Abstract
We show here the involvement of the molecular chaperone DnaK from Escherichia coli in the in vivo α-complementation of the β-galactosidase. In the dnaK756(Ts) mutant, α-complementation occurs when the organisms are grown at 30°C but not at 37 or 40°C, although these temperatures are permissive for bacterial growth. Plasmid-driven expression of wild-type dnaK restores the α-complementation in the mutant but also stimulates it in a dnaK+ strain. In a mutant which contains a disrupted dnaK gene (ΔdnaK52::Cmr), α-complementation is also impaired, even at 30°C. This observation provides an easy and original phenotype to detect subtle functional changes in a protein such as the DnaK756 chaperone, within the physiologically relevant temperature.
The α-complementation of Escherichia coli β-galactosidase, which occurs both in vivo and in vitro, is the most well-known example of protein fragment complementation in which two segments of a protein associate noncovalently to form a functional structure. In the case of β-galactosidase, which is a homotetramer, each monomer (1,023-amino-acid residue) can be divided into two segments, a small (7,500-Da) α-fragment, or α-donor peptide, and a larger α-acceptor composing the remainder of the 116-kDa monomer. The latter is also called the M15 protein because it is the product of the lacZΔM15 allele, a spontaneous partial deletion that does not disturb the open reading frame; the M15 protein, which lacks residues 11 through 41, is an inactive dimer (4 M15 →2 M152′) (10). In the presence of the α-fragment, it forms a tetramer with enzymatic activity (11, 29) according to the following multistep process determined in vitro (17): 2 M152′ + 4α→2 M152α2′→2 M152α2 → (M152α2)2.
Since the molecular chaperone HSP70, the dnaK gene product from E. coli, catalyzes specific inter- and intramolecular protein interactions (8, 14), among the myriad of functions assigned to it, it was tempting to look at its possible involvement in the α-complementation of β-galactosidase in vivo.
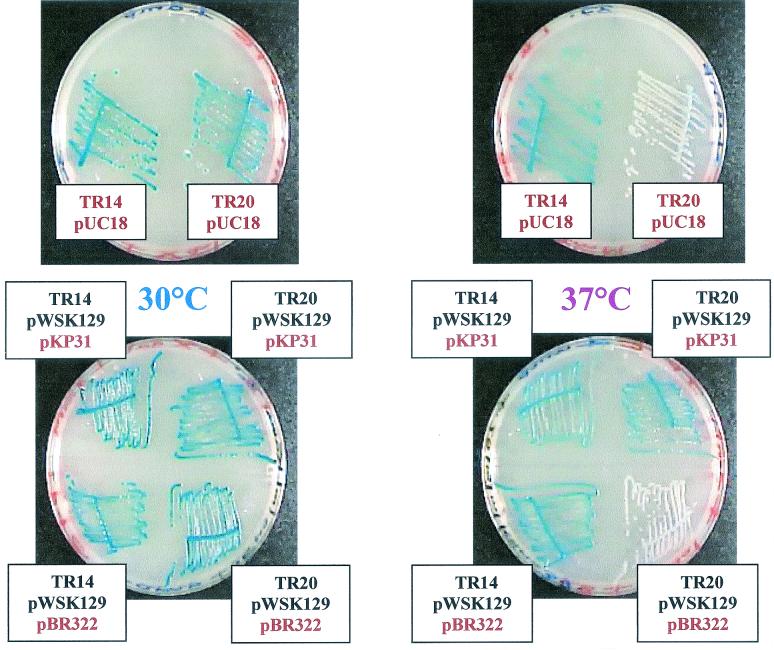
α-Complementation in two isogenic strains [dnaK+ and dnaK756(Ts)] of E. coli grown at 30, 37 and 40°C.
The dnaK756(Ts) allele (5) from strain AL19 (25) was readily transduced into the strain JM83 (lacZΔM15) (33), taking advantage of the genetic marker thr::Tn10(Tetr) adjacent to dnaK and therefore selecting the P1 transductants for tetracycline resistance at 30°C; eighteen dnaK756(Ts) and seven dnaK+ strains were obtained, in accordance with the known cotransduction frequency (around 60%). All of them were transformed to ampicillin resistance (Ampr) with plasmid pUC18 (23) and then checked for α-complementation (see below for description of methods) on plates containing ampicillin, IPTG (isopropyl-β-d-thiogalactopyranoside) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside); when grown at 37 and 40°C, all of the dnaK+ transductants gave blue colonies and all of the dnaK756(Ts) ones gave colorless (i.e., white) colonies, whereas at 30°C, both alleles produced blue colonies, showing an unambiguous correlation between the presence of the dnaK756(Ts) allele and the lack of α-complementation at 37 and 40°C (but not at 30°C).
Two strains chosen among the transductants, TR14 (lacZΔM15, dnaK+) and TR20 [lacZΔM15, dnaK756(Ts)], were transformed to either kanamycin resistance (Kanr) with plasmid pWSK129 (30) or ampicillin resistance with plasmid pUC18, which both encode the lacZ α-peptide, and the results of the α-complementation are shown in Fig. 1 and Table 1.
FIG. 1.
α-Complementation of β-galactosidase in dnaK756(Ts) and dnaK+ strains plated at 30 and 37°C. Strains TR14 (dnaK+) pUC18 and TR20 [dnaK756(Ts)] pUC18 were streaked on two LB plates containing 100 μg of X-Gal per ml, 1 mM IPTG, and 100 μg of ampicillin per ml and incubated for 48 h at 30 or 37°C. Similarly, strains TR14 pWSK129 pBR322, TR14 pWSK129 pKP31, TR20 pWSK129 pBR322, and TR20 pWSK129 pKP31 were streaked on two LB plates containing X-Gal, IPTG, ampicillin, and 25 μg of kanamycin per ml and incubated for 48 h at 30 or 37°C.
TABLE 1.
β-Galactosidase α-complementation in isolated colonies of dnaK+ and dnaK756(Ts) strains plated at 30, 37, and 40°Ca
| Strain | Plasmid | dnaK allele | Ts phenotypeb | α-Complementation in colonies grown atc:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 30°C
|
37°C
|
40°C
|
|||||||
| Blue | White | Blue | White | Blue | White | ||||
| TR14 | pUC18 | Wt | + | 100 | 0 | 99 | 1 | 99 | 1 |
| TR20 | pUC18 | 756(Ts) | − | 99 | 1 | 0 | 100 | 0 | 100 |
| TR14 | pWSK129 | Wt | + | 100 | 0 | 100 | 0 | 100 | 0 |
| TR20 | pWSK129 | 756(Ts) | − | 100 | 0 | 0 | 100 | 0 | 100 |
| TR14 | pWSK129 pBR322 | Wt | + | 100 | 0 | 100 | 0 | 100d | 0 |
| TR14 | pWSK129 pKP31 | Wt | + | 100 | 0 | 99 | 1 | 100e | 0 |
| TR20 | pWSK129 pBR322 | 756(Ts) | − | 100 | 0 | 0 | 100 | 0 | 100 |
| TR20 | pWSK129 pKP31 | 756(Ts) and wt | + | 100 | 0 | 100 | 0 | 100e | 0 |
Cultures were incubated overnight at 30°C, diluted, spread on LB plates containing IPTG, X-Gal, and the appropriate antibiotics, and incubated for 48 h at 30, 37, or 40°C.
Ts phenotype was checked by streaking an aliquot of the undiluted cultures on an LB plate at 44°C.
Five hundred colonies were screened for blue versus white color. The numbers of white and blue colonies are reported as percentages.
Colonies were pale blue.
Colonies were dark blue.
Cultures of TR14 (dnaK+) pUC18 and TR20 [dnaK756(Ts)] pUC18 were incubated overnight at 30°C in Luria-Bertani (LB) medium containing 1 mM IPTG and 100 μg of ampicillin per ml and diluted 106-fold with 10mM MgSO4. Then, 0.1 ml of the diluted bacteria was spread on three LB plates containing 100 μg of X-Gal per ml, 1 mM IPTG, and 100 μg of ampicillin per ml and incubated for 48 h at 30, 37, or 40°C. Five hundred colonies were screened for blue versus white color, and the numbers of colonies with each color were reported as percentages.
Similarly, cultures of TR14 pWSK129 and TR20 pWSK129 were incubated overnight at 30°C in LB medium containing 25 μg of kanamycin per ml, diluted as described above, spread on three LB plates containing X-Gal, IPTG, and 25 μg of kanamycin per ml, and incubated for 48 h at 30, 37, or 40°C. Likewise, cultures of TR14 pWSK129 pBR322, TR14 pWSK129 pKP31, TR20 pWSK129 pBR322, and TR20 pWSK129 pKP31 were incubated overnight at 30°C in LB medium containing X-Gal, IPTG, kanamycin, and ampicillin, diluted as described above, spread on LB plates as described in Table 1, footnote a, and incubated for 48 h at 30, 37, or 40°C. The Ts phenotype was checked by streaking an aliquot of the undiluted cultures on an LB plate at 44°C. The results are shown in Table 1.
As described above, the dnaK+ strains TR14 pWSK129 and TR14 pUC18 gave blue colonies at all the temperatures tested (30, 37, and 40°C), but the dnaK756(Ts) strains TR20 pWSK129 and TR20 pUC18 produced blue colonies only when grown at 30°C and produced white colonies after 24 h when grown at 37 and 40°C. We conclude that the lack of α-complementation in dnaK756(Ts) strains grown at 37 and 40°C is independent of the type of plasmid used, since pUC18 and pWSK129 differ in copy numbers (100 to 200 and 6 to 8, respectively), replication origins (ColE1 and pSC101), genetic markers (Ampr and Kanr), and amino acid sequences of the lacZα products (Table 2). After 48 h, all of the TR20 pWSK129 and TR20 pUC18 colonies grown at 40°C were still perfectly white, but those grown at 37°C turned to a very pale blue color, suggesting that, as far as α-complementation is concerned, the dnaK756(Ts) gene product is completely inactivated at 40°C but still has some residual activity at 37°C. On the other hand, for both strains, the growth rate is not affected by temperature (at 30, 37, and 40°C) in antibiotic-supplemented LB medium, as thermosensitive (Ts) growth of the dnaK756 strains appears only at 42 and 44°C (results not shown).
TABLE 2.
Amino acid sequences of the α-peptides found in native β-galactosidase and those encoded by the lacZα genes within plasmids pUC18 and pWSK129
| α-Peptide | Sequencea |
|---|---|
| Native | LQRRDWENPGVTQLNRLAAHPPFASWRNSEEARTDRPPSQQL |
| pUC18-encoded | mtmitnsssvpgdlpestcrhaslalawLQRRDWENPGVTQLNRLAAHPPFASWRNSEEARTDRPPSQQLrslngewrlmryfllthlcgishriwctlsticssdaa |
| pWSK129-encoded | mtmitpsaqltltkgnksxsstavaaalelvdppgcrnsisslsipstsrggpvpnspysesyyarsslaLQRRDWENPGVTQLNRLAAHPPFASWRNSEEARTDRPPSQQLrslngewdapcsgalsaagvvvtrsvtatlasalapapfaffpsflatfagfprqalnrglplgf rrfsalrhldpkkld |
α-Peptides encoded by the lacZα genes carried by the pUC18 and pWSK129 plasmids contain 110 and 190 amino acids, respectively. Those written in capital letters are the same as those found in native β-galactosidase; those written in lowercase underlined letters are translated from the polylinker sequences.
TR20 [dnaK756(Ts)] pWSK129 cells transformed to ampicillin resistance with the compatible (ColE1) plasmid pKP31 overexpressing dnaK+ (21) completely recovered their α-complementation capability when grown at 37 and 40°C but not when transformed with the empty vector pBR322 (23), confirming that the lack of α-complementation at those temperatures was due to the dnaK756 gene product and showing that dnaK756 behaves in a recessive manner towards dnaK+ for this novel phenotype (Fig. 1 and Table 1). Remarkably, a dependence of the α-complementation on the level of DnaK can also be observed in a dnaK+ strain such as TR14 pWSK129, which produced dark blue colonies when transformed with plasmid pKP31 overexpressing dnaK+ and grown at 40°C but produced pale blue colonies when transformed with the control plasmid pBR322 (Table 1). However, this dark blue color is observed only with growth at 40°C and not at 30 or 37°C (Table 1), suggesting that DnaK is limiting in strain TR14 pWSK129 pBR322 growing at 40°C, possibly because these plasmids recruit a large amount of DnaK to maintain their negative supercoiling against thermal stress (18).
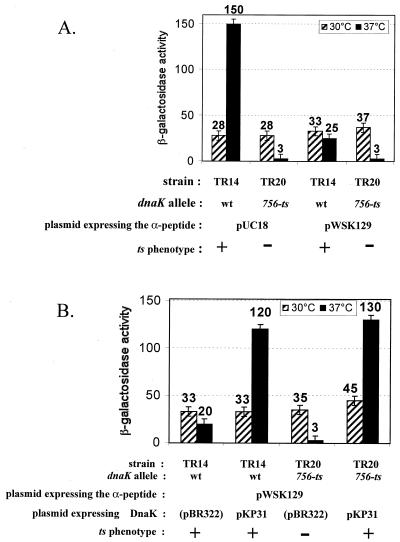
The dnaK-dependent α-complementation phenotype observed on the plates can be quantified by measuring the β-galactosidase activity in crude extracts of the different bacterial strains grown overnight in liquid cultures. The results (Fig. 2) show a clear dependence of the levels of the α-complemented β-galactosidase on the dnaK allele [the wild-type (wt) allele versus dnaK756(Ts)], the temperature (37 versus 30°C), and the overexpression or lack of overexpression of DnaK from plasmid pKP31. Almost no β-galactosidase activity is found in the dnaK756 strain (TR20) grown at 37°C (ten times less than in the same strain grown at 30°C or in the wt strain grown at 37°C) (Fig. 2A), whereas the coproduction of DnaK from plasmid pKP31 restores the β-galactosidase activity to the level found in the wt strain under the same conditions (Fig. 2B). As described in Table 1, even in the wt strain (TR14 pWSK129) grown at 37°C, the overproduction of DnaK from plasmid pKP31 leads to a sixfold increase in the α-complemented β-galactosidase level, but this level also depends on the plasmid copy number (compare TR14 pUC18 and TR14 pWSK129 in Fig. 2A), showing that the intracellular concentrations of both the α-donor peptide and DnaK are important for driving α-complementation. However, a massive production of the α-peptide (from pUC18) cannot compensate for a deficient DnaK chaperone (TR20 pUC18 at 37°C), demonstrating (i) the necessity for an active DnaK for α-complementation and (ii) the thermo-inactivation of the dnaK756 gene product at 37°C, as far as its role (direct or indirect) in α-complementation is concerned. No phenotype has so far been available to monitor such a mild-temperature-driven functional inactivation of DnaK756.
FIG. 2.
α-Complemented β-galactosidase activities in dnaK+ (TR14) and dnaK756(Ts) (TR20) strains grown at either 30 or 37°C and overproducing or not overproducing DnaK. (A) Bacteria in overnight cultures of the different strains at 30 (hatched bars) or 37°C (black bars) in LB medium containing 1 mM IPTG and the relevant antibiotics were harvested, and the β-galactosidase assay in bacterial crude extracts was performed as described previously (16). Hydrolysis of o-nitrophenyl-β-d-galactopyranoside was performed at the same temperature (30, 37, or 40°C) as that used for cultivating bacteria in order to maintain the DnaK756 chaperone in the same state (active or heat-denatured) as it was in vivo. Results are expressed as 10−1 Miller units, since the specific activity of the α-complemented β-galactosidase in a wt (dnaK+) strain is about 5% of that of a native β-galactosidase (in a fully induced lacZ+ strain). One tenth of a Miller unit under the conditions of this study corresponds to an A420 of 0.2 U/ml for 1 ml of crude extract made from a culture at a cell density corresponding to an A600 of 2 U/ml after 1 h of color production at 30°C. (B) The same experiment described in the legend for panel A was performed with strains carrying a second plasmid and overproducing (pKP31) or not overproducing (pBR322) DnaK. The Ts phenotype was checked as described in the footnote to Table 1.
α-Complementation correlates with the inducible expression of DnaK.
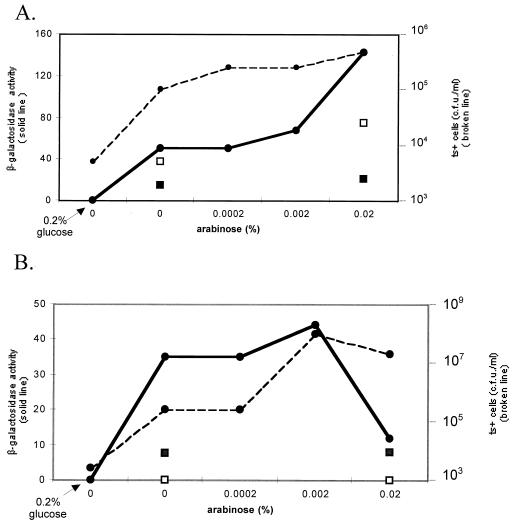
Strains TR20 [dnaK756(Ts)] pUC18 and TR20 pWSK129 were transformed at 30°C to chloramphenicol resistance by the compatible plasmid pARKJ, in which the dnaKdnaJ operon is under control of the PBAD promoter inducible by arabinose, or by the corresponding empty vector (pART3) (22). Overnight cultures of the resulting strains in the presence of the appropriate antibiotics and various concentrations of arabinose (from 0 to 0.02%) were grown at 37°C (nonpermissive conditions for α-complementation). Because of the known leakiness of the PBAD promoter, cultures of the same strains without arabinose and with 0.2% glucose, a strong repressor, were also included. Aliquots of the cultures were used for β-galactosidase activity measurements, for quantitative immunoblot analysis of DnaK (results not shown), and to determine the Ts+ phenotype of the bacteria (at 44°C). For both strains TR20 pUC18 pARKJ and TR20 pWSK129 pARKJ, the results (Fig. 3) show a good correlation between the levels of α-complemented β-galactosidase and the proportion of thermoresistant bacteria, which both increase in parallel as a function of the added concentrations of arabinose. As expected, bacteria containing the empty vector pART3 exhibited a negligible increase in thermoresistance or in β-galactosidase activity. Because the chromosomally encoded DnaK756 protein as well as the minute amount of plasmid-encoded wt DnaK is not distinguished by immunoblot analysis, the proportion of thermoresistant bacteria in the cultures was a better indication of the level of wt DnaK than was immunodetection. Figure 3 also shows that the level of expression of DnaK was already high in the absence of arabinose, but this is an expected result ascribable to the well-known leakiness of the PBAD promoter and can be reduced to zero by the addition of 0.2% glucose. Finally, an overproduction of DnaK induced by 0.02% arabinose leads, in the case of strain TR20 pWSK129 pARKJ, to a drop in α-complementation and to impaired cell growth at 44°C (Fig. 3B), but this toxic effect of overexpressed DnaK, even in dnaK+ cells, has been well documented (4, 25). The reason that this effect is not seen with the other strain (TR20 pUC18 pARKJ) (Fig. 3A) is probably related to the very different plasmid copy numbers in these two strains (100 to 200 copies of pUC18 and 6 to 8 copies of pWSK129) and, as previously mentioned, to the recruitment of a large amount of DnaK to maintain plasmid negative supercoiling in the case of the pUC18-containing strain (18).
FIG. 3.
Effect of arabinose-controlled expression of dnaKJ+ on α-complementation of β-galactosidase and on the thermoresistance phenotype of TR20 [dnaK756(Ts)] strains at 37°C. (A) Overnight cultures of strains TR20 pUC18 pARKJ (dnaKJ+) (full circles) and TR20 pUC18 pART3 (control) (squares) were grown at 37°C in LB medium containing 1 mM IPTG, 100 μg of ampicillin per ml, and 25 μg of chloramphenicol per ml with the indicated concentrations of arabinose. One of the cultures of TR20 pUC18 pARKJ lacked arabinose but contained 0.2% glucose. (B) Overnight cultures of strains TR20 pWSK129 pARKJ (dnaKJ+) (full circles) and TR20 pWSK129 pART3 (control) (squares) were grown at 37°C in LB medium containing IPTG, kanamycin, and chloramphenicol with the indicated concentrations of arabinose. One of the cultures of TR20 pWSK129 pARKJ lacked arabinose but contained 0.2% glucose. The proportion of Ts+ bacteria was measured at 44 and 30°C by plating serial dilutions of the cultures on LB plates containing the same concentrations of antibiotics and arabinose (or glucose) as those in the corresponding liquid cultures (broken lines and empty squares). The β-galactosidase assay in bacterial crude extracts was performed at 37°C (solid lines and full squares).
Immunoblot analysis of LacZΔM15 in dnaK+ and dnaK756(Ts) strains grown at 30 and 40°C, in the presence or absence of overproduced DnaK.
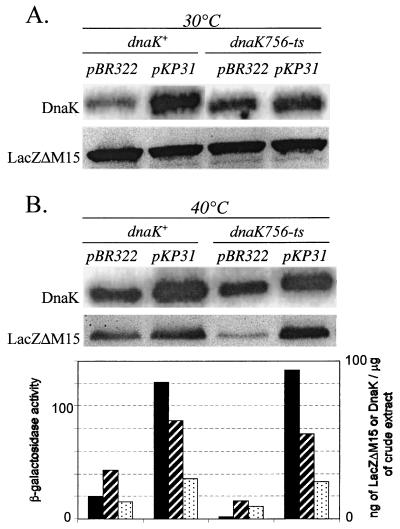
The four strains TR14 (dnaK+) pWSK129 pBR322, TR14 (dnaK+) pWSK129 pKP31, TR20 [dnaK756(Ts)] pWSK129 pBR322, and TR20 [dnaK756(Ts)] pWSK129 pKP31 were grown at both 30 and 40°C. Quantitative immunoblot analysis of bacterial crude extracts with anti-DnaK and anti-β-galactosidase antibodies was used to determine the levels of DnaK and LacZΔM15 in the eight samples (Fig. 4). As expected, plasmid pKP31, when present, produced an increase in DnaK of three to four times at both temperatures. The level of LacZΔM15 was almost the same in the four strains when grown at 30°C (Fig. 4A), but in the cultures grown at 40°C, there were significant differences depending on the dnaK allele present [dnaK+ or dnaK756(Ts)] and on the overexpression (from pKP31) or lack of overexpression (control pBR322) of DnaK (Fig. 4B). It is clear that the intracellular concentrations of LacZΔM15 correlate with those of functional DnaK in a way similar to that of the α-complemented β-galactosidase activities already measured in these strains (Fig. 2). All of these data are assembled in Fig. 4B.
FIG. 4.
Levels of DnaK, LacZΔM15, and α-complemented β-galactosidase activities in dnaK+ and dnaK756(Ts) strains grown at 30 or 40°C and overproducing or not overproducing DnaK. Overnight cultures of the four strains TR14 (dnaK+) pWSK129 pBR322, TR14 (dnaK+) pWSK129 pKP31, TR20 [dnaK756(Ts)] pWSK129 pBR322, and TR20 [dnaK756(Ts)] pWSK129 pKP31 grown in LB medium containing IPTG, kanamycin, and ampicillin at either 30 (A) or 40°C (B) were harvested. Immunoblot analysis of levels of DnaK and LacZΔM15 expression in the crude extracts of the different strains grown at the indicated temperatures was performed as described previously (23). Sonicated extracts of stationary-phase bacterial cells were centrifuged, and protein samples that were prepared by boiling aliquots of the supernatants with sodium dodecyl sulfate gel-loading buffer were resolved by electrophoresis on sodium dodecyl sulfate-polyacrylamide (8%) gels. Samples of known amounts of commercially available DnaK (Stressgen) and native β-galactosidase (Fluka) were also loaded. Serial dilutions of the samples allowed quantification of the signals in the linear range. The blots were probed with anti-E. coli HSP70 (DnaK) (Dako) or anti-native β-galactosidase (Rockland) antibodies, and in both cases reactive bands were visualized with 125I-labeled protein A (Amersham). Protein concentrations in the bands were quantified using a PhosphorImager and expressed in ng/μg of crude extract. The relative levels of LacZΔM15 (hatched bars) and DnaK (dotted bars) measured in the different strains are derived from the immunoblots shown in B. Those of the α-complemented β-galactosidase activities (black bars) in the same strains are taken from Fig. 2.
The decreased level of LacZΔM15 observed in the dnaK756(Ts) strain (TR20 pWSK129 pBR322) grown at 40°C could be either the cause or the consequence of the strong decrease in α-complemented β-galactosidase activity. Therefore, a similar immunoblot analysis of DnaK and LacZΔM15 was carried out with the four strains TR14 (dnaK+) pBR322, TR14 pKP31, TR20 [dnaK756(Ts)] pBR322, and TR20 pKP31, which do not contain the α-complementing plasmid pWSK129. In all of the cultures grown at 30 and 40°C, the intracellular concentrations of DnaK and those of LacZΔM15 (measured by quantitative immunodetection) were similar to those depicted in Fig. 4A and B; i.e., they reveal an unambiguous correlation of the relative levels of LacZΔM15 with those of DnaK (data not shown). That this correlation is independent of α-complementation therefore suggests a role for DnaK in the control of either the synthesis, the degradation, or the solubility of the LacZΔM15 protein (and perhaps that of the α-peptide too). This conclusion, however, does not preclude the possible participation of DnaK in the process of α-complementation itself.
α-Complementation in a ΔdnaK strain at 30°C.
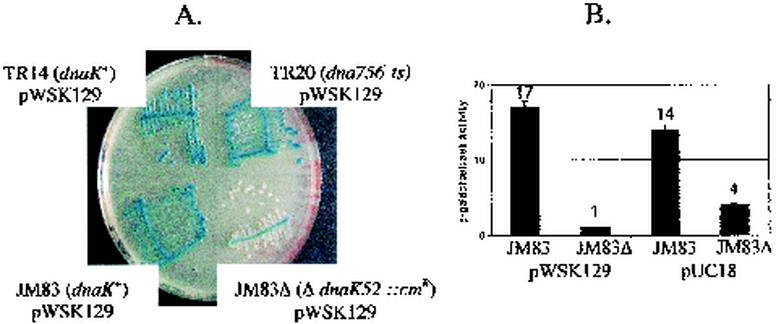
To study the possible DnaK dependence of α-complementation at 30°C, strain BB1553 (7, 21), which contains a disrupted dnaK gene (ΔdnaK52::Cmr), was used. This allele was transduced from BB1553 into the strain JM83 (lacZΔM15) (33) with selection at 30°C on LB plates containing 50 μg of chloramphenicol per ml without sodium citrate. A P1 transductant which was temperature-sensitive (at 40°C) and cold-sensitive (Cs) (at 20°C) (7) was retained and named JM83Δ. This strain, after being transformed to kanamycin resistance with plasmid pWSK129, was unable to sustain α-complementation when streaked on an LB plate containing IPTG, X-Gal, and kanamycin, since all of the isolated colonies were white after 48 h at 30°C, although the high-cell-density portion of the streak was pale blue (Fig. 5A). α-Complemented β-galactosidase activities in crude extracts of JM83Δ pWSK129 and JM83Δ pUC18 bacteria grown in liquid cultures at 30°C were negligible and strongly decreased, respectively (Fig. 5B), indicating that even at 30°C, DnaK is implicated in some way in the α-complementation of β-galactosidase in vivo.
FIG. 5.
α-Complementation of β-galactosidase in a ΔdnaK strain at 30°C. (A) Strains TR14 (dnaK+), TR20 [dnaK756(Ts)], JM83 (dnaK+), and JM83Δ (ΔdnaK52::Cmr), each carrying the plasmid pWSK129, were streaked on a plate containing X-Gal, IPTG, and kanamycin and incubated for 48 h at 30°C. As expected, JM83Δ pWSK129 displayed a chloramphenicol resistance and Cs (on LB medium with kanamycin at 20°C) and Ts phenotype (on LB medium with kanamycin at 40°C). (B) Bacteria which were grown at 30°C and reached the beginning of the stationary phase in cultures of strains JM83 pWSK129 and JM83Δ pWSK129 in LB medium containing IPTG and kanamycin and of strains JM83 pUC18 and JM83Δ pUC18 in LB medium containing IPTG and ampicillin were harvested, and the β-galactosidase was measured in bacterial crude extracts.
Conclusions.
The two main conclusions of this study are as follows. (i) With regard to the participation, directly or indirectly, of the chaperone DnaK in the α-complementation of β-galactosidase in vivo, α-complementation is strongly reduced in both mutants dnaK756(Ts) (at 37°C) and ΔdnaK (at 30°C), whereas it is stimulated in a dnaK+ strain by overproduction of plasmid-encoded DnaK at 37 or 40°C.
(ii) The functional thermo-inactivation of the chaperone DnaK756 at 37°C is revealed only by this original phenotype. The impairment in both mutants dnaK756(Ts) and ΔdnaK52::Cm is specific to the α-complementation process, since the rate of synthesis of native β-galactosidase is not affected in a dnaK756(Ts) mutant, even at 42°C (19). The same is true in the mutant ΔdnaK52::Cm at 30°C (12). However, the synthesis of β-galactosidase induced in a mutant ΔdnaK dnaJ is significantly decreased at 30 and 37°C and is abolished at 42°C (32), but this is due to the extreme instability of a positive regulator of the lac operon, namely, the cyclic AMP receptor protein (CRP), at 42°C in the absence of DnaJ (20).
The lack of α-complementation in dnaK mutants could be due to one of the following reasons. (i) Synthesis of one or both of the partners (LacZΔM15 protein and α-peptide) may be inhibited. This possibility is unlikely, since synthesis of native β-galactosidase is not affected.
(ii) The second possible reason is their accelerated degradation. Our data of immunoblot analysis of LacZΔM15 favor this hypothesis. However, degradation in a dnaK756 mutant of the nonsense lacZ-X90 allele product, a known unstable polypeptide, has been found to be three- or fourfold accelerated at 30°C relative to that in the isogenic wt strain, but by contrast, it was inhibited 20-fold at 42°C (26).
(iii) Their insolubilization or aggregation (in inclusion bodies) is also a possible explanation. Aggregation in inclusion bodies of the pre-S2-S′-β-galactosidase, a tripartite fusion protein consisting of the 55-residue pre-S2 domain and the 30-residue hydrophobic S′ domain of the hepatitis B surface antigen followed by β-galactosidase, was shown to be due only to the S′ domain of this hybrid β-galactosidase, and there is no effect of the dnaK756 mutation on the accumulation of β-galactosidase activity at 30 and 37°C. Only at 42°C does localization in an inclusion-body (insoluble) form increase (27). However, co-overproduction of plasmid-encoded DnaK-DnaJ facilitates the proper folding and assembly of the tetrameric fusion protein, leading to higher yields of active pre-S2-S′-β-galactosidase (28).
(iv) A step (tetramerization?) may be absent in the process of α-complementation itself. In yeast, LacZΔM15 has been found associated with the molecular chaperone HSP90 (1). In vitro, denatured β-galactosidase forms active tetramers upon addition of HSP70 (9). Our data on the stimulation of α-complementation in a dnaK+ background (strain TR14) by overproduction of plasmid-encoded DnaK are also in favor of a possible implication of DnaK in the process of α-complementation itself. There are many examples in which DnaK plays a role in protein quaternary structure changes, including disentanglement of the λDNA-λO-λP-DnaB preprimosomal complex found at the origin of λDNA replication (34); multimerization of the C protein, a positive regulator of bacteriophage μ late transcription (24); monomerization of the replication initiator protein RepE of the mini-F plasmid, mostly present as RepE dimers (15); monomerization of the replication initiator protein RepA of P1 plasmid, mostly present as RepA dimers (31); and dissociation of hydrophobic protein aggregates by DnaK/DnaJ/GrpE, alone or in cooperation with ClpB (3).
Previous studies on the in vitro thermodenaturation of the DnaK756 protein by biophysical techniques are in complete agreement with the data described here, since they showed that the secondary structure of DnaK756 at 20°C is similar to that of DnaK at 55°C and that DnaK756 protein starts to denature at temperatures between 35 and 40°C (2).
The observation of a role, either direct or indirect, for DnaK in α-complementation raises many questions. Which of the three genetic alterations in DnaK756 (5) is responsible for the impaired α-complementation at 37°C? This question will be easily answered, since the amino-terminal G32D and the carboxy-terminal G455D and G468D substitutions have recently been separated (5). Does DnaK act in α-complementation alone or via the general DnaK/DnaJ/GrpE pathway? This question is of particular interest regarding GrpE, since genetic alteration of residue 32 in DnaK756 was found to be solely responsible for the decreased ability of DnaK756 to bind GrpE (6). Are other chaperone machines (e.g., GroEL/GroES) implicated in α-complementation? Another important task is to determine whether DnaK also plays a role in in vitro α-complementation. If it does, dissection of the still mysterious mechanism of α-complementation (13) might become possible, and a very simple colorimetric assay for DnaK could be developed.
Finally, the phenotype described here could be a useful tool for selecting extragenic suppressors of dnaK756(Ts) at mild temperatures (37 to 40°C). It may also be profitable to extend the study made here with the dnaK756(Ts) allele to other examples of protein fragment complementation, such as the E. coli maltose binding protein, the yeast mitochondrial ribosomal protein MrpS28, the yeast phosphoglycerate kinase, and the isoleucyl-tRNA-synthetase.
Acknowledgments
This work was supported by the Centre National de le Recherche Scientifique (U.P.R. 9073) and the University of Paris-7 Denis Diderot. N.L.F. is a recipient of a fellowship from the Université Paris 7-Denis Diderot.
We thank D. Paulin for encouragement and J. Oberto for color pictures.
REFERENCES
- 1.Abbas-Terki, T., and D. Picard. 1999. α-Complemented β-galactosidase: an in vivo model substrate for the molecular chaperone heat-shock protein 90 in yeast. Eur. J. Biochem. 266:517-523. [DOI] [PubMed] [Google Scholar]
- 2.Banecki, B., M. Zylicz, E. Bertoli, and F. Tanfani. 1992. Structural and functional relationships in DnaK and DnaK756 heat-shock proteins from Escherichia coli. J. Biol. Chem. 267:25051-25058. [PubMed] [Google Scholar]
- 3.Ben-Zvi, A. P., and P. Goloubinoff. 2001. Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol. 135:84-93. [DOI] [PubMed] [Google Scholar]
- 4.Blum, P., J. Ory, J. Bauerfeind, and J. Krska. 1992. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J. Bacteriol. 174:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchberger, A., C. S. Gässler, M. Büttner, R. McMacken, and B. Bukau. 1999. Functional defects of the DnaK756 mutant chaperone of Escherichia coli indicate distinct roles for amino- and carboxyl-terminal residues in substrate and co-chaperone interaction and interdomain communication. J. Biol. Chem. 274:38017-38026. [DOI] [PubMed] [Google Scholar]
- 6.Buchberger, A., H. Schröder, J. M. Büttner, A. Valencia, and B. Bukau. 1994. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Nat. Struct. Biol. 1:95-101. [DOI] [PubMed] [Google Scholar]
- 7.Bukau, B., and G. C. Walker. 1990. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkholder, W. F., and M. E. Gottesman. 1999. Genetic evidence for the roles of molecular chaperones in E. coli metabolism, p. 105-138. In B. Bukau (ed.), Molecular chaperones and folding catalysts: regulation, cellular function and mechanisms. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 9.Freeman, B. C., and R. I. Morimoto. 1996. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 15:2969-2979. [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher, C. N., and R. E. Huber. 1997. Monomer-dimer equilibrium of uncomplemented M15 β-galactosidase from Escherichia coli. Biochemistry 36:1281-1286. [DOI] [PubMed] [Google Scholar]
- 11.Gallagher, C. N., and R. E. Huber. 1999. Stabilities of uncomplemented and complemented M15 β-galactosidase (Escherichia coli) and the relationship to α-complementation. Biochem. Cell Biol. 77:109-118. [DOI] [PubMed] [Google Scholar]
- 12.Hesterkamp, T., and B. Bukau. 1998. Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E. coli. EMBO J. 17:4818-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juers, D. H., R. H. Jacobson, D. Wigley, X. J. Zhang, R. E. Huber, D. E. Tronrud, and B. W. Matthews. 2000. High resolution refinement of β-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for α-complementation. Protein Sci. 9:1685-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund, P. A. 2001. Microbial molecular chaperones. Adv. Microb. Physiol. 44:93-140. [DOI] [PubMed] [Google Scholar]
- 15.Matsunaga, F., M. Ishiai, G. Kobayashi, H. Uga, T. Yura, and C. Wada. 1997. The central region of RepE initiator protein of mini-F plasmid plays a crucial role in dimerization required for negative replication control. J. Mol. Biol. 274:27-38. [DOI] [PubMed] [Google Scholar]
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 17.Nichtl, A., J. Buchner, R. Jaenicke, R. Rudolph, and T. Scheibel. 1998. Folding and association of β-galactosidase. J. Mol. Biol. 282:1083-1091. [DOI] [PubMed] [Google Scholar]
- 18.Ogata, Y., T. Mizushima, K. Kataoka, K. Kita, T. Miki, and K. Sekimizu. 1996. DnaK heat shock protein of E. coli maintains the negative supercoiling of DNA against thermal stress J. Biol. Chem. 271:29407-29414. [DOI] [PubMed] [Google Scholar]
- 19.Ohki, M., H. Uchida, F. Tamura, R. Ohki, and S. Nishimura. 1987. The Escherichia coli dnaJ mutation affects biosynthesis of specific proteins, including those of the lac operon. J. Bacteriol. 169:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohki, R., T. Kawamata, Y. Katoh, F. Hosoda, and M. Ohki. 1992. Escherichia coli dnaJ deletion mutation results in loss of stability of a positive regulator, CRP. J. Biol. Chem. 267:13180-13184. [PubMed] [Google Scholar]
- 21.Paek, K. H., and G. C. Walker. 1987. E. coli dnaK null mutants are inviable at high temperature. J. Bacteriol. 169:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Perez, J., C. Martinez-Caua, J. L. Barbero, and J. Gutierrez. 1995. DnaK/DnaJ supplementation improves the periplasmic production of human granulocyte-colony stimulating factor in Escherichia coli. Biochem. Biophys. Res. Commun. 210:524-529. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sand, O., L. Desmet, A. Toussaint, and M. Pato. 1995. The Escherichia coli DnaK chaperone machine and bacteriophage Mu late transcription. Mol. Microbiol. 15:977-984. [DOI] [PubMed] [Google Scholar]
- 25.Sbaï, M., and J. H. Alix. 1998. DnaK-dependent ribosome biogenesis in Escherichia coli: competition for dominance between the alleles dnaK756 and dnaK+ Mol. Gen. Genet. 260:199-206. [DOI] [PubMed] [Google Scholar]
- 26.Straus, D. B., W. A. Walter, and C. A. Gross. 1988. Escherichia coli heat shock gene mutants are defective in proteolysis. Genes Dev. 2:1851-1858. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, J. G., and F. Baneyx. 1996. Protein folding in the cytoplasm of Escherichia coli: requirements for the DnaK-DnaJ-GrpE and GroEL-GroES molecular chaperone machines. Mol. Microbiol. 21:1185-1196. [DOI] [PubMed] [Google Scholar]
- 28.Thomas, J. G., and F. Baneyx. 1996. Protein misfolding and inclusion body formation in recombinant Escherichia coli cells overexpressing heat-shock proteins. J. Biol. Chem. 271:11141-11147. [DOI] [PubMed] [Google Scholar]
- 29.Ullmann, A. 1992. α-Complementation: from protein structure to genetic engineering. Bioessays 14:201-205. [DOI] [PubMed] [Google Scholar]
- 30.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in E. coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 31.Wickner, S., D. Skowyra, J. Hoskins, and K. McKenney. 1992. DnaJ, DnaK, and GrpE heat shock proteins are required in oriP1 DNA replication solely at the RepA monomerization step. Proc. Natl. Acad. Sci. USA 89:10345-10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolska, K. I., B. Kobacz, D. Jurkiewicz, E. Bugajska, M. Kúc, and A. Józwik. 2000. Biosynthesis and secretion of several enzymes in Escherichia coli dnaK and dnaJ mutants. Microbios 101:157-168. [PubMed] [Google Scholar]
- 33.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 34.Zylicz, M., et al. 2000. Role of chaperones in replication of bacteriophage λDNA, p. 295-311. In B. Bukau (ed.), Molecular chaperones and folding catalysts: regulation, cellular function and mechanisms. Harwood Academic Publishers, Amsterdam, The Netherlands.