Abstract
Plant cell wall degradation by Clostridium cellulovorans requires the cooperative activity of its cellulases and hemicellulases. To characterize the α-l-arabinosidases that are involved in hemicellulose degradation, we screened the C. cellulovorans genomic library for clones with α-l-arabinofuranosidase or α-l-arabinopyranosidase activity, and two clones utilizing different substrates were isolated. The genes from the two clones, arfA and bgaA, encoded proteins of 493 and 659 amino acids with molecular weights of 55,731 and 76,414, respectively, and were located on neighboring loci. The amino acid sequences for ArfA and BgaA were related to α-l-arabinofuranosidase and β-galactosidase, respectively, which are classified as family 51 and family 42 glycosyl hydrolases, respectively. Recombinant ArfA (rArfA) had high activity for p-nitrophenyl α-l-arabinofuranoside, arabinoxylan, and arabinan but not for p-nitrophenyl α-l-arabinopyranoside. On the other hand, recombinant BgaA (rBgaA) hydrolyzed not only p-nitrophenyl α-l-arabinopyranoside but also p-nitrophenyl β-d-galactopyranoside. However, when the affinities of rBgaA for p-nitrophenyl α-l-arabinopyranoside and p-nitrophenyl β-d-galactopyranoside were compared, the Km values were 1.51 and 6.06 mM, respectively, suggesting that BgaA possessed higher affinity for α-l-arabinopyranose residues than for β-d-galactopyranoside residues and possessed a novel enzymatic property for a family 42 β-galactosidase. Activity staining analyses revealed that ArfA and BgaA were located exclusively in the noncellulosomal fraction. When rArfA and rBgaA were incubated with β-1,4-xylanase A (XynA), a cellulosomal enzyme from C. cellulovorans, on plant cell wall polymers, the plant cell wall-degrading activity was synergistically increased compared with that observed with XynA alone. These results indicate that, to obtain effective plant cell wall degradation, there is synergy between noncellulosomal and cellulosomal subunits.
Clostridium cellulovorans ATCC 35296 (25), an anaerobic, mesophilic, and spore-forming bacterium, produces a large extracellular polysaccharolytic multicomponent complex called the cellulosome, in which several cellulases are tightly bound to a scaffolding protein called CbpA (7). C. cellulovorans utilizes not only cellulose but also hemicelluloses, such as xylan, pectin, and several other carbon sources, for growth (7, 15, 20). Our laboratory has characterized the gene for the scaffolding protein CbpA as well as several genes necessary for the degradation of crystalline cellulose and plant cell wall polysaccharides (7, 15, 20, 26, 27).
Plant cell walls consist of a composite structure containing a complex mixture of polysaccharides such as cellulose, hemicellulose (xylan and galactomannan), pectic substances (galacturonan and arabinogalactan), and other polysaccharides (e.g., type II arabinogalactan and fucoxyloglucan) (1). l-Arabinose is a common component of several of these polysaccharides, and the arabinose residues are widely distributed in plant cell walls, where they are present in significant amounts as arabinoxylan, arabinan, and arabinogalactan (1, 29). The arabinan backbone consists of α-1,5-linked arabinofuranosyl units, and arabinoxylan consists of a β-1,4-xylopyranosyl backbone to which α-1,3-linked arabinofuranosyl units are attached (1, 2, 29). In addition, arabinogalactans are often linked covalently to protein or pectic substances in plant cell walls and are subdivided into two structurally distinct types. Type I arabinogalactans are characterized by essentially linear 1-3- or 1-4-linked chains of β-d-galactopyranose residues with 1-3- or 1-5-linked arabinofuranose side chains. Type II arabinogalactans contain highly branched structures in which β-d-galactopyranose residues are mutually joined to α-l-arabinofuranose or α-l-arabinopyranose side chains by 1-3 and 1-6 linkages (1). Complete and rapid hydrolysis of these polysaccharides requires not only β-1,4-glycosidic chain-cleaving enzymes such as β-1,4-xylanase but also the cooperation of side chain-cleaving enzymes such as α-l-arabinofuranosidase (1, 29).
α-l-Arabinofuranosidases (EC 3.2.1.55) catalyze the hydrolysis of nonreducing terminal α-l-arabinofuranosidic linkages in arabinoxylan, l-arabinan, and other l-arabinose-containing polysaccharides. These enzymes work in concert with other hemicellulases to completely degrade the hemicellulose backbone. In fact, α-l-arabinofuranosidase is applied as a tool in basic research for the elucidation of hemicellulose structure and in the industrial bioconversion of plant cell wall materials (9, 24, 29). The α-l-arabinofuranosidases from several bacteria have been characterized (2, 3, 5, 9, 14, 18, 32) and classified into three glycoside hydrolase families (GH43, GH51, and GH62) based on primary-structure similarities and hydrophobic cluster analysis (P. M. Coutinho and B. Henrissat, online data [http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html]).
To understand the hemicellulose degradation system of C. cellulovorans, we characterized two noncellulosomal α-arabinosidase genes. These noncellulosomal subunits were able to act synergistically with XynA, a key component of the C. cellulovorans cellulosome, for plant cell wall degradation.
MATERIALS AND METHODS
Bacterial strains, culture media, and plasmids.
Cultures of C. cellulovorans ATCC 35296 were grown anaerobically at 37°C in round-bottom flasks, which included 0.5% (wt/vol) birchwood xylan (Sigma) (15). Escherichia coli XL1-Blue and plasmids pBluescript SK(−) (Stratagene) and pCR2.1 (TA cloning kit; Invitrogen) served as the cloning host and vectors, respectively. Plasmid pET29b and host E. coli BL21(DE3) (Novagen) were used for production of the recombinant ArfA (rArfA) and BgaA (rBgaA) enzymes. E. coli cells were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) at 37°C.
Cloning and DNA sequencing for arfA and bgaA.
To clone genes possessing activity for α-l-arabinofuranosidase and α-l-arabinopyranosidase, the λZAPII C. cellulovorans genomic library (26) (Stratagene) was screened by overlaying with 0.7% soft agar containing 1 mM 4-methylumbelliferyl-α-l-arabinofuranoside (4-MUAf) (Sigma) or 4-methylumbelliferyl-α-l-arabinopyranoside (4-MUAp) (Sigma). The positive plaques were isolated by detection, under UV light, of fluorescent haloes surrounding recombinant plaques. To isolate full-length genes encoding each activity, phage DNA was isolated from several positive plaques by use of a Lambda mini kit (Qiagen). PCR was performed using reverse and M13-20 universal primers to amplify fragments encoding each gene, and each amplified fragment was then subcloned to pCR2.1. Positive clones were further screened for hydrolysis activity for 4-MUAf or 4-MUAp, and several colonies with activity were isolated.
DNA sequence was determined from double-stranded plasmid DNA by the dideoxy chain termination method. Sequence data were analyzed and compared using the BLAST program.
Alignment and construction of phylogenetic tree.
Amino acid sequences for several bacterial glycosyl hydrolases belonging to family 51 and family 42 were retrieved from protein databases (according to sequence accession numbers) and aligned by using CLUSTAL W (28). The phylogenetic trees were constructed by the neighbor-joining method of the Phylip program (21). Bootstrap values were calculated based on 100 replicates.
Construction of pEARF29 and pEBGA29.
Primers 5′-CTA GTC TAG ACA TTG GCT AGC TGC T-3′ (sense) and 5′-CCG CTC GAG TAG GCT TAA TTT AAG G-3′ (antisense), containing artificial XbaI and XhoI recognition sites (underlined), respectively, were used to amplify arfA fragments by PCR, and primers 5′-TAC CTA GGT GTA ATA GAT AAG GAG GA-3′ (sense) and 5′-GCC CTC GAG TTC AAC TAA TTG TAA AAC-3′ (antisense), containing artificial StyI and XhoI recognition sites, respectively, were used to amplify bgaA fragments by PCR. Each of the amplified fragments was digested with the cognate restriction enzymes and inserted between the XbaI and XhoI sites of pET29b to generate pEARF29 and pEBGA29. This plasmid provided a six-histidine tag on the C terminus.
Preparation of extracellular materials from C. cellulovorans grown on xylan medium.
Extracellular materials (cellulosomal and noncellulosomal fractions) were prepared from supernatants of cultures grown on xylan and purified according to their size and interaction with cellulose as described previously (15).
N-terminal sequencing.
The noncellulosomal fraction (10 ml) from C. cellulovorans was ultrafiltered with Ultrafree Biomax-100 membranes (Millipore) and concentrated to a volume of 1 ml. The concentrated solution was applied to a Superose 12 HR 10/30 column (Amersham Pharmacia Biotech AB) equilibrated with 50 mM sodium phosphate buffer with 150 mM NaCl (pH 7.0) by using the AKTAFPLC system (Amersham Pharmacia Biotech AB). The active fractions of α-l-arabinofuranosidase for ArfA or α-l-arabinopyranosidase for BgaA were collected and concentrated (Ultrafree Biomax-30). For N-terminal amino acid sequencing, the concentrated enzymes were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride membrane by electroblotting (Bio-Rad). The protein band was determined by staining with 0.1% Ponceau red and then cutting out the band and sequencing it by the Edman method with a model 477 protein sequencer (Applied Biosystems).
Purification of rArfA and rBgaA.
rArfA and rBgaA were purified from E. coli BL21(DE3) strains harboring pEARF29 and pEBGA29, respectively. When the culture had reached an optical density at 600 nm of 0.5 at 30°C, isopropyl-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cells were further cultivated at 30°C for 4 h. The cells were collected by centrifugation, suspended in buffer 1 (50 mM phosphate, 300 mM NaCl, 10 mM imidazole [pH 8.0]), and disrupted by sonication. The enzymes were purified by nickel affinity column chromatography (Ni-nitrilotriacetic acid agarose resins; Qiagen). Purified rArfA and rBgaA were concentrated to 1.5 to 2.0 mg/ml by ultrafiltration (Ultrafree Biomax-30; Millipore).
Construction and expression of rXynA.
Recently, we have cloned and characterized endo-1,4-β-xylanase (XynA), which belongs to glycosyl hydrolase family 11 and which is a component of C. cellulovorans cellulosomes (15, 20) (GenBank accession no. AF435978). Two primers (5′-CGAATTCGGCAACAAAAACGATCACC-3′ and 5′-CCGCTCGAGGAATGCACCATTTAACATTGT-3′), containing artificial EcoRI and XhoI recognition sites (underlined), respectively, were used to amplify full-length xynA by PCR. The amplified fragment was digested with EcoRI and XhoI and inserted into pET29b to generate pEXYNA29. This plasmid provides an S-protein-tagged and a six-histidine-tagged XynA on its N and C termini, respectively. Recombinant XynA (rXynA) was purified from E. coli BL21(DE3) harboring pEXYNA29 by using the same procedures employed for rArfA and rBgaA. To eliminate the S-protein tag sequence at the N terminus, the proteins were treated with an S-Tag thrombin purification kit (Novagen) according to the manufacturer's instructions. The eluted proteins were concentrated to 1.5 to 2.0 mg/ml by ultrafiltration (Ultrafree Biomax-30; Millipore).
Enzyme assays.
Glycosidase activity was determined based on the hydrolysis of β-nitrophenyl glycoside substrates (16). The nitrophenyl substrates used were p-nitrophenyl-α-l-arabinofuranoside (pNPAf), p-nitrophenyl-α-l-arabinopyranoside (pNPAp), p-nitrophenyl-β-d-galactopyranoside (pNPGp), o-nitrophenyl-β-d-galactopyranoside (oNPGp), p-nitrophenyl-β-d-xylopyranoside (pNPXp), p-nitrophenyl-β-d-glucopyranoside (pNPGLp), p-nitrophenyl-β-d-cellobiopyranoside (pNPCp), p-nitrophenyl-β-lactopyranoside (pNPLp), p-nitrophenyl-β-d-fucopyranoside (pNPFp), and p-nitrophenyl-α-galactopyranoside (pNPαGp) (all purchased from Sigma). pNPAf, pNPAp, and pNPGp were used routinely for α-l-arabinofuranosidase, α-l-arabinopyranosidase, and β-galactosidase assays, respectively. The glycosidase activities were measured in the presence of 1 mM concentrations of nitrophenyl substrates at 37°C in 50 mM phosphate buffer (pH 6.0) or with Britton and Robinson's universal buffer (50 mM phosphoric acid-50 mM boric acid-50 mM acetic acid; the pH was adjusted to 2 to 9 with NaOH) for 30 min, and the reaction was then terminated by addition of Na2CO3. The liberated nitrophenyl product from p- or o-nitrophenyl glycosides was determined at an absorbance at 410 nm. One unit of enzyme activity corresponds to the release of 1 μmol of nitrophenyl equivalent. Arabinoxylan (oat-spelt xylan) (Sigma), arabinan (sugar beet) (Megazyme), arabinogalactan (larch wood) (Sigma), and carboxymethylcellulose (medium viscosity; Sigma) were used as polysaccharide substrates. Corn fiber gum provided by D. B. Johnston of the U.S. Department of Agriculture (8) and corn stem powder (20) were also used as plant cell wall substrates. Enzyme activity was assayed in the presence of a 0.5% (wt/vol) concentration of each polysaccharide at 37°C for 30 min in 50 mM phosphate buffer (pH 6.0). The activity of the enzyme for polysaccharides, including plant cell walls, was determined by measuring the release of reducing sugars with the Somogyi-Nelson method (31), with l-arabinose as the standard. One unit of activity was defined as the amount of enzyme which produced 1 μmol of reducing sugar per min. Protein concentrations were determined by using a BAC protein assay kit (Pierce) with bovine serum albumin as a standard.
Activity staining.
SDS-PAGE was performed by the method of Laemmli (17). The activity staining was performed using SDS-PAGE with 10% (wt/vol) nondenaturing polyacrylamide gel. After electrophoresis, the gel was incubated for 30 min with 50 mM phosphate buffer (pH 6.0) containing 25% isopropanol at 4°C and washed twice with the same buffer without isopropanol. After being washed, the gel was incubated for 60 min at 37°C in 50 mM phosphate buffer (pH 6.0) containing 1 mM 4-MUAf or 4-MUAp. Positive bands were detected by fluorescence under UV illumination.
Determination of molecular mass.
The molecular masses of native and recombinant proteins were determined by gel filtration chromatography with a Superose 12 HR 10/30 column equilibrated with 50 mM sodium phosphate buffer with 150 mM NaCl (pH 7.0) by use of the AKTAFPLC system. The column was calibrated with an HMW gel filtration calibration kit (Amersham Pharmacia Biotech AB) composed of the proteins ferritin (molecular weight, 440,000), catalase (molecular weight, 232,000), and aldolase (molecular weight, 158,000) and with bovine serum albumin (molecular weight, 67,000) (Sigma).
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to GenBank under accession no. AY128945.
RESULTS
Cloning and nucleotide sequencing for two glycosyl hydrolase genes, arfA and bgaA.
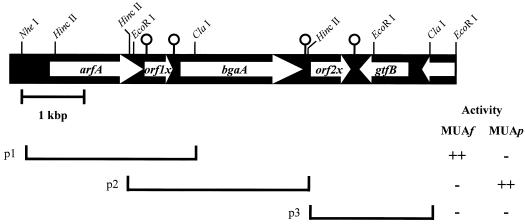
To identify α-l-arabinosidase genes in C. cellulovorans, we screened the C. cellulovorans λZAPII library by using two distinct substrates, 4-MUAf and 4-MUAp. Several positive clones coding for α-l-arabinofuranosidase and α-l-arabinopyranosidase were isolated. To obtain full-length DNA fragments coding for activity, we performed PCR with universal primers for each purified phage DNA. The amplified 2.5- and 4.6-kb fragments from each positive clone were subcloned to pCR2.1. To identify the region coding for α-l-arabinofuranosidase or α-l-arabinopyranosidase, several subclones were prepared. Figure 1 shows the physical maps of the region of the C. cellulovorans chromosome coding for the activities. As a result, the regions coding for α-l-arabinofuranosidase and α-l-arabinopyranosidase were located on a 2.7-kb fragment between the NheI and ClaI sites and on a 2.6-kb fragment between the EcoRI and HincII sites, respectively, in the amplified fragments (Fig. 1). Sequence analysis revealed that the genes coding for the α-l-arabinofuranosidase and α-l-arabinopyranosidase activities were located on neighboring loci with a hypothetical open reading frame (orf1x) between them (Fig. 1).
FIG. 1.
Restriction enzyme map of 6.9-kb fragment containing arfA and bgaA and structure of subclones. Clones coding (++) or not coding (−) for α-l-arabinofuranosidase and α-l-arabinopyranosidase activities are indicated. The activities were determined by using MUAf and MUAp as the substrates. The pin-like symbols indicate predicted palindromes. p1, 2.7-kb fragment between the NheI and ClaI sites; p2, 2.6-kb fragment between the EcoRI and HincII sites; p3, 2.0-kb fragment between the HincII and ClaI sites.
The arfA gene consists of 1,476 nucleotides encoding a protein of 492 amino acids with a predicted molecular weight of 55,731. The assigned ATG initiation codon was preceded at a spacing of 8 bp by a potential ribosomal binding sequence, GAAGGGG, which was homologous to the consensus Shine-Dalgarno sequence (10). On the other hand, bgaA, encoding α-l-arabinopyranosidase activity, was 233 bp downstream of orf1x. The open reading frame of bgaA consists of 1,977 nucleotides encoding a protein of 659 amino acids with a predicted molecular weight of 76,464. The assigned ATG initiation codon was preceded at a spacing of 6 bp by a Shine-Dalgarno sequence (AGGAGG). Two possible transcription terminators that correspond to mRNA hairpin loops were found downstream of the TAA termination codon of orf1x and bgaA (Fig. 1).
Amino acid sequences of ArfA and BgaA.
Comparison of the amino acid sequence of ArfA with sequences registered in protein databases such as SWISS-PROT revealed that mature ArfA is highly homologous to several bacterial α-l-arabinofuranosidases classified into family 51, e.g., 63% identity with AbjA of Thermobacillus xylanilyticus (GenBank accession no. CAA76421) (5), 64% identity with BH1874 of Bacillus halodurans (BAB05593), and 61% identity with ArfB of Clostridium stercorarium (AAC28125) (32).
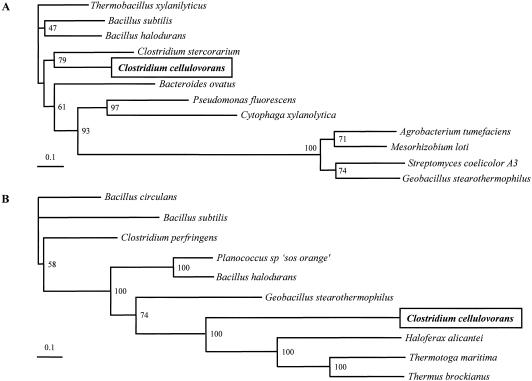
The deduced amino acid sequence of BgaA exhibited homology with bacterial β-galactosidases (EC 3.2.1.23) belonging to family 42 glycosyl hydrolases, e.g., 33% identity with BgaB of Geobacillus stearothermophilus (GenBank accession no. P19668), 33% identity with TM1195 of Thermotoga maritima (NP_229000), 31% identity with a β-galactosidase of Planococcus sp. (AAF75984) (23), and 29% identity with a β-galactosidase of Clostridium perfringens (BAA08485). To determine the evolutionary relationship between the glycosyl hydrolase families of these two proteins, we performed a phylogenetic analysis using several amino acid sequences from each family (Fig. 2). This analysis showed that ArfA of C. cellulovorans was most closely related to ArfB of Clostridium stercorarium (Fig. 2A). In addition, it appears that BgaA of C. cellulovorans is located on a deep-branched cluster and may form a separate family of β-galactosidases (Fig. 2B). The analysis also showed that C. cellulovorans BgaA was most closely related to corresponding enzymes from Thermotoga maritima, Thermus brockianus, and Haloferax alicantei (12), which are classified as halophilic archaea.
FIG. 2.
Phylogenetic tree of ArfA (A) and BgaA (B) for glycosyl family 51 and family 42, respectively. Phylogenetic trees were constructed using CLUSTAL W with the neighbor-joining method. Results from a bootstrap analysis (n = 100) are shown for the junctions. The following organisms (with GenBank accession numbers) were used as sequence sources for family 51: Thermobacillus xylanilyticus (CAA76421), Bacillus subtilis (P94552), Bacillus halodurans (BAB05593), Clostridium stercorarium (AAC28125), Bacteroides ovatus (Q59219), Pseudomonas fluorescens subsp. (AAK84947), Cytophaga xylanolytica (AAC38456), Agrobacterium tumefaciens (AAL43920), Mesorhizobium loti (BAB50453), Streptomyces coelicolor A3 (CAB86096), and Geobacillus stearothermophilus (AF159625). The following organisms were used as sequence sources for family 42: Bacillus circulans (AAA22260), Bacillus subtilis (CAB15418), Clostridium perfringens (BAA08485), Planococcus sp. ‘SOS Orange’ (AAF75984), Bacillus halodurans (BAB07420), Geobacillus stearothermophilus (P19668), Haloferax alicantei (U70664.2), Thermotoga maritima (NP_229000), and Thermus brockianus (AAD33667).
A dockerin sequence, which plays a role in cellulosome assembly by binding the various catalytic subunits to the noncatalytic scaffolding protein (CbpA), was not found in ArfA or in BgaA. Therefore, these observations indicate that ArfA and BgaA act as free subunits in the C. cellulovorans culture medium. These results are in good agreement with our previous observation that α-l-arabinosidases are found exclusively in the noncellulosomal fraction (15).
Purification and characterization of rArfA and rBgaA.
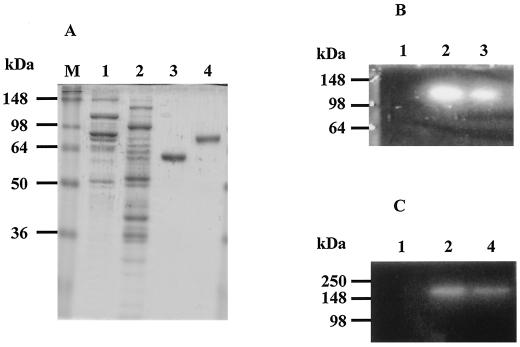
rArfA and rBgaA were purified 82- and 75-fold, respectively, from E. coli BL21 harboring pEARF29 or pEBGA29. The final preparations gave single bands on SDS-PAGE, and the molecular weights of the recombinant enzymes were estimated to be 59,000 for rArfA and 78,000 for rBgaA (Fig. 3A). This is in good agreement with the deduced molecular weights of 55,731 for ArfA and 76,464 for BgaA.
FIG. 3.
Distribution of ArfA and BgaA in the noncellulosomal fraction of C. cellulovorans. The SDS-PAGE gel was stained with Coomassie brilliant blue (A) or examined for α-l-arabinofuranosidase activity using MUAf (B) or for α-l-arabinopyranosidase activity using MUAp (C) by detection of fluorescence under UV illumination. Lane M, protein mass standard; lane 1, cellulosomal fraction from C. cellulovorans xylan-grown culture (10 or 20 μg) (15); lane 2, noncellulosomal fraction from C. cellulovorans xylan-grown culture (10 or 20 μg) (15); lane 3, purified rArfA from E. coli BL21 harboring pEARF29 (3 or 6 μg); lane 4, purified rBgaA from E. coli BL21 harboring pEBGA29 (3 or 6 μg).
Table 1 shows the substrate specificities of rArfA and rBgaA. The purified rArfA had a high specific activity for pNPAf, while activity for pNPAp and other p-nitrophenyl substrates was not detected (Table 1). In addition, hydrolyzing activity for polysaccharides such as arabinan and arabinoxylan could be detected, whereas there was less activity for arabinogalactan. The optimum pH for the activity of rArfA was 6.0 when the enzyme activity was assayed after 10 min of incubation at 37°C in Britton and Robinson's universal buffer at various pHs. rArfA was more stable at weak alkaline pHs (pH range of 8.0 to 9.0) than at neutral pH when incubated at 30°C for 12 h without a substrate. The optimum temperature for rArfA activity was found to be 40 to 50°C when the enzyme was incubated for 10 min at pH 6.0. The enzyme activity was completely lost after heating at 80°C for 20 min. The effect of different metals on the activity was determined by adding the appropriate salts (1 to 5 mM) to reaction mixtures. No effect on activity was detected after the addition of divalent cations Ca2+, Mg2+, and Mn2+; however, Cu2+, Hg2+, and Zn2+ inhibited activity. The addition of excess EDTA (10 mM) did not affect the activity, suggesting that cation cofactors were not required for the enzymatic reaction (30).
TABLE 1.
Substrate specificities of rArfA and rBgaA
| Substratea | Sp act (U/mg of protein)b
|
|
|---|---|---|
| rArfA | rBgaA | |
| pNP-α-l-arabinofuranoside | 19.05 | ND |
| pNP-α-l-arabinopyranoside | ND | 5.32 |
| pNP-β-d-xylopyranoside | ND | ND |
| pNP-β-d-glucopyranoside | ND | ND |
| pNP-β-d-cellobiopyranoside | ND | ND |
| pNP-β-d-galactopyranoside | ND | 0.51 |
| oNP-β-d-galactopyranoside | ND | <0.001 |
| pNP-β-d-fucopyranoside | ND | 0.65 |
| pNP-β-lactopyranoside | ND | <0.001 |
| pNP-α-galactopyranoside | ND | ND |
| Arabinoxylan (oat-spelt xylan) | 0.59 | <0.01 |
| Xylan (birchwood) | 0.03 | <0.01 |
| Arabinan (sugar beet) | 1.89 | <0.002 |
| Arabinogalactan (larch wood) | <0.01 | 0.07 |
| Carboxymethylcellulose | <0.001 | <0.001 |
pNP, p-nitrophenyl; oNP, o-nitrophenyl.
Enzyme activity was measured after incubation in 50 mM phosphate buffer (pH 6.0) with each substrate at 37°C for 30 min. ND, not detected.
Purified rBgaA showed hydrolysis activity against pNPAp, pNPGp, and pNPFp; however, it could not hydrolyze pNPAf, oNPGp, pNPXp, pNPGLp, pNPCp, pNPLp, and pNαGp (Table 1). In particular, rBgaA showed higher activity for pNPAp than for pNPGp. It is known that the majority of bacterial β-galactosidases belonging to family 42 has high specific activity for oNPGp (4, 11, 12, 13, 19, 23); however, rBgaA is specific for pNPGp and not for oNPGp (Table 1). These results strongly suggest that BgaA possesses novel enzymatic characteristics for a family 42 β-galactosidase. When rBgaA was incubated with polysaccharides containing nonreducing arabinofuranosyl side groups, such as arabinan and arabinoxylan, it had no detectable activity (Table 1). However, the hydrolysis activity of rBgaA for arabinogalactan could be detected, suggesting that the release of reducing sugar could be due to the effect of its β-galactosidase activity, rather than its α-l-arabinopyranosidase activity, on the β-d-galactopyranoside residues. In addition to substrate specificity, when the hydrolysis activities of rBgaA for pNPAp and pNPGp were measured, the optimum pH for both activities was found to be 6.0 and the enzyme was stable in the range of pH 6.0 to 8.0. The optimum temperature for both activities was shown to be 30 to 40°C when the enzyme was incubated for 10 min at pH 6.0; however, both enzyme activities were completely lost after heating at 50°C for 20 min. These properties of the α-l-arabinopyranosidase and β-galactosidase activities of rBgaA were similar to each other. As far as the effects of different metals are concerned, the activity pattern was similar to that observed for rArfA, i.e., no effect on either activity was detected with Ca2+, Mg2+, and Mn2+ and excess EDTA (10 mM); however, the activities were inhibited by Cu2+, Hg2+, and Zn2+ (1 to 5 mM).
To compare the substrate affinities of rBgaA for pNPAp and pNPGp, we measured the Km and Vmax values for each substrate (Table 2). The Km and Vmax values for rBgaA were 1.51 mM and 10.4 U/mg of protein, respectively, when pNPAp was used as the substrate, whereas the values when pNPGp was used were 6.06 mM and 2.5 U/mg of protein, respectively. In addition, the Km and Vmax values of rArfA were 0.71 mM and 75.8 U/mg of protein, respectively, when pNPAf was used as the substrate. These results revealed that BgaA possesses higher affinity and activity for l-arabinopyranose residues than for d-galactopyranose residues.
TABLE 2.
Kinetic parameters of rArfA and rBgaA for p-nitrophenyl substratesa
| Protein |
Km (mM)
|
Vmax (U/mg of protein)
|
||||
|---|---|---|---|---|---|---|
| pNPAf | pNPAp | pNPGp | pNPAf | pNPAp | pNPGp | |
| rArfA | 0.71 ± 0.01 | NT | NT | 75.8 ± 0.02 | NT | NT |
| rBgaA | NT | 1.51 ± 0.03 | 6.06 ± 0.01 | NT | 10.4 ± 0.01 | 2.5 ± 0.01 |
Results are means of three determinations ± standard deviations. NT, not tested.
Distribution of ArfA and BgaA in C. cellulovorans.
To elucidate the distribution of ArfA and BgaA, we carried out activity staining against cellulosomal and noncellulosomal fractions (Fig. 3B). The activities of native α-l-arabinofuranosidase and α-l-arabinopyranosidase could be detected exclusively in the noncellulosomal fraction. The activities of native ArfA and BgaA were similar to those of purified rArfA and rBgaA (Fig. 3B). In addition, we partially purified native ArfA (125-fold) and BgaA (82-fold) to confirm their N-terminal sequences in the noncellulosomal fraction. The N-terminal sequences of native ArfA and BgaA were NKLTINAQNK and RIGV, respectively, and are in good agreement with the deduced amino acid sequences. These results, including the absence of dockerins, indicate that ArfA and BgaA act as free subunits and are not associated with cellulosomes. On the other hand, the molecular masses of the active forms of purified rArfA and rBgaA were estimated to be approximately 138 and 170kDa, respectively, by gel filtration analysis. These results are in good agreement with the data for native forms of partially purified enzymes from the C. cellulovorans xylan-grown cultures, which suggest that the active forms are dimeric structures. It is known that several family 51 α-l-arabinofuranosidases and family 42 β-galactosidases of bacterial origin consist of dimeric, tetrameric, or octameric structures (3, 4, 5, 9, 13, 19, 23).
Synergistic effects between a cellulosomal subunit and noncellulosomal subunits during plant cell wall degradation.
Several synergistic effects between glycosyl hydrolases for several polysaccharides have been reported (6, 22). To determine whether or not there is a synergistic relationship between noncellulosomal and cellulosomal subunits, we measured degradation activity by using rArfA and rBgaA as noncellulosomal subunits and rXynA as the cellulosomal subunit. When rArfA and rXynA were incubated with arabinoxylan, corn fiber gum, and corn stem powder, the amount of reducing sugar released was significantly increased compared with that released with rArfA or rXynA alone, and a synergistic effect by rArfA was observed (Table 3). On the other hand, although the synergistic effects by rBgaA and rXynA could not be detected for the plant cell wall polymers, except for arabinogalactan, increases of synergistic coefficient values were observed for those substrates when rBgaA was added to reaction mixtures containing rArfA and rXynA. Of special interest is the increase of degradation activity observed with rBgaA, rArfA, and rXynA when corn fiber gum and corn stem powder were the substrates (Table 3). These results suggest that the combination of rBgaA, rArfA, and rXynA may be particularly effective for degradation of plant cell wall polymers containing galactose residues. These results also indicate that noncellulosomal subunits such as ArfA and BgaA cooperate with cellulosomal subunits to produce efficient degradation of plant cell walls.
TABLE 3.
Synergistic effects between noncellulosomal subunits and cellulosomal subunit XynA in plant cell wall degradation
| Substrate | Degradation activity (μg/ml)a
|
Synergism coefficientd
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| rArfA | rBgaA | rXynAb | rArfA and rXynA | rBgaA and rXynA | rArfA, rBgaA, and rXynA | A + X | B + X | A + B + X | |
| Arabinoxylan | 77.2 | 1.4 | 267.7 | 607.2 | 208.7 | 595.3 | 1.8 | 0.8 | 1.7 |
| Arabinogalactan | 1.5 | 8.9 | 7.0 | 7.4 | 25.7 | 20.2 | 0.9 | 1.6 | 1.2 |
| Corn fiber gum | 47.5 | 3.0 | 75.7 | 247.9 | 95.0 | 285.0 | 2.0 | 1.2 | 2.3 |
| Corn stem powderc | 8.9 | 1.5 | 20.1 | 78.7 | 26.7 | 89.1 | 2.7 | 1.2 | 2.9 |
The degradation activity is expressed as the amount of reducing sugar released per milliliter of reaction mixture. The reaction mixture was composed of 0.5% substrate (wt/vol), 50 mM phosphate buffer (pH 6.0), and a 0.2 μM concentration of each enzyme. The reaction was performed under gentle shaking conditions at 37°C for 15 h.
rXynA from C. cellulovorans endo-1,4-β-xylanase A was cloned previously (GenBank accession no. AF435978).
Corn stem powder was prepared as described previously (20).
The synergism coefficient indicates the ratio of measured to theoretical activity. Symbols: A + X, rArfA plus rXynA; B + X, rBgaA plus rXynA; A + B + X, rArfA, rBgaA, and rXynA. Results are means of three determinations.
DISCUSSION
Our recent work has suggested that C. cellulovorans might possess α-arabinosidases having activity for pNPAp as a substrate (15). To gain a better understanding of the hemicellulose degradation system of C. cellulovorans, we characterized two distinct genes, arfA and bgaA, coding for α-l-arabinofuranosidase and α-l-arabinopyranosidase, respectively.
ArfA of C. cellulovorans was highly homologous and similar to family 51 glycosyl hydrolases from several bacterial origins. The deduced amino acid sequence of BgaA indicated that it was a member of glycohydrolase family 42 and was homologous to β-galactosidases from several bacteria. However, the substrate specificity and affinity of BgaA were quite different from those of family 42 β-galactosidases from Haloferax alicantei and other bacteria. We do not know whether family 42 β-galactosidases from other bacteria involve α-l-arabinopyranosidase activity, since there is little information about α-l-arabinopyranosidases or the specificity of family 42 β-galactosidases for a wide range of substrates. It appears that C. cellulovorans BgaA possesses novel enzymatic characteristics for a family 42 enzyme that might have some potential for industrial applications.
Recently, our group reported that pectin-grown culture supernatants of C. cellulovorans can release protoplasts from cultured tobacco cells (27). We have also determined that several components of the cellulosome from pectin-grown culture supernatants show plant cell wall-degrading ability (20). Thus, our reports strongly indicate that C. cellulovorans possesses an effective ability to degrade plant cell walls. We also detected high activities of α-l-arabinofuranosidase and α-l-arabinopyranosidase corresponding to ArfA and BgaA in C. cellulovorans pectin-grown culture supernatants (data not shown). These observations suggest that ArfA and BgaA are components of the plant cell wall-degrading system of C. cellulovorans. In plant cell walls, α-l-arabinofuranosyl residues are usually present in noncellulosic polysaccharides such as xylan and type I or II arabinogalactan (1). We observed an increase in the amount of reducing sugar released by the addition of rBgaA to rArfA and rXynA in reaction mixtures containing corn fiber gum and corn stem powder. The effect of the addition of rBgaA was influenced particularly by substrates containing galactose residues, such as arabinogalactan and corn fiber gum (1, 8). Therefore, these results suggest that BgaA might function mainly as a β-galactosidase rather than an α-l-arabinopyranosidase for these substrates. Similar synergistic effects for plant cell wall degradation by β-galactosidase have been reported previously (6). On the other hand, rBgaA has activities not only for pNPAp and pNPGp but also for pNPFp (Table 1). The l-fucopyranose residues in plant cell walls have been found mainly in xyloglucan, which contains a cellulosic β-d-glucan backbone to which a single-unit α-d-xylopyranose residue is attached as a side-chain with l-fucopyranose or d-galactopyranose (1). To gain effective degradation of complex plant cell wall polymers, several glycosidases having various substrate specificities play important roles in exposing the basic components of cell walls such as cellulose and xylan and are needed to cooperate with cellulases and xylanases (1, 6, 20, 27). Thus, the α-l-arabinopyranosidase and fucopyranosidase activities of BgaA might also function to expose new hydrolysis sites for XynA, and these enzymes might play key roles in complex hemicellulosic polysaccharide degradation.
ArfA and BgaA were found exclusively in the noncellulosomal fraction (Fig. 3). So far, it has not been reported in cellulolytic clostridia and other cellulosome-producing bacteria such as Acetivibrio cellulolyticus, Bacteroides cellulosolvens, and Ruminococcus flavefaciens that side chain-degrading enzymes such as α-l-arabinofuranosidase and α-d-glucuronidase possess a dockerin domain. These observations suggest that the debranching enzymes appear either as cell-associated or free extracellular forms and might be necessary for effective degradation of highly branched hemicelluloses.
It appears that C. cellulovorans ArfA and BgaA are regulated by growth conditions. The activities of ArfA and BgaA could not be observed when C. cellulovorans was grown in medium with cellulose as a carbon source (data not shown). Recently, we have reported that C. cellulovorans was able to regulate the composition of cellulosomal and noncellulosomal subunits when grown on different carbon sources (15, 20, 27). These observations suggest that C. cellulovorans possesses a regulatory system for the degradation enzymes when extracellular substrates change from cellulose to hemicelluloses. These responses are very significant, and further studies are required to understand the conditions and mechanisms that regulate the expression and activity of hemicellulases in C. cellulovorans.
Acknowledgments
We thank David B. Johnston of the U.S. Department of Agriculture for the gift of corn fiber gum. We are also grateful to Helen Chan for skillful technical assistance and for preparation of the corn stem powder.
This research was supported in part by grant DE-DDF03-92ER20069 from the U.S. Department of Energy.
REFERENCES
- 1.Aspinall, G. O. 1980. Chemistry of cell-wall polysaccharides, p. 473-500. In J. Press (ed.), The biochemistry of plants, vol. 3. Academic Press, Inc., New York, N.Y.
- 2.Beylot, M.-H., V. A. McKie, A. G. Voragen, C. H. Doeswijk-Voragen, and H. J. Gilbert. 2001. The Pseudomonas cellulosa glycoside hydrolase family 51 arabinofuranosidase exhibits wide substrate specificity. Biochem. J. 358:607-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezalel, L., Y. Shoham, and E. Rosenberg. 1993. Characterization and delignification activity of a thermostable α-l-arabinofuranosidase from Bacillus stearothermophilus. Appl. Microbiol. Biotechnol. 40:57-62. [Google Scholar]
- 4.Coombs. J. M., and J. E. Brenchley. 1999. Biochemical and phylogenetic analysis of a cold-active β-galactosidase from the lactic acid bacterium Carnobacterium piscicola BA. Appl. Environ. Microbiol. 65:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debeche, T., N. Cummings, I. Connerton, P. Debeire, and M. J. O'Donohue. 2000. Genetic and biochemical characterization of a highly thermostable α-l-arabinofuranosidase from Thermobacillus xylanilyticus. Appl. Environ. Microbiol. 66:1734-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries. R. P., H. C. M. Kester, C. H. Poulsen, J. A. E. Benen, and J. Visser. 2000. Synergy between enzymes from Aspergillus involved in the degradation of plant cell wall polysaccharides. Carbohydr. Res. 327:401-410. [DOI] [PubMed] [Google Scholar]
- 7.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 8.Doner, W. L., D. B. Johnston, and V. Singh. 2001. Analysis and properties of arabinoxylans from discrete corn wet-milling fiber fractions. J. Agric. Food Chem. 49:1266-1269. [DOI] [PubMed] [Google Scholar]
- 9.Gilead, S., and Y. Shohan. 1995. Purification and characterization of α-l-arabinofuranosidase from Bacillus stearothermophilus T-6. Appl. Environ. Microbiol. 61:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, M. A., and R. H. Doi. 1995. Prokaryotic promoters in biotechnology. Biotechnol. Annu. Rev. 1:105-128. [DOI] [PubMed] [Google Scholar]
- 11.Gutshall, K. R., D. E Trimbur, J. J. Kasmir, and J. E. Brenchley. 1995. Analysis of a novel gene and β-galactosidase isozyme from a psychrotrophic Arthrobacter isolate. J. Bacteriol. 177:1981-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes, M. L., and M. L. Dyall-Smith. 2000. Sequence and expression of a halobacterial β-galactosidase gene. Mol. Microbiol. 36:114-122. [DOI] [PubMed] [Google Scholar]
- 13.Hung, M.-N., Z. Xia, N.-T. Hu, and B. H. Lee. 2001. Molecular and biochemical analysis of two β-galactosidases from Bifidobacterium infantis HL96. Appl. Environ. Microbiol. 67:4256-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, K. S., T. G. Lilburn, M. J. Renner, and J. A. Breznak. 1998. arfI and arfII, two genes encoding α-l-arabinofuranosidases in Cytophaga xylanolytica. Appl. Environ. Microbiol. 64:1919-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosugi, A., K. Murashima, and R. H. Doi. 2001. Characterization of xylanolytic enzymes in Clostridium cellulovorans: expression of xylanase activity dependent on growth substrates. J. Bacteriol. 183:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachke, A. H. 1988. 1,4-β-d-Xylan xylohydrolase of Sclerotium rolfsii. Methods Enzymol. 160:679-684. [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 277:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Matsuo, N., S. Kaneko, A. Kuno, H. Kobayashi, and I. Kusakabe. 2000. Purification, characterization and gene cloning of two α-l-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem. J. 346:9-15. [PMC free article] [PubMed] [Google Scholar]
- 19.Moller, P. L., F. Jorggensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular β-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 68:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method, a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz, W. H., K. Bronnenmeier, B. Krause, F. Lottspeich, and W. L. Staudenbauer. 1995. Debranching of arabinoxylan: properties of the thermoactive recombinant α-l-arabinofuranosidase from Clostridium stercorarium (ArfB). Appl. Microbiol. Biotechnol. 43:856-860. [DOI] [PubMed] [Google Scholar]
- 23.Sheridan, P. P., and J. E. Brenchley. 2000. Characterization of a salt-tolerant family 42 β-galactosidase from a psychrophilic Antarctic Planococcus isolate. Appl. Environ. Microbiol. 66:2438-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoham, Y., Z. Schwartz, A. Khasin, O. Gat, Z. Zosim, and E. Rosenberg. 1993. Delignification of wood pulp by thermostable xylanase from Bacillus stearothermophilus T-6. Biodegradation 3:207-218. [Google Scholar]
- 25.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamaru, Y., S. Ui, K. Murashima, A. Kosugi, H. Chan, R. H. Doi, and B. Liu. 2002. Formation of protoplasts from cultured tobacco cells and Arabidopsis thaliana by the action of cellulosomes and pectate lyase from Clostridium cellulovorans. Appl. Environ. Microbiol. 68:2614-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson, J. A. 1993. Molecular biology of xylan degradation. FEMS Microbiol. Rev. 104:65-82. [DOI] [PubMed] [Google Scholar]
- 30.Volkin, D. B., and A. M. Klibanov. 1989. Minimizing protein inactivation, p. 1-24. In T. E. Creighton (ed.), Protein function, a practical approach. IRL Press, Oxford, England.
- 31.Wood, W. A., and K. M. Bhat. 1988. Methods for measuring cellulose activities. Methods Enzymol. 160:87-112. [Google Scholar]
- 32.Zverlov, V. V., W. Liebl, M. Bachleitner, and W. H. Schwarz. 1998. Nucleotide sequence of arfB of Clostridium stercorarium, and prediction of catalytic residues of α-l-arabinofuranosidases based on local similarity with several families of glycosyl hydrolases. FEMS Microbiol. Lett. 164:337-343. [DOI] [PubMed] [Google Scholar]