Abstract
Two new surface layer (S-layer) proteins (SlpB and SlpD) were characterized, and three slp genes (slpB, slpC, and slpD) were isolated, sequenced, and studied for their expression in Lactobacillus brevis neotype strain ATCC 14869. Under different growth conditions, L. brevis strain 14869 was found to form two colony types, smooth (S) and rough (R), and to express the S-layer proteins differently. Under aerobic conditions R-colony type cells produced SlpB and SlpD proteins, whereas under anaerobic conditions S-colony type cells synthesized essentially only SlpB. Anaerobic and aerated cultivations of ATCC 14869 cells in rich medium also resulted in S-layer protein patterns similar to those of the S- and R-colony type cells, respectively. Electron microscopy suggested the presence of only a single S-layer with an oblique structure on the cells of both colony forms. The slpB and slpC genes were located adjacent to each other, whereas the slpD gene was not closely linked to the slpB-slpC gene region. Northern analyses confirmed that both slpB and slpD formed a monocistronic transcription unit and were effectively expressed, but slpD expression was induced under aerated conditions. slpC was a silent gene under the growth conditions tested. The amino acid contents of all the L. brevis ATCC 14869 S-layer proteins were typical of S-layer proteins, whereas their sequence similarities with other S-layer proteins were negligible. The interspecies identity of the L. brevis S-layer proteins was mainly restricted to the N-terminal regions of those proteins. Furthermore, Northern analyses, expression of a PepI reporter protein under the control of the slpD promoter, and quantitative real-time PCR analysis of slpD expression under aerated and anaerobic conditions suggested that, in L. brevis ATCC 14869, the variation of S-layer protein content involves activation of transcription by a soluble factor rather than DNA rearrangements that are typical for most of the S-layer phase variation mechanisms known.
Surface layers (S layers) are monomolecular crystalline arrays of proteinaceous subunits present in almost all archaea and all major phylogenetic groups of bacteria (26, 33). Most of the S layers are composed of subunits of a single protein or glycoprotein species capable of forming symmetrical arrays and covering the cell surface during all stages of growth. S-layer proteins commonly contain high numbers of acidic and hydrophobic amino acids but lack overall amino acid sequence homology with corresponding proteins from unrelated species. Moreover, a low pI is typical for S-layer proteins (31) except for those from different lactobacilli (5, 40) and Methanothermus fervidus (6). Diverse functions have been proposed for S layers, such as acting as molecular sieves, protective coats, molecular and ion traps, cell shape determinants, and promoters for cell adhesion and surface recognition (26). There is also increasing evidence that S-layer-carrying bacteria may use S-layer variation, by expressing alternative S-layer protein genes, for adaptation to different stress factors, such as the immune response of the host for pathogens and drastic changes in the environmental conditions for nonpathogens. In most of the characterized cases, the mechanism of S-layer variation is based on DNA rearrangements (4, 8, 26, 29). Structural features of S layers also indicate that they may offer several advantages for different biotechnological applications. For example, use of S layers as ultrafiltration membranes, matrices for the immobilization of functional molecules, and vaccine carriers has been proposed (32).
In the genus Lactobacillus, S layers have been found in many species (20, 42). However, only the S-layer protein genes from Lactobacillus brevis ATCC 8287 (40) and from the closely related species Lactobacillus acidophilus (3), Lactobacillus helveticus (7), and Lactobacillus crispatus (30) have been cloned and sequenced. The molecular masses of lactobacillar S-layer proteins are among the smallest (43 to 46 kDa) known for the S-layer proteins, and in the L. acidophilus group the proteins show sequence homology mainly in the C-terminal parts. The functions of lactobacillar S layers have remained poorly characterized. Thus far, they have been shown to function only as adhesins mediating binding to avian intestinal epithelial cells (28) or to the mammalian extracellular matrix (37). Recently, in L. brevis the S layer has been demonstrated to function as an adhesin to human epithelial cells and fibronectin (12).
We have previously characterized the S-layer protein gene (slpA) of L. brevis ATCC 8287 (40), studied its in vivo expression, which is directed by two active adjacent slpA promoters (13), and found that, in this L. brevis strain, only a single-copy S-layer protein gene can be found (40; A. Palva et al., unpublished results). Furthermore, we have demonstrated the applicability of the slpA expression and secretion signals for heterologous protein production (14, 27) and have shown that a nonadhesive lactic acid bacterium can be provided with the capacity for adhesion to human intestinal cell lines by providing it with the receptor-binding region of L. brevis ATCC 8287 SlpA (S. Åvall-Jääskeläinen et al., submitted for publication).
Our adhesion studies further indicate that the neotype strain of L. brevis, ATCC 14869, adheres more strongly to human enterocytes in vitro than L. brevis ATCC 8287, and even the binding of ATCC 14869 to pig intestinal epithelial cells was found (M. Jakava-Viljanen and A. Palva, unpublished data). To study whether the S-layer protein of ATCC 14869, like that of L. brevis ATCC 8287, mediates adhesion and whether the L. brevis S-layer proteins have structural similarity in their epithelial cell binding domains, we have started to investigate the S-layer protein of ATCC 14869 in more detail.
In this work, we have characterized two new S-layer proteins and three slp genes from L. brevis ATCC 14869 with characteristics different from those of the L. brevis ATCC 8287 SlpA protein and its gene. One of these genes, slpC, is not expressed under the growth conditions tested. The two other genes are slpB and slpD. The expression of slpD is affected by the oxygen status in ATCC 14869 cultures. Under aerobic conditions ATCC 14869 grows preferentially as rough (R) colonies and produces both SlpB and SlpD proteins, whereas under anaerobic conditions ATCC 14869 grows as smooth (S) colonies and synthesizes only the SlpB protein. Furthermore, we have shown by real-time quantitative PCR and by using a reporter gene that the regulation of slpD expression is not dependent on DNA rearrangements.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The neotype strain L. brevis ATCC 14869, obtained from the American Type Culture Collection, was cultivated in shaker flasks in MRS broth (Difco; culture was <20% of the flask volume) under aeration at 200 rpm or anaerobically in a 80% N2-20% CO2 atmosphere in an anaerobic chamber (Concept Plus; Ruskinn Technology Ltd., Leeds, United Kingdom) or on MRS agar plates (Difco) at 32°C. MRS agar plates were incubated in the anaerobic chamber (Concept Plus; Ruskinn Technology Ltd.) under the above conditions or in an ordinary incubation cabinet under normal atmosphere at 32°C for 2 days or more. The redox potential of the aerated culture was monitored with a platinum electrode (Platin Electrode BlueLine 31 RX; Schott) connected to a pH meter (model 744; Metrohm Ltd.). The electrode was calibrated with 468- and 220-mV buffer solutions (Mettler-Toledo GmbH).
Detection of S-layer proteins.
ATCC 14869 cells grown in MRS broth were harvested by centrifugation (2 min at 18,000 × g) and washed once with phosphate-buffered saline (pH 7.4). The pellet, equivalent to 1 ml of culture, was dissolved directly in 50 μl of Laemmli buffer. The polypeptides were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (19) and stained with Coomassie brilliant blue. The growth supernatants were analyzed for S-layer protein content by SDS-PAGE using concentrated fractions or directly from supernatants by Western blotting.
Isolation of S-layer proteins.
The S-layer protein bands (43 and 50 kDa) were excised from SDS-9% PAGE gel after electrophoresis and treatment of the gel with 0.5 M ice-cold KCl. The S-layer proteins were eluted from the gel by using an Electro-Eluter (model 422; Bio-Rad) according to the manufacturer's instructions. The elution was followed by dialysis against phosphate-buffered saline. Isolated S-layer proteins were used for peptide N-terminal sequence analyses and mass mapping analysis.
Peptide sequencing and mass mapping.
The N-terminal amino acid sequences of purified S-layer proteins were determined by using a gas-pulsed liquid sequencer as described previously (15). In-gel digestion with trypsin followed by mass mapping with a Biflex matrix-assisted laser desorption ionization-time of flight mass spectrometer (Bruker-Franzen Analytik, Bremen, Germany) was performed with the S-layer proteins as described by Nyman et al. (23).
Oligonucleotides and PCR.
Oligonucleotides used in this work are listed in Table 1. The oligonucleotides were purchased from the oligonucleotide synthesis service of MedProbe (Oslo, Norway). PCR was carried out with DyNAzyme II DNA polymerase and DyNAzyme EXT DNA polymerase as recommended by the manufacturer (Finnzymes). Vectorette PCRs were performed according to the manufacturer's instructions (Sigma Genosys Ltd., Cambridge, United Kingdom).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Nucleotide sequence (5′ → 3′)a | Specificityb |
|---|---|---|
| 629 | ACIAAGCGTAAYGTIAAYITIACIGG | slpB |
| 631 | TTCIACICCIGTRAAIAIIGTIGTIGC | slpB |
| 675 | CAGTTTCAACGACTGCTTCAGC | slpB |
| 730 | CAACACCATTAGCGATTGGG | slpB |
| 857 | CAACGACTAAGTCCGCTACTATGC | slpC |
| 901 | CATCGAATGTAGCACTAACTGTTGGG | slpC |
| 963 | CCAATAAGCCTTTATACTCGCAG | Intermediate region between slpB and slpC |
| 858 | GCATAGTAGCGCACTTAGTCGTTG | slpC |
| 899 | CATACTTATATCCAGTACCAGTTAGAACCG | splC |
| 946 | GATYAAGCAAGTTGGTWSTAC | slpD |
| 966 | GCTAACAATGTTGGCTGTTACCTTC | slpD |
| 973 | CGTCCGCTTTAATGAACTACGC | slpD |
| 949 | CTGATTGAGCCTTAGCAGTTGATGC | slpD |
| 1009 | GGTTCAAAAGTTGTCGCTTCTGC | slpD |
| 1010 | CCGTTGCATCACTGTTAAACTTC | slpD |
| 1156 | ATGGAGCCCACCATAACACC | slpD |
| pepi2 | AAATAAGCTTACAAACGCAGTGAAAGAATG | pepI |
| 1161 | TATTTAGGGGGATTTTTCATGGAAATTATTGAAGGA | pepI/slpD∗ |
| 1162 | AACGTCTAGAATGGAGCCCACCATAACACC | slpD |
| 1163 | TTTTCCTTCAATAATTTCCATGAAAAATCCCCCTAAATATTG | pepI/slpD∗ |
Additional nucleotide symbols used: Y, C plus T; W, A plus T; S, C plus G; I, inosine; R, A plus G.
∗, oligonucleotide used to create an exact joint between the slpD promoter and the pepI gene.
Real-time quantitative PCR.
Real-time PCR was carried out with an iCycler (Bio-Rad Laboratories) in a 50-μl reaction mixture containing 5 μl of DNA template (10 ng to 1 pg), 1 μM (each) primer, 1 U of Dynazyme II DNA polymerase (Finnzymes), 1× DNA polymerase buffer (Finnzymes), 0.2 mM (each) deoxynucleoside triphosphate, and a 1:100,000 dilution of SYBR Green I (Molecular Probes) (41). PCR amplification steps were as follows: initial denaturation at 94°C for 5 min followed by 30 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, and 85°C for 30 s. Detection of the fluorescent product at 530 nm was set at the 85°C incubation step in each cycle. Experiments were performed in triplicate. Six controls without a template were included on every plate.
Southern blotting and hybridization.
Southern blotting was carried out essentially as described earlier (36). PCR primers for the slpB- and slpD-specific probes were designed according to the amino acid sequences of the intact N termini and tryptic peptides of the SlpB and SlpD proteins. The slpB-specific probe was amplified from L. brevis ATCC 14869 chromosomal DNA with primers 629 and 631, and the slpD-specific probe was amplified with primers 946 and 966 (Table 1). The probes were labeled with digoxigenin-dUTP by using the DIG-High Prime kit according to manufacturer's instructions (Boehringer Mannheim). For hybrid detection a chemiluminescence detection kit (DIG; Boehringer Mannheim) was used.
Vectorette system.
A Vectorette library was constructed according to the instructions given by the manufacturer (Vectorette II; Sigma Genosys Ltd.). Based on the Southern blotting data, chromosomal DNA of L. brevis ATCC 14869 was extensively digested with HindIII and ligated with the HindIII Vectorette unit. The Vectorette amplicons were sequenced as described below.
DNA sequencing and sequence analyses.
Sequencing and sequence analysis of the 16S rDNA, amplified by PCR from the L. brevis ATCC 14869 DNA, were performed with a Micro Seq 500 16S rDNA bacterial sequencing kit and MicroSeq analysis software (Applied Biosystems). The nucleotide sequencing of the slp gene regions was performed by the dideoxy chain termination method of Sanger et al. (25) by using an ABI PRISM 310 genetic analyzer (Applied Biosystems) with the ABI PRISM BigDye Terminators, version 3.0, cycle sequencing kit (Applied Biosystems). Both DNA strands were sequenced. Assembly of DNA sequence data was performed with the Sequencher, version 3.0, program (Gene Codes Corporation, Ann Arbor, Mich.). The DNA and the predicted protein sequences were analyzed with the PC/GENE set of programs (release 14.0; IntelliGenetics). Homology searches of the databases were done by using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST). Alignment studies were performed on the ExPASy server by using the SIM program (http://www.expasy.org/tools/sim-prot.html). The comparison matrix used in the alignments was PAM400 (gap open penalty, 12; gap extension penalty, 4). Putative transcription terminators were searched by using mfold, version 3.1 (http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi).
Other DNA techniques.
Routine molecular biology techniques were used (24). Chromosomal DNA was isolated essentially as described before (40). L. brevis ATCC 14869 cells were transformed essentially as described by Bhowmik and Steele (1).
RNA isolation and Northern blot analysis.
L. brevis ATCC 14869 cells were grown in MRS broth at 32°C by incubating the cultures either anaerobically or under aeration (200 rpm). Cell samples were withdrawn at different growth phases for RNA isolation. Cells were collected by centrifugation in the presence of glass beads and disrupted by 90 s of cell mill homogenization (Bühler Vibrogen Cell Mill VL4; Edmund Bühler). Total RNA was isolated with the RNeasy minikit (Qiagen), and the preparation was treated with DNase (RQ1 RNase-free DNase; Promega) at 37°C for 15 min.
RNA gel electrophoresis and Northern blotting were performed essentially as described previously (10). The PCR probes for slpB, slpC, and slpD transcripts were amplified from L. brevis chromosomal DNA by using the following primer pairs: 675 and 730 for the slpB probe, 897 and 901 for the slpC probe, and 946 and 966 for the slpD probe (Table 1). Purification of PCR products was done with the QIAquick PCR purification kit (Qiagen), and labeling was done with digoxigenin-dUTP by using the DIG-High Prime kit (Boehringer Mannheim).
Primer extension analysis.
Primer extension of slpD was performed with total RNA isolated from cells grown under aeration by using an A.L.F. DNA sequencer (Amersham Pharmacia Biotech) as described previously (39) and 5 pmol of fluorescein-labeled oligonucleotide 973 (Table 1). The sequencing reaction product, obtained with primer 949 (Table 1) and the Vectorette II primer (Sigma Genosys Ltd.) by using the HindIII Vectorette II library as the template, was utilized as the marker for the 5′ end determination. The transcription start site was determined by comparing the retention time of the primer extension product with that of the sequencing reaction product.
RT-PCR.
Reverse transcription (RT) of RNA samples and PCR amplification of cDNA were done by using the RobusT RT-PCR kit (Finnzymes) as recommended by the manufacturer. Amplification products of cDNA were run on a 3% (wt/vol) MetaPhor agarose gel (BioWhittaker Molecular Applications). The absence of contaminating genomic DNA in the RNA samples used in RT-PCR was confirmed by the absence of PCR products when avian myeloblastosis virus reverse transcriptase was omitted from the reaction.
Construction of an slpD promoter-pepI reporter plasmid.
A transcriptional fusion between the slpD promoter region and the L. helveticus 53/7 proline iminopeptidase gene (pepI) (38) was constructed as follows. With the primer pair 1162 and 1163 (Table 1) the 255 bp upstream of the initiation codon of slpD was amplified by PCR from ATCC 14869 chromosomal DNA. The pepI gene, including its initiation codon and transcription terminator sequence, was amplified by PCR from plasmid pPV39 (P. Varmanen et al., unpublished data) with primers pepi2 and 1161 (Table 1). The slpD promoter and the pepI gene fragments were fused together by recombinant PCR (11, 18) and ligated as an XbaI-BglII fragment with pKT2095 (27), resulting in plasmid pKTH5117.
Aminopeptidase activity assays.
Intracellular aminopeptidase activities from L. brevis ATCC 14869 cells harboring pKTH5117 were determined as follows. Cells harvested by centrifugation were washed once with 50 mM HEPES (pH 7.0) and disrupted by cell mill homogenization for 90 s. After resuspension in HEPES buffer the cells were centrifuged and the supernatant obtained was used for aminopeptidase activity determinations as described previously (9) by using 20 mM l-proline p-nitroanilide as the substrate in 50 mM HEPES (pH 7.0).
Electron microscopy.
ATCC 14869 cells from S- and R-type colonies were grown under anaerobic or aerobic conditions, respectively, in MRS broth overnight as described above. For transmission electron microscopy the liquid cultures were centrifuged for 10 min in a bench centrifuge. The resulting cell pellets were either immediately frozen in Freon R22 for freeze-etching or chemically fixed and dehydrated as described previously (34).
Briefly, freeze-etching was performed on a BAF 400 freeze-etching machine (Balzers Union, Balzers, Liechtenstein). The fracturing temperature was −96°C, and the etching time was 1.5 min. The replicas were cleaned in 30% (wt/vol) chromium oxide and, after thorough washing with water, were mounted on 300-mesh copper grids.
For thin sectioning the wet cell pellet (∼50 mg) was suspended in 1 ml of a solution containing 2.5% glutaraldehyde, 2.5% formaldehyde, and 2% tannic acid in 0.1 M sodium cacodylate buffer, pH 7.4, and fixed for 16 h at 4°C. After five washes in cacodylate buffer the sample was postfixed for 2 h at room temperature with 1% osmium tetroxide in water, washed with water, and stained in the dark for 1 h at room temperature with 1% uranyl acetate in water. After three washes with water the sample was dehydrated in a graded ethanol series, followed by a propylene oxide exchange, and embedded in Spurr resin. Polymerization was for 24 h at 60°C, and, after curing and trimming, thin sections were cut on a Reichert Ultracut ultramicrotome (Leica-Reichert, Vienna, Austria).
Electron micrographs of both freeze-etched and thin-sectioned preparations were taken on a CM 12 electron microscope (Philips, Eindhoven, The Netherlands) at an acceleration voltage of 80 kV.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the slpB, slpC, and slpD regions are AY040846, AY040847, and AY040848, respectively.
RESULTS
Colony morphology and characterization of the S-layer protein profile.
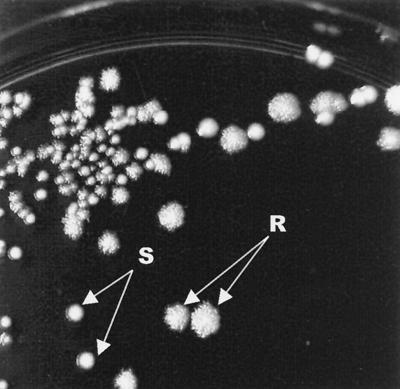
During analysis of S-layer proteins from L. brevis type strain ATCC 14869, these cells were observed to form two colony types, S and R (Fig. 1), on MRS plates when growing under aerobic conditions for 2 to 3 days, whereas under anaerobic conditions only S-type colonies were formed. When R colonies were restreaked on MRS plates and grown under aerobic condition, a few S colonies were always observed, whereas the majority of S colonies streaked from anaerobic plates to aerobic plates changed their morphology to the R type. Putative strain contamination was excluded by obtaining identical 16S rDNA sequences with L. brevis identity from the cells of both colony types (data not shown).
FIG. 1.
Strain L. brevis ATCC 14869 grown aerobically on MRS agar plates. The plate was prepared by mixing S colonies with a small amount of R colonies, and cells were grown overnight under aerobic conditions.
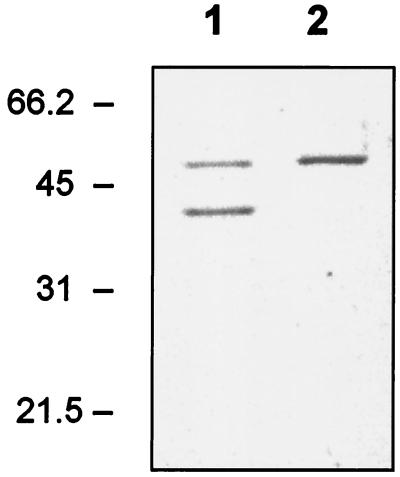
By SDS-PAGE analyses of whole ATCC 14869 cells of the R-colony type, two protein bands with apparent molecular masses of 50 and 43 kDa, representing the putative S-layer proteins, could be detected (Fig. 2, lane 1). Respective SDS-PAGE analysis of ATCC 14869 S-colony type cells resulted in essentially only one band with an apparent molecular mass of 50 kDa (Fig. 2, lane 2). The relative amount of the 50-kDa protein in the R-colony type cell sample was smaller than that in the S-colony type cell sample (Fig. 2). No significant amounts of either of the two proteins could be found in growth supernatants of the R- and S-colony type cell samples (data not shown), indicating that their synthesis was strictly controlled. When ATCC 14869 cells were propagated in MRS broth either in an anaerobic chamber or under aeration (200 rpm), S-layer protein patterns similar to those of the S- and R-colony type cells, respectively, were observed by whole-cell SDS-PAGE analyses. For anaerobically grown cells, the 50-kDa protein dominated and the amount of 43-kDa protein varied from 0 to 10% among experiments. In contrast, the ratio of 50- to 43-kDa proteins was 4:6 in cells grown under aerated conditions. Measurement of the redox potentials of ATCC 14869 cultures revealed that the difference between the anaerobic and aerated cultures was approximately 130 mV. The redox potentials of both anaerobic and aerated cultures remained substantially unchanged throughout the period represented by the growth curve (data not shown).
FIG. 2.
SDS-PAGE analysis of L. brevis ATCC 14869 cells. Lane 1, intact R-colony type cells; lane 2, intact S-colony type cells. Sizes of the molecular mass marker proteins are indicated on the left.
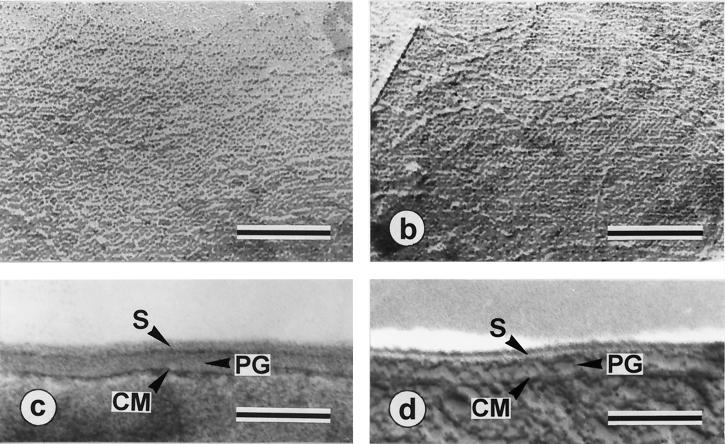
Electron microscopy of ATCC 14869.
Transmission electron microscopy, performed for cells from both colony types grown under aerated or anaerobic conditions in liquid cultures, suggested the presence of an apparently single S layer outside the cell wall structure, regardless of the mode of cultivation used (Fig. 3). By freeze-etching, the S layers from both preparations showed a delicate oblique lattice which, in either case, was partly masked by collapsed filamentous structures (Fig. 3a and b). The lattices were indistinguishable from each other and showed lattice constants of a = 8.5 nm and b = 5.0 nm for ATCC 14869 R colonies (Fig. 3a) and a = 8.4 nm and b = 5.0 nm for ATCC 14869 S colonies (Fig. 3b). In both lattices, the angle between the base vectors was approximately 80°. There was also no detectable difference in the cell wall profiles of both types of colonies, regardless of which growth conditions and preparation techniques were applied (Fig. 3c [thin sectioning] and d [freeze-etching]). In either case a single S layer was present on top of the approximately 35-nm-thick peptidoglycan layer.
FIG. 3.
Electron microscopy of L. brevis ATCC 14869 cells. (a and b) Freeze-etched preparations of the S-layer protein of ATCC 14869 cells grown from R-type colonies in MRS broth under aerated conditions (a) and ATCC 14869 cells grown from S-type colonies in MRS broth under anaerobic conditions (b), showing identical oblique lattices. (c and d) Ultrathin section of an ATCC 14869 R-type cell (c) and cross-fracture of a freeze-etched ATCC 14869 S-type cell (d), showing that only a single S-layer protein is present on the cell surface. Abbreviations: S, S layer; PG, peptidoglycan; CM, cytoplasmic membrane. Bars, 100 nm.
Isolation and sequencing of the slp genes.
The 50- and 43-kDa S-layer protein bands were extracted from SDS-PAGE gel and electroeluted, followed by the N-terminal sequencing of the intact proteins and tryptic peptides. The obtained N-terminal sequences of the intact 50- and 43-kDa proteins wereKSAAKVTSDKVLTTDATKRNVNLTGTNAIYSKPGTVGAK and QVGSTNLEPTTAYIR, respectively. Degenerate PCR oligonucleotides (Table 1) were designed according to the N-terminal sequences by utilizing the codon usage determined for the L. brevis ATCC 8287 S-layer protein (40), and the PCR products obtained were sequenced and used as probes in Southern hybridizations with different restriction enzyme digestions. Both probes detected a chromosomal DNA fragment size of approximately 5 kb with HindIII digestions (results not shown), suggesting the use of HindIII-cut chromosomal DNA for analysis of the upstream and downstream regions of the slp genes. Therefore, HindIII-cut chromosomal DNA was ligated with HindIII Vectorette units, and PCR primers specific for the isolated gene and the Vectorette system were used to amplify and sequence the slp gene regions.
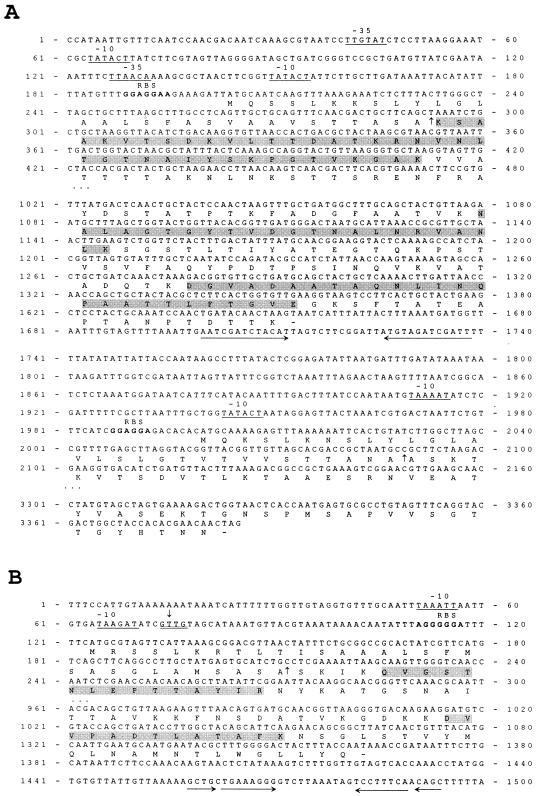
Sequence analysis of the slp genes.
In the amino acid sequence predicted from the open reading frame expected to encode the 50-kDa S-layer protein, the amino acid sequences of the intact N terminus and tryptic peptides of this protein could be found (Fig. 4), and the associated gene was designated slpB. Downstream of the slpB sequence, another putative slp gene with the capacity for encoding an approximately 49-kDa protein was found and designated slpC. The DNA sequence and coding capacity of slpC did not, however, correspond to the N-terminal sequence and size of the 43-kDa S-layer protein synthesized by the R-colony type cells. Therefore, to isolate the third gene, slpD, encoding the 43-kDa S-layer protein, specific primers were designed according to the available nucleotide sequences of slpD and used for isolation of its up- and downstream regions from the Vectorette library.
FIG. 4.
Relevant parts of the nucleotide sequences from the L. brevis ATCC 14869 slpB and slpC genes (A) and slpD gene (B) and deduced amino acid sequences. The predicted −10 and −35 regions for promoters are underlined. Vertical arrows pointing up, signal peptide cleavage sites; horizontal arrows, putative transcription terminators of the slpB and slpD genes. RBS, putative ribosome-binding site. The 5′ end of slpD mRNA (vertical arrow pointing down) is double underlined. The N-terminal and internal amino acid sequences of the intact SlpB and SlpD proteins and their tryptic peptides are shaded.
The slpB gene was found to consist of 1,449 nucleotides and to have the capacity to encode a polypeptide of 483 amino acids. A signal sequence of 30 amino acids was found from the deduced amino acid sequence, and the molecular mass of the mature SlpB was calculated to be 47,968 kDa, which is in good agreement with the molecular mass of the 50-kDa protein estimated by SDS-PAGE (Fig. 2). Two putative promoter regions and a ribosome-binding site were identified from the gene sequence (Fig. 4A). Furthermore, a putative rho-independent type transcription terminator sequence (ΔG = −92.88 kJ/mol) could be recognized 44 nucleotides downstream of the stop codon of the slpB gene (Fig. 4A), suggesting that the slpB gene is monocistronic.
The slpC gene, separated from slpB by 347 nucleotides, was found to consist of 1,383 nucleotides with a capacity for encoding a polypeptide of 461 amino acids. A predicted signal sequence of 30 amino acids was identified from the deduced slpC-encoded polypeptide, SlpC, indicating that the slpC gene has a coding capacity for a mature protein of 45,793 kDa. Upstream of the translation start codon of slpC, only two putative −10 regions for a promoter (Fig. 4A) and a possible ribosome-binding site (Fig. 4A) were recognized. No obvious rho-independent type transcription terminator could be distinguished downstream of the stop codon of slpC. The DNA sequencing of the slpB-slpC region directly from PCR fragments that were amplified from chromosomal DNA, separately isolated from the S- and R-colony type cells, revealed no differences in the sequences (data not shown).
The open reading frame of slpD was shown to consist of 1,239 nucleotides and to encode a putative polypeptide, SlpD, of 413 amino acids. From the deduced amino acid sequence of SlpD, a signal sequence of 30 amino acids was identified, and the molecular mass of the mature SlpD was calculated to be 41,571 kDa. This is in good agreement with the molecular mass of the 43-kDa protein estimated by SDS-PAGE (Fig. 2). From the upstream region of slpD, only two putative −10 regions for a promoter (Fig. 4B) and a possible ribosome-binding site (Fig. 4B) could be distinguished. A putative rho-independent type transcription terminator sequence (ΔG = −65.27 kJ/mol) was identified approximately 92 nucleotides downstream of the stop codon of slpD (Fig. 4B), suggesting that the slpD gene is also monocistronic. Furthermore, Southern blotting and PCR experiments, carried out to test whether the slpD gene is located on the same DNA region as slpB and slpC, revealed that slpD was not closely linked to the slpB-slpC gene region (data not shown).
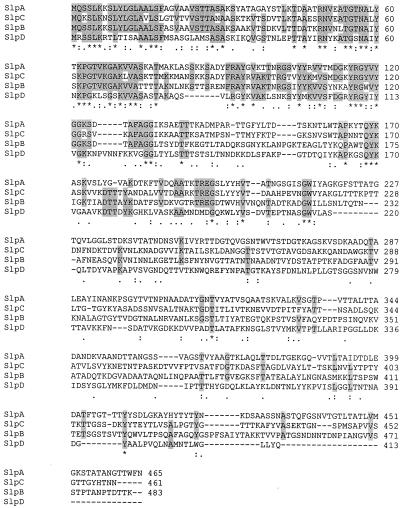
The deduced amino acid sequences of all these L. brevis ATCC 14869 Slp proteins were shown to possess typical features of S-layer proteins. All mature ATCC 14869 Slp proteins were found to have a high content of amino acids with hydroxyl groups (SlpB, 25.2%; SlpC, 32.3%; SlpD, 24.8%) and to lack cysteine residues. They were also found to possess a high content of hydrophobic amino acid residues (SlpB, 34.6%; SlpC, 33.8%; SlpD, 36.8%). The CHARGPRO program of PC/GENE predicted that the isoelectric points of the ATCC 14869 Slp proteins would also be high (SlpB, 9.99; SlpC, 10.12; SlpD, 10.08), which is a typical feature of S-layer proteins of the genus Lactobacillus (5). The SIM program revealed clear similarity between the S-layer proteins of L. brevis ATCC 14869 and SlpA from L. brevis ATCC 8287. However, the levels of identity were significantly lower than would have been expected from two strains belonging to same species (Table 2). Comparison of the four L. brevis Slp proteins revealed that the signal peptides and the N-terminal parts of the mature proteins had the highest similarity, whereas the C-terminal regions were very dissimilar (Fig. 5), unlike what is found for L. acidophilus, L. crispatus, and L. helveticus, where the C termini are highly conserved (35). Among the L. brevis S-layer proteins, ATCC 14869 SlpD was found to be the most distantly related and, interestingly, the closest relatedness was between the L. brevis ATCC 8287 SlpA protein and the predicted product of the silent ATCC 14869 slpC gene (Fig. 5). The amino acid sequence identities between the Slp proteins from L. brevis ATCC 14869 and those from other Lactobacillus species (L. acidophilus SlpA and SlpB, L. helveticus SlpH1, and L. crispatus CbsA, CbsB, SlpNA, and SlpNB) were negligible (under 17%).
TABLE 2.
Homology comparisonsa of the deduced amino acid sequences of the different L. brevis ATCC 14869 S-layer proteins
| L. brevis strain/S protein | % Identical amino acids in L. brevis GRL62
|
||
|---|---|---|---|
| SlpB | SlpC | SlpD | |
| ATCC 14869/SlpB | 35.4 | 25.9 | |
| ATCC 14869/SlpC | 35.4 | 29.5 | |
| ATCC 14869/SlpD | 25.9 | 29.5 | |
| ATCC 8287/SlpA | 30.3 | 38.2 | 23.7 |
Alignment studies were performed on the ExPASy server by using the SIM program. The comparison matrix used in the alignments was PAM400 (gap open penalty, 12; gap extension penalty, 4).
FIG. 5.
Alignment of the SlpA, SlpB, SlpC, and SlpD protein sequences. Asterisks, colons, and dots, identical, conserved, and semiconserved amino acid residues, respectively, in all sequences in the alignment. The regions with the highest identity are shaded.
Expression of the slpB, slpC, and slpD genes and primer extension analysis of slpD.
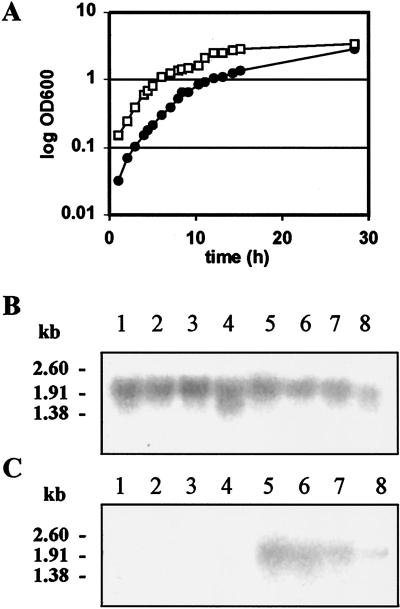
Northern blot analyses were performed to investigate expression of all the three L. brevis ATCC 14869 slp genes under aerated and anaerobic growth conditions. Under both conditions, the slpB-specific probe detected an approximately 2.0-kb transcript (Fig. 6B), confirming the monocistronic nature of slpB transcripts. The slpB promoter region (Fig. 4A) was shown to have a nearly 100% DNA sequence identity with the tandem promoter region of L. brevis ATCC 8287 slpA (40). The signals obtained with the slpB probe were almost equally strong in all the RNA samples isolated from different growth stages (Fig. 6B), indicating that slpB mRNA is very stable.
FIG. 6.
Expression of the L. brevis slpB and slpD genes as a function of growth. L. brevis cells were grown in MRS broth at 32°C. (A) Growth curves of R- (□) and S (•)-colony type cells. R-colony type cells were grown under aeration (200 rpm), and S-colony type cells were grown anaerobically. OD600, optical density at 600 nm. (B and C) Northern blot analysis of total RNA isolated from S-colony cells after 8.4, 13.2, 15.1, and 28.4 h of growth (lanes 1 to 4, respectively) and from R-colony cells after 4.5, 8, 11.1, and 28.4 h of growth (lanes 5 to 8, respectively). Probes used in panels B and C were specific to slpB and slpD, respectively. Numbers on the left refer to an RNA ladder (Promega) used as the size marker.
No slpC transcripts were obtained in Northern blots when the slpC-specific probe was hybridized with total RNA isolated from cells grown under either aerated or anaerobic conditions (data not shown). However, RT-PCR analyses with the slpC-specific primers (857 and 899; Table 1) and slpB- and slpC-specific primers (963 and 858; Table 1) gave PCR products (results not shown). The RT-PCR products most likely, however, represented a minor fraction of slp transcripts since the Northern blots could reveal neither slpC-specific transcripts nor read-through mRNAs from slpB. Furthermore, to ensure that small amounts of the slpC gene products are not synthesized and masked under the SlpB protein, the protein band of 50 kDa was isolated from SDS-PAGE gel and analyzed by mass mapping (data not shown). All of the peptides analyzed matched with the SlpB sequence, suggesting that slpC indeed was not expressed under the growth conditions tested.
The slpD-specific probe detected an approximately 1.9-kb transcript only from total RNA isolated from cells grown under aerated conditions (Fig. 6C). The size of slpD transcripts indicated that the slpD gene is also monocistronic. The Northern blotting result (Fig. 6C) furthermore indicated that slpD transcripts are more labile at the stationary phase than the other slp transcripts. For determination of the transcription start site of the slpD gene, a primer extension analysis was performed with primer 973 (Table 1). With the primer extension result, the transcription start site could be localized between 50 to 47 nucleotides upstream of the start codon of slpD (Fig. 4B).
Real-time quantitative PCR analysis.
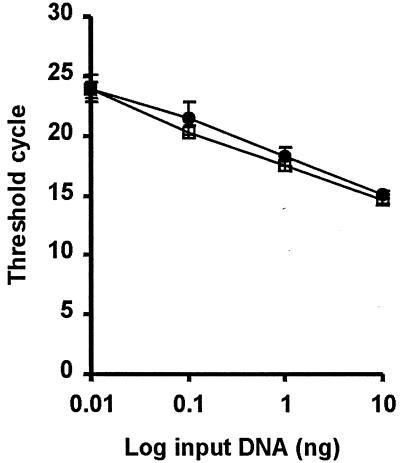
Phase variation mechanisms of S-layer proteins described so far are essentially based on DNA rearrangements (4, 8, 26, 29). To study whether the activation of slpD gene expression takes place by chromosomal rearrangement, a real-time quantitative PCR analysis was performed. To establish a standard calibration curve, serially diluted chromosomal DNA, isolated from cells grown under either aerated or anaerobic conditions, was amplified with primers 1009 and 1010 (Table 1). These primers amplified a 674-bp internal fragment of the slpD gene. The amount of this PCR product should be constant under both growth conditions even if slpD expression is regulated by rearrangement of a chromosomal fragment. Indeed, when the primer pair 1009 and 1010 was used, the threshold cycle (Ct) was achieved at the same time in each template dilution in both cell types (results not shown). Thus, for further analysis the standard calibration curve was based on the amplification of ATCC 14869 DNA from anaerobically grown cells with the primer pair 1009 and 1010. The standard calibration curve obtained with this chromosomal DNA was linear over a range of 4 log10 (correlation coefficient, 0.987) (results not shown).
For studying the possible chromosomal rearrangement during slpD gene expression, primers 949 and 1156 (Table 1) were used. These primers amplified an approximately 500-bp fragment spanning the slpD promoter region and 247 bp from the structural gene slpD. A linear relationship over a range of 3 log10 between the input DNA and the Ct values in cells from both cultivations was observed (correlation coefficients, 0.996 and 0.995) (Fig. 7). These curves were nearly identical, indicating that the regulation of slpD gene expression is not likely to occur by chromosomal rearrangement.
FIG. 7.
Quantitative real-time PCR analyses of the slpD gene of L. brevis ATCC 14869 cells grown under aerated or anaerobic conditions. Chromosomal DNA isolated from both cultivations was serially diluted 10-fold and amplified by PCR with primer pair 949 and 1156, spanning the slpD promoter region. Each symbol represents the average result ± standard deviation of triplicate PCR amplifications for each dilution. Symbols: □, chromosomal DNA isolated from anaerobically grown cells; •, chromosomal DNA from aerated cells.
Expression of an L. helveticus pepI reporter gene under the control of the slpD promoter.
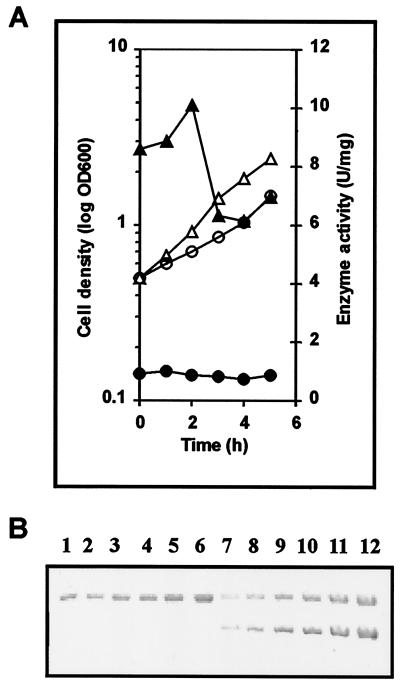
To address the possibility that a soluble transcription regulator controls slpD gene expression, the slpD promoter (PslpD) region was fused with an L. helveticus pepI gene (38), chosen as the reporter gene because it encodes proline iminopeptidase (PepI) activity that can be easily assayed in lactobacilli. The PslpD-pepI fusion was cloned into low-copy-number plasmid vector pKTH2095 (27). The resulting construct, pKTH5117, was used to transform L. brevis ATCC 14869. The resulting recombinant L. brevis strain, GRL1033, was cultivated under aerated and anaerobic conditions, and the aminopeptidase activity of PepI and synthesis of the Slp proteins were measured as a function of growth (Fig. 8). The results showed that under aerated conditions, where the SlpD protein was synthesized, the specific PepI activity increased up to 15-fold in the exponential growth phase, followed by a decrease on the onset of the stationary phase (Fig. 8). This is in agreement with the level of slpD transcripts in Northern analysis. PCR analysis confirmed that no plasmid integration took place during the experiment (data not shown). These results strongly indicate that the variation of the S-layer protein content in L. brevis ATCC 14869 indeed occurs by a mechanism involving activation of transcription by a soluble factor.
FIG. 8.
Expression of the proline iminopeptidase I gene (pepI) of L. helveticus under the control of the L. brevis ATCC 14869 slpD promoter (pKTH5117) under aerated or anaerobic conditions as a function of growth. L. brevis ATCC 14869 cells harboring pKTH5117 were grown in MRS broth at 32°C either under aeration or anaerobically until the optical density at 600 nm (OD600) was 0.5, after which samples were withdrawn at 1-h intervals for enzyme activity and cell density measurements. (A) Growth curves and specific PepI activities, measured as proline p-nitroanilide-hydrolyzing activity of L. brevis ATCC 14869 cells. One unit is defined as 1 μmol of substrate (l-proline p-nitroanilide) hydrolyzed min−1. Shown are growth curves (▵ and ○) and PepI activities (▴ and •) of the aerated (triangles) and anaerobically grown (circles) cells. (B) SDS-PAGE of the whole cells of the growth experiments of panel A. Lanes 1 to 6, synthesis of L. brevis ATCC 14869 SlpB protein; lanes 7 to 12, synthesis of SlpB and SlpD proteins under anaerobic and aerated growth conditions, respectively. Sampling time points were as in panel A.
DISCUSSION
In this work, we have characterized two new S-layer proteins and three slp genes from L. brevis ATCC 14869. The proteins have distinct differences from the S-layer proteins from L. brevis ATCC 8287 and other lactobacilli previously described.
One of the distinctive characteristics of L. brevis ATCC 14869 observed was its ability to alter colony morphology under different growth conditions, a phenomenon not earlier described for this L. brevis neotype strain. When growing under anaerobic conditions, ATCC 14869 produced only S-type colonies, whereas under aerobic conditions preferentially R-type colonies were formed. Furthermore, the colony morphology was reversible depending on the growth conditions. In association with the morphological change of colonies, a switch of the slp gene type expressed was also observed. ATCC 14869 S-type colonies expressed the 50-kDa SlpB S-layer protein on their surfaces, whereas ATCC 14869 R-type colonies produced both SlpB and 43-kDa SlpD S-layer proteins (Fig. 2). Since the mechanism of this regulation is as yet uncharacterized, it is not clear whether the variation of the colony forms and that of slp gene expression are connected by the same regulon or whether they are independent events. Furthermore, it cannot yet be predicted whether the change of redox potential or amount of dissolved oxygen has a direct effect on the induction of slpD expression or whether some other changes under these conditions affect the control of slpD transcription. Most likely, a stress-related cascade of events may be involved. It is feasible that this same response in the S-layer protein content may be caused by other environmental factors as well.
Electron microscopy of ATCC 14869 cells grown in liquid cultures under either anaerobic or aerated conditions and synthesis of the SlpB protein or both SlpB and SlpD, respectively, showed only a single S layer (Fig. 3). The S layers of both types of cells were indistinguishable by electron microscopy. Thus, the electron microscopy data do not explain the difference in the colony morphology but suggest that the S-layer structures are not directly involved. It is not known at present whether the change of the colony morphology is a consequence of the change in the S-layer content or whether it is an independent event induced under the same environmental conditions as the induction of SlpD expression. On the basis of this study, the location of SlpD cannot be clearly elucidated. Treatments with 6 M LiCl released only one-half of the SlpD content under conditions where SlpB was fully released (data not shown), suggesting different interactions with the cell wall material. In a whole-cell enzyme-linked immunosorbent assay with SlpB and SlpD antisera, both SlpB and SlpD appear to be accessible on the cell surface (unpublished data). Thus, it is conceivable that part of the SlpD subunits form a chimeric S layer with the SlpB protein.
The genetic organization of the three ATCC 14869 slp genes is also distinct from that described so far for other lactobacilli. Immediately downstream of the functional ATCC 14869 slpB gene another gene, slpC, was found; this gene was shown to be silent under the growth conditions tested. From L. brevis ATCC 8287, we have been able to find only one S-layer protein gene, slpA (40; Palva et al., unpublished results). In L. acidophilus and the closely related L. crispatus, two S-layer protein genes, an active and a silent one, have been identified (5, 30). Furthermore, the L. acidophilus slpA and slpB genes have been shown to be located on a 6-kb fragment, and an inversion of this fragment controls their expression (4), but conditions that would induce this change have not been established.
In addition to the L. acidophilus group, the phase variation of S layers based on DNA arrangement has been shown for Campylobacter and Geobacillus (formerly Bacillus) stearothermophilus (22). In pathogenic S-layer-carrying species, such as Campylobacter fetus (2), S-layer variation, via the expression of different slp genes as a response to environmental factors, is an essential strategy to avoid host defense systems and maintain virulence. Environmental factors, such as oxygen content, have also been shown to affect the expression of S-layer protein genes sbsA and sbsB in G. stearothermophilus (16, 17). G. stearothermophilus SbsA is produced under oxygen-limiting conditions but is irreversibly replaced by SbsB in the presence of an oxygen supply (16, 17). This variation in S layer is due to DNA rearrangements of the sbs genes between the chromosome and a megaplasmid naturally present in G. stearothermophilus (29). The role of oxygen and possibly other factors in inducing the switch of the sbs genes remains, however, to be elucidated. Another type of mechanism to modify the S-layer protein content of the cell has been described for Bacillus anthracis (21). B. anthracis encodes two S-layer protein types, Sap and EA1, that are colocalized at the cell surface when EA1 synthesis starts in the stationary phase. EA1 synthesis eventually replaces the Sap S layer, which is released to the growth medium. This developmental S-layer switch appears to be controlled by the Sap protein, which is a transcriptional repressor of the eag gene (21). In our study, Northern analyses indicated that slp gene expression in ATCC 14869 is regulated at the transcriptional level. This was also confirmed by expressing reporter protein PepI under the control of the slpD promoter. Furthermore, we showed by quantitative real-time PCR analysis of the chromosomal DNA from cells grown under aerated or anaerobic conditions that there are no conformational changes on the known promoter region of slpD. When SlpD is produced, the amount of SlpB is lowered and also the level of slpB transcripts is somewhat decreased. However, there appears to be no marked repression of slpB transcription, although it might be partly masked by the long half-lives of slpB transcripts. Thus, this suggests that the control mechanism of ATCC 14869 slp gene expression is different from all the S-layer variation mechanisms described so far. The variation of the S-layer protein content takes place by a unique mechanism involving activation of slpD transcription at the exponential growth phase by a soluble factor as a result of an environmental change in L. brevis ATCC 14869. How this response is triggered and what are all the participating components remain to be elucidated.
Acknowledgments
This work was supported by the Academy of Finland.
We thank Ilkka Palva for valuable discussions and critical reading of the manuscript, Anja Osola, Esa Pohjolainen, Harald Mayer, and Andrea Scheberl for skillful technical assistance, and Nisse Kalkkinen for amino acid sequencing and mass mapping.
REFERENCES
- 1.Bhowmik, T., and J. L. Steele. 1993. Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ32. J. Gen. Microbiol. 139:1-7.8450303 [Google Scholar]
- 2.Blaser, M. J., E. Wang, M. K. Tummuru, R. Washburn, S. Fuimoto, and A. Labigne. 1994. High frequency S-layer protein variation in Campylobacter fetus revealed by sapA mutagenesis. Mol. Microbiol. 14:521-532. [DOI] [PubMed] [Google Scholar]
- 3.Boot, H. J., C. P. A. M. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boot, H. J., C. P. A. M. Kolen, and P. H. Pouwels. 1996. Interchange of the active and silent S-layer protein genes of Lactobacillus acidophilus by inversion of the chromosomal slp segment. Mol. Microbiol. 21:799-809. [DOI] [PubMed] [Google Scholar]
- 5.Boot, H. J., and P. H. Pouwels. 1996. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21:1117-1123. [DOI] [PubMed] [Google Scholar]
- 6.Bröckel, G., M. Behr, S. Fabry, R. Hensel, H. Kaudewitz, E. Biendl, and H. König. 1991. Analysis and nucleotide sequence of the genes encoding the surface-layer glycoprotein of the hyperthermophilic methanogens Methanothermus fervidus and Methanothermus sociabilis. Eur. J. Biochem. 199:147-152. [DOI] [PubMed] [Google Scholar]
- 7.Callegari, M. L., B. Riboli, J. W. Sanders, P. S. Cocconcelli, J. Kok, G. Venema, and L. Morelli. 1998. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology 144:719-726. [DOI] [PubMed] [Google Scholar]
- 8.Dworkin, J., and M. J. Blaser. 1996. Generation of Campylobacter fetus S-layer protein diversity utilizes a single promoter on an invertible DNA segment. Mol. Microbiol. 19:1241-1253. [DOI] [PubMed] [Google Scholar]
- 9.El Soda, M., and M. Desmazeaud. 1982. Les peptide hydrolases des lactobacilles du groupe Thermobacterium. I. Mise en évidence de ces activités chez Lactobacillus helveticus, L. acidophilus, L. lactis et L. bulgaricus. Can. J. Microbiol. 28:1181-1188. [PubMed] [Google Scholar]
- 10.Hames, B., and S. Higgins. 1985. Nucleic acid hybridization: a practical approach. IRL, Washington, D.C.
- 11.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols. Academic Press, San Diego, Calif.
- 12.Hynönen, U., B. Westerlund-Wikström, A. Palva, and T. K. Korhonen. 2002. Fibronectin-binding function in the SlpA surface protein of Lactobacillus brevis. J. Bacteriol. 184:3360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahala, M., and A. Palva. 1997. In vivo expression of the Lactobacillus brevis S-layer gene. J. Bacteriol. 179:284-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahala, M., and A. Palva. 1999. The expression signals of the Lactobacillus brevis slpA gene direct efficient heterologous protein production in lactic acid bacteria. Appl. Microbiol. Biotechnol. 51:71-78. [DOI] [PubMed] [Google Scholar]
- 15.Kalkkinen, N., and C. Tilgmann. 1988. A gas-pulsed-liquid-phase sequencer constructed from a Beckman 890D instrument by using Applied Biosystems delivery and cartridge blocks. J. Protein Chem. 7:242-243. [Google Scholar]
- 16.Kuen, B., U. B. Sleytr, and W. Lubitz. 1994. Sequence analysis of the sbsA gene encoding the 130-kDa surface-layer protein of Bacillus stearothermophilus strain PV72. Gene 45:115-120. [DOI] [PubMed] [Google Scholar]
- 17.Kuen, B., A. Koch, E. Asenbauer, M. Sára, and W. Lubitz. 1997. Molecular characterization of the Bacillus stearothermophilus PV72 S-layer gene sbsB induced by oxidative stress. J. Bacteriol. 179:1664-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kylä-Nikkilä, K., M. Hujanen, M. Leisola, and A. Palva. 2000. Metabolic engineering of Lactobacillus helveticus CNRZ32 for production of pure l-(+)-lactic acid. Appl. Environ. Microbiol. 66:3835-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Masuda, K., and T. Kawata. 1983. Distribution and chemical characterization of regular arrays in the cell walls of strains of the genus Lactobacillus. FEMS Microbiol. Lett. 20:145-150. [Google Scholar]
- 21.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 22.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petrol reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51:433-446. [DOI] [PubMed] [Google Scholar]
- 23.Nyman, T. A., S. Matikainen, T. Saraneva, I. Julkunen, and N. Kalkkinen. 2000. Proteome analysis reveals ubiquitin-conjugating enzymes to be a new family of interferon-alpha-regulated genes. Eur. J. Biochem. 267:4011-4019. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savijoki, K., M. Kahala, and A. Palva. 1997. High level heterologous protein production in Lactococcus and Lactobacillus using a new secretion system based on the Lactobacillus brevis S-layer signals. Gene 186:255-262. [DOI] [PubMed] [Google Scholar]
- 28.Schneitz, C., L. Nuotio, and K. Lounatmaa. 1993. Adhesion of Lactobacillus acidophilus to avian intestinal epithelial cells mediated by the crystalline bacterial cell surface layer (S-layer). J. Appl. Bacteriol. 74:290-294. [DOI] [PubMed] [Google Scholar]
- 29.Scholz, H. C., E. Riedmann, A. Witte, W. Lubitz, and B. Kuen. 2001. S-layer variation in Bacillus stearothermophilus PV72 is based on DNA rearrangements between the chromosome and the naturally occurring megaplasmids. J. Bacteriol. 183:1672-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sillanpää, J., B. Martínez, J. Antikainen, T. Toba, N. Kalkkinen, S. Tankka, K. Lounatmaa, J. Keränen, M. Höök, B. Westerlund-Wikström, P. H. Pouwels, and T. K. Korhonen. 2000. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 182:6440-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sleytr, U. B., and P. Messner. 1983. Crystalline surface layers on bacteria. Annu. Rev. Microbiol. 37:311-339. [DOI] [PubMed] [Google Scholar]
- 32.Sleytr, U. B., and M. Sára. 1997. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 15:20-26. [DOI] [PubMed] [Google Scholar]
- 33.Sleytr, U. B., and T. J. Beveridge. 1999. Bacterial S-layers. Trends Microbiol. 7:253-260. [DOI] [PubMed] [Google Scholar]
- 34.Sleytr, U. B., P. Messner, and D. Pum. 1988. Analysis of crystalline bacterial surface layers by freeze-etching, metal shadowing, negative staining and ultrathin sectioning. Methods Microbiol. 20:29-60. [Google Scholar]
- 35.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of Lactobacillus acidophilus ATCC 4356: identification and characterization of domains responsible for S-layer protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 36.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 37.Toba, T., R. Virkola, B. Westerlund, Y. Björkman, J. Sillanpää, T. Vartio, N. Kalkkinen, and T. K. Korhonen. 1995. A collagen binding S-layer protein in Lactobacillus crispatus. Appl. Environ. Microbiol. 61:2467-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varmanen, P., T. Rantanen, and A. Palva. 1996. An operon from Lactobacillus helveticus composed of a proline iminopeptidase gene (pepI) and two genes coding for putative members of the ABC transporter family of proteins. Microbiology 142:3459-3468. [DOI] [PubMed] [Google Scholar]
- 39.Vesanto, E., K. Savijoki, T. Rantanen, J. L. Steele, and A. Palva. 1994. An X-prolyl dipeptidyl aminopeptidase (pepX) gene from Lactobacillus helveticus. Microbiology 141:3067-3075. [DOI] [PubMed] [Google Scholar]
- 40.Vidgrén, G., I. Palva, R. Pakkanen, K. Lounatmaa, and A. Palva. 1992. S-layer protein gene of Lactobacillus brevis: cloning by polymerase chain reaction and determination of the nucleotide sequence. J. Bacteriol. 174:7419-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittwer, C. T., M. G. Hermann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 22:130-131, 134-138. [DOI] [PubMed] [Google Scholar]
- 42.Yasui, T., K. Yoda, and T. Kamiya. 1995. Analysis of S-layer proteins of Lactobacillus brevis. FEMS Microbiol. Lett. 133:181-186. [DOI] [PubMed] [Google Scholar]