Abstract
Nidogens are highly conserved proteins in vertebrates and invertebrates and are found in almost all basement membranes. According to the classical hypothesis of basement membrane organization, nidogens connect the laminin and collagen IV networks, so stabilizing the basement membrane, and integrate other proteins. In mammals two nidogen proteins, nidogen-1 and nidogen-2, have been discovered. Nidogen-2 is typically enriched in endothelial basement membranes, whereas nidogen-1 shows broader localization in most basement membranes. Surprisingly, analysis of nidogen-1 gene knockout mice presented evidence that nidogen-1 is not essential for basement membrane formation and may be compensated for by nidogen-2. In order to assess the structure and in vivo function of the nidogen-2 gene in mice, we cloned the gene and determined its structure and chromosomal location. Next we analyzed mice carrying an insertional mutation in the nidogen-2 gene that was generated by the secretory gene trap approach. Our molecular and biochemical characterization identified the mutation as a phenotypic null allele. Nidogen-2-deficient mice show no overt abnormalities and are fertile, and basement membranes appear normal by ultrastructural analysis and immunostaining. Nidogen-2 deficiency does not lead to hemorrhages in mice as one may have expected. Our results show that nidogen-2 is not essential for basement membrane formation or maintenance.
Tissues employ basement membranes (BM) as specialized sheet-like extracellular matrices which are instrumental for tissue compartmentalization and maintenance of cell phenotypes, but they also may act as a source of morphogenetic stimuli for development and tissue remodeling (8, 10, 52). The major BM components are laminins, collagen IV, perlecan, and nidogen. Nidogen, also referred to as entactin, represents a highly conserved protein in vertebrates and invertebrates and is found in almost all BM (25). A large body of in vitro data has guided concepts about the deposition, supramolecular organization, maintenance, and dynamics of BM and nourished our views about the in vivo functions of individual BM proteins (54). Invertebrates, including Caenorhabditis elegans, possess only a single nidogen gene (14), whereas in mammals two nidogens have been discovered and named nidogen-1 and nidogen-2, or entactin-1 and entactin-2, respectively (6, 19, 21, 23). Nidogen-1 (150 kDa) and nidogen-2 (200 kDa) are elongated molecules composed of three globular domains (G1, G2, and G3) connected by a flexible, protease-sensitive link and a rigid rod-like domain (11, 19, 21). In vitro binding affinities of both nidogens for collagen IV and perlecan are nearly identical, whereas the binding affinities of human nidogen-2 to mouse laminin-1 and the corresponding high-affinity nidogen binding module, the laminin-type epidermal growth factor-like module γ1III4 of the laminin γ1 chain, is on the order of 2 to 3 magnitudes lower compared to human or mouse nidogen-1 (21). In contrast, mouse nidogen-2 binds to the γ1III3-5 fragment of the laminin γ1 chain, which contains the γ1III4 module, with only about 10-fold lower affinity than mouse nidogen- 1, and thus significantly (10- to 100-fold) better than human nidogen-2 (44). In vitro data have shown that recombinant nidogen-1 interacts through different binding sites with all three main BM components—laminin, collagen IV, and perlecan—and can mediate the formation of ternary complexes between laminin and collagen IV (11). This was interpreted to mean that nidogen may be a key component for BM assembly, connecting the laminin and collagen networks and integrating other BM components (54).
During embryonic development in mice, nidogen-1 can be detected in preimplantation embryos (7), and both nidogens share overlapping expression patterns at later developmental stages and in adult tissues (19, 21, 32, 33, 44). Enhanced localization in endothelial BM has been recognized as a distinguishing feature of nidogen-2 expression as opposed to a more ubiquitous and even distribution of nidogen-1 in BM. In addition, nidogen-2 shows distinct binding to endostatin and tropoelastin in vitro and colocalizes with both in elastic fibers of vessel walls (30, 46). Data from in vitro angiogenesis studies indicate that laminin and its complex with nidogen play an important modulatory role in angiogenesis (38). Furthermore, a strong interference of capillary formation is observed in in vitro angiogenesis assays upon the addition of antibodies recognizing laminin or nidogen (12). In this respect it is tempting to speculate about specific roles of laminin-nidogen interactions, in particular those of nidogen-2, in blood vessel formation and homeostasis.
Several other functional assays have revealed important roles for nidogens in the development of several organ systems. For example, addition of antibodies that interfere with nidogen-1 binding to laminin induced a distortion of the BM and perturbed epithelial morphogenesis of embryonic kidney, lung, and submandibular glands in organ culture experiments (9, 16). Recently genetic experiments have described C. elegans mutations causing the loss of its nidogen gene. It was shown that nidogen is nonessential and not required for normal collagen IV localization in BM (17) or for BM formation in general but resulted in alterations in axonal patterning in the worm (18). This suggests that the nidogens may have nonstructural functions. Similarly, nidogen-1 gene knockout mice are viable and produce normal BM. This has led to the suggestion that the function of nidogen-1 may be compensated for by the natural presence and/or de novo up-regulation of nidogen-2 (35). In contrast to the surprisingly mild phenotype of nidogen deficiencies described so far, genetic deletion of the LAMC1 gene coding for the laminin γ1 chain eliminates the laminin network and prevents BM formation, resulting in early embryonic lethality (50). Interestingly, genetic elimination of a single laminin-nidogen binding module γ1III4 within the LAMC1 gene interferes with nidogen deposition in embryoid bodies (27) and with kidney and lung development in mice (57).
To gain a more complete understanding of nidogens and the nidogen laminin interaction in mice, we report here (i) the cloning, organization, and chromosomal location of the mouse nidogen-2 gene and (ii) the molecular and biochemical, as well as the genetic and histological, analyses of a nidogen-2 gene mutation in a mouse line recovered by the secretory gene trap approach (34). Our data show that mice homozygous for the gene trap insertion are viable and nidogen-2 deficient but without ultrastructural overt alterations in BM.
MATERIALS AND METHODS
Isolation and characterization of the mouse nidogen-2 gene.
Genomic clones of the mouse nidogen-2 gene were obtained from genomic mouse 129/Sv libraries (catalog no. 946309; Stratagene; supplied by T. Doetschman, University of Cincinnati, Cincinnati, Ohio) using a 32P-labeled HindIII/KpnI restriction fragment (nucleotide [nt] 1 to 2387) of the human nidogen-2 cDNA (21) and a 1.9-kb EcoRI/SacI restriction fragment of a mouse nidogen-2 partial cDNA clone, pEST-3.1 (expressed sequence tag [EST]-cDNA; EMBL GenBank 937317; GenBank EST Division, Merck/Wash U project [supplied by the Resource Center GHGP, Berlin, Germany]) as probes. pEST-3.1 encodes sequences comparable to nt 1983 to 4778 of the published mouse nidogen-2 cDNA (19). DNA inserts of recombinant λ−phage clones and their nidogen-2-positive overlapping DNA fragments were characterized by restriction endonuclease mapping and Southern blot hybridization and subcloned into plasmid vector pBluescript II KS (Stratagene). A 1.5-kb genomic region between λ−phage clones 4-72 and 3-1 which was not covered by recombinant λ phages was isolated by PCR-cloning experiments using 129/Sv genomic DNA and forward primer ND-38 (5′-AAGGACTTGCCATCGACCAC-3′, exon 17) and reverse primer ND-54 (5′-ATGCTACACAGTCCAGCCTCC-3′, exon 18). The 2.7-kb amplification product was cloned by a TOPO-TA-cloning kit pCR2.1-TOPO (Invitrogen), and the DNA sequence of PCR clone pEx17/18 confirmed the missing region. The nucleotide sequences of exons, adjacent intron sequences, and untranslated 5′- and 3′-flanking regions of the gene were determined from overlapping plasmids together with M13, M13rev, T3, T7, and nidogen-2 internal specific primers using the ABI Prism Big Dye terminator cycle sequencing ready reaction kit and an ABI 373A automated DNA sequencer (Perkin-Elmer/Applied Biosystems). Sequence analyses were performed with the programs of the Genetics Computer Group (University of Wisconsin, Madison) package. Intron sizes were determined by DNA sequence analysis, PCR analysis using exon-specific primers, and/or restriction endonuclease mapping. The sequences of 15 DNA stretches encoding the complete sequence of all 21 translated exons of the nidogen-2 gene and their adjacent exon-intron sequences have been deposited in the GenBank/EMBL/DDBJ databases. The accession numbers and the corresponding genomic exon and intron sequences are listed below. Partial intron sequences are labeled with an asterisk: AJ428498, exon 1-intron 1-exon 2-intron 2∗; AJ428499, intron 2∗-exon 3-intron 3∗; AJ428500, intron 3∗-exon 4-intron 4∗; AJ428501, intron 4∗-exon 5-intron 5-exon 6-intron 6∗; AJ428502, intron 6∗-exon 7-intron 7∗; AJ428503, intron 7∗-exon 8-intron 8∗; AJ428504, intron 8∗-exon 9-intron 9-exon 10-intron 10∗; AJ428505, intron 10∗-exon 11-intron 11∗; AJ428506, intron 11∗-exon 12-intron 12∗; AJ428507, intron 12∗-exon 13-intron 13∗; AJ428508, intron 13∗-exon 14-intron 14∗; AJ428509, intron 14∗-exon 15-intron 15∗; AJ428510, intron 15∗-exon 16-intron 16-exon 17-intron 17∗; AJ428511, intron 17∗-exon 18-intron 18∗; AJ428512, intron 18∗-exon 19-intron 19-exon 20-intron 21-exon 21.
Chromosomal assignment of the nidogen-2 gene.
The chromosomal localization of the nidogen-2 gene was determined by radiation hybrid mapping using the T31 radiation hybrid screening panel (28). Primer pair P8-P9 (P8, 5′-ACAATAGGTGGACGGTAG-3′; P9, 5′-AGACAGTTCAGCAGGTAAG-3′) located in intron 1 of nidogen-2 was used to test by PCR genomic DNA from 96 of the 100 T31 radiation hybrid clones for the presence of a specific 223-bp fragment. Genomic DNAs were amplified by standard PCR protocols (denatured at 94°C for 1 min, annealed at 55°C for 1 min, and extended at 72°C for 2 min for 40 cycles, with a final extension at 72°C). The results were tabulated and analyzed by submission to The Jackson Laboratory T31 Mouse Radiation Hybrid Database(http://www.jax.org/resources/documents/cmdata/rhmap/rhsubmit.html) (42).
Generation of mutant mice.
Mouse embryonic stem cells (E14Tg2a.4, derived from the 129/Ola mouse strain) were electroporated with the secretory trap vector pGT1.8TM as described previously (49). By direct sequencing of cDNAs obtained by rapid amplification of cDNA ends (RACE) (56), we identified an embryonic stem (ES) cell line (GST011) expressing a fusion transcript composed of gene trap vector sequences joined to the entactin-2/nidogen-2 endogenous gene (19). The generation of chimeric mice was performed as described previously (34, 49). To genotype animals at weaning, dot blots of DNA prepared from tail biopsy specimens were initially probed with vector (βgeo) sequences (3). The mutant nidogen-2 allele and mouse strain was named Nid2GSF(pGT1.8TM)011Ska.
Identification and cloning of the gene trap vector insertion site.
Restriction endonuclease mapping and Southern blot experiments identified a 345-bp PstI/KpnI DNA fragment containing exon 4 (probe A) and a 436-bp EcoRI/PstI DNA fragment from intron 4 (probe B) as probes which distinguish wild-type- and mutant-specific genomic restriction fragments and map the insertion site of the gene trap vector in intron 4. PCR amplifications using Taq polymerase and standard reaction buffer (Sigma) were carried out to clone the adjacent genomic regions of the insertion site in mutant alleles. The primer pairs P1-ND101 and N6-ND70 were designed to generate insertion site-specific amplicons: P1, 5′-AGCTACACCGAGGACAGTTTCC-3′, exon 4; ND70, 5′-CTGCTTGCTAAACTTTTCACC-3′, intron 4; N6, 5′-ATATTGCTGAAGAGCTTGGCG-3′, gene trap-vector-SV40-poly(A); ND101, 5′-TGTGTGTGTGAAAATGAGATGG-3′, gene trap vector-engrailed intron. The following PCR conditions were applied. PCR samples were initially denatured at 94°C for 4 min, followed by 10 cycles of amplification (94°C for 1 min, 60°C for 2 min with 1°C increment per cycle, 72°C for 4 min), a further 25 cycles of amplification (94°C for 1 min, 56°C for 2 min, 72°C for 4 min), and a final 10-min extension at 72°C. PCR amplification products were separated through agarose gel electrophoresis, and DNA fragments were cloned by a TOPO-TA-cloning kit pCR2.1-TOPO (Invitrogen) according to the supplier's instructions.
Genotype analysis.
Wild-type and mutant nidogen-2 alleles were assessed by Southern blot hybridization and/or PCR from DNA isolated from mouse tail biopsy specimens and from yolk sacs of embryos. For Southern blot analysis, 10 μg of genomic DNA was digested with PstI and analyzed using radiolabeled probe A (see above). PCR assays were performed with primer pair P1-ND70 wild-type allele-specific (1.1-kb) and with primer pair ND79-ND70 mutant allele-specific (0.7-kb) sequences. For PCR primer sequences P1 and ND70, see above; the sequence of ND79 is 5′-CAACGATCAAGGCGAGTTAC-3′ (gene trap vector-specific primer).
Northern blot analysis.
Total RNA was prepared from mouse tissues by extraction in Tri-reagent (Sigma). Poly(A)+ RNA was recovered from total RNA with an RNA Polytex-Kit (Qiagen). RNA was fractionated with 2.2 M formaldehyde-1% agarose gel electrophoresis, transferred to nitrocellulose membranes (Protran; Schleicher & Schuell), and independently hybridized with the following gene-specific 32P-labeled cDNA fragments: mouse nidogen-2, SacI/HindIII fragment (nt 4417 to 5094) (19); mouse nidogen-1, EcoRI/BamHI fragment (nt 1 to 1014) (23); E. coli lacZ gene, 840-bp BamHI/ClaI fragment or mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 1.2-kb fragment (Clontech). Blots were washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% sodium dodecyl sulfate at room temperature for 30 min and then at 60°C for 1 h and exposed to Biomax X-ray film (Kodak) for autoradiography. Blots were stripped of radioactivity before consecutive rehybridizations.
Tissue extraction and protein analysis.
Tissues from various organs were extracted following a previously established protocol (45). Protein extracts were used for immunoblotting and radioimmunoassays. Western blot experiments and immunodetection of proteins were performed as described previously (57). Equal protein loadings of lanes were controlled by staining the membranes with Ponceau S (Sigma) before staining with antisera. Polyclonal rabbit antisera directed against either nidogen-1 (diluted 1:1,000) (11) or nidogen-2 (diluted 1:400) (21) were used, and detection was done using a horseradish peroxidase-conjugated swine anti-rabbit antibody (diluted 1:1,200; DAKO) followed by development with the ECL reagent (Amersham Pharmacia Biotech).
Radioimmunoassays with different specific rabbit antisera were used for the quantitation of individual proteins and established protocols were followed (55). All samples were analyzed at three to four different dilutions, and the average values of individual extracts showed a standard deviation of less than 20%. The assay for laminin γ1 chain was based on recombinant mouse laminin γ1 fragment γ1III3-5 (24), and the assays for nidogens were based on recombinant mouse nidogen-1 (11) and mouse nidogen-2 (44). The reference inhibitors (nidogen-1 or nidogen-2, respectively) showed a steep dose-response profile over a 20-fold range of concentrations, with half-maximal inhibition being achieved at 200 pM. No inhibition was observed with more than 1,000-fold excess of the other nidogen isoform. The same steep inhibition profiles were also observed with the tissue extracts obtained using dilutions of 1:10 to 1:1,000.
Histology and immunofluorescence experiments.
Histology and immunofluorescence experiments were performed as described previously (57). For immunolocalization of different proteins, rabbit antisera were prepared against recombinant mouse nidogen-1 (11), mouse and human nidogen-2 (21, 44), mouse laminin γ1 chain domain IV/V (unpublished), perlecan domain V (4), and collagen IV extracted from the mouse EHS tumor (20). The antibodies were purified by affinity chromatography on the antigen used for immunization. A purified rat monoclonal antibody (JF4) against nidogen-1 has been recently characterized (41). The nidogen-1- and nidogen-2-specific antibodies do not show cross-reactivity with nidogen-2 or nidogen-1, respectively. They were used at 4 or 10 μg/ml (JF4) for immunostaining. Cy2- or Cy3-conjugated secondary antibodies were used at dilutions recommended by the manufacturer (Jackson Immunodiagnostic Labs).
Transmission electron microscopy.
Organs of adult control (nidogen-2+/+, nidogen-2+/−) and homozygous mutant (nidogen-2−/−) mice were dissected. For ultrastructural investigation, 2-mm3 tissue fragments were isolated. Fixation, preparation, and staining have been previously described (30, 31).
RESULTS
Cloning, exon-intron organization, and chromosomal location of the murine nidogen-2 gene.
The mouse nidogen-2 gene locus was isolated as overlapping DNA inserts of λ phage clones from mouse 129/Sv genomic libraries by using cDNA probes specific for nidogen-2 (19, 21) and of a genomic PCR clone generated by using 129/Sv genomic DNA as template and primers specific for exons 17 and 18 of the nidogen-2 gene (Fig. 1A and B). The characterization of the gene was performed by restriction endonuclease mapping, Southern blotting, and DNA sequence analyses. DNA sequences were identified for all translated exons, its exon-intron borders, and introns as well as 5′- and 3′-untranslated regions by sequencing and alignment of genomic and nidogen-2 cDNA sequences (19). The obtained DNA sequences from overlapping subclones of the nidogen-2 gene locus and its sequence analyses resulted in 15 noncontiguous DNA stretches comprising the complete DNA sequences of all 21 translated exons adjacent to exon-intron borders and of 6 introns, as well as partial DNA sequences of the remaining 14 introns, and 5′- and 3′-untranslated sequences. These sequences have been deposited in the GenBank/EMBL/DDBJ databases (for accession numbers see Materials and Methods). The complete nidogen-2 gene contains 21 translated exons spanning approximately 60 kb from the translational start-site to the polyadenylation signal sequence (Fig. 1A and Table 1). All exon-intron borders follow the consensus sequences of U2-type GT-AG splice sites (48), and translated exon sequences range in size from 11 to 728 bp, while the length of the 20 flanking introns varies between 119 bp and approximately 14 kb (Table 1). The 4,209-bp translated sequences of the nidogen-2 gene encode 1,403 amino acids, including a characteristic 30-residue signal peptide. Comparisons of the amino acid sequences deduced from genomic sequences with the nidogen-2 cDNA (19) show a very high sequence identity of 99.35%, but we noticed changes at nine amino acid positions as follows (note: amino acids according to Kimura et al. [19] are shown in parentheses): 251-S (F), 367-A (V), 375-P (H), 386-S (W), 496-S (F), 651-R (C), 810-K (E), 876-L (F), and 1151-(F). In addition, two DNA repeats of a 64-nt (nt 4192 to 4255 and nt 4256 to 4319) and a 185-nt (nt 4358 to 4542 and nt 4543 to 4727) sequence, detected in the 3′-untranslated region of the cDNA sequence published by Kimura et al. (19) are only present as single sequences in our genomic sequences. The additional sequences of the cDNA represent repeats of coding (nt 4192 to 4255) and noncoding (nt 4358 to 4542) cDNA sequences. Our sequence analysis of EST-cDNA clone pEST-3.1 encoding mouse nidogen-2 sequences (see Materials and Methods) and an independent mouse nidogen-2 full-length cDNA sequence by Salmivirta et al. (44) confirmed the genomic sequences in the corresponding amino acid positions and untranslated sequences.
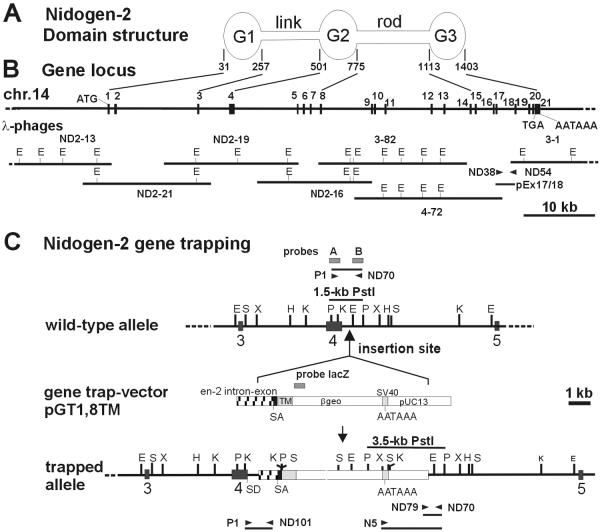
FIG. 1.
Domain structure (A) and gene locus (B) of murine nidogen-2, and characterization of the gene trap vector insertion site in the mutant nidogen-2 allele (C). (A) Schematic drawing of the domain structure of nidogen-2. Lines define the exons encoding the predicted globular domains G1, G2, and G3 and the connecting elements, the flexible link and the rigid rod. Numbers refer to amino acid positions of corresponding domain borders. (B) The organization of the murine nidogen-2 gene is shown. The filled boxes representing the 21 translated exons are drawn to scale. The exon numbers are indicated on the top of the boxes. The translational start and stop codon (ATG and TGA) and the poly(A) adenylation signal AATAAA are indicated. The lines at bottom represent the inserts of overlapping λ phages and genomic PCR clone pEX17/18 carrying the complete nidogen-2 gene. Only the EcoRI restriction sites are indicated on the phage maps. (C) The schematic drawings represent the wild-type and trapped alleles of nidogen-2. The vertical arrow indicates the insertion site of the gene trapping vector pGT1.8TM within intron 4. The positions of the probes (probes A, B, and lacZ) used in genomic Southern blot analysis are indicated as boxes. The positions of the primers used in PCR assays are depicted by horizontal arrowheads. Key DNA restriction fragments (wild-type, 1.5-kb PstI fragment; mutant, 3.5-kb PstI fragment) detected by Southern blot and DNA fragments (wild-type, 1.1 kb; mutant, 0.7 kb) generated by PCR assays for genotyping mice are shown as horizontal bars. Abbreviations: E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SalI; X, XbaI; βgeo, β-galactosidase-neomycin fusion gene; en-2, engrailed 2 gene; SA, splice acceptor site; SD, splice donor site; TM, transmembrane domain of CD4.
TABLE 1.
Gene organization and splice junction sites of the mouse nidogen-2 genea
| Exon | Protein domain/modulee | Exon size (bp) | Splice donor | Intron | Splice acceptor | Intron sizec | Codon phase | Amino acid interrupted codon |
|---|---|---|---|---|---|---|---|---|
| 1 | SP/G1 | 228b | C TAC gtaagt | 1 | tagtag GTG G | 675 bp (Seq) | 0 | Y76/V77 |
| 2 | G1 | 309 | A GAG gtaagt | 2 | ccacag CTG A | ≈14 kb (R) | 0 | E179/L180 |
| 3 | G1 | 233 | AC CA gtaagt | 3 | tcttag G CTA | ≈4.5 kb (R) | 2 | Q257 |
| 4 | Link | 728 | CAA G gtaagt | 4 | cttcag TC TTC | ≈9 kb (R) | 1 | V500 |
| 5 | G2/EGF1 | 150 | GAA G gtgatt | 5 | tttcag GG GCA | 593 bp (Seq) | 1 | G530 |
| 6 | G2 | 246 | ACA G gtgagc | 6 | ctgcag GT GCT | ≈1 kb (R) | 1 | G632 |
| 7 | G2 | 201 | TCA G gtatgt | 7 | ttacag TC GTG | ≈1.5 kb (R) | 1 | V699 |
| 8 | G2 | 231 | GAA G gtatgt | 8 | acatag TG GAC | ≈3.5 kb (R) | 1 | V776 |
| 9 | Rod/EGF2 | 144 | GTG G gtgaga | 9 | gcctag AT GTC | 463 bp (Seq) | 1 | D824 |
| 10 | Rod/EGF3 | 129 | ATC T gtgagt | 10 | ttccag TG ATC | ≈1.5 kb (PCR) | 1 | L867 |
| 11 | Rod/EGF4 | 144 | TCT G gtgagt | 11 | ctgcag AT GTT | ≈5.5 kb (PCR) | 1 | D915 |
| 12 | Rod/EGF5 | 119 | TCT G gtaagt | 12 | cctcag AC ACG | ≈2 kb (R) | 1 | D955 |
| 13 | Rod/TY1 | 234 | CCA G gtgagc | 13 | tcttag AG CCC | ≈4 kb (PCR) | 1 | D1038 |
| 14 | Rod/TY2 | 222 | GCA T gtaagt | 14 | aaacag GC ATA | ≈0.9 kb (PCR) | 1 | C1112 |
| 15 | G3/YWTD1 | 170 | G CAT gtaaac | 15 | tcaaag GGC T | ≈2.5 kb (PCR) | 0 | H1168/G1169 |
| 16 | G3/YWTD2 | 130 | TCA G gtcagc | 16 | ttccag GT TTG | 415 bp (Seq) | 1 | G1212 |
| 17 | G3/YWTD3 | 172 | GA GG gttagc | 17 | ccacag C AAC | ≈2.5 kb (PCR) | 2 | G1269 |
| 18 | G3/YWTD4 | 158 | GCA G gtaata | 18 | taaaag GA ACC | ≈1.7 kb (PCR) | 1 | G1322 |
| 19 | G3/YWTD5 | 124 | GG AG gtgagc | 19 | tcctag G GAT | 849 bp (Seq) | 2 | R1363 |
| 20 | G3/YWTD6 | 113 | ACA G gtgagt | 20 | ctacag GA AGA | 119 bp (Seq) | 1 | G1401 |
| 21 | G3 | 11b | ||||||
| KAG gtragtood | Yyyoyag Good |
Exon sequences are in capital letters; intron sequences are in lowercase letters.
The lengths of the 5′ and 3′ untranslated region in exon 1 and exon 21 are not included.
Intron sizes were determined either by the complete intron sequence (Seq), PCR analyses with primers specific for flanking exons (PCR), or restriction endonuclease mapping (R).
Consensus for U2-type GT-AG introns (48).
Abbreviations of protein modules: EGF, epidermal growth factor-like module; TY, thyroglobulin type-I-like module; LDLR, LDL-receptor-like YWTD module (51); SP, signal peptide.
The predicted modular structure of the nidogen-2 protein as deduced from genomic coding exons or cDNA sequences and/or identified by structural studies (19, 21) is characterized by three globular protein domains—G1, G2, and G3—and their connecting elements, a flexible link and a rigid rod. We have localized these domains containing several typical extracellular protein modules (2) to specific exons of the mouse nidogen-2 gene (Table 1 and Fig. 1A and B). The overall exon-intron organization of the mouse nidogen-2 gene is similar to the modular structure of the nidogen-1 genes in humans and mice (6, 62). Our nidogen-2 gene expression study by Northern blotting of poly(A)+ RNA from adult tissue indicates the expression of a single approximately 5.5-kb mRNA transcript in all tissues examined (Fig. 3A), whereas in the study by Kimura et al. (19) at least two bands for nidogen-2 were detected.
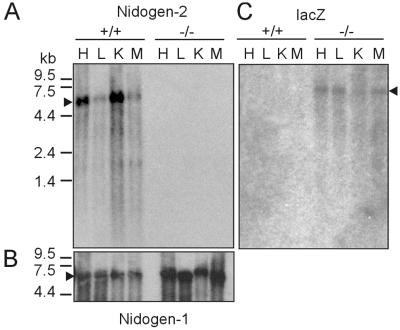
FIG. 3.
Northern blot analysis of RNA isolated from heart, lung, kidney, and skeletal muscle from wild-type and mutant (−/−) nidogen-2 mice. (A) Poly(A)+ RNA fractionated by agarose gel electrophoresis (4 μg/lane) and transferred to nylon membranes was hybridized to nidogen-2-specific probe C. (B) After stripping of the radioactivity, the same blot was rehybridized with a nidogen-1-specific probe and a GAPDH-specific probe (not shown). (C) In a parallel blot hybridization was performed using a lacZ-specific probe. The markers on the left represent an RNA ladder (GIBCO-BRL). Abbreviations: H, heart; K, kidney; L, lung; M, skeletal muscle.
Computer analysis (TRANSFAC and MatInspector [40]) of the 480-bp 5′-untranslated region flanking the translational start codon revealed several high-scoring matches for transcription factor binding sequences, as follows: CAAT (nt −107 to −97, nt −129 to −117), SP1 (nt −71 to −58, nt −78 to −64, nt −264 to −252), Gfi1 (nt −138 to −115), Ik2 (nt −78 to −68, nt −394 to −382), and c-Myb (nt −465 to −449). The analyzed sequence carrying several putative SP1 and CAAT recognition sites lacked a TATA box-like sequence characteristic of housekeeping promoters. In the 3′-untranslated region of the nidogen-2 gene the putative polyadenylation signal AATAAA, already identified in the cDNA (19), and 14 bp further downstream the poly(A) adenylation site were found.
The chromosomal localization of the nidogen-2 gene was determined by radiation hybrid mapping using the T31 radiation hybrid screening panel (28). Based upon linkage to adjacent STS markers (58), the nidogen-2 gene was assigned to chromosome 14, between the microsatellite markers D14Mit12 (1.7 cM) and D14Mit171 (2.5 cM). This is consistent with the physical analysis of nidogen-2 on the mouse genomic map generated by the Ensembl Project (http://www.ensembl.org/Mus_musculus), which places it within 100 kb of D14Mit249 (2.5 cM).
Identification and cloning of a gene trap mutation in the nidogen-2 gene.
Recently, a modified gene trap approach, the secretory trap, was successfully applied in ES cells en route to identifying the in vivo function of large numbers of genes encoding secreted proteins and membrane proteins in mice (34). In this genetic screen, ES cell lines harboring insertions of the secretory trap vector were first characterized by direct sequencing of cDNAs obtained by rapid amplification of cDNA ends (5′ RACE). Next, 60 lines of mice were generated from selected ES cell lines and bred for phenotype analysis. One such line, GST011, was initially identified as an insertion in an unknown gene that later proved to match the entactin- 2/nidogen-2 cDNA (19) once its sequence was deposited in the public database (accession number NM_008695). A comparison of the 283-bp partial 5′-RACE sequences shows almost 100% identity to murine nidogen-2 cDNA sequence, except for a gap of 38 bp in the RACE sequence (data not shown), and we mapped the 3′-end of the RACE sequence to the nidogen-2 cDNA nt 1502 (amino acid Q-469) (19) and to the exon 4-intron 4 border in the mouse nidogen-2 gene (Table 1) by DNA sequence alignments. This suggested that the insertion of the gene trapping vector may have occurred in intron 4 and fusion transcripts harbor exon 1 to exon 4 sequences of nidogen-2 representing the G1 domain and the link region. ES cell line GST011 was used to generate chimeric males that transmitted the mutant allele to their progeny. The presence of the gene-trapped mutant allele in mice was initially identified by a lacZ-specific dot blot assay (3). The putative mutant nidogen-2 allele was named Nid2GSF(pGT1.8TM)011Ska. We will use a superscript plus sign for the wild-type nidogen-2 allele and a superscript minus sign for the mutant nidogen-2 Nid2GSF(pGT1.8TM)011Ska allele to simplify the writing of the genotypes in the remainder of the text.
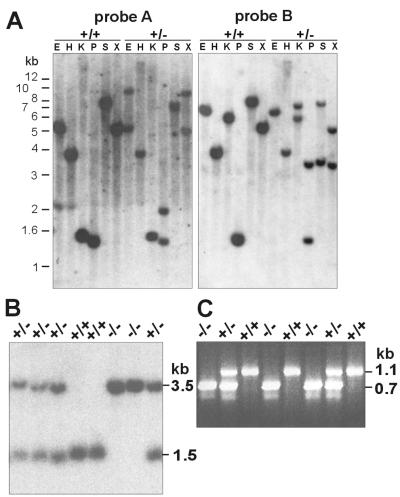
Molecular characterization of the trapped nidogen-2 allele resulted in the cloning of sequences surrounding the insertion site of the gene trap vector and provided us with a probe for reliable genotyping of heterozygous and homozygous mutant offspring of heterozygous intercrosses (Fig. 1 and 2). First we mapped the insertion of the gene trap vector in intron 4 close to the exon 4-intron 4 border by restriction endonuclease mapping and Southern blot analyses with exon 4- and intron 4-specific genomic DNA probes (probes A and B) and genomic DNA from wild-type and nidogen-2+/− mice (Fig. 1C and 2A). We next determined the DNA sequences of the adjacent regions of the insertion site by cloning and DNA sequence analysis of genomic PCR products amplified by primer pairs P1-ND101 and N6-ND70 (Fig. 1C and data not shown). Analysis of DNA sequences revealed that the gene trap vector insertion event occurred 414 bp downstream of the exon 4-intron 4 border and resulted in the deletion of flanking sequences of the linearized gene trap vector DNA, 0.75 kb of the engrailed 2 intron and 0.9 kb of the plasmid vector pUC13 (Fig. 1C and 2A and data not shown). Mice heterozygous for the mutant nidogen-2 gene were identified by Southern blot analysis of PstI-digested tail DNA or PCR assays (Fig. 2B and C). Heterozygous mice appeared normal and were indistinguishable from their wild-type littermates. To obtain homozygous animals for the nidogen-2 mutation, heterozygous mice were intercrossed.
FIG. 2.
Characterization of the gene trap vector insertion site by Southern blotting (A) and genotype analyses by Southern blotting (B) and PCR (C). (A) Genomic DNA digested with different restriction endonucleases was analyzed by hybridization with probes A and B. The probes detect in genomic DNA derived from wild-type (+/+) mice one single band, whereas in genomic DNA derived from heterozygous mutant (+/−) in most cases two specific bands corresponding to both alleles are readily recognized. (B) Southern blots of genomic DNA digested with the restriction endonuclease PstI were analyzed by hybridization with probe B. The 1.5-kb band derived from the wild-type allele and the 3.5-kb band specific for the mutant allele are indicated on the right. Genotypes with the expected specific hybridization patterns of wild-type (+/+), heterozygous (+/−) and homozygous (−/−) mutant mice are shown. (C) PCR analysis of genomic DNA as template and with primers P1, ND70, and ND79. The 1.1-kb band, the product of primers P1 and ND70, represents the wild-type allele (+/+); the 0.7-kb band, the product of primers ND79 and ND70, represents the mutant allele (−/−). The markers on the left represent a 1-kb ladder (GIBCO-BRL). Abbreviations: E, EcoRI; H, HindIII; K, KpnI; P, PstI; S, SalI; X, XbaI.
Nidogen-2 homozygous mutant mice are viable, and secretory gene trapping vector insertion causes nidogen-2 deficiency.
Interbreeding of heterozygous mice yielded 321 viable offspring. Offspring genotypes were identified by Southern blot analysis of PstI-digested tail DNA or PCR assays (Fig. 2B and C) with genotype ratios at the following approximate Mendelian ratios: 67 +/+ (21%), 174 +/− (54%), and 80 −/− (25%). Homozygous animals were born and appeared phenotypically normal compared to their wild-type or heterozygous littermates. Adult homozygous mutant mice did not show any gross anatomical abnormality and proved to be fertile.
Next we analyzed nidogen-2 RNA and protein levels in adult wild-type and mutant mice to confirm that the gene trap insertion represents a phenotypic null allele leading to a nidogen-2-deficiency. Northern blots of RNA from heart, lung, kidney, and skeletal muscle with DNA probes specific for nidogen-1, nidogen-2 or βgeo were performed (Fig. 3). In poly(A)+RNA from wild-type and nidogen-2−/− tissue we detected nidogen-1 mRNA of approximately 6 kb at comparable levels (Fig. 3B), whereas wild-type nidogen-2 mRNA of approximately 5.5 kb was present in wild-type tissue and absent in mutant tissue (Fig. 3A). The generation of putative fusion transcripts with a predicted size of approximately 6.5 kb encoding gene trap vector sequences and nidogen-2 sequences (exon 1 to 4) was identified in poly(A)+ RNA from nidogen-2−/− tissue by a lacZ-specific probe (Fig. 3C).
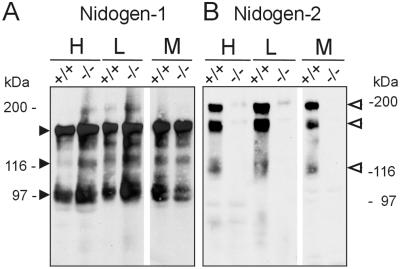
The functional consequence of the nidogen-2 mutation was further characterized by Western blot experiments (Fig. 4) and sensitive and quantitative radioimmunoassays with protein extracts from different tissues (heart, lung, kidney, and skeletal muscle) of adult mice using antisera detecting specifically nidogen-1, nidogen-2, or laminin γ1 chain (Table 2 and data not shown). In Western blot experiments nidogen-1-specific antiserum identified protein bands at 150, 130, and 100 kDa characteristic for full-length nidogen-1 and its proteolytic fragments (39) in wild-type and nidogen-2−/− extracts (Fig. 4A). In parallel, nidogen-2-specific antiserum was applied to the same sets of protein extracts. While this antiserum detects a major band at approximately 200 kDa accompanied by smaller and variable amounts of a 160- to 170-kDa band typical for nidogen-2 (21) in wild-type extracts, no comparable bands were detectable in extracts from tissues of homozygous mutant mice (Fig. 4B). We further quantitated the levels of nidogens and their major binding partner the laminin γ1 chain carrying the well-characterized nidogen binding domain (24) in protein extracts from tissue of adult mice. We established radioimmunoassays for reliable quantitation of antigens in biological samples in the picomolar range (55). The analyses of extracts from kidney and skeletal muscle are summarized in Table 2. In wild-type, nidogen-2+/−, and nidogen-2−/− extracts, similar high amounts of nidogen-1 were detected in both tissues while the amounts of nidogen-2 were up to 22- or 32-fold lower in kidney and 46- or 70-fold lower in skeletal muscle compared to nidogen-1 in wild-type or nidogen-2+/− extracts, respectively. No significant changes in the amounts of the laminin γ1 chain were observed in the corresponding tissues for each genotype. Most importantly, in nidogen-2−/− extracts nidogen-2 could not be detected, indicating a content of less than 1% compared to wild-type nidogen-2 content. Taken together our RNA and protein analyses of tissues from control and homozygous nidogen-2 mutant mice show that the gene trap mutation in intron 4 of the mutant nidogen-2 allele causes a phenotypic null allele and a nidogen-2 deficiency in mice.
FIG. 4.
Western blot analysis of protein extracts of heart, lung and skeletal muscle from normal (+/+) and mutant (−/−) mice. Extracted protein (60 μg) was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to Immobilon membranes, and stained for nidogens with antiserum to nidogen-1 (A) and antiserum to nidogen-2 (B). (A) Both wild-type and nidogen-2 (−/−) extracts contain nidogen-1-specific bands of 150, 130, and 100 kDa (closed arrowheads) in apparently normal and comparable amounts. (B) The nidogen-2 (−/−) mice lack the nidogen-2-specific bands (open arrowheads). The gels were calibrated by globular proteins, denoted in the margins. Abbreviations: H, heart; L, lung; M, skeletal muscle.
TABLE 2.
Quantitation of nidogen-1, nidogen-2, and laminin γ1 chain in tissue extracts by radioimmunoassaya
| Genotypea | Amt of protein (pmol/g[wet weight]) in:
|
|||||
|---|---|---|---|---|---|---|
| Kidney
|
Skeletal muscle
|
|||||
| Nidogen-1 | Nidogen-2 | Laminin γ1 | Nidogen-1 | Nidogen-2 | Laminin γ1 | |
| +/+ | 777 ± 160 | 36 ± 6.7 | 50.2 ± 12.4 | 732 ± 72 | 16 ± 6.4 | 95 ± 30 |
| +/− | 703 ± 192 | 22.2 ± 4.4 | 48.2 ± 17.4 | 735 ± 137 | 10.5 ± 3 | 98 ± 19 |
| −/− | 639 ± 46 | <0.3 | 44.6 ± 8.8 | 679 ± 82 | <0.1 | 90 ± 8 |
+/+, +/−, and −/− refer to nidogen-2 genotypes, and amounts of proteins are shown as means ± standard deviations based on independent measurements of single extracts of each tissue and genotype (n = 5).
Normal histology and BM formation in nidogen-2-deficient tissues.
We performed gross histological analysis on formalin fixed and paraffin embedded tissues of a variety of organs from adult nidogen-2-deficient mice and control littermates (data not shown). The skin, lung, and vascular system were of particular interest, since in vitro nidogen-2-tropoelastin binding studies and immunogold colocalization studies have been suggestive that nidogen-2 may be involved in the assembly and/or linking of elastic fibers to the surface of surrounding cells (46). Furthermore, the strong binding of nidogen-2 to endostatin in vitro and its colocalization in elastic fibers in vessel walls implicated it to be functional in vivo (30). However, we did not observe any obvious histological changes in tissues of adult control and nidogen-2-deficient mice at the age of 2 to 9 months (data not shown).
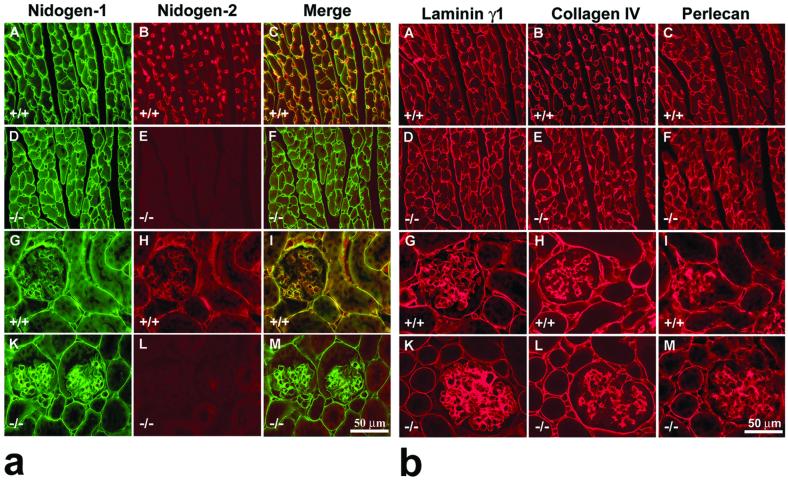
To assess the formation of BM, molecular composition and localization of the main BM proteins in these organs, including heart (Fig. 5a, A to F and Fig. 5b, A to F), kidney (Fig. 5a, panels G to M and 5b, panels G to M), and skeletal muscle (data not shown) frozen and/or paraffin tissue sections, were analyzed either by indirect single and/or double immunofluorescence techniques for nidogen-1 and nidogen-2 (Fig. 5a), as well as for the main BM proteins laminin γ1 chain, collagen IV, and perlecan (Fig. 5b). The typical feature of nidogen-2 protein localization in tissue section of wild-type animals could be observed in cardiac muscle (Fig. 5a, panel B) and striated muscles (data not shown). Nidogen-2-specific antibodies weakly stained the BM surrounding the sarcomeres and cardiomyocytes, while the endothelial BM of blood vessels were typically stained much more strongly (Fig. 5a, panels B and C, and data not shown). In all examined tissues of homozygous mutant mice no BM-specific signal was observed for nidogen-2 (Fig. 5a, panels E and L, and data not shown). However, nidogen-1 appeared abundant in most BM and showed indistinguishable staining pattern and intensities in the control (Fig. 5a, panels A and G) and mutant (Fig. 5a, panels D and K) animals. In all examined tissues of homozygous mutant mice, the staining for laminin γ1 chain, collagen IV, and perlecan was unchanged compared with that of control littermates, as shown in Fig. 5b.
FIG. 5.
Indirect immunofluorescence microscopy of cardiac muscle (A to F) and kidney (G to M) from adult control (+/+) and nidogen-2 (−/−) mice. (a) Double-staining immunofluorescence was applied to sections of cardiac muscle and kidney using rat monoclonal antibodies to mouse nidogen-1 (A, D, G, and K) (green signal) and affinity-purified antibodies to nidogen-2 (B, E, H, and L) (red signal). (C, F, I, and M) Both color channels were merged to demonstrate codistribution (yellow signal) of both immunofluorescence staining signals in the corresponding section. (A and D) Prominent nidogen-1-specific staining is shown around cardiomyocytes and capillaries in both wild-type (A) and mutant (D) sections. (B) In control sections, staining for nidogen-2 was typically prominent around capillaries, whereas the staining intensities around cardiomyocytes are much weaker. This aspect is well demonstrated by showing the merged images (C). (E) In the tissue section of the mutant heart, no nidogen-2-specific staining was observed. (G to I) In the sections of wild-type kidney, a close colocalization of both nidogens was mainly demonstrated in the BM zones of the proximal and distal tubuli, glomeruli, the Bowman's capsule, and blood vessels. In the tissue section of the mutant kidney (K), nidogen-1 staining is similar to nidogen-1 immunofluorescence in the wild-type section (G), whereas no nidogen-2-specific staining was observed in panel L. Bar, 50 μm. (b) Single immunofluorescence microscopy of cardiac muscle (A to F) and kidney (G to M) from adult control (+/+) and nidogen-2 (−/−) mice was applied on consecutive sections using rabbit antisera as primary antibodies recognizing BM proteins laminin γ1 (A, D, G, and K), collagen IV (B, E, H, and L), or perlecan (C, F, I, and M). Cy3-conjugated secondary antibodies were used to visualize specific stainings. Prominent staining for all three BM proteins is shown in wild-type and mutant sections. Immunofluorescence staining intensities in control and mutant tissues appear to be maintained at equal levels. Bar, 50 μm.
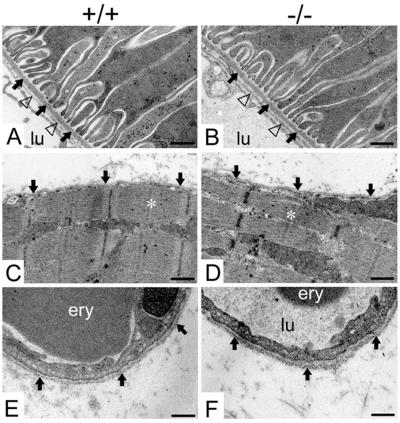
We next examined whether nidogen-2 deficiency resulted in ultrastructural changes in BM of various organs, including kidney, skeletal muscle, and heart. In our analyses, we detected normal-looking BM, and there were no evident alterations in the cellular architecture due to the absence of nidogen-2 (Fig. 6). We recognized slight variations in the thickness of BM of each genotype, e.g., endothelial BM of blood capillaries, which were in the range that is routinely observed.
FIG. 6.
Electron micrographs of ultrathin sections of the kidney cortex (A and B), the hind limb soleus muscle (C and D), and capillaries (E and F) from adult wild-type (A, C, and E) and homozygous mutant (B, D, and F) animals. (A and B) The BM of a proximal tubule is marked by arrows, and the endothelial BM is indicated by triangles. (C and D) The BM of the myocyte is marked by arrows, and a sarcomere is marked by a white asterisk. (E and F) Arrows point to the BM of a capillary of the soleus muscle. Abbreviations: ery, erythrocyte; lu, capillary lumen. Bars, 0.25 μm (A to D) and 0.2 μm (E and F).
DISCUSSION
BM deposition and organization have been suggested to depend on the formation of laminin and the collagen IV networks, which are linked together by nidogen (54). For some time, it was assumed that there is only one nidogen in the vertebrate genome until human and mouse nidogen-2 cDNAs were discovered (19, 21). Both nidogen proteins have a similar modular structure, and the in vitro binding properties for major BM proteins have demonstrated a comparable binding to perlecan and collagen IV while the affinity of recombinant human and mouse nidogen-2 for mouse laminin γ1 chain was 100- to 1,000-fold and 10-fold lower, respectively, compared to human and mouse nidogen-1 (21, 44). To assess the in vivo roles of nidogens, nidogen-1 gene knockout mice have been generated and, unexpectedly, are viable and produce normal BM (35). In this study it was suggested that the loss of nidogen-1 function may be compensated for by the natural presence and/or de novo up-regulation of nidogen-2.
Our present studies described the cloning, gene organization, and chromosomal localization of the mouse nidogen-2 gene and identified a gene trap insertion in the nidogen-2 gene as a phenotypic null allele leading to nidogen-2-deficient mice. We have shown that the mouse nidogen-2 gene locus is approximately 60 kb and is similar in size to the 65-kb mouse nidogen-1 gene locus (6), whereas the human nidogen-1 gene spans 90 kb (62). The nidogen-2 gene comprises 21 translated exons, whereas nidogen-1 is encoded by 20 exons. Comparisons of the proteins as well as the genes for both nidogens characterize their modular structures (6, 62). Nidogen-2 has a modular structure similar to that of nidogen-1, except that it possesses one additional TY module in the rod and lacks an epidermal growth factor-like module at the COOH-terminal end of the G3 domain (19, 21). In both genes exons are organized in a domain-specific manner, and both nidogens contain several typical extracellular protein modules (2) which can be localized to single exons. The presence of exons interrupted by codon phase 1 introns in the two major domains G2 and G3 in both nidogen genes supports the concept of exon shuffling and duplication of epidermal growth factor- and TY-like modules that may have played an important role in vertebrate nidogen evolution (6, 36). In this work the mouse nidogen-2 gene was assigned to chromosome 14, whereas the nidogen-1 gene is localized on chromosome 13 (15). Thus, the different chromosomal locations of nidogen genes will facilitate the generation of nidogen-1 nidogen-2 double-mutant mice by simple cross-breeding of the corresponding mouse lines.
Toward understanding the in vivo roles of nidogen-2 our isolation and molecular characterization of a mutant allele of the nidogen-2 gene has identified the gene trap insertion site in intron 4 of the nidogen-2 gene. Our data from genetic, biochemical, and molecular analyses show that homozygous mutant mice are viable and nidogen-2 deficient. This work presents further evidence that the secretory gene trapping method can access all classes of proteins targeted to the secretory pathway, and most importantly, can efficiently mutate trapped genes and create null alleles in mice in both gene-based and phenotype-driven screens (34). The histological and ultrastructural analyses of tissues from different organs performed in the study show the formation of BM in the absence of nidogen-2, and there are no indications of alterations in BM. Although nidogen-2 shows a prominent localization in endothelial BM of capillaries in striated and cardiac muscle, we have not observed any overt vascular abnormality or hemorrhagic phenotype in normal physiological situations as one may have expected. Interestingly, it has been reported that deletion of the laminin α4 chain of laminin-8 (α4:β1:γ1) and laminin-9 (α4:β2:γ1), both of which carry a laminin γ1 chain, leads to impaired microvessel maturation and hemorrhages during embryonic and neonatal period (53). With respect to large blood vessels it is noteworthy that nidogen-2 and fibulin-2 can bind to endostatin and tropoelastin in vitro and both colocalize with endostatin in elastic fibers of vessel walls. Interestingly, nidogen-1 does not show this distinct binding property and expression profile. Since the collagen XVIII-derived angiogenesis inhibitor endostatin has no affinity for tropoelastin, its localization to aortic media may be explained by binding to either nidogen-2 and/or fibulin-2 (30, 46). Therefore, nidogen-2 may function in modulating the endostatin reservoir in the vessel wall and/or in the supramolecular organization of elastic fibers. In nidogen-2-null mice under normal physiological conditions we have not observed overt signs of the abnormal angiogenic alterations which may take place in the injured aorta. With respect to interactions among nidogens, tropoelastin, and fibulins, it is interesting that fibulin-1-deficient mice show vascular abnormalities (22) and fibulin-5-null mice exhibit severe elastic fiber defects in the skin, lung, and vasculature. These studies identify fibulin-5 as an essential determinant of elastic fiber organization (37, 60). Despite the fact that nidogen-2 binds strongly to tropoelastin (46) and shows cell binding properties in vitro (21, 44) our histological analyses have not revealed pathological changes in the elastic fiber organization and/or connection to adjacent cellular components. We cannot, however, exclude changes which may manifest in aged mice.
In nidogen-1-null mice, nidogen-2 staining is increased in certain BM, particularly in cardiac and skeletal muscle, where it normally has only a limited expression (35). We have not observed a similar compensatory increase in the level of nidogen-1, particularly in endothelial BM of nidogen-2-null mice. Our quantitative studies comparing the content of nidogens and laminin γ1 chain in different tissues from wild-type and nidogen-2-deficient mice by highly sensitive radioimmunoassays (55) document the dominant expression of nidogen-1 (in the range of 30- to 70-fold) over nidogen-2 protein in control tissues and the absence of nidogen-2 in tissues from homozygous mutant nidogen-2 mice. Furthermore, the amount of nidogen-1 in these tissues is 7.5- to 14-fold higher than laminin γ1 independent of the genotype of mice. Since unbound nidogen-1 appears to be highly protease sensitive (26), it is suggestive that excess of nidogen-1 over laminin γ1 may be in part derived from its binding to nonprocessed forms of laminin-5 γ2 chain as has been recently shown in vitro (47). Thus, this newly identified interaction may also take place in vivo and may be functionally different from the nidogen-laminin γ1 interaction.
Interestingly, a C. elegans mutation was described recently in which the loss of its single nidogen gene did not interfere with BM formation but resulted in alterations in axonal patterning (18). This suggests that the nidogens may have nonstructural functions. This will have to be further considered in the analysis of mice with either single or double mutations for nidogen-1 and nidogen-2, in particular with respect to subtle neurological changes.
Targeting the LAMC1 gene coding for the ubiquitous laminin γ1 chain causes the absence of the laminin network and prevents BM formation. This mutation results in early embryonic lethality due to the failure of endoderm differentiation (50). In addition, it has been shown that the loss of less-ubiquitous laminin subunit α5, α3, or β2 causes an intrinsically weaker BM, affecting different tissues (29, 43, 59). As nidogen-1-null and nidogen-2-null animals each on their own appear to develop normally without any evidence of structural alterations or deformity in the BM, we conclude that the lack of nidogen-1 or nidogen-2 has no effect upon laminin deposition. Interestingly, the loss of perlecan results in a less-stable BM in certain tissues and defects in cartilage development during embryonic development (1, 5). Therefore, our studies and those by Murshed et al. (35) suggest that individual nidogens are dispensable for the retention of perlecan in the BM. Together these results suggest that nidogen-1 and nidogen-2 have no structural role in the BM, that their function in BM formation is redundant with other proteins, or that in the absence of one the other can fully compensate.
The distinction between the biological roles of nidogen binding modules of the laminin γ1 chain and nonprocessed form of laminin-5 γ2 chain may be achieved genetically by module-specific gene mutations in ES cells and mice. This is feasible as it is shown for the deletion of the nidogen binding module γ1III4 of the LAMC1 gene compromising kidney and lung development in mice (57). The in vivo relevance of nidogens' interactions to other proteins, e.g., collagen IV, perlecan and fibulins, is less clear, but in mice deficient for both nidogens we may observe important roles for nidogens in cartilage development, where perlecan is important (1, 5), or in matrix assembly of BM, especially in blood vessel BM, which express fibulins (13, 22, 37, 61).
Taken together, our studies of nidogen-2-deficient mice and those of the nidogen-1 gene knockout (35) have prompted us to generate nidogen-1-nidogen-2 double-mutant mice, and preliminary analyses indicate a perinatal lethal phenotype (B. L. Bader and S. Nedbal, Max-Planck-Institute for Biochemistry, Martinsried, Germany, and R. Nischt and N. Smyth, University of Cologne, Cologne, Germany, unpublished data). Now, crosses of nidogen mutant mice with mouse lines lacking the binding sites on relevant ligands (57) are possible and will help to genetically dissect the in vivo roles of nidogens and their multiple interactions. Perhaps these experiments will reveal unexpected functions of nidogens in the BM network and may provide a more focused view on how nidogens work as organizers of BM formation.
Acknowledgments
We thank Viola Westerbarkey for technical assistance and Markus Keller and other members of the Bader Laboratory for their helpful comments.
This work and Sabine Nedbal were supported by the grant Ba 1327/3-1 from the priority program SPP-1086 of the Deutsche Forschungsgemeinschaft (DFG) to Bernhard L. Bader.
REFERENCES
- 1.Arikawa-Hirasawa, E., H. Watanabe, H. Takami, J. R. Hassell, and Y. Yamada. 1999. Perlecan is essential for cartilage and cephalic development. Nat. Genet. 23:354-358. [DOI] [PubMed] [Google Scholar]
- 2.Bork, P., A. K. Downing, B. Kieffer, and I. D. Campbell. 1996. Structure and distribution of modules in extracellular proteins. Q. Rev. Biophys. 29:119-167. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, J., and W. C. Skarnes. 1999. Gene trapping in mouse embryonic stem cells. Methods Mol. Biol. 97:123-138. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. C., T. Sasaki, W. Göhring, Y. Yamada, and R. Timpl. 1997. The C-terminal domain V of perlecan promotes β1 integrin-mediated cell adhesion, binds heparin, nidogen and fibulin-2 and can be modified by glycosaminoglycans. Eur. J. Biochem. 250:39-46. [DOI] [PubMed] [Google Scholar]
- 5.Costell, M., E. Gustafsson, A. Aszodi, M. Mörgelin, W. Bloch, E. Hunziker, K. Addicks, R. Timpl, and R. Fässler. 1999. Perlecan maintains the integrity of cartilage and some basement membranes. J. Cell Biol. 147:1109-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durkin, M. E., U. M. Wewer, and A. E. Chung. 1995. Exon organization of the mouse entactin gene corresponds to the structural domains of the polypeptide and has regional homology to the low-density lipoprotein receptor gene. Genomics 26:219-228. [DOI] [PubMed] [Google Scholar]
- 7.Dziadek, M., and R. Timpl. 1985. Expression of nidogen and laminin in basement membranes during mouse embryogenesis and in teratocarcinoma cells. Dev. Biol. 111:372-382. [DOI] [PubMed] [Google Scholar]
- 8.Ekblom, P. 1993. Basement membranes in development, p. 359-378. In D. H. Rohrbach and R. Timpl (ed.), Molecular and cellular aspects of basement membranes. Academic Press, New York, N.Y.
- 9.Ekblom, P., M. Ekblom, L. Fecker, G. Klein, H.-Y. Zhang, Y. Kadoya, M.-L. Chu, U. Mayer, and R. Timpl. 1994. Role of mesenchymal nidogen for epithelial morphogenesis in vitro. Development 120:2003-2014. [DOI] [PubMed] [Google Scholar]
- 10.Ekblom, P., M. Durbeej, and M. Ekblom. 1996. Laminin isoforms in development, p. 185-216. In P. Ekblom and R. Timpl (ed.), The laminins. Harwood Academic Publishers, Chur, Switzerland.
- 11.Fox, J. W., U. Mayer, R. Nischt, M. Aumailley, D. Reinhardt, H. Wiedemann, K. Mann, R. Timpl, T. Krieg, J. Engel, and M.-L. Chu. 1991. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 10:3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, D. S., and H. K. Kleinman. 1997. Regulation of capillary formation by laminin and other components of the extracellular matrix, p. 317-359. In I. D. Goldberg and E. M. Rosen (ed.), Regulation of angiogenesis. Birkhäuser Verlag, Basel, Switzerland. [DOI] [PubMed]
- 13.Hungerford, J. E., G. K. Owens, W. S. Argraves, and C. D. Little. 1996. Development of aortic vessel wall as defined by vascular smooth muscle and extracellular matrix markers. Dev. Biol. 178:375-392. [DOI] [PubMed] [Google Scholar]
- 14.Hutter, H., B. E. Vogel, J. D. Plenefisch, C. R. Norris, R. B. Proenca, J. Spieth, C. Guo, S. Mastwal, X. Zhu, J. Scheel, and E. M. Hedgecock. 2000. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science 287:989-994. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, N. A., M. J. Justice, D. J. Gilbert, M.-L. Chu, and N. G. Copeland. 1991. Nidogen/entactin (Nid) maps to the proximal end of mouse chromosome 13 linked to beige (bg) and identifies a new region of homology between mouse and human chromosomes. Genomics 9:401-403. [DOI] [PubMed] [Google Scholar]
- 16.Kadoya, Y., K. Salmivirta, J. F. Talts, K. Kadoya, U. Mayer, R. Timpl, and P. Ekblom. 1997. Importance of nidogen binding to laminin γ1 for branching epithelial morphogenesis of the submandibular gland. Development 124:683-691. [DOI] [PubMed] [Google Scholar]
- 17.Kang, S. H., and J. M. Kramer. 2000. Nidogen is not essentiell and not required for normal collagen IV localization in Caenorhabditis elegans. Mol. Biol. Cell 11:3911-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S., and W. G. Wadsworth. 2000. Positioning of longitudinal nerves in C. elegans by nidogen. Science 288:150-154. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, N., T. Toyoshima, T. Kojima, and M. Shimane. 1998. Entactin-2: a new member of basement membrane protein with high homology to entactin/nidogen. Exp. Cell Res. 241:36-45. [DOI] [PubMed] [Google Scholar]
- 20.Kleinman, H. K., M. L. McGarvey, L. A. Liotta, P. Gehron Robebey, K. Tryggvarson, and G. R. Martin. 1982. Isolation and characterization of IV procollagen, laminin and heparan sulfate proteoglycan from EHS sarcoma. Biochemistry 21:6188-6193. [DOI] [PubMed] [Google Scholar]
- 21.Kohfeldt, E., T. Sasaki, W. Göhring, and R. Timpl. 1998. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 282:99-109. [DOI] [PubMed] [Google Scholar]
- 22.Kostka, G., R. Giltay, W. Bloch, K. Addicks, R. Timpl, R. Fässler, and M.-L. Chu. 2001. Perinatal lethality and endothelial cell abnormalities in several compartments of fibulin-1-deficient mice. Mol. Cell. Biol. 20:7025-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mann, K., R. Deutzmann, M. Aumailley, R. Timpl, L. Raimondi, Y. Yamada, T.-C. Pan, D. Conway, and M.-L. Chu. 1989. Amino acid sequence of mouse nidogen, a multidomain basement membrane protein with binding activity for laminin, collagen IV and cells. EMBO J. 8:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer, U., R. Nischt, E. Pöschl, K. Mann, K. Fukuda, M. Gerl, Y. Yamada, and R. Timpl. 1993. A single EGF-like motif of laminin is responsible for high affinity nidogen binding. EMBO J. 125:1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer, U., and R. Timpl. 1994. Nidogen, a versatile binding protein of basement membranes, p. 389-416. In P. D. Yurchenco, D. Birk, and R. P. Mecham (ed.), Extracellular matrix assembly and structure. Academic Press, Orlando, Fla.
- 26.Mayer, U., K. Zimmermann, K. Mann, D. Reinhardt, R. Timpl, and R. Nischt. 1995. Binding properties and protease stability of recombinant human nidogen. Eur. J. Biochem. 227:681-686. [DOI] [PubMed] [Google Scholar]
- 27.Mayer, U., E. Kohfeldt, and R. Timpl. 1998. Structural and genetic analysis of laminin-nidogen interaction. Ann. N. Y. Acad. Sci. 857:130-142. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy, L. C., J. Terrett, M. E. Davis, C. J. Knights, A. L. Smith, R. Critcher, K. Schmitt, J. Hudson, N. K. Spurr, and P. N. Goodfellow. 1997. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 7:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miner, J. H., J. Cunningham, and J. R. Sanes. 1998. Roles for laminin in embryogenesis: exencephaly, syndactyly, and placentopathy in mice lacking the laminin α5 chain. J. Cell Biol. 143:1713-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miosge, N., T. Sasaki, and R. Timpl. 1999. Angiogenesis inhibitor endostatin is a distinct component of elastic fibers in vessel walls. FASEB J. 13:1743-1790. [DOI] [PubMed] [Google Scholar]
- 31.Miosge, N., S. Heinemann, A. Leissling, C. Klenczar, and R. Herken. 1999. Ultrastructural triple localization of laminin-1, nidogen-1, and collagen type IV helps elucidate basement membrane structure in vivo. Anat. Rec. 254:382-388. [DOI] [PubMed] [Google Scholar]
- 32.Miosge, N., F. Quondamatteo, C. Klenczar, and R. Herken. 2000. Nidogen-1: expression and ultrastructural localization during the onset of mesoderm formation in the early mouse embryo. J. Histochem. Cytochem. 48:229-237. [DOI] [PubMed] [Google Scholar]
- 33.Miosge, N., F. Köther, S. Heinemann, E. Kohfeld, R. Herken, and R. Timpl. 2000. Ultrastructural colocalization of nidogen-1 and nidogen-2 in murine kidney basement membranes. Histochem. Cell Biol. 113:115-124. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, K. J., K. I. Pinson, O. G. Kelly, J. Brennan, J. Zupicich, P. Scherz, P. A. Leighton, L. V. Goodrich, X. Lu, B. J. Avery, P. Tate, K. Dill, E. Pangilinan, P. Wakenight, M. Tessier-Lavigne, and W. C. Skarnes. 2001. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 28:241-249. [DOI] [PubMed] [Google Scholar]
- 35.Murshed, M., N. Smyth, N. Miosge, J. Karolat, T. Krieg, M. Paulsson, and R. Nischt. 2000. The absence of nidogen 1 does not affect murine basement membrane formation. Mol. Cell. Biol. 20:7007-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakae, H., M. Sugano, Y. Ishimori, T. Endo, and T. Obinata. 1993. Ascidian entactin/nidogen. Implication of evolution by shuffling two kinds of cysteine-rich motifs. Eur. J. Biochem. 213:11-19. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, T., P. R. Lozana, Y. Ikeda, Y. Iwanga, A. Hinek, S. Minamisawa, C.-F. Cheng, K. Kobuke, N. Dalton, Y. Takada, K. Tashiro, J. Ross, Jr., T. Honjo, and K. R. Chien. 2002. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 415:171-175. [DOI] [PubMed] [Google Scholar]
- 38.Nicosia, R. F., E. Bonanno, M. Smith, and P. Yurchenco. 1994. Modulation of angiogenesis in vitro by laminin-entactin complex. Dev. Biol. 164:197-206. [DOI] [PubMed] [Google Scholar]
- 39.Paulsson, M., R. Deutzmann, M. Dziadek, H. Nowack, R. Timpl, S. Weber, and J. Engel. 1986. Purification and properties of intact and degraded nidogen obtained from a tumor basement membrane. Eur. J. Biochem. 166:11-19. [DOI] [PubMed] [Google Scholar]
- 40.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ries, A., W. Göhring, J. W. Fox, R. Timpl, and T. Sasaki. 2001. Recombinant domains of mouse nidogen and their binding to basement membrane proteins and monoclonal antibodies. Eur. J. Biochem. 268:5119-5128. [DOI] [PubMed] [Google Scholar]
- 42.Rowe, L. B., M. E. Barter, and J. T. Eppig. 2000. Cross-referencing radiation hybrid data to the recombination map: lessons from mouse chromosome 18. Genomics 69:27-36. [DOI] [PubMed] [Google Scholar]
- 43.Ryan, M. C., K. Lee, Y. Myashita, and W. G. Carter. 1999. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J. Cell Biol. 145:1309-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salmivirta, K., J. F. Talts, M. Olsson, T. Sasaki, R. Timpl, and P. Ekblom. Binding of mouse nidogen-2 to basement membrane components and cells and its expression in embryonic and adult tissues suggest complementary functions of the two nidogens. Exp. Cell Res., in press. [DOI] [PubMed]
- 45.Sasaki, T., K. Mann, G. Murphy, M.-L. Chu, and R. Timpl. 1996. Different susceptibilities of fibulin-1 and fibulin-2 to cleavage by matrix metalloproteinases and other tissue proteases. Eur. J. Biochem. 240:427-434. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki, T., W. Göhring, N. Miosge, W. R. Abrams, J. Rosenbloom, and R. Timpl. 1999. Tropoelastin binding to fibulins, nidogen-2 and other extracellular matrix proteins. FEBS Lett. 460:280-284. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki, T., W. Göhring, K. Mann, C. Brakebusch, Y. Yamada, R. Fässler, and R. Timpl. 2001. Short arm region of laminin-5 γ2 chain: structure, mechanisms of processing and binding to heparin and proteins. J. Mol. Biol. 314:761-763. [DOI] [PubMed] [Google Scholar]
- 48.Sharp, P. A., and C. B. Burge. 1997. Classification of introns: U2-type or U12-type. Cell 91:875-879. [DOI] [PubMed] [Google Scholar]
- 49.Skarnes, W. C., J. E. Moss, S. M. Hurtley, and R. S. Beddington. 1995. Capturing genes encoding membrane and secretory proteins important for mouse development. Proc. Natl. Acad. Sci. USA 92:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth, N., H. S. Vatansever, P. Murray, M. Meyer, C. Frie, M. Paulsson, and D. Edgar. 1999. Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to the failure of endoderm differentiation. J. Cell Biol. 144:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer, T. A. 1998. An extracellular β-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 283:837-862. [DOI] [PubMed] [Google Scholar]
- 52.Streuli, C. 1996. Basement membrane as a differentiation and survival factor, p. 185-216. In P. Ekblom and R. Timpl (ed.), The laminins. Harwood Academic Publishers, Chur, Switzerland.
- 53.Thyboll, J., J. Kortesmaa, C. Renhai, R. Soininen, L. Wang, A. Iivanainen, L. Sorokin, M. Risling, Y. Cao, and K. Tryggvason. 2002. Deletion of the laminin α4 chain leads to impaired microvessel maturation. Mol. Cell. Biol. 22:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timpl, R., and J. C. Brown. 1996. Supramolecular assembly of basement membranes. Bioessay 18:123-132. [DOI] [PubMed] [Google Scholar]
- 55.Timpl, R., and L. Risteli. 1982. Radioimmunoassays in studies of connective tissue proteins, p. 199-235. In H. Furthmayr (ed.), Immunochemistry of the extracellular matrix, vol. I. Methods. CRC Press, Boca Raton, Fla.
- 56.Townley, D. J., B. J. Avery, B. Rosen, and W. C. Skarnes. 1997. Rapid sequence analysis of gene trap integrations to generate a resource of insertional mutations in mice. Genome Res. 7:293-298. [DOI] [PubMed] [Google Scholar]
- 57.Willem, M., N. Miosge, W. Halfter, N. Smyth, I. Jannetti, E. Burghart, R. Timpl, and U. Mayer. 2002. Specific ablation of the nidogen-binding site in the laminin γ1 chain interferes with kidney and lung development. Development 129:2711-2722. [DOI] [PubMed] [Google Scholar]
- 58.Van Etten, W. J., R. G. Steen, H. Nguyen, A. B. Castle, D. K. Slonim, B. Ge, C. Nusbaum, G. D. Schuler, E. S. Lander, and T. J. Hudson. 1999. Radiation hybrid map of the mouse genome. Nat. Genet. 22:384-387. [DOI] [PubMed] [Google Scholar]
- 59.Xu, H., P. Christmas, X. R. Wu, U. M. Wewer, and E. Engvall. 1994. Defective muscle basement membrane and lack of M-laminin in the dystrophic dy/dy mouse. Proc. Natl. Acad. Sci. USA 91:5572-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanagisawa, H., E. C. Davis, B. C. Starcher, T. Ouchi, M. Yanagisawa, J. A. Richardson, and E. N. Olson. 2002. Fibulin-5 is an elastin-binding protein essential for elastic fibre development. Nature 415:168-171. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, H.-Y., R. Timpl, T. Sasaki, M.-L. Chu, and P. Ekblom. 1996. Fibulin-1 and fibulin-2 expression during organogenesis in the developing mouse embryo. Dev. Dyn. 205:348-364. [DOI] [PubMed] [Google Scholar]
- 62.Zimmermann, K., S. Hoischen, M. Hafner, and R. Nischt. 1995. Genomic sequences and structural organization of the human nidogen gene (NID). Genomics 27:245-250. [DOI] [PubMed] [Google Scholar]