Abstract
An important feature of Escherichia coli pathogenesis is an ability to withstand extremely acidic environments of pH 2 or lower. This acid resistance property contributes to the low infectious dose of pathogenic E. coli species. One very efficient E. coli acid resistance system encompasses two isoforms of glutamate decarboxylase (gadA and gadB) and a putative glutamate:γ-amino butyric acid (GABA) antiporter (gadC). The system is subject to complex controls that vary with growth media, growth phase, and growth pH. Previous work has revealed that the system is controlled by two sigma factors, two negative regulators (cyclic AMP receptor protein [CRP] and H-NS), and an AraC-like regulator called GadX. Earlier evidence suggested that the GadX protein acts both as a positive and negative regulator of the gadA and gadBC genes depending on environmental conditions. New data clarify this finding, revealing a collaborative regulation between GadX and another AraC-like regulator called GadW (previously YhiW). GadX and GadW are DNA binding proteins that form homodimers in vivo and are 42% homologous to each other. GadX activates expression of gadA and gadBC at any pH, while GadW inhibits GadX-dependent activation. Regulation of gadA and gadBC by either regulator requires an upstream, 20-bp GAD box sequence. Northern blot analysis further indicates that GadW represses expression of gadX. The results suggest a control circuit whereby GadW interacts with both the gadA and gadX promoters. GadW clearly represses gadX and, in situations where GadX is missing, activates gadA and gadBC. GadX, however, activates only gadA and gadBC expression. CRP also represses gadX expression. It does this primarily by repressing production of sigma S, the sigma factor responsible for gadX expression. In fact, the acid induction of gadA and gadBC observed when rich-medium cultures enter stationary phase corresponds to the acid induction of sigma S production. These complex control circuits impose tight rein over expression of the gadA and gadBC system yet provide flexibility for inducing acid resistance under many conditions that presage acid stress.
The acidic pH of the human stomach is a daunting environment for any pathogen. After a meal is ingested, the gastric pH can fall to 2 or less, an acidification sufficient to kill most enteric pathogens. However, commensal and pathogenic strains of Escherichia coli have evolved three systems of acid resistance that will, upon induction, protect the organism from pH 2 environments for hours (2, 11, 12). This period is long enough for the organism to breach the gastric acid barrier and gain entrance to the less acidic intestinal environment. The most potent of the three acid resistance systems requires extracellular glutamate to function and is induced either in moderately acid environments (pH 5 to 6) during exponential growth or upon entry into stationary phase at any pH. Extracellular glutamate is not required, however, for induction. The glutamate-dependent acid resistance system includes two isoforms of glutamate decarboxylase encoded by gadA and gadB that convert intracellular glutamate to γ-amino butyric acid (GABA) and consume a proton in the process. In addition, a dedicated antiporter, GadC, is believed necessary to export the GABA end product out of the cell while simultaneously importing more glutamate as the substrate, although this has not yet been demonstrated. The genes gadB and gadC form the gadBC operon, while gadA maps to a different location. How this system actually enables acid resistance is not known, but one proposal is that it affects intracellular pH.
Regulation of the system is extremely complex. Earlier work from this and other laboratories has shown that gadA and gadBC expression is regulated by separate sigma S (σS)-dependent and -independent pathways even though only a single transcriptional start site has been identified for either operon (1, 2, 4). The alternative sigma factor σS, encoded by rpoS, is used to express a regulon of stress-regulated genes classically associated with entry into stationary phase (6, 7). The σS-dependent pathway for gad expression is evident in complex-medium-grown cells, whereas σS-independent regulation occurs in minimal-glucose-grown cells. The cyclic AMP (cAMP) receptor protein (CRP) and the nucleoid protein H-NS have been implicated as potent repressors of gad expression in complex Luria-Bertani (LB) media (1, 4). In addition, a 20-bp conserved DNA sequence, called the GAD box, is located upstream from the gadBC and gadA promoters and is required for induction (1). Outside this one area, almost no homology is apparent between the two promoters.
The gadX (yhiX) gene, located downstream of gadA and cotranscribed with it, encodes an AraC- and XylS-like regulator implicated in the control of the gadAX and gadBC loci (8, 18, 19). Tramonti et al. (20) recently showed that gadX was an RpoS-dependent gene and is itself acid induced. GadX was originally ascribed to have both positive and negative regulatory function, depending on the environmental conditions tested (18). However, another gene encoding an AraC-like regulatory protein with possible influence over gadA and gadBC expression was recently discovered as a result of gene array analysis (D. Tucker and T. Conway, submitted for publication). The gene, gadW (yhiW), is located downstream of gadX (D. Tucker and T. Conway, unpublished data). GadW is 60 and 30% homologous to GadX in the DNA binding and dimerization domains, respectively. Previous work describing GadX as both a positive and negative regulator did not consider the potential influence of the gadW product (18), and the gadW array studies did not directly examine effects on protein levels or explore a potential collaboration with GadX. So with the discovery of GadW, it became necessary to reexamine the role of GadX as well as the influence of GadW over gadA and gadBC expression.
We have taken a direct genetic approach to better define the roles of GadX and GadW in controlling this system. The results indicate that GadW mediates control of the gadA and gadBC genes by repressing gadX and, under some situations, by activating gadA and gadBC. CRP represses the system largely by inhibiting the production of RpoS, the sigma factor primarily responsible for gadX expression. Finally, in contrast to reports using a different background, repression by H-NS was independent of gadX. The overlapping control of gadX by CRP, RpoS, and GadW makes E. coli highly attuned to situations that can lead to lethal acid stress.
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. The media used included minimal E medium containing 0.4% glucose (EG) (21) and complex LB medium buffered with either 100 mM morpholinepropanesulfonic acid (MOPS [pH 8]) or 100 mM morpholineethanesulfonic acid (MES [pH 5.5]). Antibiotics were used at the following concentrations: ampicillin, 60 μg/ml; kanamycin, 25 μg/ml; tetracycline, 20 μg/ml; and chloramphenicol, 30 μg/ml. All strains were grown at 37°C with aeration.
TABLE 1.
Bacterial strains and plasmids in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| EK227 | K-12, wild type, λ− F− | A. C. Matin |
| EK344 | (GE1050) Δcrp::Cm | 16 |
| EK395 | Δcya-1400::Km λ− e14−relA1 spoT1 thi-1 | J. Kaper |
| EK432 | Δ(lacA-lacZ)515(::Cat) | 3 |
| EK426 | K-12 (MG1655) ΔgadX::Km | T. Conway |
| EK437 | hns-206oppC506::Tn10 | C. Higgens |
| EK441 | K-12 (MG1655) ΔgadW::Km | T. Conway |
| EK442 | K-12 (MG1655) ΔgadXW::Km | T. Conway |
| EK540 | TOPO 10 F−mcrA Δ(mrr-hsdRMS-merBC) φ80 lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| EK541 | BL21 Star(DE3): F−ompT hsdB (rB− mB−) dcm rne-131 | Invitrogen |
| EF362 | K-12 (EK227) rpoS::Tn10 | 2 |
| EF528 | K-12 (EK227) Δcrp::Cm | EK227 × EK344 |
| EF615 | EK298 trpDC::putPA1303-Km-gadA::lacZ(o) (−165 to +788) | 2 |
| EF646 | Δcya-1400::Km/pT18-Zip × pT25-Zip | 14 |
| EF663 | EK298 trpDC::putPA1303-Km-gadA::lacZ(o) (−51 to +788) | 2 |
| EF666 | EK298 trpDC::putPA1303-Km-gadA::lacZ(o) (−86 to +788) | 2 |
| EF678 | K-12 (EK227) Δcrp::Cm rpoS::Tn10 | EF528 × EF362 |
| EF757 | K-12 (EK227) ΔgadX::Km | EK227 × EF426 |
| EF825 | K-12 (EK227) Δ(lacA-lacZ)::Cat | EK227 × EK432 |
| EF828 | K-12 (EK227) ΔgadX | EF757 × pCP20 |
| EF830 | K-12 (EK227) ΔgadX Δ(lacA-lacZ)::Cat | EF828 × EK432 |
| EF833 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−165 to +788) Δ(lacA-lacZ)::Cat | EF825 × EF615 |
| EF839 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−165 to +788) ΔgadX Δ(lacA-lacZ)::Cat | EF830 × EF615 |
| EF861 | K-12 (EK227) ΔgadW::Km | EK227 × EK441 |
| EF863 | K-12 (EK227) ΔgadXW::Km | EK227 × EK442 |
| EF864 | K-12 (EK227) ΔgadW::Km Δcrp::Cm | EF528 × EK441 |
| EF865 | K-12 (EK227) ΔgadXW::Km Δcrp::Cm | EF528 × EK442 |
| EF906 | K-12 (EK227) ΔgadX Δcrp::Cm | EF828 × EF528 |
| EF907 | K-12 (EK227) ΔgadW | EF861 × pCP20 |
| EF908 | K-12 (EK227) ΔgadXW | EF863 × pCP20 |
| EF911 | K-12 (EK227) ΔgadW Δ(lacA-lacZ)::Cat | EF907 × EK432 |
| EF912 | K-12 (EK227) ΔgadXW Δ(lacA-lacZ)::Cat | EF908 × EK432 |
| EF919 | Δcya-1400::Km/pT18 × pT25 | EK395 × pT18 × pT25 |
| EF920 | Δcya-1400::Km/pMF497/pMF498 | This study |
| EF921 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−165 to +788) ΔgadW Δ(lacA-lacZ)::Cat | EF911 × EF615 |
| EF922 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−51 to +788) ΔgadW Δ(lacA-lacZ)::Cat | EF911 × EF663 |
| EF923 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−86 to +788) ΔgadW Δ(lacA-lacZ)::Cat | EF911 × EF666 |
| EF928 | K-12 (EK227) Δcya::Km | EK227 × EK395 |
| EF929 | K-12 (EK227) ΔgadXW::Km hns::Tn10 Δcrp::Cm | EF865 × EK437 |
| EF931 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−51 to +788) Δ(lacA-lacZ)::Cat | EF825 × EF663 |
| EF932 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−86 to +788) Δ(lacA-lacZ)::Cat | EF825 × EF666 |
| EF933 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−165 to +788) ΔgadXW Δ(lacA-lacZ)::Cat | EF912 × EF615 |
| EF934 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−51 to +788) ΔgadXW Δ(lacA-lacZ)::Cat | EF912 × EF663 |
| EF935 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−86 to +788) ΔgadXW Δ(lacA-lacZ)::Cat | EF912 × EF666 |
| EF948 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−51 to +788) ΔgadX Δ(lacA-lacZ)::Cat | EF830 × EF663 |
| EF949 | K-12 (EK227) trpDC::putPA1303-Km-gadA::lacZ(o) (−86 to +788) ΔgadX Δ(lacA-lacZ)::Cat | EF830 × EF663 |
| EF957 | K-12 (EK227) hns-206 oppC506::Tn10 | EK227 × EK437 |
| Plasmids | ||
| pT18 | pBluescript II KS containing T18 C terminus of B. pertussis cyaA | 9 |
| pT25 | pACYC184 containing T25 N terminus of B. pertussis cyaA | 9 |
| pT18-Zip | pT18 containing leucine zipper domain | 9 |
| pT25-Zip | pT25 containing leucine zipper domain | 9 |
| pCP20 | Flp recombinase | 3 |
| pMF497 | gadW-′cyaA in pT18 | This study |
| pMF498 | cyaA′-gadX in pT25 | This study |
| pMF499 | pET102/D-TOPO containing Trx-GadW-His6 | |
| pMF506 | cyaA′-gadW in pT25 | This study |
| pMF507 | yhiX-′cyaA in pT18 | This study |
| pET102/D-TOPO | His-Patch thioredoxin fusion vector | Invitrogen |
Genetic and molecular procedures.
Phage P1 transduction, transformation with CaCl2, and electroporation were performed by standard methods (13). General DNA manipulations were carried out as described earlier (17). Deletion of the kanamycin resistance cassette in the gadX, gadW, and gadXW mutants was carried out by the method reported by Datsenko and Wanner (3). The list of oligonucleotide primers used is given in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence |
|---|---|
| Oligo-201 | 5′-CAGCAATGTTTGGGCGATTTTTATTAC-3′ |
| Oligo-202 | 5′-GATAATTCAGGAGACACAGAATGCG-3′ |
| Oligo-233 | 5′-CTCGTCGACACGTGAATCGAGTAGTTC-3′ |
| Oligo-340 | 5′-CTATGCAATCACTACATGGGAATT-3′ |
| Oligo-341 | 5′-CTCTATAATCTTATTCCTTCCGCAG-3′ |
| Oligo-377 | 5′-GGAGTTCGAAATGGACCAGAAG-3′ |
| Oligo-378 | 5′-AGTTTCGGGTGATCGCTGAG-3′ |
| Oligo-379 | 5′-GGTGTCGACTATGAACCTGCTTCCCATCGACTAC-3′ |
| Oligo-406 | 5′-TGCGCCAAAACGTGAATCGAGTAGTTC-3′ |
| Oligo-407 | 5′-TCGCACCAAAACGTGAATCGAGTAGTT-3′ |
| Oligo-414 | 5′-ATACATATGACTCATGTCTGCTCGGTGAT-3′ |
| Oligo-415 | 5′-TAAGAATCCATTCAAATGCGAAATATGTCA-3′ |
| Oligo-444 | 5′-AAGCTGCAGGGATGCAATCACTACATGGGAATT-3′ |
| Oligo-445 | 5′-TCAGGATCCCTATAATCTTATTCCTTCCGCAGA-3′ |
| Oligo-446 | 5′-AAGGGGCCCCATGCAATCACTACAT-3′ |
| Oligo-447 | 5′-TCAAAGCTTATTAATCTTATTCCTTC-3′ |
| Oligo-448 | 5′-AAGCTGCAGGGATGACTCATGTCTG-3′ |
| Oligo-449 | 5′-ATGGGATCCTCAGGAAAAGGTACCT-3′ |
| Oligo-450 | 5′-AAGGGGCCCCATGACTCATGTCTGCT-3′ |
| Oligo-451 | 5′-ATGAAGCTTATGGAAAAGGTACCT-3′ |
| Oligo-458 | 5′-AATAAGATTATAGAGTTTTACT-3′ |
| Oligo-459 | 5′-CATGAGTCATGATTATCCCTTA-3′ |
| Oligo-465 | 5′-CACCATGACTCATGTCTGCTCG-3′ |
| Oligo-466 | 5′-GGAAAAGGTACCTGGCGAATG-3′ |
| Oligo-507 | 5′-CAATACGCAAACCGCCTCTCC-3′ |
| Oligo-508 | 5′-AGTGAATCCGTAATCATGGTCATA-3′ |
Western blot analysis.
Strains were grown at 37°C in media containing the required antibiotics as indicated. At an optical density of 600 nm (OD600) of 0.4 (log phase) or 3.8 (late stationary phase), cells were collected by centrifugation and then were resuspended in 0.01% sodium dodecyl sulfate (SDS) solution. Protein concentrations were determined using Bio-Rad Protein Assay reagent. Samples (5 μg of protein) were mixed with 2× SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer (125 mM Tris [pH 7.0], 20% glycerol, 10% β-mercaptoethanol, 6% SDS, and 0.2% bromophenol blue), boiled for 5 min, and were then separated by PAGE on 10% Tris-HCl Criterion gels (Bio-Rad). After semidry electrophoretic transfer of proteins onto polyvinylidene difluoride membranes (Millipore Co., Bedford, Mass.), the membranes were incubated overnight with 5% powdered milk in Tris-buffered saline-Tween buffer (150 mM NaCl, 10 mM Tris-HCl [pH 8.0], and 0.05% [vol/vol] Tween 20) to block nonspecific protein interactions. The membranes were probed with one of the following primary antibodies: rat anti-GAD (2), mouse anti-RpoS, and rabbit anti-GadX (kindly provided by S. Shin and J. Kaper), followed by the appropriate monoclonal secondary antibodies coupled to peroxidase (Sigma): anti-rat (1:10,000), anti-mouse (1:3,000), and anti-rabbit (1:4,000). Antibody-tagged protein bands on the probed membranes were detected using an enhanced chemiluminescence Western blot detection kit (Amersham) (2).
Northern blot and primer extension analyses.
Total RNA was extracted from log-phase cultures (OD600 = 0.4; 2 × 108 cells per ml) grown under both alkaline and acidic conditions in LB medium using the RNeasy kit (Qiagen). RNA concentrations were determined by measuring the optical densities at 260 and 280 nm. Aliquots of RNA (5 μg) were denatured at 65°C for 15 min and were separated by electrophoresis through a denaturing formaldehyde-agarose (1.2%) gel as described previously (17). The RNA was then transferred onto a positively charged nylon membrane (Amersham-Pharmacia) and was baked at 80°C for 2 h. The membranes were probed with a 1.4-kb gadA and gadB probe or a 0.827-kb gadX probe generated by PCR using oligo-377 and -378 or oligo-340 and -341, respectively. Probes were labeled with [α-32P]dCTP (Amersham) by using a random-primed DNA-labeling kit (Boehringer Mannheim). The gadA and gadB probe corresponds to the entire open reading frame of gadA or gadB and hybridizes to both gadA and gadB. As a control, the membranes were also hybridized with a 23S rRNA probe (oligo-379) that was end labeled with [γ-32P]ATP.
For primer extension analysis of the gad transcriptional start sites, 1 pmol each of the primers for gadA (oligo-406) and gadB (oligo-407) was 5′ end labeled with T4 polynucleotide kinase (Promega) and [γ-32P]ATP. Reverse transcription of gadA or gadBC mRNA was performed using the Qiagen Omniscript reverse transcriptase (RT) kit. Sequencing reactions were carried out using the GIBCO BRL double-stranded DNA cycle-sequencing system and were run in parallel with the cDNA transcripts to map the 5′ ends of gadA or gadBC mRNA. The promoter and beginning regions of gadA and gadB genes were used as the templates in the sequencing reactions.
RT-PCR.
The RNA used for RT-PCR was extracted using TRIzol Reagent (GIBCO BRL) per the manufacturer's recommendation from cells grown to log phase (OD600 = 0.4) in LB medium at pH 8 and 5.5 and in EG medium at pH 7.7 and 5.5 during log phase. RT-PCR was performed with the Superscript One-Step RT-PCR kit (GIBCO BRL). cDNA for gadX was prepared using oligo-340 and oligo-341. cDNA for gadW was prepared using oligo-414 and oligo-415. To detect potential gadX gadW cotranscripts, the pairs oligo-340 (forward primer) and oligo-415 (reverse primer) and oligo-458 and oligo-459 were used to amplify the entire gadX gadW region and the shorter intergenic region between gadX and gadW, respectively. Reaction conditions were designed as follows: cDNA synthesis and predenaturation, 1 cycle of 55°C for 30 min and 94°C for 2 min. PCR amplification involved 40 cycles of denaturation (94°C for 15 s), annealing (55°C for 30 s), extension (72°C for 1 min), and one final extension cycle of 72°C for 10 min. Control PCRs were performed using DNA to confirm primer efficacy. RNA purity was confirmed by performing the reaction in the absence of RT.
Purification of Trx-GadW-His6.
The K-12 gadW-coding region was amplified with oligo-465 and oligo-466 by using Pwo proofreading polymerase (Invitrogen), and the resulting fragment was cloned into the pET102/D-TOPO vector containing the His-Patch thioredoxin leader (pET Directional Expression Kits; Invitrogen). To enable directional cloning, the forward primer, oligo-465, was designed to contain 5′ CACC at the 5′ end, which pairs with the overhang sequence GTGG in the vector. The reverse primer, oligo-466, starts with the codon preceding the gadW stop codon (TGA) so that the cloned PCR product will produce an in-frame fusion with the C-terminal His tag. The resulting plasmid, pMF499, was transformed into TOPO 10 cells and, from there, into BL21 Star(DE3) for expression. The BL21 strain containing pMF499 was grown in 1 liter of LB medium containing 100 μg of ampicillin/ml at 30°C with shaking. When the OD600 of the culture reached 0.55, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added and the culture was incubated for an additional 3 h at 30°C. The cells were harvested by centrifugation and were resuspended overnight in 60 ml of lysis buffer (6 M guanidine, 0.1 M NaH2PO4, 0.01 mM Tris-HCl, and 5 mM imidazole) with shaking at 4°C. Insoluble cell debris was removed by centrifugation (8,000 × g, 4°C for 15 min), and the supernatant was passed through a 0.45-μm-pore-size filter. The filter-sterilized solution was subsequently incubated with 5 ml of ProBond resin (Invitrogen) at room temperature for 1 h, after which the resuspension was transferred to a Poly-Prep Chromatography column (Bio-Rad). The column was washed with 50 ml of washing buffer (8 M urea, 6 M guanidine, 0.1 M NaH2PO4, 0.01 mM Tris-HCl, and 5 mM imidazole), and the protein was eluted with the column buffer (8 M urea, 0.1 M NaH2PO4, 0.01 mM Tris-HCl, and 250 mM imidazole [pH 4.9]). Three-milliliter samples of the three highest-protein-content fractions were combined and desalted through a PD-10 Sephadex column (Amersham Pharmacia). The protein was eluted with desalting buffer (10 mM NaH2PO4 [pH 6.8], 0.1 mM EDTA, and 0.2 M NaCl), and glycerol was added to a final concentration of 50% for storage. This procedure purified Trx-GadW-His6 to homogeneity based on Coomassie-stained SDS-PAGE (data not shown). Western blot analysis also failed to detect any GadX in these preparations.
EMSA.
The ability of His-tagged GadW protein to bind the gadA and gadB promoters was tested using the electrophoretic mobility shift assay (EMSA). The promoter target fragments were amplified with oligo-201 and oligo-233 (gadA) and oligo-202 and oligo-233 (gadB). The PCR-generated gadA promoter fragment extends from bp −164 to +78, while the gadB promoter extends from bp −203 to +78 relative to the transcriptional start sites. Both fragments include the 20-bp GAD box region implicated in gad expression. Both fragments were end labeled with [γ-32P]ATP by T4 polynucleotide kinase. A PCR fragment of lacZ promoter (oligo-507 and oligo-508) was used as a probe for nonspecific DNA binding. Radiolabeled DNA probes (5,000 cpm) were incubated with GadW fusion protein at room temperature for 30 min in 30 μl of binding buffer (20 mM HEPES [pH 8.0], 5 mM MgCl2, 50 mM potassium glutamate, 0.01 mM EDTA, 1 mM NaH2PO4, 20 mM NaCl, 1 mM dithiothreitol, 30 μg of bovine serum albumin/ml, and 30 μg of salmon sperm DNA/ml). Samples were loaded onto 5% Tris-borate-EDTA nondenaturing ready gel (Bio-Rad) and were electrophoresed at room temperature in 0.5% Tris-borate-EDTA buffer with 0.2% glycerol. The gels were dried and exposed to X-Omat Kodak film at −70°C for 3 h.
Bacterial two-hybrid system.
The two-hybrid system used to demonstrate in vivo interaction between GadX and GadW is based upon the reconstitution of adenylate cyclase activity of Bordetella pertussis CyaA protein (9). Plasmid pT25 is a derivative of pACYC184 that encodes the N-terminal T25 fragment of CyaA (amino acids 1 to 224; CyaA′) with a multicloning site at the C terminus. Expression is controlled by the lacUV5 promoter. Plasmid pT18 is a derivative of pBluescript II KS (Stratagene), compatible with pT25, that encodes the C-terminal T18 fragment of CyaA (amino acids 225 to 339; ′CyaA) with the multicloning site of pBluescript II KS located at the N terminus. Functional reconstitution of adenylate cyclase activity is achieved if each Cya fragment is fused in frame to one of two interacting proteins. Adenylate cyclase activity was monitored in an E. coli strain deficient in endogenous adenylate cyclase (cya). The gadW gene was PCR amplified (Pwo polymerase) from EK227 using oligo-450 and oligo-451, which contain ApaI and HindIII sites, respectively. The ApaI and HindIII sites were used to directionally clone the insert into pT18. This created plasmid pMF497, in which the N terminus of the gadW open reading frame was fused in frame with the ′Cya fragment of pT18. The gadW stop codon was omitted from oligo-451 to allow read-through into the T18 fragment of Cya to generate a GadW-′Cya hybrid protein. A CyaA′-GadW hybrid was then constructed by amplifying gadW with oligo-448 and oligo-449 engineered with PstI and BamHI sites, respectively. Insertion of the PCR fragment into pT25 cut with PstI and BamHI created an in-frame CyaA′-GadW fusion (pMF506).
The gadX open reading frame was cloned in frame into the pT25 vector as follows: the gadX gene was PCR amplified from E. coli K-12 (EK227) with the proofreading Pwo polymerase by using oligo-444, which has a PstI site, and oligo-445, which was engineered to contain a BamHI site. The oligonucleotides were designed so that the insert would be in frame with CyaA′ and contain a stop codon after gadX. The PCR product was gel purified (Geneclean II kit; Bio 101), digested with PstI and BamHI, and ligated with T4 ligase into similarly digested pT25, resulting in plasmid pMF498. A GadX-′CyaA hybrid was then constructed by amplifying gadX with oligo-446 and oligo-447 engineered with ApaI and HindIII sites, respectively. Insertion of the PCR fragment into pT18 cut with ApaI and HindIII created an in-frame GadX-′CyaA fusion (pMF507). Potentially complementing pairs of these plasmids were transformed into the cya-deficient strain EK395, and the expression of chromosomal lacZ was monitored as an indicator of cAMP production resulting from potential interactions between pairs of Cya fusion protein. All cultures were grown in LB broth in the presence of chloramphenicol, ampicillin, or both as needed, and 0.5 mM IPTG at 30°C with aeration (250 rpm). β-Galactosidase activity was measured as described by Miller (13).
RESULTS
The gadX and gadW genes constitute separate transcriptional units.
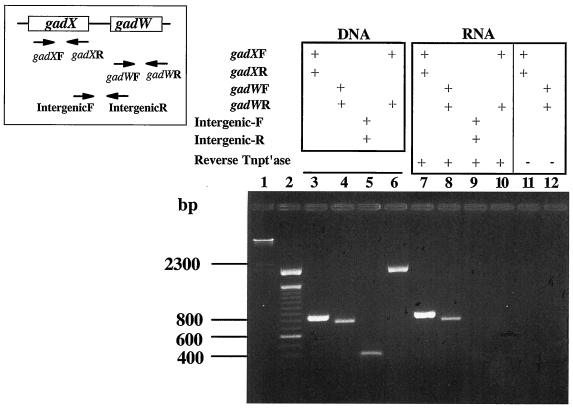
Since the gadW gene is located downstream of gadA and gadX, it was necessary to determine if independent promoters drive expression of each gene before analyzing the individual roles of GadX and GadW in controlling gadA and gadBC expression. If gadX and gadW are transcribed separately, then individual mutations in these genes could be examined without fear of polar effects. RT-PCR was chosen to identify whether these genes form an operon. As shown in Fig. 1, primers designed to detect the individual messages amplified both gadX and gadW (lanes 7 and 8). However, primers designed to amplify a potential intercistronic message failed (Fig. 1, lane 9), indicating that gadX and gadW do not form an operon. Thus, gadX and gadW mutants can be examined separately for their effects on gadA and gadBC expression.
FIG. 1.
RT-PCR analysis of gadX and gadW transcripts. Inset, left, represents the gadX and -W genes and the location of primers used for RT-PCR. gadX forward, oligo-340; gadX reverse, oligo-341; gadW forward, oligo-414; gadW reverse, oligo-415; intergenic forward, oligo-458; and intergenic reverse, oligo-459. PCRs with and without RT (Reverse Tntp′ase) were run, and products were separated on 2% agarose gels. Lane 1, HindIII-cut lambda DNA; lane 2, 100-bp ladder; conditions for lanes 3 to 12 are shown above the gel. RNA used was from log-phase (pH 5.5), LB-grown EK227 cells. Similar results were obtained using RNA from cells grown at pH 8 and minimal-glucose-grown cells (data not shown).
GadW inhibits activation of gadA and gadBC by GadX.
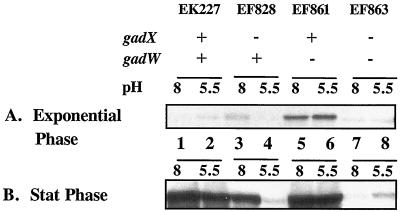
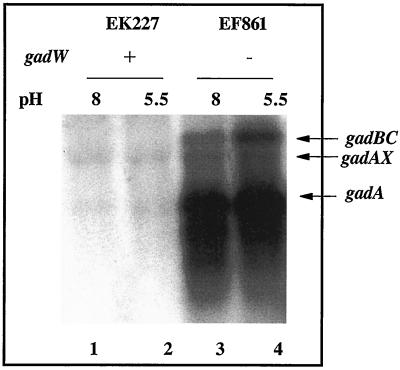
Western blot results presented in Fig. 2 illustrate that gadA and gadBC are normally repressed in cells growing exponentially in complex LB media (Fig. 2A, lanes 1 and 2). However, when GadW was removed through mutation, GadX activated gadA and gadBC expression under acid or alkaline conditions (Fig. 2A, lanes 5 and 6 versus lanes 7 and 8). Thus, GadW is a negative regulatory protein that inhibits the activation of gadA and gadBC by GadX. Figure 3 presents evidence reinforcing this conclusion from Northern blot analyses that examined both the gadA and gadBC transcripts. The results indicate that GadX is not both a positive and negative regulator as previously reported (18). Strain EF861, possessing GadX but lacking GadW, highly expressed the gadA and gadBC messages under both acid and alkaline conditions. Thus, GadX serves as an activator of gadA and gadBC, while GadW can inhibit activation.
FIG. 2.
Effects of GadX and GadW on pH-regulated gadA and gadBC expression in exponential- and stationary-phase cells grown in LB. Western blot analysis of extracts prepared from cultures grown to mid-log (OD600 = 0.4) (A) or stationary (Stat) (OD600 = 3.8) (B) phase in LB medium buffered to the pH values indicated. Strains and genotypes are indicated in the figure. Whole-cell proteins were extracted by boiling in SDS, and 5 μg of protein was loaded per sample on SDS-10% PAGE gels. Blots were probed with polyclonal anti-GadA and -B antibody.
FIG. 3.
Northern blot analysis of gadW effects on gadA and gadBC message. Cells were grown to log phase in buffered LB medium. RNA was extracted, and 5 μg was electrophoresed in formaldahyde-agarose gels and probed with a 1.4-kb gadA and -B probe that can detect gadAX, gadA, and gadBC transcripts. Strains and relevant genotypes are indicated in the figure.
Interestingly, Fig. 2A and B (lanes 3 and 4 versus lanes 7 and 8) also indicate that the opposite scenario is true: that GadX can prevent GadW induction of gadA and gadBC. It seems that, while either GadX or GadW can induce gadA and gadBC, when both AraC-like regulators are present, they inhibit each other's function.
GadW represses expression of gadX.
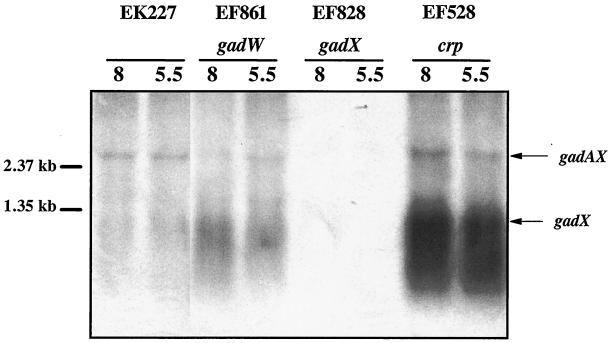
The gadX gene is expressed from two promoters, PgadA and PgadX (20). Since GadW inhibits GadX-dependent expression of gadA and gadBC and since a putative GAD box exists upstream of the gadX promoter, we hypothesized that GadW might repress gadX expression. Figure 4 presents Northern blot data that confirmed this idea. In mid-log-phase cells, gadX is not expressed well at either pH (EK227) but is expressed three- to fourfold higher in a gadW mutant (EF861). This increased expression appears to come from the gadX promoter, not the gadA promoter, since the level of gadAX transcript did not increase. It is significant that the same gadW mutant also overexpressed the gadA transcript under the same conditions (Fig. 3), suggesting that either the gadAX cotranscript is processed to separate the two RNA molecules or that there is a terminator between gadA and gadX that prevents most transcripts originating at PgadA from extending into gadX.
FIG. 4.
Northern blot analysis of GadW effects on gadX message. Cells were grown to log phase in buffered LB medium. RNA was extracted; 5 μg was electrophoresed in formaldahyde-agarose gels and probed with an 827-bp radiolabled gadX probe that can detect gadAX and gadX transcripts. Strains and relevant genotypes are indicated in the figure. pH is given immediately over the blot.
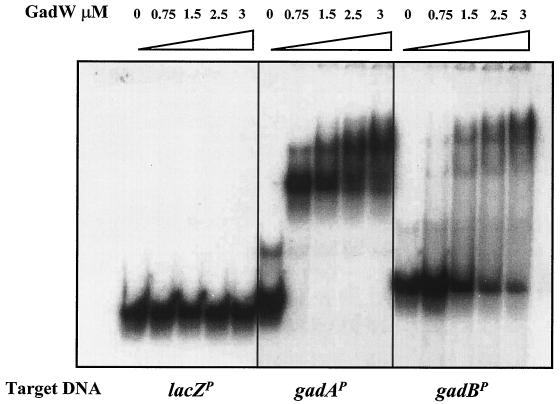
Purified GadW binds the gadA and gadBC promoters.
The gadX and gadW products are both implicated as regulators of gadA and gadBC expression (8, 18-20) (Tucker and Conway, unpublished). EMSAs previously demonstrated that a purified MalE-GadX hybrid protein directly binds a 447-bp promoter fragment of gadA encompassing the 20-bp GAD box (18). We have now purified a thioredoxin-His-tagged GadW fusion protein suitable for conducting similar DNA binding assays (see Materials and Methods). The results presented in Fig. 5 illustrate that GadW binds to both the gadA (242 bp) and gadBC (281 bp) promoters but not to the unrelated lacZ promoter. The purified GadW protein was not contaminated with GadX, based on Western blot analysis (data not shown). The results suggest that GadW can act directly on the gadA and gadBC promoter regions in vivo. The slightly larger band seen in lane 1 of the second panel in Fig. 5 is an artifact of the PCR. It was not present on other GadW EMSA gels. The multiple shifted bands suggest that GadW may bind as a multimer to these promoters.
FIG. 5.
GadW binds to the gadA and gadB promoters. EMSA conditions were described in Materials and Methods. Radiolabeled promoter fragments (5,000 cpm) were incubated with purified Trx-GadW-His6 protein for 30 min at 25°C and were electrophoresed through a 5% nondenaturing polyacrylamide gel.
The conserved 20-bp cis-acting regulatory region is required for GadX and GadW control.
A 20-bp conserved sequence (TTAGGATTTTGTTATTTAAA) located between −53 and −73 bp from the gadA and gadBC transcriptional start sites was previously identified as being required for gadA and gadBC expression (1). To determine if this 20-bp cis-acting GAD box region was required for GadX or -W control of gadA and gadBC expression, gadX and gadW mutations were tested for effects on the expression of a series of gadA-lacZ fusions in which different portions of the gadA promoter were deleted. Strains carrying the entire region from −164 to +788 or a truncation that retains the region from −85 to +788, both of which contain the cis-acting GAD box sequence, were subjected to normal control by GadX and GadW (Fig. 6A and B). In contrast to the fusions containing the GAD box, fusions starting at −51 and thus lacking the 20-bp sequence were devoid of X or W control (Fig. 6C).
FIG. 6.
Effects of promoter deletions on GadX and GadW regulation of gadA. Cells were grown to exponential phase in LB medium buffered to pH 8 with 100 mM MOPS or to pH 5.5 with 100 mM MES and were examined for β-galactosidase activity (Miller units). (A) gadA-lacZ fusions; (B) gadA-lacZ fusions; and (C) gadA-lacZ fusions. The length of the gadA promoter region (in nucleotides) is given along top of panels. Wild type (WT), EF833, EF932, and EF931; ΔX, EF839, EF949, and EF948; ΔXW, EF933, EF935, and EF934; and ΔW, EF921, EF923, and EF922.
Another unusual aspect of GadW is seen in Fig. 6A and B. Whereas removing GadW allowed GadX to activate gadA-lacZ expression at both growth pH values, removing GadX allowed W to activate expression only at pH 8. This is consistent with what was observed in the Western blots, both in log-phase cultures (Fig. 2A, lanes 3 and 4) and more dramatically in stationary-phase cultures (Fig. 2B, lanes 3 and 4). The data are consistent with a model in which GadW binds the gadA and -B promoters best at pH 8. This binding could inhibit GadX activity. However, when GadX is missing, GadW could inadvertently activate gadA and gadBC expression.
GadX and GadW form homodimers in vivo.
An important characteristic of many AraC-like regulators is that they form homodimers via an N-terminal dimerization domain (5, 15). Since GadX and -W are predicted to be AraC-like regulators, a two-hybrid approach was taken to demonstrate whether or not either of them dimerizes in vivo. The genes encoding GadX and GadW were fused to the B. pertussis CyaA T18 and T25 fragments as described in Materials and Methods. The T18 and T25 fusion pairs were then placed into a Δcya mutant of E. coli. If the T25-GadX and GadX-T18 hybrids dimerize in vivo, the two CyaA fragments will be brought together, reconstituting an active adenylate cyclase. Adenylate cyclase activity was then monitored by measuring the expression of the cAMP-dependent lacZ gene. The same approach was taken to test GadW dimerization. As shown in Table 3, the two-GadX-hybrid fusions (EF974) increased lacZ expression 55-fold over that in control strains (EF919), while the two-GadW-hybrid fusions (EF975) yielded a 45-fold increase. The results indicate that GadX and GadW form homodimers in vivo.
TABLE 3.
Two-hybrid analysis of GadX and GadW in vivo interactions
| Strain | Relevant genotype | β-Galactosidase activitya |
|---|---|---|
| EK395 | Δcya | 60 |
| EF919 | EK395/pT25/pT18 | 65 |
| EF646 | EK395/pT25-Zip/pT18-Zip | 1,570 |
| EF1017 | EK395/pT25/pT18-gadX | 62 |
| EF1018 | EK395/pT25/pT18-gadW | 72 |
| EF1019 | EK395/pT25-gadX/pT18 | 68 |
| EF1020 | EK395/pT25-gadW/pT18 | 62 |
| EF974 | EK395/pT25-gadX/pT18-gadX | 3,600 |
| EF975 | EK395/pT25-gadW/pT18-gadW | 2,880 |
| EF920 | EK395/pT25-gadX/pT18-gadW | 318 |
| EF973 | EK395/pT25-gadW/pT18-gadX | 246 |
Strains were grown to stationary phase (22 h) at 30°C in LB medium with 0.5 mM IPTG and were assayed for β-galactosidase (Miller units).
We also noticed that GadX and GadW can form heterodimers in vivo, although this was a much weaker interaction than that of homodimers. Table 3 illustrates that the X-Cya and W-Cya hybrid proteins (EF920 and EF973) generated four- to fivefold-higher levels of β-galactosidase than did controls. The significance of heterodimer formation is not yet apparent.
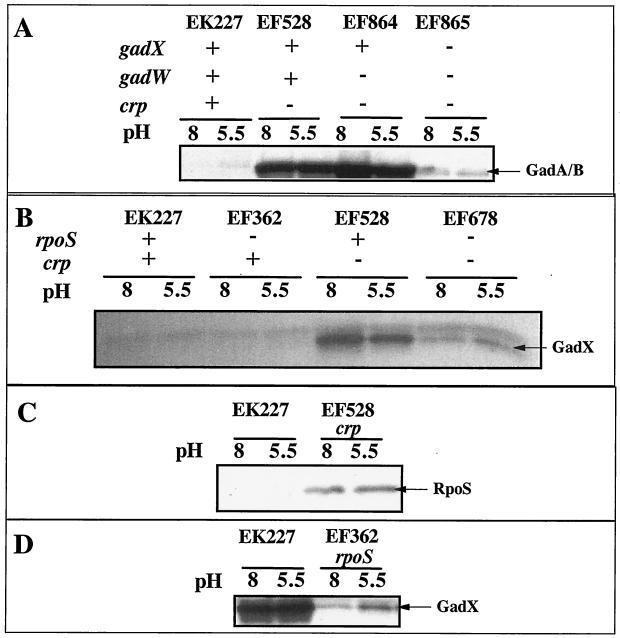
CRP regulates gadX.
Previous studies have shown that CRP is a potent negative regulator of gadA and gadBC expression in log-phase, LB-grown cells (1). Figure 7A illustrates that a crp mutation caused overexpression of gadA and gadBC (EF227 versus EF528) and that overexpression required GadX (EF864 versus EF865). This suggested that CRP might repress gadA and gadBC indirectly by regulating gadX expression. The Northern blot in Fig. 4 (EF528) and the Western blot in Fig. 7B indicate that CRP does repress gadX expression at the RNA level. The crp mutation caused elevated gadX message and protein (compare EF528 to EK227). A crp mutation did not affect expression of gadW (data not shown).
FIG. 7.
Western blot analysis of crp and rpoS effects on GAD, RpoS, and GadX protein levels. Cells were grown in LB medium with MES, pH 5.5, or LB medium with MOPS, pH 8, to log phase (A to C) or to stationary phase (D). Strains used and relevant genotypes are indicated in the figure.
As reported previously, expression of gadX is principally driven by the RpoS sigma factor (Fig. 7D, stationary phase), and it is known that CRP represses rpoS expression (Fig. 7C, exponential phase) (10, 20). Because of this, the effect of a crp mutation on GadX production was tested in an rpoS mutant background (Fig. 7B). The results indicate that CRP repression of gadX occurs primarily, but not completely, through its control over rpoS (Fig. 7B, compare EF528 and EF678). The rpoS crp mutant (EF678) still produced elevated levels of GadX compared to the rpoS mutant (EF362). The data, in sum, suggest that the majority of CRP control over gadA and gadBC occurs through its control over RpoS, which, in turn, is needed to maximally express gadX.
Acid induction of gadA and gadBC in complex media is due to the timing of RpoS production.
In complex media, GAD production is acid induced as cells enter stationary phase but occurs in equivalent amounts by late stationary phase. We asked whether pH control could be due to altered levels of RpoS rather than through GadX sensing some parameter of pH. Cells were grown to late log phase (OD600 = 1.2) in LB medium with MES, pH 5.5, and LB medium with MOPS, pH 8. Under these conditions GAD is observed in the pH 5.5 but not the pH 8 culture. Western blot analysis of RpoS revealed that the levels of this sigma factor were higher at pH 5.5 than at pH 8, suggesting that pH control of GAD production in complex media was indirect, through RpoS effects on GadX production (Fig. 8). The growth rates of the pH 5.5 and pH 8 cultures were equivalent (ca. 30 min). Thus, the accelerated production of RpoS under acidic conditions was not the result of an early entry into stationary phase but was a consequence of low pH.
FIG. 8.
Low-pH and growth phase induction of glutamate decarboxylase and RpoS in rich media. EK227 was grown to different optical densities in LB media buffered to pH 5.5 or 8. The inset illustrates the growth under both conditions measured as OD600. Extracts were probed by Western blot analysis as described in the Fig. 2 legend. (A) Detection of RpoS; (B) detection of glutamate decarboxylase (GAD).
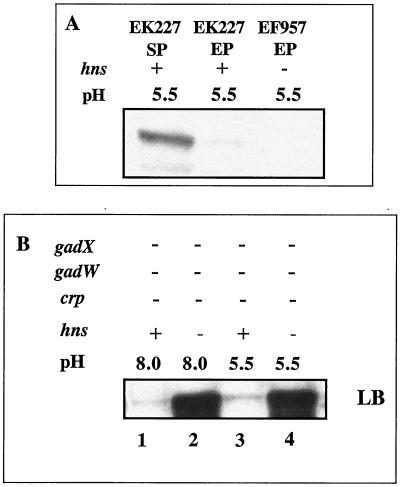
H-NS conditionally represses gadA and gadBC expression independent of GadX.
Previous reports utilizing E. coli strain W3110 convincingly showed that H-NS can also indirectly repress gadA and gadBC expression by inhibiting RpoS production (4, 20). However, we could not reproduce those results with the Stanford University wild-type K-12 strain in which CRP was the main global regulator (Fig. 9A). We did find that a crp gadX gadW mutant (EF865), which produces almost no glutamate decarboxylase when grown to exponential phase in LB medium, would make copious amounts of GAD once an hns mutation was introduced (Fig. 9B, compare lane 1 to lane 2 and lane 3 to lane 4). This confirms that H-NS will repress gadA and gadBC; however, this control did not require gadX as reported for the W3110 strain and was not even evident in a gadX+ culture (data not shown).
FIG. 9.
Effects of H-NS on gadA and gadBC expression. Cells were grown in complex, buffered LB media to exponential phase. Western blot analysis using anti-GadA and -B antibody was performed as described in the Fig. 2 legend. For panel A, all cells were gadA and gadBC+ gadX+ gadW+. For panel B, all cells lacked GadX, GadW, and CRP (hns+, lanes 1 and 3, EF865; hns mutant, lanes 2 and 4, EF929).
DISCUSSION
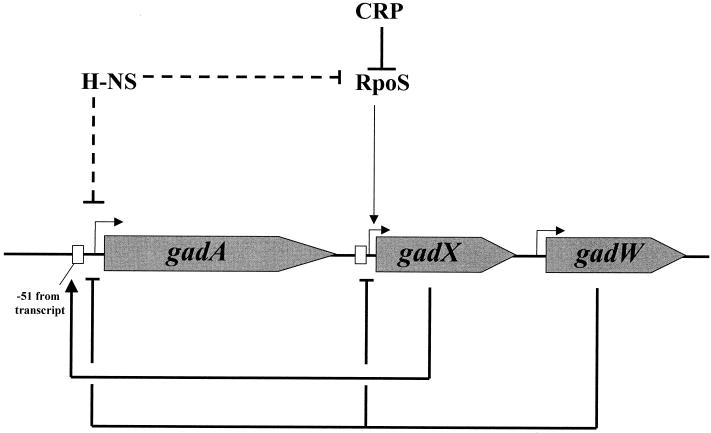
The glutamic acid decarboxylase and putative GABA:glutamate antiporter system plays a prominent role in E. coli acid resistance. While it might seem that regulation of this system should be straightforward with acid pH inducing expression and alkaline pH preventing induction, regulation of the gadA and gadBC operons is much more complex. Several reports, some seemingly contradictory, have invoked various roles for CRP and H-NS as repressors, σS and σ70 as principal sigma factors, and an AraC-like regulator, GadX, as both inducer and repressor. The data presented in this study concern another AraC-like regulatory protein, GadW, and its collaborative role with GadX in controlling this system prior to and during entry into stationary phase. The study also examines CRP-mediated control of this system. The results add more clarity to the complex model of gadA and gadBC expression and its regulation. Figure 10 summarizes the basic control circuits mapped for this system.
FIG. 10.
Model of the GadX and -W and CRP-RpoS control circuit. GadX activates expression from PgadA, while GadW inhibits expression (in the presence of GadX) from PgadA and PgadX. Transcription of the activator gene gadX is largely dependent on the RpoS sigma factor, tying expression of the system to various stress conditions that increase RpoS levels. Expression is also influenced by growth on carbohydrates through CRP, which dampens expression of RpoS, an effect more evident in LB-grown cells, where cAMP levels are high, than in cells grown on glucose, where cAMP levels are low. H-NS is also reported to inhibit gadX expression but appears capable of inhibiting gadA and gadBC expression independently of gadX control.
An earlier report describing GadX as a repressor at pH 8 but an activator at pH 5.5 can now be partly explained (18). The activator-repressor model was based on data obtained from cells grown in minimal glucose media. The repressor effect can also be seen, although less dramatically, in the LB-grown cells represented in Fig. 2A. More GadA and -B protein is seen at pH 8 in the gadX mutant (Fig. 2A, lane 3) than is found in the wild type (Fig. 2A, lane 1), originally suggesting that GadX was a repressor at pH 8. The inducer effect at pH 5.5 can be seen in Fig. 2B, where the gadX mutant (Fig. 2B, lane 4) produces less GadA and -B protein than does the wild type (Fig. 2B, lane 2), suggesting that GadX was an activator at pH 5.5. However, when GadW is missing, GadX is really only a positive regulator (Fig. 2A, lanes 5 and 6). Repression at pH 8 appears to be due to an effect of pH on GadW function (Fig. 2A and B, lanes 3 and 4).
The fact that GadX and GadW can each independently activate gadA and gadBC under certain conditions indicates that both proteins are capable of binding to the gadA and gadBC promoter regions, a conclusion supported in vitro for both GadX (18, 20) and GadW (this report). The data also shows that GadW regulates the gadX promoter (Fig. 4). The effects of GadW on gadA and gadBC expression are not solely through its impact on gadX expression, however, since GadW can affect gadA and gadBC expression even in the absence of GadX (Fig. 2A and B, compare lanes 3 and 4 with lanes 7 and 8). An interesting question is why does GadW, in the absence of GadX, activate gadA and gadBC only at pH 8? One model, discussed briefly above, is that GadW might bind gadA and gadBC promoters best when cells are grown at pH 8, which either prevents GadX from binding or attenuates GadX activation. Alternatively, GadW may not strongly bind the gadA and gadBC promoter regions if GadX is already bound to them. In this model, GadW, under ordinary circumstances, might primarily temper GadX activity by repressing gadX.
The transcriptional organization of the gadA locus involves three promoters, one upstream of gadA, one upstream of gadX, and one upstream of gadW. Transcription from PgadA can produce a cotranscript with gadX, but transcription from PgadX does not yield a cotranscript with gadW (20) (this report). However, mutations that affect expression from PgadA have a large effect on gadA message levels but almost no effect on the gadAX cotranscript (Fig. 3 and 4 for gadW effects and Fig. 4 for crp effects). This suggests either that gadA and gadX are usually transcribed separately and only occasionally form cotranscripts or that the gadAX cotranscript is subject to vigorous processing that separates the two messages.
Consistent with their annotation as AraC-like regulators, GadX and GadW will readily form homodimers in vivo (5). The finding that these proteins also appear capable of forming heterodimers may provide insight into the GadX and -W control circuit. Heterodimers could be incorporated into a model that explains GadW inhibition of GadX activity, although equally plausible alternative hypotheses involving competitive DNA binding are possible. Many AraC-like regulators bind ligands that alter their function (5), but whether or not GadX or GadW binds specific cytoplasmic ligands remains unknown.
Expression of gadA and gadBC in LB media is principally dependent on RpoS. A recent study, confirmed here, demonstrated that expression of gadX is RpoS dependent (20). In that same study, gadX was uncoupled from RpoS control by placing gadX under the control of a Tn5-lacO promoter-operator element. When gadX was expressed from this promoter, the gadA and gadBC genes were transcribed even in log-phase (pH 7.4), LB-grown cells, a condition ordinarily devoid of gadA and gadBC expression. This finding suggested that the RpoS effects on gadA and gadBC expression are indirect, occurring due to an RpoS requirement for gadX expression. The result also suggests that the gadA and gadBC promoters are themselves RpoS independent. The fact that expression of gadA and gadBC in minimal glucose media is mostly RpoS independent and GadX independent supports that idea (data not shown). However, the result also suggests that another gadA and gadBC induction pathway exists, one independent of GadX.
We have also found that CRP represses gadX expression, which explains the derepressive effect that crp mutations have on gadA and gadBC expression. CRP appears to negatively regulate gadX expression by virtue of its effects on RpoS production. The developing model for the control circuit is that, when E. coli is actively growing under conditions where cAMP levels are high (e.g., exponential growth in media like LB medium), CRP will repress gadX by inhibiting RpoS production. GadW also represses gadX under this condition. As cells approach stationary phase, RpoS levels rise, causing increased expression of gadX. GadX levels increase and overcome repression of gadA and gadBC by GadW. Cells grown fermentatively on glucose would have lower cAMP levels and less CRP-dependent repression of the system.
The pH control of this system, at least in rich, undefined media, also appears tied to the pH control of RpoS production. Evidence supporting this model includes the fact that gadA and gadBC expression in this media requires RpoS and that the levels of glutamate decarboxylase and of RpoS are induced earlier in pH 5.5 cultures entering stationary phase than in pH 8 cultures. It has previously been shown that pH influences translation of rpoS message and degradation of RpoS protein in Salmonella enterica (14, 22).
The nucleoid protein H-NS has also been identified as a repressor for this system (4, 8, 23). A recent report has demonstrated that H-NS may work to repress the gadA and gadBC genes by repressing gadX (20). However, our results, using a different genetic background, indicate that H-NS still affects gadA and gadBC expression in a gadXW mutant. Thus, H-NS must also act on gadA and gadBC independently of gadX. Another inconsistency between the two studies is that an hns mutation in our K-12 strain did not relieve CRP-dependent repression of gadA and gadBC. Thus, our results argue that CRP, not H-NS, is a master regulator of glutamate-dependent acid resistance. The reason(s) for these discrepancies is not apparent and may be strain dependent.
The question remains: why does the cell expend so much energy to regulate the gad genes? At present count there are two repressors (H-NS and CRP), one activator (GadX), one repressor-activator (GadW), and two sigma factors focused on controlling this system. This complex regulatory network must reflect the importance that E. coli places on surviving transient exposures to extreme acid stress, ensuring that the system is in place under any environmental condition that could lead to acid stress. Numerous questions remain regarding the regulatory interactions that take place between these many factors.
Acknowledgments
This work was supported by the National Institutes of Health (R01-GM61147 to J.W.F. and R01-AI48945-01 to T.C.) and the U.S. Department of Agriculture (97-35201-4751 to J.W.F.).
We are grateful to J. Kaper and S. Shin for helpful discussions and for providing GadX antibody. We also thank J. Audia for critically reading the manuscript.
REFERENCES
- 1.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 2.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 5.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 7.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 8.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 9.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σs subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 11.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Moreno, M., J. P. Audia, S. M. Bearson, C. Webb, and J. W. Foster. 2000. Regulation of sigma S degradation in Salmonella enterica var typhimurium: in vivo interactions between sigma S, the response regulator MviA(RssB) and ClpX. J. Mol. Microbiol. Biotechnol. 2:245-254. [PubMed] [Google Scholar]
- 15.Porter, M. E., and C. J. Dorman. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salles, B., and G. M. Weinstock. 1989. Interaction of the CRP-cAMP complex with the cea regulatory region. Mol. Gen. Genet. 215:537-542. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 19.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 22.Webb, C., M. Moreno, M. Wilmes-Riesenberg, R. Curtiss III, and J. W. Foster. 1999. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 34:112-123. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida, T., T. Yamashino, C. Ueguchi, and T. Mizuno. 1993. Expression of the Escherichia coli dimorphic glutamic acid decarboxylases is regulated by the nucleoid protein H-NS. Biosci. Biotechnol. Biochem. 57:1568-1569. [DOI] [PubMed] [Google Scholar]