Abstract
P13 is a chromosomally encoded 13-kDa integral outer membrane protein of the Lyme disease agent, Borrelia burgdorferi. The aim of this study was to investigate the function of the P13 protein. Here, we inactivated the p13 gene by targeted mutagenesis and investigated the porin activities of outer membrane proteins by using lipid bilayer experiments. Channel-forming activity was lost in the p13 mutant compared to wild-type B. burgdorferi, indicating that P13 may function as a porin. We purified native P13 to homogeneity by fast performance liquid chromatography and demonstrated that pure P13 has channel-forming activity with a single-channel conductance in 1 M KCl of 3.5 nS, the same as the porin activity that was lost in the p13 mutant. Further characterization of the channel formed by P13 suggested that it is cation selective and voltage independent. In addition, no major physiological effects of the inactivated p13 gene could be detected under normal growth conditions. The inactivation of p13 is the first reported inactivation of a gene encoding an integral outer membrane protein in B. burgdorferi. Here, we describe both genetic and biophysical experiments indicating that P13 in B. burgdorferi is an outer membrane protein with porin activity.
Lyme disease is caused by infection with the spirochete Borrelia burgdorferi sensu lato and is transmitted by Ixodes ticks (18, 58). Like gram-negative bacteria, B. burgdorferi contains an inner membrane, an outer membrane, and a periplasmic space including the murein sacculus (4). However, a number of studies have revealed major differences between the B. burgdorferi cell envelope structure and membrane composition and those of gram-negative bacteria (7, 23, 24, 47, 63). The flagella are localized to the periplasmic space (4), and B. burgdorferi has been shown to lack lipopolysaccharide (59). Proteins associated with the Borrelia outer membrane are mostly lipoprotein (46, 54). Another important difference is that a large portion of the membrane lipids in B. burgdorferi consist of glycolipids, which comprises only galactose as a monosaccharide constituent (13, 32). These glycolipids are absent in gram-negative bacteria such as Escherichia coli (13). Only a few integral membrane proteins have been identified and characterized for B. burgdorferi (17, 43, 45, 56).
Proteins embedded in bacterial membranes fulfill a number of tasks that are crucial for bacterial cells, such as solute and protein transport, signal transduction, and interaction with other cells (1). Substances can be taken into bacteria by diffusion through the outer membrane via porin proteins (9) or via receptor-mediated uptake (41). Porins are located in the outer membrane, where they often form trimers of three identical subunits, allowing the diffusion of molecules across lipid bilayer membranes (8, 34). Their structure, location, and diversity confer on them multiple functions, including those of general diffusion pores and substrate-specific channels (1, 8). Porin loops are potential targets for adhesion to other cells (14), to bacteriophages (64), and to bactericidal compounds (52). In addition, porins can also activate complement (2, 40) and insert into eukaryotic host cellular membranes; an example of the latter is PorB of pathogenic Neisseria strains (50).
Variation of the loop structure as a mechanism to escape immune pressure (33) or modulation of porin expression in response to the pressure of antibiotics (48) are survival strategies used by some pathogenic bacteria. The presence of porins in other spirochetes has been investigated. Tromp1 of Treponema pallidum and OmpL1 from pathogenic Leptospira kirschneri are reported to have porin activity (15, 53). The major surface protein (Msp) of the oral spirochete Treponema denticola has been shown to form very large conductance ion channels and to form oligomers on the bacterial surface (27). This protein also binds to cell surface receptors and forms channels in the cytoplasmic membranes of epithelial cells (39). Borrelia spirochetes have an extremely limited metabolic capacity but a complex lifestyle, allowing them to persist in warm-blooded animals and arthropod hosts. Therefore, they need to efficiently use nutrients in these different environments. Hence, the few integral membrane proteins present in the B. burgdoferi outer membrane could act as channel-forming proteins. Two porins have been characterized for B. burgdorferi, Oms28 (56) and Oms66 (P66) (57). The Oms66 (P66) porin has been further shown to act as an adhesin and to bind to β3-integrin (22, 25). Porin activities have also been found in Borrelia hermsii (54).
In an earlier study, a 13-kDa integral outer membrane protein, P13, of B. burgdorferi sensu lato was characterized (43). P13 was first identified when monoclonal antibodies (MAbs) binding to a 13-kDa protein inhibited the growth of a B. burgdorferi strain lacking several of the major outer surface lipoproteins (51). P13 is processed at the amino (N) terminus, where the first 19 amino acids are cleaved off, most likely by signal peptidase I, and at the carboxyl (C) terminus by the removal of 28 amino acids (42, 43). Furthermore, it was recently shown that the N terminus of P13 was blocked for Edman degradation by pyroglutamylation (42).
In the present investigation, we studied the function of integral outer membane protein P13. The p13 gene was inactivated by allelic exchange; when the mutant was compared to an isogenic wild-type strain by use of a lipid bilayer assay, porin activity was found to be lost from the mutant. A fast performance liquid chromatography (FPLC) system (Pharmacia Biotech, Piscataway, N.J.) with a MonoQ column was used to purify P13 from B. burgdorferi, and the pure protein formed pores in a planar bilayer assay. Thus, we have shown by use of both genetic and biochemical analyses that the P13 protein of B. burgdorferi has channel-forming activity. The results presented in this study suggest that this protein forms an unusual porin structure compared to those of other known outer membrane porins. Furthermore, the structure and the substrate specificity of the P13 channel remain to be determined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study were as follows. B. burgdorferi B31 (ATCC 35210), the prototype strain of B. burgdorferi sensu stricto, was originally isolated from a tick collected on Shelter Island, N.Y. (18). Strain B31-A, a high-passage, noninfectious clone of B31, was used for gene inactivation experiments (16). B. burgdorferi N40 is an infectious strain isolated from an Ixodes tick (5, 6). Strain B313 was derived from B31 by cultivation in the presence of antibodies against the major outer surface proteins (51). Bacteria were grown in BSK-H (Sigma, St. Louis, Mo.) or BSK-II (3a) supplemented with 6% rabbit serum at 32 or 34°C. Single colonies were obtained as described elsewhere (49).
Construction of plasmid pYMP13.
A recombinant plasmid was constructed to inactivate the B. burgdorferi p13 gene. A 1.3-kb fragment containing 215 bp of the p13 gene and upstream DNA was amplified by PCR with an Expand high-fidelity PCR system (Roche, Mannheim, Germany) from the chromosomal DNA of strain B31 by using primers z1-PstI and z2-XbaI (Table 1 and Fig. 1). After restriction enzyme digestion of the PCR product, the fragment was cloned into vector pUC18 (New England Biolabs, Beverly, Mass.). A 1.4-kb fragment containing 265 bp of the p13 gene and downstream sequence was amplified by using primers z3-XbaI and z4-BamHI (Table 1 and Fig. 1) and ligated downstream of the first fragment in the same vector. The following reaction conditions were used: 94°C for 30 s, 45°C for 1 min, and 72°C for 1 min for 5 cycles and 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min for 35 cycles. The 1.3-kb PflaB-kanamycin resistance fragment from plasmid pJLB12a was used to inactivate the p13 gene (16). This fragment was amplified by using primers KNa1 and KNa1rev containing XbaI sites (Table 1 and Fig. 1) and 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min. After XbaI digestion, the kanamycin resistance fragment was ligated into the p13 gene. To prevent the introduction of an ampicillin resistance gene into B. burgdorferi, the p13-containing fragment was recloned into pOK12, a 2.1-kb low-copy-number plasmid containing a kanamycin resistance gene (62). An XhoI site was identified 921 bp upstream of the p13 gene, and the p13::kan fragment was excised from vector pUC18 by restriction with XhoI and BamHI and ligated into pOK12. The resulting plasmid was called pYMP13.
TABLE 1.
Oligonucleotides (5′-3′) used in this study
| Oligo- nucleotide | Sequencea | Location |
|---|---|---|
| z1-PstI | ATCCTGCAGTTTATACTATTAAAGATC | Upstream of p13 |
| z2-XbaI | CCTTCTAGAATATCTCCTTGAGCAAAG | p13 |
| z3-XbaI | TTTCTAGATCAAAGCGCTTGATGGTATTAC | p13 |
| z4-BamHI | ATGAAAGGATCCATATAAAATTACCTGG | Downstream of p13 |
| z12b | GGGAGAAAATGATGAATTTGTCG | p13 |
| z43b | TTTAAAGAAAAAGAGGGATATAC | p13 |
| KNa1 | TCTAGATGTCTGTCGCCTCTTGTGGCT | PflaB upstream of kan |
| KNa1rev | TCTAGAGGCGAATGAGCTAGCGCCGTC | Downstream of kan |
| KNaB | CCATGTTGGAATTTAATCGCGG | kan |
Restriction sites are in boldface type.
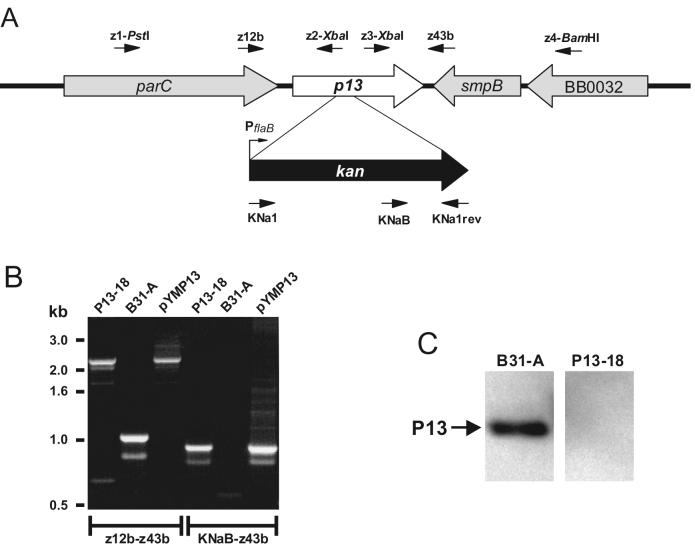
FIG. 1.
Insertional inactivation of P13. (A) Organization of the p13 gene and flanking genes on the B. burgdorferi B31 chromosome and insertion of the kan gene cassette. Arrows indicate relative positions of oligonucleotides used. The diagram is not drawn to scale. (B) PCR analysis of strain B31-A and kanamycin-resistant mutant P13-18 with primer pairs z12b-z43b and KNaB-z43b (Table 1). (C) Western blot analysis of P13-18 (mutant) and B31-A (wild type) with polyclonal antiserum against P13.
Allelic exchange mutagenesis and screening of B. burgdorferi transformants.
Preparation of competent B. burgdorferi, electroporation, and plating of transformants were done as described previously (60). Briefly, for each transformation, 20 μg of plasmid DNA in 5 μl of water was used. After electroporation, the bacteria were resuspended in 5 ml of BSK-H and incubated overnight at 34°C. The transformed bacteria were plated in solid BSK-II medium containing 200 μg of kanamycin per ml. Plates were incubated at 34°C with 1% CO2 until colonies were visible.
Kanamycin-resistant colonies were screened for allelic exchange at the p13 locus by PCR (Fig. 1). Individual colonies were picked with sterile toothpicks and added to tubes containing a PCR mixture, and oligonucleotides 1 and 9 (kan5′+NdeI and pOK.7+NheI) described by Bono et al. (16) were used to detect the kan gene. A number of potential transformant clones were chosen for further analysis by using primer z12b, primer z43b spanning the p13 gene, and primer pair KNaB-z43b amplifying a portion of the kan gene and a portion of the p13 gene (Fig. 1). PCR was carried out for 35 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 1 min. To obtain clonal mutants, individual colonies were aspirated with a sterile Pasteur pipette and grown in 5 ml of BSK-H with 200 μg of kanamycin per ml.
Determination of the plasmid profile.
Total DNAs from B31-A and a p13 knockout mutant were prepared by using a Wizard genomic DNA purification kit (Promega, Madison, Wis.), and plasmid contents were determined by PCR as previously described by Elias et al. (28).
Protein electrophoresis, immunoblotting, and antiserum.
For gel electrophoresis, proteins were heated to 85°C for 5 min in Novex 2× Tricine-sodium dodecyl sulfate (SDS) sample buffer (Invitrogen, Carlsbad, Calif.) or to 70°C for 10 min in Novex 4× NuPAGE sample buffer and separated through Tricine-10 to 20% polyacrylamide gradient gels or through 4 to 12% NuPAGE bis-Tris gels by using a Novex XCell Sure Lock electrophoresis cell (Invitrogen). Total B. burgdorferi proteins were prepared by growing cells to stationary phase, harvesting cells by centrifugation, and washing cells twice in phosphate-buffered saline. Proteins in peak FPLC fractions were concentrated by precipitation with a 2.5-fold excess of ice-cold ethanol. Precipitated proteins were pelleted by centrifugation at 13,000 × g for 40 min at 4°C and resuspended in sample buffer. Proteins were stained with Coomassie blue R-250 (Sigma). For immunoblotting, proteins were transferred to a polyvinylidene difluoride membrane (PALL Corporation, Ann Arbor, Mich.) and probed with antibodies. Polyclonal antibodies used against P13 were described before (43). MAb 15G6, which recognizes P13, was previously described by Sadziene et al. (51). Bound antibodies were detected by using peroxidase-conjugated anti-rabbit or anti-mouse antibodies (DAKO A/S, Glostrup, Denmark) and enhanced chemiluminescence reagents according to the manufacturer's instructions (Amersham Pharmacia Biotech, Buckinghamshire, England).
Separation of the outer membrane proteins of B. burgdorferi and purification of P13.
Outer membrane fractions of B. burgdorferi strains B313, B31-A, P13-18 (a clone with an inactivated p13 gene), and N40 were prepared as described elsewhere (37). P13 was purified by preparative SDS-polyacrylamide gel electrophoresis (PAGE) as previously described (43). For purification of native P13, the outer membrane proteins of strain B313 were applied to a MonoQ column in conjunction with an FPLC system. The column was washed with 15 ml of 0.4% lauryl-dimethyl-amine-oxide (LDAO; Sigma) buffered in 10 mM Tris HCl (pH 8.0), and 12 separate 0.5-ml flowthrough fractions were collected. Proteins bound to the column were eluted by using a 0 to 1 M linear NaCl gradient containing 0.4% LDAO buffered in 10 mM Tris HCl (pH 8.0).
Chemical cross-linking.
Proteins of intact high-passage B. burgdorferi strain B313 were cross-linked by a modification of a procedure described elsewhere (55). Approximately 1.3 × 108 freshly harvested and washed spirochetes were suspended in 350 ml of 0.1 M sodium phosphate buffer (pH 6.8). Formaldehyde (Riedel-de Haën, Seelze, Germany) was added to a final concentration of 1%, and the samples were incubated for 30 min at room temperature. The cells were washed once in cold 0.1 M sodium phosphate buffer (pH 6.8) and centrifuged, and the cell pellets were solubilized in 4× NuPAGE sample buffer and subjected to electrophoresis and then immunoblot analysis.
Planar lipid bilayer assay.
The methods used for black lipid bilayer experiments have been described elsewhere (10). The instrumentation consisted of a Teflon chamber with two compartments separated by a thin wall and connected by a small circular hole with an area of 0.2 mm2. The membranes were formed from a 1% (wt/vol) solution of diphytanoyl phosphatidylcholine (PC) (Avanti Polar Lipids, Alabaster, Ala.) in n-decane. The porin-containing protein solutions were highly diluted in 1% Genapol (Fluka, Buchs, Switzerland) and added to the aqueous phase either immediately before membrane formation or after the membrane had turned black. All salt solutions were used unbuffered and had a pH of about 6. The membrane current was measured with a pair of Ag-AgCl electrodes with salt bridges switched in series with a voltage source and a highly sensitive current amplifier (Keithley 427). The voltage dependence of the porin channels was checked as described elsewhere (36) by using membrane potentials as high as −150 to +150 V. Zero-current membrane potential measurements were performed by establishing a salt gradient across membranes containing 100 to 1,000 channels as described earlier (11). Zero-current membrane potentials were measured with a high-impedance electrometer (Keithley 617).
RESULTS
Construction of the p13 mutant.
In order to investigate the function of P13, we inactivated the p13 gene by targeted insertion of a kanamycin resistance gene cassette (16) (Fig. 1A). P13 maps at positions 32089 to 31553 on the linear chromosome of B. burgdorferi B31 and is transcribed as a monocistronic mRNA (29, 43). To confirm that the p13 gene was inactivated by allelic exchange, we amplified a chromosomal region spanning the kan gene and flanking sequences (Fig. 1A). We also used primers binding inside the kan gene and downstream of the kan gene insertion site (Fig. 1A). The PCR results were compatible with allelic exchange occurring only at the p13 locus (Fig. 1B). Using primers z12b and z43b, covering the kan gene and flanking sequences (Fig. 1A), we amplified a DNA fragment of the same size from both the p13 mutant and control plasmid pYMP13 but amplified a smaller fragment from B31-A (Fig. 1B). To further confirm that the kan gene was inserted into the chromosome of the mutant, a primer (KNaB) that binds within the kan gene cassette and another primer (z43b) downstream of the kan gene insertion site were used for amplification (Fig. 1A). A PCR product was obtained from the mutant but not from B31-A, as expected (Fig. 1B). The PCR results also showed that the kan gene was inserted in the same direction as the inactivated gene (Fig. 1A and B).
Clone P13-18 was grown to late exponential phase and analyzed for P13 expression. Western blot analysis with polyclonal rabbit serum against P13 provided additional evidence for the inactivation of the p13 gene (Fig. 1C). Total protein from B31-A was used as a positive control and, as expected, analysis of the total protein content of P13-18 revealed no P13 protein (Fig. 1C). Thus, we could successfully inactivate the gene for the integral outer membrane protein P13 in B. burgdorferi. Interestingly, no obvious defect of the mutant was found under normal growth conditions.
Porin activities in the outer membrane fraction.
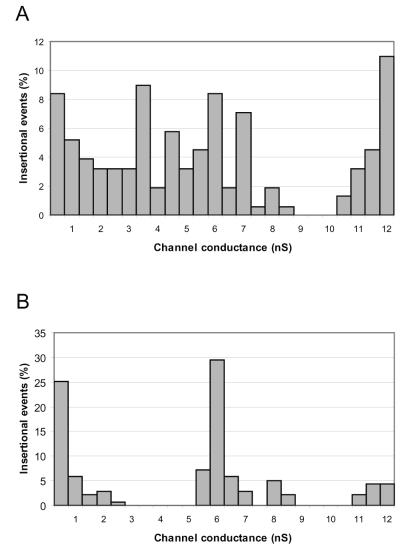
In a recent study, P13 was shown to be an integral membrane protein (43). To investigate whether P13 has porin activity, the channel-forming activities of outer membrane proteins from P13-18, B31-A, and infectious strain N40 were compared. Outer membrane proteins were enriched from B. burgdorferi strain B313 as previously described (37). Proteins were solubilized in 1% Genapol to a concentration of 0.05 μg/μl, diluted 100,000-fold, and investigated for channel-forming activities in planar lipid bilayer experiments. Strain P13-18, with an inactivated p13 gene, exhibited the same pore-forming activities as B31-A, with the exception of an activity of about 3.5 nS that was lost in the mutant (Fig. 2). The same numbers of insertional events were measured for both the p13 mutant and B31-A. However, the smaller number of events observed for the 12-nS pore in the total outer membrane extract of the p13 mutant than in that of B31-A may be due to relative differences in the amounts of certain proteins present in the outer membrane fractions. Another possible explanation for the reduction in the 12-nS channel-forming activity is that the inactivation of p13 lowers the level of expression of the protein responsible for the 12-nS activity or that the lack of P13 in the membrane fraction lowers the reconstitution of the other channel in the bilayer experiments. It is also noteworthy that the frequency of channel reconstitution in lipid bilayer membranes does not necessarily reflect the relative amounts of different porins expressed in the bacteria. Channel frequency in lipid bilayer measurements is also dependent on how efficiently a particular porin is reconstituted in the lipid bilayer; i.e., channel frequency is dependent on the porin’s channel-forming activity. We therefore have provided genetic evidence that 13-kDa protein P13 is a B. burgdorferi porin. In addition, strains B31-A and N40 showed similar single-channel conductance activities, ranging from 0.3 to 12 nS, with main activities of approximately 0.6, 2, 3.5, 6, and 12 nS (data not shown).
FIG. 2.
Porin activities of outer membrane proteins. Outer membrane proteins from B31-A (wild type) (A) and P13-18 (mutant) (B) in PC-n-decane membranes were analyzed. The total numbers of insertional events were 143 for B31-A and 139 for P13-18. The aqueous phase contained 1 M KCl. The temperature was 20°C, and the applied voltage was 20 mV.
Plasmid contents of B31-A and P13-18.
In vitro propagation of B. burgdorferi strains can result in the loss of plasmids; therefore, monitoring of plasmid content is essential for studying the phenotype of a derived mutant. To determine whether the p13 mutant and background strain B31-A had the same plasmid profiles, PCR was performed as described elsewhere (28). Both B31-A and P13-18 harbored linear plasmids lp17, lp28-2, lp28-3, lp38, lp54, and lp56 and circular plasmids cp26, cp32-1, cp32-3, cp32-4, cp32-7, cp32-8, and cp32-9 (data not shown). These results strongly indicated that the loss of channel-forming activity observed in the p13 null mutant was due to the inactivation of the p13 gene alone.
P13 purification and oligomer formation.
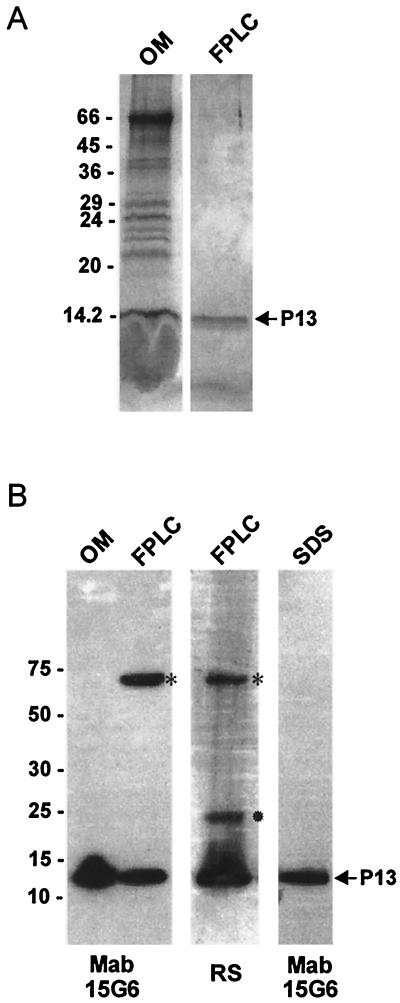
To purify the P13 protein, a membrane fraction from B. burgdorferi B313 corresponding to approximately 100 μg of total protein was solubilized in 2% LDAO, and proteins were subsequently purified by FPLC (Fig. 3). The P13 protein was eluted from the MonoQ column in the flowthrough fractions (Fig. 3) and was confirmed to be P13 by SDS-PAGE, staining with Coomassie blue R-250, and immunoblotting with MAb 15G6 (Fig. 4). P13 was present in flowthrough fractions 2 to 5. Pure P13, without detected contamination with other outer membrane proteins, was eluted from the column in fraction 4 (Fig. 4A). The amount of protein in fraction 4 containing P13 corresponded to approximately 10% (approximately 10 μg) of the total protein in the outer membrane fraction. Immunoblotting with MAb 15G6, demonstrating reactivity to FPLC-purified P13, revealed the formation of a P13 oligomer of about 60 kDa (Fig. 4B). Additionally, in FPLC-purified P13, a dimer form was detected by use of polyclonal antibodies (Fig. 4B). This result could be explained by masking of the epitope recognized by MAb 15G6 in the dimer form. Furthermore, the P13 oligomer was not detected in the outer membrane fraction, a result which could be explained by the presence during outer membrane preparation of the detergent n-octyl-β-d-glucopyranoside (OGP), which could prevent P13 oligomer formation. A dimer form of P13 was also detected in chemical cross-linking experiment with formaldehyde (data not shown). P13 purified by preparative SDS-PAGE does not form oligomers, as indicated by immunoblotting with the same MAb (Fig. 4B). The discrepancy in the SDS sensitivity of the P13 oligomer between P13 purified by preparative SDS-PAGE and P13 purified by FPLC and separated by SDS-PAGE could be caused by less influence of SDS on the latter form.
FIG. 3.
FPLC MonoQ purification of P13. Shown is a chromatogram of FPLC-purified outer membrane proteins derived from B. burgdorferi B313. Detergent-solubilized membrane proteins were loaded on a MonoQ column and eluted as described in Materials and Methods. The arrow indicates the fraction containing pure P13. The solid line showing an increase indicates the concentration of NaCl for a given fraction volume (indicated on the x axis).
FIG. 4.
Analysis of the P13 protein. (A) Analysis by SDS-PAGE and Coomassie blue R-250 staining of the membrane fraction (OM) of B. burgdorferi B313 and FPLC-purified P13 (FPLC). (B) Immunoblot analysis of the membrane fraction (OM) of B. burgdorferi B313, FPLC-purified P13 (FPLC), and preparative SDS-PAGE-purified P13 (SDS) probed with either MAb 15G6 or polyclonal rabbit serum (RS). ✹, possible dimer form; *, oligomer form. Molecular mass standards in kilodaltons are indicated at the left.
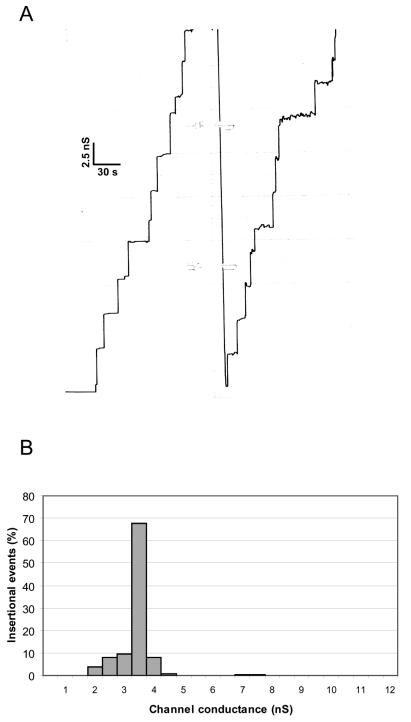
The addition of approximately 60 ng of pure P13 protein to lipid bilayer membranes composed of PC-n-decane showed that it had channel-forming activity, and the membrane conductance increased in a step-like fashion (Fig. 5A). Control experiments with the detergent LDAO alone revealed that these conductance steps were specific for the presence of P13. Most of the conductance steps were directed upward, and closing steps were only rarely observed at a transmembrane potential of 20 mV. These results indicated a long life span of the channels formed by P13. Figure 5B shows a histogram of 211 conductance steps in 1 M KCl at a membrane potential of 20 mV. The most frequent value for the single-channel conductance of P13 was 3.5 nS, but we observed some other conductance steps of 7.0 to 7.5 nS that could be explained by the formation of more than one channel simultaneously. It is noteworthy that the P13-containing fraction had a high specific activity, since a 100,000-fold dilution of the protein yielded 211 individual stepwise insertion events over a time period of approximately 45 min in the lipid bilayer assay (Fig. 5A). P13 was found to be sensitive to SDS and therefore became inactive after purification by preparative SDS-PAGE. However, it was possible to regain some channel-forming activity when the protein was incubated in 1% Genapol at 4°C (data not shown). Together with the results of a previous computer analysis of the P13 protein (43), the results presented here suggest that the P13 protein forms an unusual porin structure that does not fit into the architecture shown for other outer membrane porins.
FIG. 5.
Porin activity associated with FPLC-purified P13. (A) Single-channel conductance observed for P13 in PC-n-decane membranes. The fraction containing FPLC-purified P13 was added to a lipid bilayer composed of PC bathed in 1 M KCl at a concentration of about 10 ng/ml. (B) Histogram of the individual single-channel conductance events observed for the purified P13 porin. The average single-channel conductance for 211 single-channel events was 3.5 nS.
Single-channel analysis of P13.
P13 was found to be permeable to a variety of different ions. Table 2 shows the single-channel conductance of the P13 channel in the presence of different salt solutions. Changing the cations by replacing potassium with lithium resulted in an alteration that had a substantial influence (3.9-fold) on the conductance of the P13 channel. The influence of the anions (Cl− and CO3COO−) on the single-channel conductance, on the other hand, was less pronounced (1.8-fold) (Table 2); this result is consistent with the assumption that the P13 channel is cation selective. Alternatively, the single-channel conductance seen with different salts followed more or less the mobility sequence of the ions in the aqueous phase. This means that the P13 channel is wide and filled with water and has a small field strength on its interior face. Table 2 also shows the average single-channel conductance as a function of the KCl concentration in the aqueous phase. Despite the possible cation selectivity described above, we observed a 1:1 relationship between conductance and KCl concentration; this result is consistent with the absence of a binding site for ions within the channel and the absence of point charge effects.
TABLE 2.
Average single-channel conductance of the P13 channel in various salt solutionsa
| Salt | Salt concn (M) | Single-channel conductance (nS) | Hydrated cation radius (nm) |
|---|---|---|---|
| LiCl | 1.0 | 0.9 | 0.216 |
| KCl | 0.1 | 0.35 | 0.110 |
| 0.3 | 1.4 | ||
| 1.0 | 3.5 | ||
| 3.0 | 9.0 | ||
| KCO3COO− | 1.0 | 2.0 | 0.110 |
Membranes were formed from 1% PC dissolved in n-decane. The applied voltage was 20 mV, and the temperature was 20°C. The average single-channel conductance was calculated from at least 50 single events. The radii of the hydrated cations were calculated by using the Stokes equation (21) and the limiting molar conductivity of the various cations (61).
Purified P13 is cation selective and voltage independent.
For analysis of the ion selectivity of P13, we performed zero-current membrane potential experiments. After the incorporation of 100 to 1,000 porin channels into the PC membrane, the KCl concentration on one side of the membrane was raised from 0.1 to 0.5 M by the addition of 3 M KCl. The zero-current membrane potential was measured 3 min after the gradient was established. The more dilute side was always positive, indicating preferential movement of the cations through the channel; i.e., the channel was cation selective. The zero-current membrane potential was 14 mV under these conditions (from 0.1 to 0.5 M KCl). Analysis with the Hodgkin-Katz equation (11) suggested that anions could permeate the channel because the cation/anion permeability ratio was 2.1 for KCl. Together with the single-channel conductance data, this slight selectivity suggests that cations and anions can enter the channel and that the ion selectivity most likely is created by an excess of negatively charged residues within the channel, which can also create cation selectivity in a giant conductance porin channel (38). Additionally, the P13 channel exhibited no voltage dependence even at voltages as high as ±150 V (data not shown). This means that P13 did not show voltage-dependent closure.
DISCUSSION
In the present study, we provide the first evidence via gene inactivation for the presence of a possible porin in the outer membrane of B. burgdorferi. Porins form water-filled channels through the outer membrane and allow the diffusion of hydrophilic substrates in and out of cells (8, 9). Porins may also function as virulence determinants (3, 14, 31). It was shown in earlier studies that a 13-kDa integral membrane protein (P13) in B. burgdorferi is surface exposed, processed at both ends, and blocked for Edman degradation by a pyroglutamic acid modification (42, 43). In this study, we further investigated the function of the 13-kDa protein by inactivating the p13 gene by targeted mutagenesis (16, 60) and determined the channel-forming activities of the outer membrane proteins of the mutant (Fig. 1 and 2). Since the p13 knockout mutant, P13-18, did not display any porin activity of about 3.5 nS, we assumed that P13 is a channel-forming protein.
Due to the genetic instability of the B. burgdorferi genome during in vitro cultivation, we compared the plasmid contents of control strain B31-A and the p13 knockout mutant. As expected, both strains contained the same plasmids. Thus, the loss of porin activity in strain P13-18 is caused by insertional inactivation of the p13 gene and not by differences in plasmid content.
Purifying P13 from detergent-extracted membrane proteins by FPLC with a MonoQ column (Fig. 3 and 4) and testing porin activity in a planar bilayer assay showed that the purified P13 protein displayed a channel-forming activity of 3.5 nS (Fig. 5). This channel-forming activity was the same as that lost in the p13 knockout mutant. The only porins previously described for B. burgdorferi are Oms28 and Oms66 (P66), with single-channel conductance activities of 0.6 and 10.7 nS, respectively (56, 57). Our experiments showed that at least two or three additional channels may exist in the outer membrane fraction of B. burgdorferi, where P13 is responsible for the 3.5-nS activity (Fig. 2A and 5). The finding of additional porin activities that have not been discovered before could be explained by differences between the methods used to prepare outer membrane proteins or genetic differences in the strains investigated.
The p13 gene is a member of paralogous gene family 48, which include seven additional members situated on different linear plasmids (20, 29). Two members of this family are regarded as pseudogenes, and one gene contains a frameshift. Sequence similarities between the deduced amino acid sequences for P13 and four other family 48 members (BBA01, BBH41, BBI31, and BBQ06) suggest that they also may function as porins (20, 29). The channel-forming activity of P13 may be different from the possible channel-forming activities of the other paralogous proteins. It is therefore possible that some of the additional porin activities seen in the membrane fraction of B31-A are caused by some of the family 48 members. However, one of the paralogous genes (bbi31) is situated on linear plasmid lp28-4, which is missing in B31-A; hence, the BBI31 protein cannot be responsible for any channel-forming activity in the experiments. Furthermore, the other members may not be expressed at all in cultured B. burgdorferi or may not be present in the membrane protein fraction used in the lipid bilayer experiments. Further investigations are needed to determine the role of the other members of family 48 in channel formation in B. burgdorferi membranes.
Our attempts to show porin activity of P13 purified by preparative SDS-PAGE failed because of the sensitivity of P13 to SDS. Although we could detect reconstitution of porin into the lipid bilayer and regain some porin activity after incubation of SDS-PAGE-purified P13 in 1% Genapol at 4°C, we were not able to obtain clear single-channel activity (data not shown). Most enterobacterial porins are resistant to SDS; however, it was shown earlier that some porins can become inactive after treatment with SDS, such as Tsx of E. coli (12). It is noteworthy that after treatment of FPLC-purified P13 with SDS, a defective channel-forming activity pattern similar to that seen with P13 obtained from preparative SDS-PAGE and incubated in Genapol was observed (data not shown).
Secondary structure analysis indicates that the P13 monomer does not contain a sufficient number of β-barrel structures to form a monomeric channel. Therefore, it is likely that the protein functions as an oligomer. Cross-linking experiments with formaldehyde revealed that P13 could form dimers (data not shown). Oligomer formation was also observed in an immunoblot with FPLC-purified P13 and monoclonal antibodies directed against P13 (Fig. 4B). This presumably means that P13 does not fit the known porin structure, which forms trimers of three identical polypeptides containing three channels. The method used could not reveal the exact size of the P13 oligomer but suggested that the protein complex is probably formed by four or five monomers of P13. Another possibility is that P13 can form a heterodimer mixture together with another protein(s). This possibility has to be further investigated. In addition, we observed that the P13 oligomer was not detected by immunoblotting of outer membrane proteins with MAb 15G6. This finding could be explained by the presence during outer membrane preparation of the detergent OGP, which could prevent P13 oligomer formation. However, P13 channel-forming activity was detected in the outer membrane fraction, presumably because of drastic dilution of OGP and treatment of the proteins with 1% Genapol, which aids in the formation of the native porin structure during lipid bilayer experiments.
The P13 protein showed no voltage dependence and was cation selective, with a cation/anion permeability ratio of 2.1. This cation selectivity is not based on point charges at the channel mouth, since the single-channel conductance was a linear function of the cation concentration, as Table 2 clearly indicates. Although it was cation selective, the P13 channel also allowed anion diffusion because of some influence of the replacement of chloride by acetate on the single-channel conductance (Table 2). These results suggest that the P13 channel is wide and filled with water. A comparison with the single-channel conductance of E. coli porins suggests that it has at least the same permeability and the same diameter as the E. coli porins (9). However, further studies are required to elucidate the substrate for the 3.5-nS channel of B. burgdorferi. It is known that in addition to general diffusion pores that discriminate between solutes primarily on the basis of size and charge, the bacterial outer membrane may also contain channels with specificity for certain substrates, such as phosphate or carbohydrates (8, 9).
The specific role of P13 porin activity in B. burgdorferi metabolism or pathogenesis remains to be determined. However, we demonstrated that inactivation of the p13 gene is not lethal for B. burgdorferi during in vitro cultivation, indicating that this gene is most likely required for the survival of B. burgdorferi inside the tick or mammalian host. Interestingly, inactivation did not significantly affect the growth of the spirochetes in liquid medium. Since inactivation was done with the high-passage, noninfectious strain B31-A, the relevance for the pathogenesis of the P13 protein cannot be tested in an animal model. However, lipid bilayer experiments with detergent-solubilized outer membrane proteins from avirulent clone B31-A and virulent strain N40 revealed that these strains contain similar porin activities, including P13 porin activity (data not shown). These results indicate that genes coding for detected porins are present on the chromosome or on plasmids that are stably maintained during in vitro cultivation. This is the case for oms66 (p66) and p13, which are present on the chromosome, and oms28, which is present on the 54-kb linear plasmid.
Structural analysis of the subunits of conventional E. coli outer membrane proteins, such as OmpA, OmpF, and LamB, reveal that these proteins contain between 8 and 18 β sheets that form a β-barrel structure spanning the outer membrane (26, 30, 44) and providing only for OmpF and LamB enough space for a transmembrane channel (8). Thus, the results of this study demonstrating channel formation by B. burgdorferi P13 suggest that this protein forms an unusual structure different from those of other known outer membrane porins because of its low molecular mass and unusual C-terminal processing. Gram-negative bacterial porins form trimers in the outer membrane with a minimum molecular mass of 30 kDa for the monomer (8, 34). A computer analysis of the P13 amino acid sequence shows that this protein does not contain pronounced or sufficient amounts of β sheets to form channels following conventional pore formation mechanisms; instead, three hydrophobic α helices are predicted to span the membrane (43). Additionally, crystallographic studies of TolC in E. coli have revealed that this channel-forming protein also lacks the common structural elements of channel-forming proteins. In the TolC protein, three subunits each contribute to four β strands that form a single 12-strand β barrel, and 12 α helices pack in an antiparallel fashion to form a hollow cylinder that spans the space between the outer and the inner membranes (19, 35). These findings show that unconventional structures for channel formation may exist; however, further structural analyses are needed to elucidate the novel channel structure formation of P13.
Borrelia spirochetes have an unusual outer membrane structure and have been shown to have a limited metabolic capacity, meaning that they need to efficiently take up nutrients from the environment. Presumably, a large number of transporters are needed to move essential compounds from extracellular sources across the inner membrane. However, it is still unknown how these compounds are transported to the periplasm. We suggest that porins, which form channels in the outer membrane, are very important for the survival strategy of B. burgdorferi, as this obligate parasite can inhabit both ticks and mammals. Thus, we believe that to obtain more complete knowledge of the biology of Borrelia spirochetes, it is important to understand and investigate the physical properties of outer membrane proteins, such as porins.
Acknowledgments
This study was supported by Swedish Medical Research Council grants 07922 and 13564, Swedish Council for Forestry and Agricultural Research grant 23.0161, Symbicom AB, the Medical Science Council, the Royal Academy of Science (Olof Ahlöfs Foundation), a Federation of European Microbiology Societies fellowship, the J. C. Kempe Foundation, and the Deutsche Forschungsgemeinschaft (Be 865/10).
We thank Abdallah Elias, Dorothee Grimm, Philip Stewart, and Kit Tilly for help during mutant construction; Elke Maier for help with the lipid bilayer experiments; and Alan G. Barbour for providing MAb 15G6 and B. burgdorferi N40.
REFERENCES
- 1.Achouak, W., T. Heulin, and J. M. Pages. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Alberti, S., G. Marques, S. Hernandez-Alles, X. Rubires, J. M. Tomas, F. Vivanco, and V. J. Benedi. 1996. Interaction between complement subcomponent C1q and the Klebsiella pneumoniae porin OmpK36. Infect. Immun. 64:4719-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., W. M. Johnson, and F. G. Rodgers. 1999. Identification and characterisation of a cytotoxic porin-lipopolysaccharide complex from Campylobacter jejuni. J. Med. Microbiol. 48:139-148. [DOI] [PubMed] [Google Scholar]
- 3a.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50:381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barthold, S. W., D. S. Beck, G. M. Hansen, G. A. Terwilliger, and K. D. Moody. 1990. Lyme borreliosis in selected strains and ages of laboratory mice. J. Infect. Dis. 162:133-138. [DOI] [PubMed] [Google Scholar]
- 6.Barthold, S. W., K. D. Moody, G. A. Terwilliger, P. H. Duray, R. O. Jacoby, and A. C. Steere. 1988. Experimental Lyme arthritis in rats infected with Borrelia burgdorferi. J. Infect. Dis. 157:842-846. [DOI] [PubMed] [Google Scholar]
- 7.Belisle, J. T., M. E. Brandt, J. D. Radolf, and M. V. Norgard. 1994. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J. Bacteriol. 176:2151-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benz, R. 2001. Porins—structure and function, p. 227-246. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 9.Benz, R. 1994. Solute uptake through bacterial outer membranes, p. 397-423. In R. Hackenbek and J.-M. Ghuysen (ed.), Bacterial cell wall. Elsevier, Amsterdam, The Netherlands.
- 10.Benz, R., K. Janko, W. Boos, and P. Lauger. 1978. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta 511:305-319. [DOI] [PubMed] [Google Scholar]
- 11.Benz, R., K. Janko, and P. Lauger. 1979. Ionic selectivity of pores formed by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta 551:238-247. [DOI] [PubMed] [Google Scholar]
- 12.Benz, R., A. Schmid, C. Maier, and E. Bremer. 1988. Characterization of the nucleosid-specific binding site inside the Tsx channel of Escherichia coli outer membrane: reconstitution experiments with lipid bilayer membranes. Eur. J. Biochem. 176:699-705. [DOI] [PubMed] [Google Scholar]
- 13.Berg, S., Y. Östberg, S. Bergström, and Å. Wieslander. 2001. Functional analysis of a lipid galactosyltransferase from Lyme disease Borrelia spirochetes, p. 161-186. In The glucolipid pathway in Acholeplasma laidlawii. VMC-KBC, Umeå, Sweden.
- 14.Bernardini, M. L., M. G. Sanna, A. Fontaine, and P. J. Sansonetti. 1993. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect. Immun. 61:3625-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco, D. R., C. I. Champion, M. M. Exner, H. Erdjument-Bromage, R. E. Hancock, P. Tempst, J. N. Miller, and M. A. Lovett. 1995. Porin activity and sequence analysis of a 31-kilodalton Treponema pallidum subsp. pallidum rare outer membrane protein (Tromp1). J. Bacteriol. 177:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunikis, J., L. Noppa, and S. Bergström. 1995. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol. Lett. 131:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 19.Calladine, C. R., A. Sharff, and B. Luisi. 2001. How to untwist an alpha-helix: structural principles of an alpha-helical barrel. J. Mol. Biol. 305:603-618. [DOI] [PubMed] [Google Scholar]
- 20.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 21.Castellan, G. W. 1983. The ionic current in aqueous solutions, p. 769-780. In G. W. Castellan (ed.), Physical chemistry. Addison-Wesley, Reading, Mass.
- 22.Coburn, J., W. Chege, L. Magoun, S. C. Bodary, and J. M. Leong. 1999. Characterization of a candidate Borrelia burgdorferi β3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926-940. [DOI] [PubMed] [Google Scholar]
- 23.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 93:7973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox, D. L., and J. D. Radolf. 2001. Insertion of fluorescent fatty acid probes into the outer membranes of the pathogenic spirochaetes Treponema pallidum and Borrelia burgdorferi. Microbiology 147:1161-1169. [DOI] [PubMed] [Google Scholar]
- 25.Defoe, G., and J. Coburn. 2001. Delineation of Borrelia burgdorferi p66 sequences required for integrin α(IIb)β(3) recognition. Infect. Immun. 69:3455-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutzler, R., Y. F. Wang, P. Rizkallah, J. P. Rosenbusch, and T. Schirmer. 1996. Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway. Structure 4:127-134. [DOI] [PubMed] [Google Scholar]
- 27.Egli, C., W. K. Leung, K. H. Muller, R. E. Hancock, and B. C. McBride. 1993. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect. Immun. 61:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 30.Garavito, R. M., and J. P. Rosenbusch. 1980. Three-dimensional crystals of an integral membrane protein: an initial x-ray analysis. J. Cell Biol. 86:327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haines, K. A., L. Yeh, M. S. Blake, P. Cristello, H. Korchak, and G. Weissmann. 1988. Protein I, a translocatable ion channel from Neisseria gonorrhoeae, selectively inhibits exocytosis from human neutrophils without inhibiting O2- generation. J. Biol. Chem. 263:945-951. [PubMed] [Google Scholar]
- 32.Hossain, H., H. Wellensiek, R. Geyer, and G. Lochnit. 2001. Structural analysis of glycolipids from Borrelia burgdorferi. Biochimie 83:683-692. [DOI] [PubMed] [Google Scholar]
- 33.Jelfs, J., R. Munro, E. Wedege, and D. A. Caugant. 2000. Sequence variation in the porA gene of a clone of Neisseria meningitidis during epidemic spread. Clin. Diagn. Lab. Immunol. 7:390-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 35.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig, O., V. De Pinto, F. Palmieri, and R. Benz. 1986. Pore formation by the mitochondrial porin of rat brain in lipid bilayer membranes. Biochim. Biophys. Acta 860:268-276. [DOI] [PubMed] [Google Scholar]
- 37.Magnarelli, L. A., J. F. Anderson, and A. G. Barbour. 1989. Enzyme-linked immunosorbent assays for Lyme disease: reactivity of subunits of Borrelia burgdorferi. J. Infect. Dis. 159:43-49. [DOI] [PubMed] [Google Scholar]
- 38.Maier, E., G. Polleichtner, B. Boeck, R. Schinzel, and R. Benz. 2001. Identification of the outer membrane porin of Thermus thermophilus HB8: the channel-forming complex has an unusually high molecular mass and an extremely large single-channel conductance. J. Bacteriol. 183:800-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathers, D. A., W. K. Leung, J. C. Fenno, Y. Hong, and B. C. McBride. 1996. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect. Immun. 64:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merino, S., M. M. Nogueras, A. Aguilar, X. Rubires, S. Alberti, V. J. Benedi, and J. M. Tomas. 1998. Activation of the complement classical pathway (C1q binding) by mesophilic Aeromonas hydrophila outer membrane protein. Infect. Immun. 66:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikaido, H. 1992. Porins and specific channels of bacterial outer membranes. Mol. Microbiol. 6:435-442. [DOI] [PubMed] [Google Scholar]
- 42.Nilsson, C., H. J. Cooper, K. Håkansson, A. G. Marshall, Y. Östberg, M. Lavrinovicha, and S. Bergström. 2002. Characterization of the P13 membrane protein of Borrelia burgdorferi by mass spectrometry. J. Am. Soc. Mass. Spectrom. 13:295-299. [DOI] [PubMed] [Google Scholar]
- 43.Noppa, L., Y. Östberg, M. Lavrinovicha, and S. Bergström. 2001. P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect. Immun. 69:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 45.Probert, W. S., K. M. Allsup, and R. B. LeFebvre. 1995. Identification and characterization of a surface-exposed, 66-kilodalton protein from Borrelia burgdorferi. Infect. Immun. 63:1933-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radolf, J. D., K. W. Bourell, D. R. Akins, J. S. Brusca, and M. V. Norgard. 1994. Analysis of Borrelia burgdorferi membrane architecture by freeze-fracture electron microscopy. J. Bacteriol. 176:21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radolf, J. D., M. S. Goldberg, K. Bourell, S. I. Baker, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 63:2154-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regelink, A. G., D. Dahan, L. V. Moller, J. W. Coulton, P. Eijk, P. Van Ulsen, J. Dankert, and L. Van Alphen. 1999. Variation in the composition and pore function of major outer membrane pore protein P2 of Haemophilus influenzae from cystic fibrosis patients. Antimicrob. Agents Chemother. 43:226-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudel, T., A. Schmid, R. Benz, H.-A. Kolb, L. F., and T. F. Meyer. 1996. Modulation of translocatable pathogenic Neisseria porin (PorB) by cytosolic ATP/GTP of target cells: parallels between intracellular pathogen accomodation and mitochondrial endosymbiosis. Cell 85:391-402. [DOI] [PubMed] [Google Scholar]
- 51.Sadziene, A., D. D. Thomas, and A. G. Barbour. 1995. Borrelia burgdorferi mutant lacking Osp: biological and immunological characterization. Infect. Immun. 63:1573-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sallmann, F. R., S. Baveye-Descamps, F. Pattus, V. Salmon, N. Branza, G. Spik, and D. Legrand. 1999. Porins OmpC and PhoE of Escherichia coli as specific cell-surface targets of human lactoferrin. Binding characteristics and biological effects. J. Biol. Chem. 274:16107-16114. [DOI] [PubMed] [Google Scholar]
- 53.Shang, E. S., M. M. Exner, T. A. Summers, C. Martinich, C. I. Champion, R. E. Hancock, and D. A. Haake. 1995. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 63:3174-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shang, E. S., J. T. Skare, M. M. Exner, D. R. Blanco, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1998. Isolation and characterization of the outer membrane of Borrelia hermsii. Infect. Immun. 66:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skare, J. T., B. M. Ahmer, C. L. Seachord, R. P. Darveau, and K. Postle. 1993. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J. Biol. Chem. 268:16302-16308. [PubMed] [Google Scholar]
- 56.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 178:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skare, J. T., T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, J. Bunikis, S. Bergström, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1997. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect. Immun. 65:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 59.Takayama, K., R. J. Rothenberg, and A. G. Barbour. 1987. Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 55:2311-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilly, K., A. F. Elias, J. L. Bono, P. Stewart, and P. Rosa. 2000. DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol. 2:433-442. [PubMed] [Google Scholar]
- 61.Trias, J., and R. Benz. 1993. Characterization of the channel formed by the mycobacterial porin in lipid bilayer membranes. Demonstration of voltage gating and of negative point charges at the channel mouth. J. Biol. Chem. 268:6234-6240. [PubMed] [Google Scholar]
- 62.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 63.Walker, E. M., L. A. Borenstein, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1991. Analysis of outer membrane ultrastructure of pathogenic Treponema and Borrelia species by freeze-fracture electron microscopy. J. Bacteriol. 173:5585-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, S., H. Ding, J. Seah, K. Wu, Y. Chang, K. S. Chang, M. F. Tam, and W. Syu. 1998. Characterization of a phage specific to hemorrhagic Escherichia coli O157:H7 and disclosure of variations in host outer membrane protein OmpC. J. Biomed. Sci. 5:370-382. [DOI] [PubMed] [Google Scholar]