activities are not consistent with GABA-specific induction. Instead, slower growth correlates with higher activities. BLS1 (gabC nac) consistently had about twice the activities of SR1 (gabC) with aspartate or GABA as the carbon source, which suggests that Nac impairs expression during carbon-limited growth.
TABLE 6.
Transaminase and dehydrogenase activities and rates of carbon-limited growth
| Strain and genotype | Carbon sourcea | Activity (nmol min−1 mg of protein−1) ± SE (no. of determinations)
|
Avg doubling time (min) ± SEb | |
|---|---|---|---|---|
| Transaminase | Dehydrogenase | |||
| SR6 (wild type) | Glucose | 25.7 ± 1.4 (3) | 28.5 ± 3.5 (3) | 84 ± 11 |
| SR8 (gabC) | Glucose | 34.1 ± 6.1 (3) | 29.2 ± 5.1 (3) | 78 ± 6.4 |
| W3110 (wild type) | Glycerol | 38.8 ± 11 (3) | 32.7 ± 4.3 (3) | 104 ± 2.7 |
| SR1 (gabC) | Glycerol | 110 ± 17 (3) | 80.6 ± 9.4 (3) | 100 ± 3.7 |
| BLS1 (gabC nac) | Glycerol | 121 ± 6.9 (3) | 71.9 ± 5.2 (3) | 116 ± 9.1 |
| BLS2 (gabC rpoS) | Glycerol | 13.9 ± 0.47 (3) | 10.5 ± 0.29 (3) | 130 ± 7.8 |
| W3110 (wild type) | GABA | NGc | NG | NG |
| SR1 (gabC) | GABA | 738 ± 50 (3) | 434 ± 69 (3) | 236 ± 2.7 |
| BLS1 (gabC nac) | GABA | 1,500 ± 51 (3) | 779 ± 47 (3) | 245 ± 14 |
| BLS2 (gabC rpoS) | GABA | NG | NG | NG |
| W3110 (wild type) | Aspartate | 47.2 ± 7.2 (3) | 36.3 ± 4.9 (4) | 256 ± 3.3 |
| SR1 (gabC) | Aspartate | 1,000 ± 30 (3) | 534 ± 43 (2) | 310 ± 14 |
| BLS1 (gabC nac) | Aspartate | 1,700 ± 67 (3) | 961 ± 61 (3) | 344 ± 3.2 |
| BLS2 (gabC rpoS) | Aspartate | 63.2 ± 6.7 (3) | 33.1 ± 1.4 (3) | 336 ± 13 |
Cells were grown with the indicated carbon source at 0.4% and 0.2% (NH4)2SO4.
Doubling times of at least three cultures are shown.
NG, no growth.
It has been reported that stationary-phase gab expression requires σS (1, 37). Our results show that gab expression during carbon-limited growth also requires σS. BLS2 (gabC rpoS) could not utilize GABA as a carbon source. Furthermore, BLS2 (gabC rpoS), compared to SR1 (gabC), had 8- and 30-fold less transaminase and dehydrogenase activities with glycerol and aspartate, respectively, as the carbon sources (Table 6).
gab operon transcripts and regulatory sites.
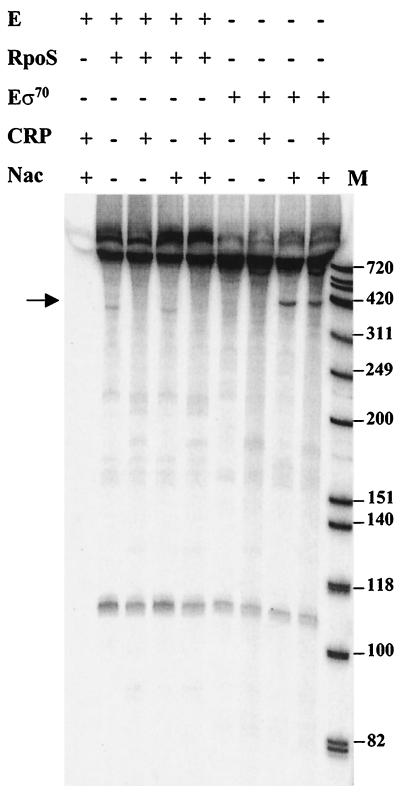
To confirm the results of the mutant analysis and enzyme assays, we analyzed transcription with purified core RNA polymerase (E), σ70, σS, CRP, and Nac. We used purified Nac from K. aerogenes, which is stable, unlike Nac from E. coli, which is unstable (11, 25). The template, plasmid pGabp, produced a transcript of about 400 bases, but only with Eσ70 and Nac (see the two lanes next to the markers in Fig. 4). The plasmid produces a discrete transcript because of a strong transcriptional terminator downstream from the promoter region. Eσ70 did not direct synthesis of this transcript by itself or with cAMP and CRP. cAMP and CRP had no effect on Nac-dependent transcription. EσS did not generate a similar transcript by itself or with either CRP or Nac. The 110- and 720-base transcripts serve as internal controls and can presumably be synthesized by either Eσ70 or EσS. The transcripts probably correspond to a regulatory RNA of plasmid replication and the bla transcript, respectively. The purified EσS complex was active because it synthesizes these transcripts and initiates transcription from the astCADBE regulatory region (17).
FIG. 4.
Requirements for transcription in vitro from the gabD promoter. The arrow indicates the location of the transcript that requires Eσ70 and Nac. M, molecular size markers (sizes are indicated in bases on the right).
Primer extension analysis of the in vitro transcript indicated a start site 101 bases from the structural gene (Fig. 5). (We had great difficulty detecting primer extension products from RNA extracted from cells, presumably because of extensive degradation.) Nac sites in K. aerogenes have a consensus of ATA(A/G)-N3-(A/T)NT(C/G)GTAT and are centered 64 or 44 bases from the transcription start site (11, 14, 27). There is a potential Nac-binding site that is centered 44 bases from the transcription start site preceding gabD (Fig. 6), which is consistent with the location of a known Nac-binding site.
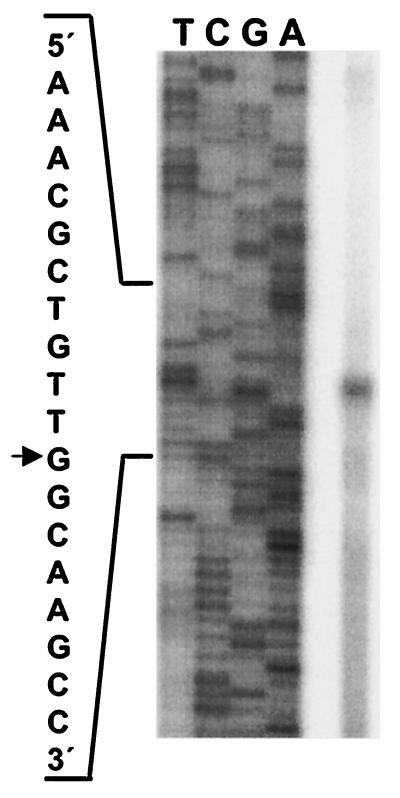
FIG. 5.
Primer extension from the in vitro transcription product. The arrow indicates the initiating nucleotide. The sequence of the entire region is shown in Fig. 6.
FIG. 6.
Nucleotide sequence of the gabD promoter region. The coordinates refer to the transcription start site. The RNA polymerase −10 and −35 regions are underlined. The arrows above the sequence indicate possible GabC-binding half-sites. The dashed arrows indicate one mismatch with the t-GTa consensus for GntR-like proteins. The solid arrows show a potential palindromic GabC site that has no mismatches in the downstream half-site and two mismatches in the upstream half-site. There are also potential half-sites with one mismatch centered at +45, +58, +96, and +108 that are not shown.
GabC is a member of the FadR subfamily of the GntR family of bacterial regulators, which are generally repressors (32). The extremes of the protected regions for characterized GntR-like proteins are −80 and +28 with respect to the transcription start site (6, 10, 18, 21, 28). A consensus operator site is an inverted repeat with t-GTa half-sites (capital letters indicate highly conserved bases) separated by one to three bases (32). Assuming that GabC binds a similar sequence, there are four potential half-sites with no or one mismatch from −80 to +28 (Fig. 6). A consensus half-site (no mismatches) overlaps the −10 RNA polymerase-binding region and has an adjacent half-site with two mismatches (Fig. 6).
The intercistronic gabT-gabP region contains 237 bases and three repetitive extragenic palindromic (REP) elements. Nonetheless, we considered the possibility that a promoter precedes the third gene of the operon, gabP. Primer extension indicated a transcript with a 5′ end that was 88 bases upstream of gabP, which was detectable only from SR1 (Ruback and Reitzer, unpublished). A binding site for σ70-containing RNA polymerase was not apparent from the region upstream of this transcript. We also constructed a fusion of this region to a promoterless lacZ gene on the chromosome, but expression was always less than twice the background and unregulated (Ruback and Reitzer, unpublished). Considering the polar effect of an insertion in gabT on gabC, we conclude that no promoter precedes gabP. The primer extension results probably indicate an RNA-processing site.
DISCUSSION
The gabDTPC operon.
Our results, combined with previously published results, suggest an operon structure for the gab genes and that four genes form the gabDTPC operon. The evidence that the first three genes are members of the gab operon is unambiguous. The GabT transaminase and GabD dehydrogenase activities change coordinately (Tables 3 and 4; Fig. 3 and 4). Furthermore, mutations in gabD have been previously shown to be polar and reduce synthesis of the GABA permease, i.e., GabP (23). Finally, microarray analysis indicates coordinate expression of gabD, gabT, and gabP (50).
The evidence that gabC is also a member of this operon is not as strong but reasonably convincing. The strongest evidence is that a strain with a polar insertion in gabT mimics the phenotype of a gabC mutant with respect to gabD expression. It is unusual for a repressor to be encoded within the operon that it regulates, so we considered alternative interpretations. If it is proposed that gabC is not part of the gab operon, then it becomes necessary to explain the differential effect of the insertion in gabT versus an in-frame deletion of gabT on GabD activity (Table 4). The only viable explanation that accounts for this difference is that polarity on gabP expression affects regulation. This could occur if GabP itself has a regulatory function in addition to its transport function or if impairment of transport affects accumulation of an inducer. The first possibility seems unlikely, although it is certainly not impossible. We can eliminate the second possibility, since gab operon expression does not require either GABA or a product of GABA catabolism. Therefore, the simplest explanation for the effect of the gabT lesion on GabD activity is polarity on gabC expression. The only uncertainty in this conclusion is the absence of evidence of a four-gene gab operon transcript. We tried to obtain evidence of this transcript but could not see discrete bands in a Northern blot. Even if we had observed gab operon transcripts, our expectation would be that the Northern blot would confirm the microarray analysis, which did not indicate gabC coregulation with other genes of the gab operon (50). This would be a reasonable expectation because transcripts of regulatory genes (e.g., gabC) are usually less abundant than those for structural genes (e.g., gabD and gabT). The apparent discoordinate level of gabC transcripts could result from less frequent transcription of the distal gabC gene or greater susceptibility to degradation. In any case, we consider the genetic evidence to be more reliable than the biochemical evidence and therefore conclude that gabC is the fourth gene of the gab operon.
In contrast, our results argue that ygaF, the gene preceding gabD, is not part of the gab operon. First, a deletion of ygaF had no effect on catabolism of GABA or any other nitrogen source, which implies that the function of YgaF is not related to the function of gab operon products. Second, transcription with purified proteins indicated that a promoter with the appropriate regulation and regulatory sites precedes gabD. Finally, microarray analysis did not provide evidence of coregulation of ygaF with the gab operon (50), which in this case is considered meaningful because the ygaF transcript would be expected to be as abundant as those for other enzymes, i.e., GabD and GabT.
Control of the gab operon.
Nac mediates the induction of the gab operon by nitrogen limitation. Nac also activates codBA (b0336-0337), a putative polyamine transport/catabolic operon (b1440-1444), yedL (b1932), nupC (b2393), and fklB-cycA (b4207-4208) and represses serA (b2913) and gltBDF (b3212-3214) (50). However, the phenotype of E. coli nac mutants tends to be subtle (25). The failure to utilize GABA as a nitrogen source is its most distinctive phenotype.
GabC does not obviously respond to a specific inducer. GabC is in the FadR subfamily of the GntR family of transcriptional regulators (32). At least one other member of the family, MatR, which regulates malonate metabolism in Rhizobium leguminosarum, also fails to respond to a specific inducer (21). Therefore, the absence of specific induction for these proteins has a precedent.
gab expression shows a nitrogen source modulation, and GabC reduces this modulation. This function could explain why gabC is a member of the gab operon since a modulatory function would become important only after the operon is expressed. Nitrogen source modulation of Ntr gene expression is also evident for activation of astCADBE (17, 38), a minimal glnAp2 promoter (39), and ygjG (C. Pybus and L. Reitzer, unpublished data) and repression of gltBDF (12). The mechanism of this modulation is not known and may vary from gene to gene. For example, ArgR mediates arginine-specific stimulation of astCADBE expression and this control may be unique among Ntr genes (17). Control of Nac specific activity is probably not involved in nitrogen source-specific modulation, since intracellular metabolites in K. aerogenes do not regulate its activity (40). However, the instability of E. coli Nac has prevented examination of the effects of metabolic intermediates on its activity (25).
In addition to expression during nitrogen-limited growth, we also observed gab expression during carbon-limited growth, which required both σS and inactivation of gabC. Several groups have observed gab expression in environments that are not nitrogen limited (1, 37, 44, 48). When examined, such expression requires σS and always involves strains with insertions in the gab operon (1, 37, 44, 48). We propose that the common factor is loss of GabC, since we could not detect significant σS-dependent expression in wild-type cells during either carbon-limited growth (Table 6) or stationary phase (B. Schneider, H. Kasbarian, and L. Reitzer, unpublished data). However, loss of GabC is not sufficient for expression since we could not reconstitute σS-dependent gab expression. cAMP, growth rate, cell density, osmolarity, and starvation have been shown to affect σS-dependent gene expression (20). cAMP and its receptor protein did not stimulate expression (Fig. 4). Instead, our results suggest that a growth rate factor, such as guanosine tetraphosphate, may be required, which has been shown to be the case for other σS-dependent genes (19).
The function of the gab operon and polyamine metabolism.
Our results show that the gab operon is required for GABA catabolism but does not contribute to the catabolism of any other nitrogen source. The exclusivity of nitrogen source catabolism contrasts with the nonspecificity of induction; i.e., all nitrogen sources induce. One explanation for this discrepancy is that nitrogen source catabolism is not the only function of the gab operon. Examination of the pattern of genes expressed during nitrogen limitation suggests a possible alternate function for the gab operon. Nitrogen limitation induces few catabolic enzymes, and surprisingly, most metabolize compounds that are related to putrescine. Nitrogen limitation induces b1440-1444 (putative role in both polyamine transport and catabolism), gabDTPC (GABA catabolism, which could remove products of putrescine catabolism), and ygjG (a putrescine-specific transaminase) (50; Pybus and Reitzer, unpublished). Nitrogen limitation also activates the astCADBE operon (17, 38), whose products metabolize both arginine and ornithine away from putrescine synthesis.
Putrescine is the major polyamine in E. coli (5, 45). Polyamines have been implicated in protein and nucleic acid elongation rates, translational fidelity, chromosomal structure, and other crucial functions (5). Putrescine content is higher in faster-growing cells, and loss of RpoS diminishes putrescine pools (47). The putrescine content of cells in high-ionic-strength media is also low (45). There is no agreement on the mechanisms that control the intracellular putrescine concentration, and it is likely that several mechanisms are involved. It has been argued that feedback inhibition controls putrescine synthesis (46), and the opposite has also been argued (5). It has been proposed that putrescine export controls intracellular putrescine (13, 16), but putrescine is not excreted in certain media (45). Putrescine catabolism has not been considered as a possible homeostatic control mechanism, but only because polyamine catabolism has not been extensively studied. The pattern of Ntr gene expression suggests that several catabolic Ntr enzymes act in concert to diminish putrescine levels during nitrogen-limited growth. If this is true, then enzymes of the Ntr response not only assimilate ammonia and scavenge nitrogen but also control polyamine homeostasis. Such a function would account for the seemingly nonspecific Ntr induction of the gabDTPC operon.
Acknowledgments
National Institute of General Medical Sciences grant GM47965 and National Science Foundation grants MCB-9723003 and MCB-0077904 supported this work.
We acknowledge R. Bender and D. Friedman (University of Michigan), F. Blattner (University of Wisconsin), and P. Rather (Case Western Reserve University) for supplying strains.
REFERENCES
- 1.Baca-DeLancey, R. R., M. M. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsch, K., A. von Johnn-Marteville, and A. Schulz. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J. Bacteriol. 172:7035-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, R. H., D. R. Morris, and P. Coffino. 1992. Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol. Rev. 56:280-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiRusso, C. C., T. L. Heimert, and A. K. Metzger. 1992. Characterization of FadR, a global transcriptional regulator of fatty acid metabolism in Escherichia coli. Interaction with the fadB promoter is prevented by long chain fatty acyl coenzyme A. J. Biol. Chem. 267:8685-8691. [PubMed] [Google Scholar]
- 7.Dover, S., and Y. S. Halpern. 1974. Genetic analysis of the γ-aminobutyrate utilization pathway in Escherichia coli K-12. J. Bacteriol. 117:494-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dover, S., and Y. S. Halpern. 1972. Utilization of γ-aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J. Bacteriol. 109:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, T. 1992. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J. Bacteriol. 174:245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, Y., and Y. Miwa. 1989. Identification of an operator sequence for the Bacillus subtilis gnt operon. J. Biol. Chem. 264:4201-4206. [PubMed] [Google Scholar]
- 11.Goss, T. J., and R. A. Bender. 1995. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J. Bacteriol. 177:3546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goss, T. J., A. Perez-Matos, and R. A. Bender. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183:6607-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbst, E. J., R. H. Weaver, and D. L. Keister. 1958. The Gram reaction and cell composition: diamines and polyamines. Arch. Biochem. Biophys. 75:171-177. [DOI] [PubMed] [Google Scholar]
- 14.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahane, S., R. Levitz, and Y. S. Halpern. 1978. Specificity and regulation of γ-aminobutyrate transport in Escherichia coli. J. Bacteriol. 135:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashiwagi, K., and K. Igarashi. 1988. Adjustment of polyamine contents in Escherichia coli. J. Bacteriol. 170:3131-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiupakis, A. K., and L. Reitzer. 2002. ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli. J. Bacteriol. 184:2940-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koo, J. H., I. H. Cho, and Y. S. Kim. 2000. The malonate decarboxylase operon of Acinetobacter calcoaceticus KCCM 40902 is regulated by malonate and the transcriptional repressor MdcY. J. Bacteriol. 182:6382-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of σS. J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 20.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σS subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 21.Lee, H. Y., J. H. An, and Y. S. Kim. 2000. Identification and characterization of a novel transcriptional regulator, MatR, for malonate metabolism in Rhizobium leguminosarum bv. trifolii. Eur. J. Biochem. 267:7224-7230. [DOI] [PubMed] [Google Scholar]
- 22.Mandecki, W., M. A. Hayden, M. A. Shallcross, and E. Stotland. 1990. A totally synthetic plasmid for general cloning, gene expression and mutagenesis in Escherichia coli. Gene 94:103-107. [DOI] [PubMed] [Google Scholar]
- 23.Metzer, E., R. Levitz, and Y. S. Halpern. 1979. Isolation and properties of Escherichia coli K-12 mutants impaired in the utilization of γ-aminobutyrate. J. Bacteriol. 137:1111-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Muse, W. B., and R. A. Bender. 1998. The nac (nitrogen assimilation control) gene from Escherichia coli. J. Bacteriol. 180:1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niegemann, E., A. Schulz, and K. Bartsch. 1993. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Arch. Microbiol. 160:454-460. [DOI] [PubMed] [Google Scholar]
- 27.Pomposiello, P. J., B. K. Janes, and R. A. Bender. 1998. Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein. J. Bacteriol. 180:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 29.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reitzer, L. J. 1996. Sources of nitrogen and their utilization, p. 380-390. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, second ed. ASM Press, Washington, D.C.
- 31.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, second ed. ASM Press, Washington, D.C.
- 32.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 33.Rothstein, D. M., G. Pahel, B. Tyler, and B. Magasanik. 1980. Regulation of expression from the glnA promoter of Escherichia coli in the absence of glutamine synthetase. Proc. Natl. Acad. Sci. USA 77:7372-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudd, K. E. 1998. Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol. Mol. Biol. Rev. 62:985-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schellhorn, H. E., J. P. Audia, L. I. Wei, and L. Chang. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 180:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider, B. L., A. K. Kiupakis, and L. J. Reitzer. 1998. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180:4278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider, B. L., S. P. Shiau, and L. J. Reitzer. 1991. Role of multiple environmental stimuli in control of transcription from a nitrogen-regulated promoter in Escherichia coli with weak or no activator-binding sites. J. Bacteriol. 173:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaibe, E., E. Metzer, and Y. S. Halpern. 1985. Metabolic pathway for the utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J. Bacteriol. 163:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 43.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tabor, H., and C. W. Tabor. 1969. Formation of 1,4-diaminobutane and of spermidine by an ornithine auxotroph of Escherichia coli grown on limiting ornithine or arginine. J. Biol. Chem. 244:2286-2292. [PubMed] [Google Scholar]
- 47.Tweeddale, H., L. Notley-McRobb, and T. Ferenci. 1998. Effect of slow growth on metabolism of Escherichia coli, as revealed by global metabolite pool (“metabolome”) analysis. J. Bacteriol. 180:5109-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zaboura, M., and Y. S. Halpern. 1978. Regulation of γ-aminobutyric acid degradation in Escherichia coli by nitrogen metabolism enzymes. J. Bacteriol. 133:447-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]