Abstract
The chromatin structure of the pea plastocyanin gene (PetE) was examined at three different transcriptional states by investigating the acetylation states of histones H3 and H4 and the nuclease accessibility of the gene in pea roots, etiolated shoots, and green shoots. The acetylation states of histones associated with different regions of PetE were analyzed by chromatin immunoprecipitation with antibodies specific for acetylated or nonacetylated histone H3 or H4 tails, followed by polymerase chain reaction quantification. Comparison of pea tissues indicated that histone hyperacetylation was associated with increased PetE transcription in green shoots. Moreover, hyperacetylation of both histones H3 and H4 was targeted to the enhancer/promoter region in green shoots, suggesting that only specific nucleosomes along the gene were modified. Time-course digestions of nuclei with micrococcal nuclease and DNaseI indicated that the enhancer/promoter region was more resistant to digestion in the inactive gene in pea roots than was the same region in the active gene in shoots, whereas the transcribed region of PetE was digested similarly among the tissues. This finding indicates that transcription is accompanied by changes in the nuclease accessibility of the enhancer/promoter region only. Moreover, these results indicate that the changes in nuclease accessibility are organ specific, whereas histone hyperacetylation is light dependent, and they suggest that changes in nuclease accessibility precede histone hyperacetylation during PetE activation.
INTRODUCTION
Plastocyanin is a 10-kD copper protein that transfers electrons from cytochrome f to the primary donor P700 of the photosystem I reaction center in the photosynthetic electron transfer chain. In pea, the plastocyanin gene (PetE) is a single-copy intronless nuclear gene that is expressed in a light-induced and organ-specific manner (Last and Gray, 1989, 1990). Pea PetE contains an upstream enhancer element (−444 to −177 with respect to the start codon) that activates the expression of reporter genes, directed by minimal cauliflower mosaic virus 35S, patatin, or PetE promoters, in the leaves and roots of stable transgenic tobacco and potato plants (Sandhu et al., 1998). The enhancer element is able to increase reporter gene expression by as much as 40-fold, thereby representing one of the strongest plant enhancers characterized to date (Pwee and Gray, 1993; Sandhu et al., 1998). At least two lines of evidence suggest that the enhancer element increases transcription through the modulation of chromatin structure: (1) the enhancer element fails to increase reporter gene expression when the same constructs are introduced transiently into plant cells, suggesting that the enhancer requires a chromatin context to increase transcription (J.S. Sandhu and J.C. Gray, unpublished data); and (2) the enhancer element interacts strongly with pea HMG-I/Y and HMG-1 proteins (Pwee et al., 1994; Webster et al., 1997), which are “architectural” chromosomal proteins that maintain chromatin in a conformation favorable for transcription (Grosschedl et al., 1994; Grasser, 1998).
Chromatin structure affects transcription through nucleosomes, the basic structural units of chromatin in eukaryotic cells (Brownell and Allis, 1996; Wolffe and Hayes, 1999). Each nucleosome is composed of two turns of DNA wound around a histone octamer containing two molecules each of H2A, H2B, H3, and H4 and is linked to the next nucleosome by linker DNA. Mutations that change the lysine residues in the N termini of histone H3 or H4 or that alter the levels of nucleosomes in yeast change the transcriptional activities of genes, indicating that nucleosomes affect transcription through histone acetylation and nucleosome positioning (Durrin et al., 1991; Fisher-Adams and Grunstein, 1995; Wyrick et al., 1999).
Acetylation of histones involves the transfer of acetyl groups from acetyl-CoA to the ɛ-amino groups of K9 or K14 of histone H3, or K5, K8, K12, or K16 of histone H4 by histone acetyltransferases (HATs). Hyperacetylated nucleosomes are correlated with the potential for transcription because both active and inducible genes are generally associated with hyperacetylated histone H3 or H4 (reviewed in Struhl, 1998). Conversely, the inactive X chromosomes in human and mouse, the transcriptionally silent telomeric and heterochromatic regions in human chromosomes, and the silent loci in yeast are generally associated with hypoacetylated or nonacetylated histone H3 or H4 (Braunstein et al., 1993; Jeppesen and Turner, 1993; O'Neill and Turner, 1995). A direct link between histone hyperacetylation and transcription has been established through the characterization of the following: (1) transcriptional coactivators, which require their HAT activities for transcription activation (Kuo et al., 1998; Wang et al., 1998); (2) the viral oncoprotein E1A, which represses transcription by inhibiting the HAT activities of transcription regulators (Chakravarti et al., 1999); and (3) transcription repressors, which require the deacetylase activities of histone deacetylase complexes to function (Bird and Wolffe, 1999; Brehm et al., 1999; Kao et al., 2000). Histone acetylation is also important in nucleolar dominance in Brassica napus, in which silent rRNA genes become active in the presence of histone deacetylase inhibitors. This finding suggests that histone acetylation plays a role in the regulation of RNA polymerase I transcription in plants (Chen and Pikaard, 1997).
The positioning of nucleosomes on promoters is inhibitory to transcription in that it prevents transcription factors from binding to the promoter sites (Venter et al., 1994; Tyler and Kadonaga, 1999). Transcriptional activation of these promoters, therefore, requires the active repositioning of nucleosomes. Consistent with this is the characterization of a variety of chromatin-remodeling factors in yeast and metazoans, such as the SWI/SNF, ISWI, and NURF complexes, which are capable of promoting gene activation by moving nucleosomes (Tyler and Kadonaga, 1999; Whitehouse et al., 1999).
In plants, chromatin structure also influences transcription. New regions of DNaseI hypersensitivity upstream of the maize Adh1 and Adh2 genes (Ashraf et al., 1987; Paul et al., 1987), the maize Shrunken gene (Frommer and Starlinger, 1988), the maize P gene (Lund et al., 1995), the pea RbcS genes (Görz et al., 1988), and the Arabidopsis Adh gene (Vega-Palas and Ferl, 1995; Paul and Ferl, 1998) are formed in vivo as the expression of the genes increases. These induced hypersensitivity sites suggest that the nucleosome arrays in these regions are disrupted upon transcription and are likely to be the binding sites of transcription factors (Gross and Garrard, 1988). In wheat, DNaseI preferentially digests transcriptionally active sequences, suggesting that these sequences assume open chromatin structures (Spiker et al., 1983). Moreover, the nucleosome present on the β-phaseolin promoter is removed upon gene activation in transgenic tobacco (Li et al., 1998), and nucleosomes are partly responsible for the closed conformation of a silent, methylated A1 transgene in petunia (van Blokland et al., 1997). Therefore, nucleosome positioning and arrangement also may play an active role in plant gene transcription regulation.
Despite these experiments, little information is available on the chromatin structure of regions across a plant gene and its effects on transcription. We have examined the distribution of histone acetylation states and the nuclease accessibility of PetE at the single gene level to investigate whether changes in chromatin structure are involved in the regulation of PetE transcription. Short DNA regions of 140 to 607 bp were analyzed across PetE to identify regions whose chromatin structure changes with transcription. Our results indicate that the structural changes are not random but are targeted to short defined regions in a highly specific manner. Moreover, changes in nuclease accessibility can be correlated with the absence or presence of transcription, whereas modification of histone acetylation states is correlated with the rate at which transcription occurs.
RESULTS
Transcription of PetE Is Higher in Green Pea Shoots Than in Etiolated Shoots and Is Absent in Roots
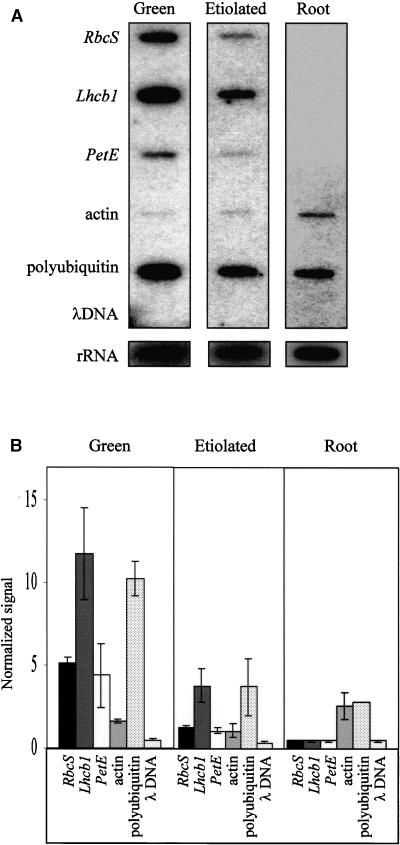
In pea, the accumulation of PetE transcripts is shoot specific and light induced (Last and Gray, 1989, 1990). Thus, the regulation of PetE expression at the transcriptional level was examined by performing run-on assays with nuclei isolated from green pea shoots, etiolated shoots, and roots (Figure 1A). In nuclei from green shoots, high levels of 32P-labeled transcripts were produced from the photosynthesis genes RbcS, Lhcb1, and PetE. These genes were transcribed at much lower levels in etiolated shoots. Gallagher and Ellis (1982) and Sagar et al. (1988) previously investigated the transcription of pea RbcS and Lhcb1 with nuclear run-on assays and demonstrated that these two genes are transcribed at a basal level in dark-grown pea shoots. To compare the relative transcription rates of the photosynthesis genes in etiolated and green shoots, the amount of transcript obtained from the genes was normalized to the amount of rRNA transcript to account for loading differences between experiments; the results are presented in Figure 1B. In green shoots, RbcS was transcribed to levels approximately fivefold higher, Lhcb1 to levels fourfold higher, and PetE to levels threefold higher than in etiolated shoots. Thus, like that of RbcS and Lhcb1, the transcription of PetE was activated in light.
Figure 1.
Run-On Transcription of PetE in Isolated Pea Nuclei.
Run-on assays were performed with nuclei prepared from shoots of 7-day-old pea seedlings grown in light or dark or from roots of 7-day- old pea seedlings grown in light. The nuclei were incubated with α-32P-labeled UTP, ATP, CTP, GTP, phosphocreatine, and creatine phosphokinase at 30°C for 25 min before RNA extraction.
(A) DNA slot blots hybridized with 32P-labeled RNA synthesized in isolated nuclei. The slot blots were exposed to x-ray films for 7 days to obtain signals for the photosynthesis, actin, and polyubiquitin genes and for 2 days to obtain signals for the rRNA genes.
(B) Quantification of hybridization signals. The hybridization signals obtained for the genes were normalized to rRNA transcription, and the averages determined were from three independent run-on experiments. The error bars indicate ±se.
In contrast to the shoot tissues, no transcripts were detected for the photosynthesis genes in pea roots. Hence, the organ-specific expression of RbcS, Lhcb1, and PetE is controlled at the transcriptional level in pea. Consistent with this result, transgenic tobacco plants that contain the uidA reporter gene fused to the PetE promoter show β-glucuronidase (GUS) activity in shoots but not in roots (Pwee and Gray, 1993).
Green shoots, etiolated shoots, and roots thereby represent three different transcriptional states of PetE: an activated state, a basal state, and an inactive state. The chromatin structure of the gene in these three transcriptional states was analyzed next.
The Enhancer, Promoter, and Transcribed Regions of PetE Are Associated with Hyperacetylated Histone H4 in Green Shoots
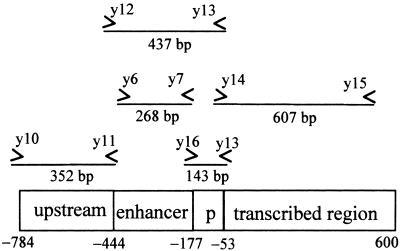
As shown in Figure 2, pea PetE can be divided into four functionally defined regions. The gene possesses an enhancer of 268 bp (−444 to −177), a minimal promoter of 124 bp (−176 to −53), and a transcribed region containing a coding sequence of 504 bp with no introns (Last and Gray, 1989). The enhancer region increases expression by as much as 40-fold when fused upstream to the minimal PetE promoter and GUS reporter gene (Pwee and Gray, 1993; Sandhu et al., 1998). The minimal promoter is able to confer cell-specific expression by directing a low level of reporter gene expression in chloroplast-containing cells (Pwee and Gray, 1993). Transgenic work has also indicated that the transcribed region is required for the light-regulated expression of PetE (Helliwell et al., 1997). Pea PetE also possesses an upstream region (−784 to −445) that does not influence reporter gene expression in transgenic tobacco plants (Pwee and Gray, 1993). Therefore, we analyzed the chromatin structure of the upstream, enhancer, promoter, and transcribed regions of PetE.
Figure 2.
Scheme of Pea PetE.
The locations of primers and the sizes of PCR fragments generated by each primer set are indicated. p, promoter.
Components of transcriptional activators have been shown to preferentially acetylate lysines in either histone H3 or H4 (reviewed in Grunstein, 1997; Strahl and Allis, 2000). Therefore, the acetylation states of both histones were analyzed to determine whether they were involved in PetE transcription. To accomplish this, we performed chromatin immunoprecipitation (ChIP), a method that investigates whether a DNA sequence and a protein of interest interact (Braunstein et al., 1993). Nuclei extracted from pea tissues were first treated with formaldehyde to cross-link proteins to their contiguous DNA sequences and then sonicated to fragment the chromatin material into an average length of 300 to 400 bp. Next, the sheared chromatin was incubated with antibodies specific for acetylated or nonacetylated histone H3 or H4 for coimmunoprecipitation of DNAs that were associated with the proteins. As controls, the precipitation step was also performed in the absence of antibody and with nonimmune rabbit serum. The immunocomplexes were heated at 65°C for 5 hr to reverse the cross-links, and the DNA released was analyzed by polymerase chain reaction (PCR).
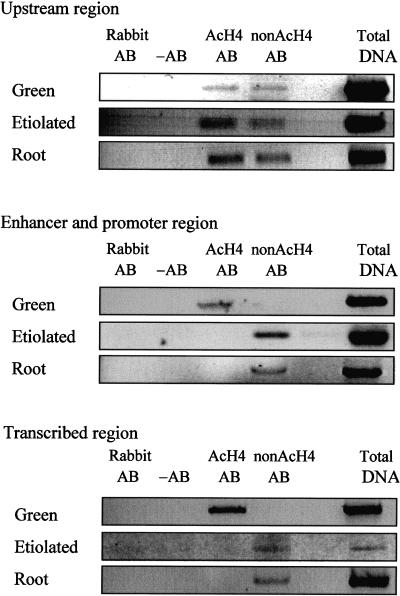
The enhancer/promoter (−492 to −56) and transcribed regions of PetE (−103 to +504) were enriched in the hyperacetylated H4 antibody immunoprecipitates from green shoots but were depleted in the immunoprecipitates from etiolated shoots and roots (Figure 3). This finding suggests that these regions were associated with hyperacetylated histone H4 in green shoots and with hypoacetylated histone H4 in etiolated shoots and roots. However, to eliminate the possibility that the absence of these DNA sequences in the immunoprecipitates from etiolated shoots and roots was due to decreased protein–DNA cross-linking efficiencies or decreased accessibility of nucleosomes to antibodies in these tissues, we included an antibody that recognizes nonacetylated histone H4 in our ChIP experiment. The incubation of the etiolated shoot and root chromatin with this antibody coprecipitated the enhancer/promoter and transcribed sequences, confirming that these regions were associated with hypoacetylated histone H4 in both tissues (Figure 3). Thus, upon greening, the histone H4 present on the enhancer/promoter and transcribed regions of PetE in etiolated shoots was selectively hyperacetylated. The histone H4 present on the upstream region (−800 to −449) of PetE possessed a mixture of acetylated and nonacetylated lysine residues in all three tissues (Figure 3), indicating that the acetylation status of the region remained unchanged in relation to the transcriptional activity of PetE.
Figure 3.
Acetylation States of Histone H4 Associated with Pea PetE.
Nuclei isolated from green shoots, etiolated shoots, and roots were cross-linked, sonicated, and immunoprecipitated with antibodies specific for acetylated histone H4 (AcH4 AB) or nonacetylated histone H4 (nonAcH4 AB). The immunoprecipitates were analyzed for the presence of DNA with PCR. The upstream (−800 to −449), enhancer/promoter (−492 to −56), and transcribed (−103 to +504) regions of PetE were examined. The lanes labeled Total DNA contain the products of PCR performed with chromatin solution before immunoprecipitation. The immunoprecipitation step was also performed without antibody (−AB) and with nonimmune rabbit serum (Rabbit AB). Similar results were obtained for three replicates of ChIP experiments.
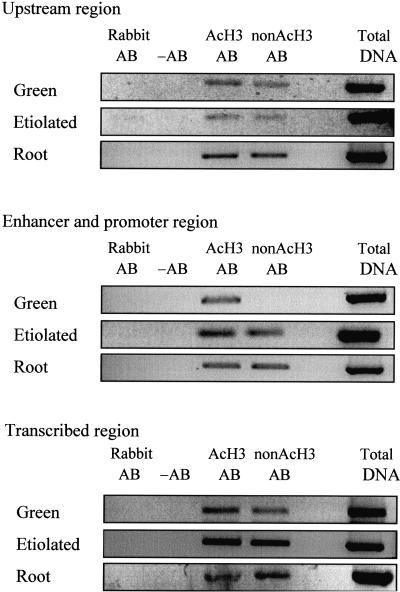
The Enhancer and Promoter of PetE Are Associated with Hyperacetylated Histone H3 in Green Shoots
ChIPs performed with antibodies against acetylated or nonacetylated histone H3 indicated that only the enhancer/promoter region of PetE contained hyperacetylated histone H3 in green shoots (Figure 4). The hyperacetylation of histone H3, therefore, was targeted to the enhancer/promoter region. This is in contrast to histone H4, which was hyperacetylated in nucleosomes present on both the enhancer/promoter and transcribed regions. Thus, the regions of PetE that are associated with hyperacetylated H3 and H4 are different in green shoots.
Figure 4.
Acetylation States of Histone H3 Associated with Pea PetE.
Nuclei were cross-linked, sonicated, and immunoprecipitated with antibodies specific for acetylated histone H3 (AcH3 AB) or nonacetylated histone H3 (nonAcH3 AB). The immunoprecipitates were analyzed for the presence of DNA with PCR. The upstream (−800 to −449), enhancer/promoter (−492 to −56), and transcribed (−103 to +504) regions of PetE were examined. The lanes labeled Total DNA contain the products of PCR performed with chromatin solution before immunoprecipitation. The immunoprecipitation step was also performed without antibody (−AB) and with nonimmune rabbit serum (Rabbit AB). Similar results were obtained for three replicates of ChIP experiments.
In etiolated shoots and roots, histone H3 associated with the upstream, enhancer/promoter, and transcribed regions contained a mixture of acetylated and nonacetylated lysine residues (Figure 4). Thus, the acetylation patterns of both histones H3 and H4 associated with PetE are similar in etiolated shoots and roots. In green shoots, the histone H3 associated with the upstream and transcribed regions also contained a mixture of acetylated and nonacetylated lysines. Therefore, the overall histone H3 acetylation patterns in the upstream and transcribed regions were similar among the three tissues. In contrast, the histone H4 acetylation pattern in the transcribed region in green shoots was very different from that in etiolated shoots and roots.
Nuclease Accessibility of PetE
After investigating histone acetylation states, we proceeded to examine the nuclease accessibility of regions of PetE as a means of assessing the chromatin structure of the gene. We examined the rates of degradation of short regions (140 to 607 bp) of PetE in green shoots, etiolated shoots, and roots by micrococcal nuclease and DNaseI. Both endonucleases possess different DNA digestion characteristics and exhibit different sequence specificities (Hörz and Altenburger, 1981; Simpson, 1998). Micrococcal nuclease preferentially attacks the internucleosomal linker regions but not the DNA sequences that are wrapped around the nucleosome. In contrast, DNaseI cleaves the DNA wrapped around the nucleosome readily and at positions where the minor groove faces away from the nucleosome (Simpson, 1998). Because the digestion of DNA by both enzymes is influenced by the presence of nucleosomes, the digestion data for both enzymes should complement each other to reflect the nucleosome structure along PetE.
The digestions were performed by incubating nuclei from green shoots, etiolated shoots, and roots with the endonucleases. A time-course experiment was performed for each digestion, and DNA was isolated from the various time points. The amounts of the upstream, enhancer, promoter, and transcribed regions present at each time point were determined by quantitative PCR and normalized to the amount of DNA present at time 0. A control reaction without endonuclease was included in the digestion experiment to assess the presence of inherent nuclease activities in the nuclei.
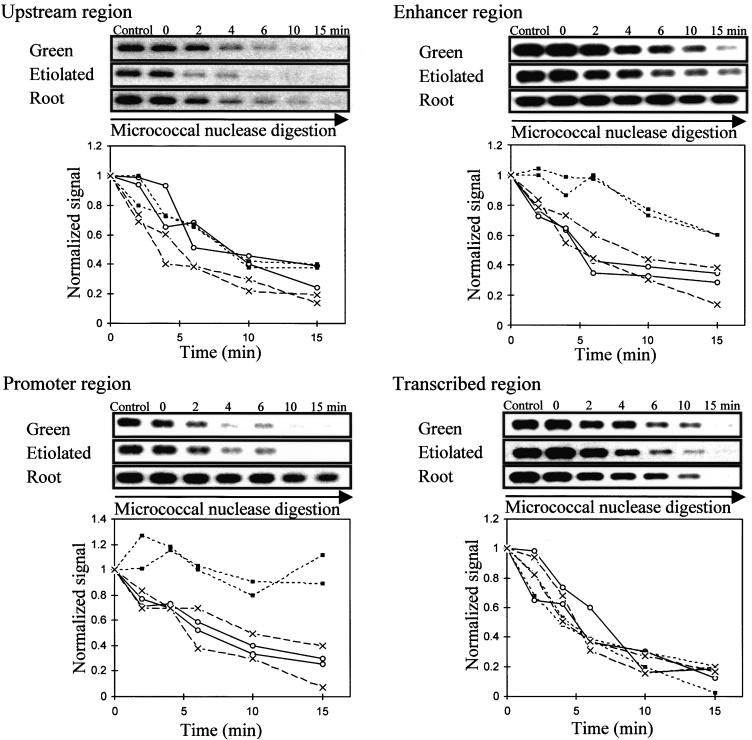
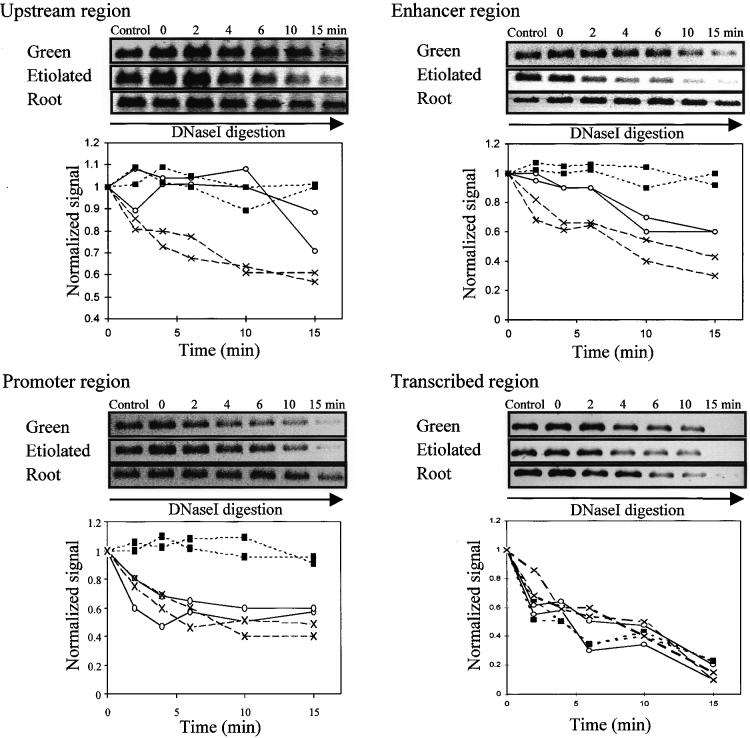
The rates of degradation of the transcribed region of PetE by both micrococcal nuclease and DNaseI were similar in green shoot, etiolated shoot, and root nuclei (Figures 5 and 6). The accessibility of this region to protein factors was thus similar among the three pea tissues. In contrast, the promoter (−195 to −56) was degraded at a slower rate in pea roots than was the same sequence in green and etiolated shoots (Figures 5 and 6). This finding indicates that the promoter possessed a relatively closed conformation in pea roots. A closed conformation in the promoter has also been observed in many transcriptionally inactive genes in animal systems and has been proposed to be responsible for the inactive state of the genes by preventing transcription factors from binding to their cognate DNA sites (Paranjape et al., 1994). In the pea PetE gene, the relative inaccessibility of the promoter region was also correlated with the silent state of the gene.
Figure 5.
Accessibility of Regions of PetE to Micrococcal Nuclease.
Nuclei were incubated with micrococcal nuclease for the times indicated. Control represents a reaction without added micrococcal nuclease. The amount of DNA sequence present at each time was determined by PCR. The PCR products were separated on an agarose gel, transferred onto GeneScreen Plus membrane, probed with 32P-labeled PCR products, exposed to x-ray film, and quantified. The upstream (−800 to −449), enhancer (−444 to −177), promoter (−195 to −56), and transcribed (−103 to +504) regions of PetE were examined. For each region of PetE, PCR products obtained for each time are shown at the top, and degradation curves for two independent digestion experiments for each pea organ are shown at the bottom. The amount of DNA present at each time was normalized to that at time 0 and plotted against time to compare degradation rates in green shoots (open circles), etiolated shoots (crosses), and roots (closed squares).
Figure 6.
Accessibility of Regions of PetE to DNaseI.
Nuclei were incubated with DNaseI for the times indicated. Control represents a reaction without added DNaseI. The amount of DNA sequence present at each time was determined by PCR. The PCR products were separated on an agarose gel and stained with ethidium bromide, and the fluorescence of the bands was quantified. The upstream (−800 to −449), enhancer (−444 to −177), promoter (−195 to −56), and transcribed (−103 to +504) regions of PetE were examined. For each region of PetE, PCR products obtained for each time are shown at the top, and degradation curves for two independent digestion experiments for each pea organ are shown at the bottom. The amount of DNA present at each time was normalized to that at time 0 and plotted against time to compare degradation rates in green shoots (open circles), etiolated shoots (crosses), and roots (closed squares).
Like the promoter, the pea PetE enhancer (−444 to −177) was largely inaccessible to both micrococcal nuclease and DNaseI in pea roots (Figures 5 and 6). This also indicates a closed conformation, which may be expected to discourage the binding of transcriptional factors and thereby prevent transcription. Comparing green and etiolated shoots, the rates of degradation of the enhancer region were similar in micrococcal nuclease digestions but different in DNaseI digestions. The enhancer sequence was degraded more rapidly by DNaseI in etiolated shoots than in green shoots. This suggests that non-nucleosomal proteins may be bound to the enhancer region in green shoots because digestion by DNaseI, but not by micrococcal nuclease, is also affected by the presence of non-nucleosomal proteins (Gross and Garrard, 1988).
Both the micrococcal nuclease and DNaseI digestion experiments indicated that the upstream region was more accessible in etiolated shoots than in green shoots and roots (Figures 5 and 6), suggesting that the upstream region possessed a more open conformation in etiolated shoots. Comparing the digestion data obtained for all four regions among the tissues, it was observed that the chromatin structure of PetE is very open in etiolated shoots.
The digestion experiments were also performed at a higher nuclease concentration, and similar results were obtained (data not shown). We also investigated the rates at which the four regions of PetE were digested after DNA binding proteins were removed. Total genomic DNA isolated from green shoot, etiolated shoot, and root nuclei was treated with phenol to remove proteins. Similar time-course digestion experiments with micrococcal nuclease and DNaseI were performed with the naked DNA. The rates of digestion of the naked DNA by both nucleases were similar among the three tissues for all regions (data not shown). This further indicated that the differences observed in the digestion rates of the upstream, enhancer, and promoter regions in intact nuclei were due to the different degrees to which these regions were accessible to proteins and not to DNA modifications.
DISCUSSION
Hyperacetylation of Nucleosomes Is Associated with High Transcription Rates
Using ChIP with a panel of antisera that recognize acetylated and nonacetylated histone tails, we mapped the distribution of histone acetylation across PetE. The results show that histones H3 and H4 are hyperacetylated in green shoots, where the transcription is high, but not in etiolated shoots or roots, where there is basal or no transcription. This finding suggests that histone hyperacetylation is correlated with increased transcription. This is consistent with the observation that HATs are components of mammalian and yeast transcriptional coactivator complexes (Struhl, 1998; Cheung et al., 2000). These coactivator complexes activate transcription through histone hyperacetylation; mutations that abolish HAT activities also prevent gene activation (Kuo et al., 1998; Wang et al., 1998). In addition, the human basal transcription factor TFIID and the yeast elongator complex also possess HAT activities (Kornberg and Lorch, 1999; Wittschieben et al., 1999). Transcriptional activation of PetE by recruitment of coactivator complexes, basal transcriptional machineries, and elongator complexes may be expected to result in histone hyperacetylation, as observed in green pea shoots.
In etiolated shoots and pea roots, reduced PetE transcription is associated with histone H4 hypoacetylation (Figure 7). This has been similarly observed in animal systems, where histone deacetylation results in transcriptional repression (Kornberg and Lorch, 1999; Turner, 2000). Furthermore, histone deacetylase complexes associate with repressor proteins to repress transcription (Luo et al., 1998; Kornberg and Lorch, 1999). Targeting of Arabidopsis homologs of yeast HD2 and RPD3 histone deacetylases to promoters of reporter genes also results in transcriptional repression (Wu et al., 2000a, 2000b). It is therefore likely that the hypoacetylated states of histone H4 present on PetE in etiolated shoots and roots are maintained by histone deacetylase complexes. It is interesting to note that histone H3 proteins on PetE are not completely deacetylated in etiolated shoots and roots. Hence, the histone deacetylase complexes involved may specifically deacetylate histone H4 to repress transcription.
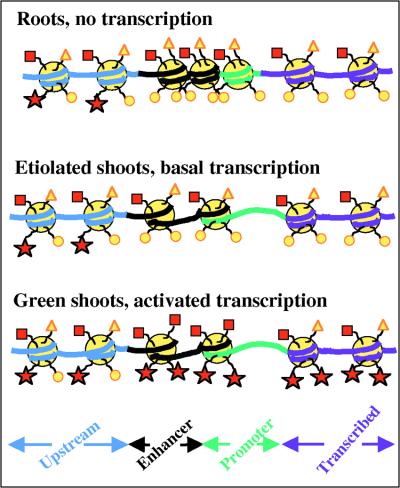
Figure 7.
Scheme for the Chromatin Structure of PetE.
The figure shows a proposed model of the histone acetylation states and nucleosome arrangement over the four analyzed regions of PetE in pea roots, etiolated shoots, and green shoots. Large yellow circles, nucleosomes; red squares, acetylated lysines in histone H3 tails; yellow triangles, nonacetylated lysines in histone H3 tails; red stars, acetylated lysines in histone H4 tails; small yellow circles, nonacetylated lysines in histone H4 tails.
In green pea shoots, the hyperacetylated histones H3 and H4 that are associated with the enhancer/promoter and transcribed regions of PetE may also function to increase transcription. There are currently two models proposed for the roles of hyperacetylated histones H3 and H4 in transcriptional activation (Wolffe and Hayes, 1999). Hyperacetylated histones may increase transcription by altering the structure of chromatin to facilitate the entry of transcription factors. Acetylation of lysine residues neutralizes the positive charges of histones, weakens histone-DNA contacts, and facilitates the displacement of nucleosomes by transcription factors (Lee et al., 1993; Puig et al., 1998). Histone hyperacetylation can also weaken nucleosome interconnections and inhibit the formation of condensed chromatin structures (Luger and Richmond, 1998; Wolffe and Hayes, 1999). Alternatively, hyperacetylated histones H3 and H4 can recruit transcription factors by functioning as signals for proteins (Strahl and Allis, 2000; Turner, 2000). Because HATs possess bromodomains that interact specifically with acetylated lysines in histone H3 and H4 tails, hyperacetylated histones may also directly recruit transcriptional coactivator complexes or anchor the complexes to genes to increase transcription (Dhalluin et al., 1999; Krajewski, 2000). It can thus be envisaged that in pea shoots, the initial recruitment of coactivator complexes to PetE involves sequence-specific transcription factors (Korzus et al., 1998; Winston and Allis, 1999; Strahl and Allis, 2000). However, once histone hyperacetylation occurs, transcription machineries are assembled more rapidly because the hyperacetylated histones can also recruit transcription factors or stabilize the coactivator complexes on chromatin. This would maintain the high PetE transcription rates in green shoots.
The observation that histone H3 is hyperacetylated only in the enhancer/promoter region of PetE in green shoots suggests that histone acetylation is not a random process but is targeted to nucleosomes present on specific DNA sequences. Considering the fact that 180 bp of DNA wraps around a nucleosome, the enhancer/promoter region (437 bp) is likely to contain two nucleosomes. Moreover, comparing the histone H3 and H4 acetylation patterns of nucleosomes present on the upstream (352 bp), enhancer/promoter (437 bp), and transcribed (607 bp) regions in green shoots, it appears that the overall histone acetylation state can change over a region of two to three nucleosomes in a specific manner. This may suggest that the modification of histone acetylation states is specific to individual nucleosomes. Indeed, targeted modification of a few nucleosomes on a gene also has been reported in mammalian systems (Kuo et al., 1998; Rundlett et al., 1998). The yeast transcriptional coactivator Gcn5p acetylates only the nucleosomes present on the promoters of reporter genes to increase transcription (Kuo et al., 1998). The yeast repressor RPD3 also selectively deacetylates the histone H4 associated with the promoter regions of UME6-regulated genes to repress transcription (Rundlett et al., 1998). The histones H3 and H4 on the interferon β promoter are specifically acetylated by the human p300/CBP HAT during viral infection (Parekh and Maniatis, 1999). Targeted histone hyperacetylation in the enhancer/promoter region in the animal genes and pea PetE can direct transcription factors to these regions either by altering the chromatin structure or by serving as signals to transcription factors. This appears to be an extremely efficient system for maintaining high transcriptional rates, as transcription factors would not be recruited to other parts of the gene.
The observation that the histone H4 tails but not the histone H3 tails are hyperacetylated in the transcribed region of the active PetE gene in green pea shoots suggests that histone H3 hyperacetylation in this region is not required for elevated transcription. Hyperacetylated histones H3 and H4 may therefore affect transcription differently. Consistent with this, histones H4 and H3 have been shown to have different functions in transcription: (1) deletion of the N terminus of histone H4 decreases the induction of GAL1 promoter by 20-fold, whereas deletion of the N terminus of histone H3 activates the promoter by threefold (Durrin et al., 1991; Fisher-Adams and Grunstein, 1995); (2) point mutations in the N terminus of histone H4 reactivate the expression of telomeric regions to a greater extent than deletion in the N terminus of histone H3 (Thompson et al., 1994); (3) in GAL1 hyperactivation, the N terminus of histone H3 regulates transcription through the upstream UAS regulatory sequence, whereas the N terminus of histone H4 acts through the promoter (Wan et al., 1995). Histone H3 and H4 tails may contact DNA differently or may obstruct the entry of transcription factors to different extents. Hyperacetylation of histones H3 and H4 may thus change the structure of chromatin differently. Alternately, the acetylation states of histones H3 and H4 may form a unique code for transcription factors (Strahl and Allis, 2000; Turner, 2000). Hence, in green pea shoots, histone H3 and H4 hyperacetylation in the enhancer/promoter region of PetE may serve to recruit transcription factors or alter the local chromatin structure to facilitate the entry of proteins, whereas hyperacetylated histone H4 on the transcribed region inhibits the formation of regular nucleosome arrays and facilitates the elongation process of transcription.
Unlike the enhancer/promoter and transcribed regions, the acetylation patterns of histones H3 and H4 in the upstream region remain relatively unchanged in the different transcriptional states of PetE. This suggests that the enhancer/ promoter and transcribed regions, but not the upstream region, participate actively in increasing the expression of PetE through histone acetylation. Consistent with this observation, the enhancer can increase transcription from the minimal cauliflower mosaic virus 35S, PetE, or patatin promoter (Sandhu et al., 1998), and the transcribed region is able to increase GUS expression in a light-dependent manner (Helliwell et al., 1997), whereas the upstream region has no effect on reporter expression in transgenic plants (Pwee and Gray, 1993).
An explanation for the observed histone H4 acetylation patterns along PetE can be suggested from the current model of histone H4 deposition onto chromatin (reviewed in Grunstein, 1997). Newly synthesized histone H4 proteins are deposited onto replicating chromatin in the diacetylated isoform in fly, frog, protist, and human (Dimitrov et al., 1993; Sobel et al., 1995). Depending on the transcription state of the gene, the histone H4 tails are then progressively hyperacetylated or hypoacetylated (Lee et al., 1993). Extrapolating this model to plants suggests that the histone H4 proteins present on newly replicated PetE are diacetylated. The acetyl groups on histone H4 would next be selectively removed in the enhancer, promoter, and transcribed regions of PetE, leaving the upstream region with diacetylated histone H4. The hypoacetylated state of histone H4 would be maintained in etiolated shoots and roots, where there is little or no transcription. However, upon illumination, the histone H4 present in nucleosomes on the enhancer, promoter, and transcribed regions would be hyperacetylated to recruit transcriptional activators. This proposal cannot be extended to histone H3 because the acetylated sites in newly synthesized histone H3 are less conserved (Sobel et al., 1995).
Nuclease Accessibility of PetE
Nucleosomes positioned over cis-elements inhibit transcription by preventing transcription factors from accessing their binding sites (Paranjape et al., 1994; Niu et al., 1996). The nuclease experiments performed here investigated directly the accessibility of various regions of PetE to protein factors. Because micrococcal nuclease and DNaseI were used, the digestion data are likely to reflect the nucleosome structure of these regions (Simpson, 1998). The promoter region of PetE was less accessible to digestion in roots than was the same region in shoots, suggesting that the promoter possesses a closed conformation in roots. This closed conformation is also correlated with a lack of basal transcription and hence is likely to suppress transcription in roots by preventing the binding of transcription factors. Consistent with our observation, the silent β-phaseolin promoter is closely associated with nucleosomes in transgenic tobacco plants (Li et al., 1998), whereas the active Adh promoter contains loosely bound nucleosomes in Arabidopsis (Vega-Palas and Ferl, 1995).
The chromatin conformation of parts of a gene other than the promoter and its effects on transcription have been little examined in animal or plant systems. Here, we examined the upstream, enhancer, and transcribed regions of PetE with the same set of nuclease digestions. Like the promoter, the pea PetE enhancer region is inaccessible to nucleases in roots. The closed conformation of the enhancer region may further act to prevent the binding of transcription factors and thereby decrease transcription. In contrast, the upstream and transcribed regions possess similar nuclease accessibilities in the active PetE in shoots and the inactive PetE in roots, suggesting that the accessibility of these regions to protein factors is unlikely to be important in transcription initiation.
The similar nuclease accessibility of the transcribed region of PetE in roots and shoots suggests that nucleosomes are not mobilized from these regions during transcription. This is consistent with results obtained from fruit fly hsp70 and rat rRNA genes, in which the transcribed regions possess comparable amounts of histones in active and inactive genes (Belikov et al., 1993; Mutskov et al., 1996). Furthermore, nucleosomes also are present in the coding region of the active Adh in Arabidopsis (Vega-Palas and Ferl, 1995). Therefore, RNA polymerase is likely to transcribe through nucleosomes in plant genes, as has been proposed for animal genes.
Although the accessibility of the promoter/enhancer region of PetE to micrococcal nuclease and DNaseI is similar in green and etiolated shoots, the transcription level of PetE is manyfold higher in green shoots. Thus, an open structure in the enhancer/promoter region in etiolated shoots is not sufficient for high transcription rates. This finding indicates that other factors are important for transcriptional activation. Chromatin conformation may act to determine the presence or absence of transcription and is likely to affect the assembly of basal transcription complexes. Indeed, nucleosomes inhibit the binding of the basal transcription factors TATA box binding protein and transcription factor IID to their recognition sites (Imbalzano et al., 1994; Ryan et al., 1998). Nucleosome remodeling on the β-phaseolin promoter potentiates transcription, and other transcription factors are necessary for activation (Li et al., 1999). In contrast, histone acetylation is part of a different mechanism that acts to fine-tune transcription rates through the recruitment of transcription factors, as discussed above.
Chromatin Structure of PetE
Using the results from ChIP and the nuclease accessibility experiments to indicate the likely presence of nucleosomes, we can suggest a model for the chromatin structure of PetE in different transcriptional states (Figure 7). The results indicate that chromatin structure undergoes targeted changes in short, defined regions of the gene as the transcriptional state of the gene changes. In particular, the chromatin structure of the enhancer/promoter region appears to be the most dynamic, because it undergoes changes in histone H3 and H4 acetylation states and in nuclease accessibility. This is perhaps not surprising because the enhancer and promoter are important DNA regions where transcription factors bind. This model does not propose that the chromatin structure of PetE in each of the pea tissues is static, as illustrated, but suggests a predominant structure for PetE in each tissue. It is expected that intermediate structures will form as transcription changes.
The data in this article clearly show that the chromatin structure of PetE, as assessed by histone acetylation and nuclease accessibility, is different in different transcriptional states. Moreover, this work provides evidence that the two processes, histone acetylation and nucleosome organization, function to regulate the transcription of a single gene. We have shown by nuclear run-on assays that the transcription of PetE is organ specific and light induced. The accessibility of the PetE promoter/enhancer region appears to be regulated in an organ-specific manner, and histone acetylation occurs in green but not in etiolated shoots. Hence, the organ-specific regulation of PetE transcription is likely to involve modifications in nucleosome organization, whereas light induction requires histone acetylation. Thus, each mechanism regulates a different aspect of PetE transcription. It is now of interest to isolate and characterize the protein complexes involved in these two processes to increase our understanding of the control of transcription by chromatin structure.
METHODS
Nuclei Extraction
Roots or green shoots were harvested from pea (Pisum sativum cv Progress No. 9) grown in Levington M3 potting compost (Scotts, Suffolk, UK) under a 16-hr-light (450 μmol photons m−2 sec−1) and 8-hr-dark regimen at 22 to 30°C for 10 days. Etiolated shoots were harvested from pea plants grown in total darkness for 10 days. The pea tissues were washed with water to remove soil before extraction.
Nuclei were isolated from 100 g of tissue as described by Hatton and Gray (1999). Briefly, tissues were homogenized with a Polytron homogenizer and a PTA20SM head (Kinematica, Lucerne, Switzerland) in 600 mL of cold nuclear extraction buffer (0.44 M sucrose, 25 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 2.5% Ficoll 400, 5% dextran, 0.5% Triton X-100, 2 mM spermine, 2.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin), filtered through four layers of muslin cloth, and centrifuged at 1500g for 10 min. The nuclei were suspended in 20 mL of nuclear extraction buffer without spermine, layered onto four 10-mL 30/60% Percoll step gradients (gradients contained 0.44 M sucrose, 25 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.5% Triton X-100, 2.5 mM DTT, 0.5 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin), and centrifuged at 1000g for 30 min. The pellet was next washed once with nuclear extraction buffer without spermine, once with NSB (50% glycerol, 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 2.5 mM DTT, 0.5 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin), and suspended in 1 mL of NSB. All steps were performed at 4°C. For run-on assays, the nuclei were prepared as described above, but the Percoll gradient step was omitted.
Transcription in Isolated Nuclei
Transcription reactions (400 μL) containing 200 μL of nuclei, 0.1 mM ammonium sulfate, 4 mM MgCl2, 0.3 μM phosphocreatine, 1 mM DTT, 100 ng (0.025 unit) of creatine phosphokinase (Sigma), 480 units of RNasin (Promega), 5 mM each of ATP, CTP, and GTP, and 100 μCi of α-32P-UTP (3000 Ci/mmol) were incubated at 30°C for 25 min. After the addition of 80 μg of yeast tRNA (Boehringer Mannheim) and 20 units of RNase-free DNaseI (Boehringer Mannheim), the reaction mixtures were incubated at 30°C for another 10 min. An aliquot of 50 μL of 100 mM Tris-HCl, pH 7.6, 50 mM EDTA, 10% SDS, and 10 μL of proteinase K (5 mg/mL, predigested at 37°C for 20 min) was added next, and the reaction mixtures were incubated at 42°C for 30 min. After extraction with phenol:chloroform, the labeled RNA was precipitated overnight with 0.1 volume of diethyl pyrocarbonate–treated 3 M sodium acetate and 2 volumes of ethanol at −20°C. After washing once with 70% ethanol, the RNA pellet was dissolved in 120 μL of diethyl pyrocarbonate–treated TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) and used for hybridization. Slot blots were prepared using 2 μg of plasmids and GeneScreen Plus membrane according to the manufacturer's instructions (DuPont–New England Nuclear Research Products). The plasmids used contained pea Lhcb1 (Cline et al., 1989), pea RbcS (Anderson and Smith, 1986), pea PetE (Last and Gray, 1989), tobacco actin gene Tac9 (Thangavelu et al., 1993), pea polyubiquitin gene (Watts and Moore, 1989), and pea rRNA gene cluster (Jorgensen et al., 1987). Hybridization was performed as described by Helliwell et al. (1997). The blots were exposed to x-ray film and scanned with a Molecular Dynamics (Sunnyvale, CA) 300S laser scanning densitometer. Alternately, the blots were exposed to phosphor screens and scanned with a Molecular Dynamics STORM 840 phosphorimager.
Chromatin Immunoprecipitation
Nuclei in NSB (1 mL) were fixed with 1% formaldehyde at 22°C for 15 min immediately after extraction. The formaldehyde concentration was adjusted from a 37% solution (Sigma). Glycine (150 μL of 1 M) was added, and the incubation was allowed to proceed for another 5 min. Nuclei were collected by centrifugation, suspended in 500 μL of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin), and sonicated three times for 10 sec on ice with an MSE Soniprep 150 (Fisher Scientific, Loughborough, UK). Between sonications, the solution was incubated for 5 min on ice. The sonicated solution was then centrifuged at 11,600g for 10 min at 4°C. The supernatant was collected and diluted 10-fold with IP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1 mM PMSF, 0.5 μg/mL antipain, and 0.5 μg/mL leupeptin) to give a chromatin solution at the correct ionic concentration for immunoprecipitation.
Chromatin solution (1 mL) was combined with the following amounts of rabbit antisera: 10 μL of anti-acetylated H4 (Serotec, Oxford, UK), 10 μL of anti-nonacetylated H4 (Serotec), 10 μL of anti-acetylated H3 (Upstate Biotechnology, Lake Placid, NY), or 30 μL of anti-nonacetylated H3 (Upstate Biotechnology). The solutions were incubated overnight at 4°C on a rotation wheel. After the addition of 0.66 μg of sonicated bacteriophage λ DNA and 20 μL of protein A–Sepharose beads (50% slurry in TE and 0.1% BSA), the chromatin solutions were incubated for another 2 hr at room temperature on the rotation wheel. The beads were washed twice with 500 μL of TSE-150 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl), once with 500 μL of LNDET (0.25 M LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and twice with 500 μL of TE. The immunocomplexes were eluted from the beads with 500 μL of 1% SDS and 0.1 M NaHCO3, mixed with 2.5 μL of 5 M NaCl, and heated at 65°C for 5 hr to reverse the formaldehyde cross-linkages. After the addition of 2 volumes of ethanol, the samples were incubated at 4°C overnight. The precipitates were collected by centrifugation, dissolved in 100 μL of TE, treated with 1.5 μL of proteinase K (18.6 mg/mL) for 2 hr at 42°C, and extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and chloroform. After precipitation with ethanol in the presence of 5 μg of glycogen, the purified DNA pellets were suspended in 10 μL of TE and analyzed by polymerase chain reaction (PCR). Three replicates of chromatin immunoprecipitation (ChIP) were performed on different nuclei preparations from each pea tissue to analyze the acetylation states of histones associated with PetE.
Treatment of Nuclei with Endonucleases
Endonuclease digestions were performed in reaction mixtures containing 320 μL of nuclei in NSB, 480 μL of 2 × nuclease buffer (30 mM NaCl and 8 mM CaCl2), 160 μL of Tris buffer (50 mM Tris-HCl, pH 8.0, and 5 mM MgCl2), and the endonuclease (0.2 unit/mL for micrococcal nuclease [Worthington] or 0.3 unit/mL for DNaseI [Boehringer Mannheim]). Before the addition of the endonuclease, two 120-μL aliquots were removed from the reaction mixture for the non-nuclease control and for the zero time point. After addition of the endonuclease, the digestion mixture and the non-nuclease control were incubated at 37°C. Aliquots of 120 μL were taken from the digestion mixture at 2, 4, 6, 10, and 15 min after endonuclease addition, mixed immediately with phenol, and placed on ice. After extraction with phenol and phenol:chloroform:isoamyl alcohol (25:24:1), the DNA was precipitated with ethanol, dried, suspended in 10 μL of TE, and analyzed by quantitative PCR.
Quantitative PCR
The amounts of DNA present in the samples from endonuclease digestion experiments and ChIP were determined by quantitative PCR. Primers were designed based on the pea PetE sequence reported by Last and Gray (1989). The primer pairs used are as follows: the upstream region (y10, 5′-AGTGTAGAATTGGACATATGC-3′; y11, 5′-CGTTTAGCTTGCATTATGCC-3′); the enhancer region (y6, 5′-AGCTTAGTTAATCATGTTAAACAAC-3′; y7, 5′-CAGAGTGCGTGGGCTTAAAGTGG-3′); the promoter region (y16, 5′-CTTTAAGCCCACGCA-CTCTGTGG-3′; y13, 5′-AATCGAAAGAGAGACACACAGAAGG-3′); the enhancer and promoter regions (y12, 5′-GATGGAGCTCATTATAATTTGAATGG-3′; y13, 5′-AATCGAAAGAGAGACACACAGAAGG-3′); and the transcribed region (y14, 5′-TTATCATCCATTCTA-TAAAATCACC-3′; y15, 5′-ATTAACAGTGACTTGTCCAACC-3′). The PCR reactions were first performed with various dilutions of the template DNA to ensure that the amount of DNA used and the PCR conditions were within the quantitative range of the amplification reaction. Typically, a threefold dilution was used for the nuclease digestion experiments and a twofold dilution was used for the ChIP experiments. For PCR of chromatin solution before immunoprecipitation, fivefold dilution of samples was used. PCR was performed in a 50-μL reaction volume containing DNA template, 2.5 units of Taq polymerase (Bioline, UK), 5 μL of 10 × reaction buffer supplied by the manufacturer of the enzyme, 400 μM each of dATP, dTTP, dCTP, and dGTP, 1 μM of each primer, and 2 mM MgCl2. The PCR started with an incubation at 95°C (10 min), followed by 22 cycles of 55°C (1 min), 72°C (1 min), and 95°C (1 min), and ended with an incubation step at 72°C (10 min). The PCR fragments were resolved by electrophoresis on a 2% agarose gel, stained with ethidium bromide, and quantified with an Alpha Imager 1200 (Alpha Innotech Corp., San Leandro, CA). Alternately, the fragments were blotted onto GeneScreen Plus membrane (DuPont–New England Nuclear Research Products), probed with 32P-labeled PCR products, and exposed to x-ray film, and the bands were scanned with a Molecular Dynamics 300S laser scanning densitometer.
Acknowledgments
We thank Dr. James A. Sullivan for help with nuclear run-on experiments. Y.L.C. was supported by funds from Advanced Technologies (Cambridge Ltd.) and by an Overseas Research Student award. A.P.C.B. was supported by a research grant from the Biotechnology and Biological Sciences Research Council (UK).
References
- Anderson, S., and Smith, S.M. (1986). Synthesis of the small subunit of ribulose-bisphosphate carboxylase from genes cloned into plasmids containing the SP6 promoter. Biochem. J. 240, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf, M., Vasil, V., Vasil, I.K., and Ferl, R.J. (1987). Chromatin structure at the 5′ promoter region of the maize Adh2 gene and its role in gene regulation. Mol. Gen. Genet. 208, 185–190. [Google Scholar]
- Belikov, S.V., Belgovsky, A.I., Preobrazhenskaya, O.V., Karpov, V.L., and Mirzabekov, A.D. (1993). Two non-histone proteins are associated with the promoter region and histone H1 with the transcribed region of active hsp-70 genes as revealed by UV-induced DNA-protein crosslinking in vivo. Nucleic Acids Res. 21, 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression— belts, braces and chromatin. Cell 99, 451–454. [DOI] [PubMed] [Google Scholar]
- Braunstein, M., Rose, A.B., Holmes, S.G., Allis, D.C., and Broach, J.R. (1993). Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Brehm, A., Nielsen, S.J., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J., and Kouzarides, T. (1999). The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 18, 2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell, J.E., and Allis, D.C. (1996). Special HATs for special occasions: Linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 6, 176–184. [DOI] [PubMed] [Google Scholar]
- Chakravarti, D., Ogryzko, V., Kao, H.-Y., Nash, A., Chen, H., Nakatani, Y., and Evans, R.M. (1999). A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chen, Z.J., and Pikaard, C.S. (1997). Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 16, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, W.L., Briggs, S.D., and Allis, C.D. (2000). Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12, 326–333. [DOI] [PubMed] [Google Scholar]
- Cline, K., Fulsom, D.R., and Viitanen, P.V. (1989). An imported thylakoid protein accumulates in the stroma when insertion into thylakoids is inhibited. J. Biol. Chem. 264, 14225–14232. [PubMed] [Google Scholar]
- Dhalluin, C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A.K., and Zhou, M.-M. (1999). Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Dimitrov, S., Almouzni, G., Dasso, M., and Wolffe, A.P. (1993). Chromatin transitions during early Xenopus embryogenesis: Changes in histone H4 acetylation and in linker histone type. Dev. Biol. 160, 214–227. [DOI] [PubMed] [Google Scholar]
- Durrin, L.K., Mann, R.K., Kayne, P.S., and Grunstein, M. (1991). Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Fisher-Adams, G., and Grunstein, M. (1995). Yeast histone H4 and H3 N-termini have different effects on the chromatin structure of the GAL1 promoter. EMBO J. 14, 1468–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer, W.B., and Starlinger, P. (1988). DNase I hypersensitive sites in the 5′-region of the maize Shrunken gene in nuclei from different organs. Mol. Gen. Genet. 212, 351–359. [DOI] [PubMed] [Google Scholar]
- Gallagher, T.F., and Ellis, R.J. (1982). Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J. 1, 1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görz, A., Schäfer, W., Hirasawa, E., and Kahl, G. (1988). Constitutive and light-induced DNaseI hypersensitive sites in the rbcS genes of pea (Pisum sativum). Plant Mol. Biol. 11, 561–573. [DOI] [PubMed] [Google Scholar]
- Grasser, K.D. (1998). HMG1 and HU proteins: Architectural elements in plant chromatin. Trends Plant Sci. 3, 260–265. [Google Scholar]
- Gross, D.S., and Garrard, W.T. (1988). Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197. [DOI] [PubMed] [Google Scholar]
- Grosschedl, R., Giese, K., and Pagel, J. (1994). HMG domain proteins: Architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10, 94–100. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hatton, D., and Gray, J.C. (1999). Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J. 18, 417–429. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Webster, C.I., and Gray, J.C. (1997). Light-regulated expression of the pea plastocyanin gene is mediated by elements within the transcribed region of the gene. Plant J. 12, 499–506. [DOI] [PubMed] [Google Scholar]
- Hörz, W., and Altenburger, W. (1981). Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 9, 2643–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano, A.N., Kwon, H., Green, M.R., and Kingston, R.E. (1994). Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370, 481–485. [DOI] [PubMed] [Google Scholar]
- Jeppesen, P., and Turner, B.M. (1993). The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74, 281–289. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Cuellar, R.E., Thompson, W.F., and Kavanagh, T.A. (1987). Structure and variation in ribosomal RNA genes of pea. Characterisation of a cloned rDNA repeat and chromosomal rDNA variants. Plant Mol. Biol. 8, 3–12. [DOI] [PubMed] [Google Scholar]
- Kao, H.-Y., Downes, M., Ordentlich, P., and Evans, R.M. (2000). Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14, 55–66. [PMC free article] [PubMed] [Google Scholar]
- Kornberg, R.D., and Lorch, Y. (1999). Twenty-five years of the nucleosome, fundamental particle of eukaryote chromosome. Cell 98, 285–294. [DOI] [PubMed] [Google Scholar]
- Korzus, E., Torchia, J., Rose, D.W., Xu, L., Kurokawa, R., Mclnerney, E.M., Mullen, T.-M., Glass, C.K., and Rosenfeld, M.G. (1998). Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279, 703–707. [DOI] [PubMed] [Google Scholar]
- Krajewski, W.A. (2000). Histone hyperacetylation facilitates chromatin remodelling in a Drosophila embryo cell-free system. Mol. Gen. Genet. 263, 38–47. [DOI] [PubMed] [Google Scholar]
- Kuo, M.-H., Zhou, J., Jambeck, P., Churchill, M.E.A., and Allis, D.C. (1998). Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12, 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last, D.I., and Gray, J.C. (1989). Plastocyanin is encoded by a single-copy gene in the pea haploid genome. Plant Mol. Biol. 12, 655–666. [DOI] [PubMed] [Google Scholar]
- Last, D.I., and Gray, J.C. (1990). Synthesis and accumulation of pea plastocyanin in transgenic tobacco plants. Plant Mol. Biol. 14, 229–238. [DOI] [PubMed] [Google Scholar]
- Lee, D.Y., Hayes, J.J., Pruss, D., and Wolffe, A.P. (1993). A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72, 73–84. [DOI] [PubMed] [Google Scholar]
- Li, G., Chandler, S.P., Wolffe, A.P., and Hall, T.C. (1998). Architectural specificity in chromatin structure at the TATA box in vivo: Nucleosome displacement upon the β-phaseolin gene activation. Proc. Natl. Acad. Sci. USA 95, 4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Bishop, K.J., Chandrasekharan, M.B., and Hall, T.C. (1999). β-Phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid–mediated gene activation. Proc. Natl. Acad. Sci. USA 96, 7104–7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K., and Richmond, T.J. (1998). The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8, 140–146. [DOI] [PubMed] [Google Scholar]
- Lund, G., Das, O.P., and Messing, J. (1995). Tissue-specific DNase I-sensitive sites of the maize P gene and their changes upon epimutation. Plant J. 7, 797–807. [PubMed] [Google Scholar]
- Luo, R.L., Postigo, A.A., and Dean, D.C. (1998). RB interacts with histone deacetylase to repress transcription. Cell 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Mutskov, V.J., Russanova, V.R., Dimitrov, S.I., and Pashev, I.G. (1996). Histones associated with non-nucleosomal rat ribosomal genes are acetylated while those bound to nucleosome-organised gene copies are not. J. Biol. Chem. 271, 11852–11857. [DOI] [PubMed] [Google Scholar]
- Niu, X., Adams, C.C., Workman, J.L., and Guiltinan, M.J. (1996). Binding of the wheat basic leucine zipper protein EmBP-1 to the nucleosomal binding sites is modulated by nucleosome positioning. Plant Cell 8, 1569–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, L.P., and Turner, B.M. (1995). Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape, S.M., Kamakaka, R.T., and Kadonaga, J.T. (1994). Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63, 265–297. [DOI] [PubMed] [Google Scholar]
- Parekh, B.S., and Maniatis, T. (1999). Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell 3, 125–129. [DOI] [PubMed] [Google Scholar]
- Paul, A.-L., and Ferl, R.J. (1998). Permeabilized Arabidopsis protoplasts provide new insight into the chromatin structure of plant alcohol dehydrogenase genes. Dev. Genet. 22, 7–16. [DOI] [PubMed] [Google Scholar]
- Paul, A.-L., Vasil, V., Vasil, I.K., and Ferl, R.J. (1987). Constitutive and anaerobically induced DNase-I-hypersensitive sites in the 5′ region of the maize Adh1 gene. Proc. Natl. Acad. Sci. USA 84, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O.M., Belles, E., Lopez-Rodas, G., Sendra, R., and Tordera, V. (1998). Interaction between N-terminal domain of H4 and DNA is regulated by the acetylation degree. Biochim. Biophys. Acta 1397, 79–90. [DOI] [PubMed] [Google Scholar]
- Pwee, K.-H., and Gray, J.C. (1993). The pea plastocyanin promoter directs cell-specific but not full light-regulated expression in transgenic tobacco plants. Plant J. 3, 437–449. [DOI] [PubMed] [Google Scholar]
- Pwee, K.-H., Webster, C.I., and Gray, J.C. (1994). HMG protein binding to an A/T-rich positive regulatory region of the pea plastocyanin gene promoter. Plant Mol. Biol. 26, 1907–1920. [DOI] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Suka, N., Turner, B.M., and Grunstein, M. (1998). Transcriptional repression of UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 381–385. [DOI] [PubMed] [Google Scholar]
- Ryan, M.P., Jones, R., and Morse, R.H. (1998). SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol. Cell. Biol. 18, 1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar, A.D., Horwitz, B.A., Elliott, R.C., Thompson, W.F., and Briggs, W.R. (1988). Light effects on several chloroplast components in norflurazon-treated pea seedlings. Plant Physiol. 88, 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, J.S., Webster, C.I., and Gray, J.C. (1998). A/T-rich sequences act as quantitative enhancers of gene expression in transgenic tobacco and potato plants. Plant Mol. Biol. 37, 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, R.T. (1998). Chromatin structure and analysis of mechanisms of activators and repressors. Methods Companion Methods Enzymol. 15, 283–294. [DOI] [PubMed] [Google Scholar]
- Sobel, R.E., Cook, R.G., Perry, C.A., Annunziato, A.T., and Allis, D.C. (1995). Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA 92, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker, S., Murray, M.G., and Thompson, W.F. (1983). DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc. Natl. Acad. Sci. USA 80, 815–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, D.C. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Thangavelu, M., Belostotsky, D., Bevan, M.W., Flavell, R.B., Rogers, H.J., and Lonsdale, D.M. (1993). Partial characterization of the Nicotiana tabacum actin gene family: Evidence for pollen-specific expression of one of the gene family members. Mol. Gen. Genet. 240, 290–295. [DOI] [PubMed] [Google Scholar]
- Thompson, J.S., Ling, X., and Grunstein, M. (1994). Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature 369, 245–247. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- Tyler, J.K., and Kadonaga, J.T. (1999). The “dark side” of chromatin remodelling: Repressive effects on transcription. Cell 99, 443–446. [DOI] [PubMed] [Google Scholar]
- van Blokland, R., ten Lohuis, M., and Meyer, P. (1997). Condensation of chromatin in transcriptional regions of an inactivated plant transgene: Evidence for an active role of transcription in gene silencing. Mol. Gen. Genet. 257, 1–13. [DOI] [PubMed] [Google Scholar]
- Vega-Palas, M.A., and Ferl, R.J. (1995). The arabidopsis Adh gene exhibits diverse nucleosome arrangements within a small DNaseI-sensitive domain. Plant Cell 7, 1923–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, U., Svaren, J., Schmitz, J., Schmid, A., and Hörz, W. (1994). A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13, 4848–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J.S., Mann, R.K., and Grunstein, M. (1995). Yeast histone H3 and H4 N termini function through different GAL1 regulatory elements to repress and activate transcription. Proc. Natl. Acad. Sci. USA 92, 5664–5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Liu, L., and Berger, S.L. (1998). Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, F.Z., and Moore, A.L. (1989). Nucleotide sequence of a full- length cDNA clone encoding a polyubiquitin gene from Pisum sativum. Nucleic Acids Res. 17, 10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, C.I., Packman, L.C., Pwee, K.-H., and Gray, J.C. (1997). High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J. 11, 703–715. [DOI] [PubMed] [Google Scholar]
- Whitehouse, I., Flaus, A., Cairns, B.R., White, M.F., Workman, J.L., and Owen-Hughes, T. (1999). Nucleosome mobilisation catalysed by the yeast SWI/SNF complex. Nature 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Winston, F., and Allis, C.D. (1999). The bromodomain: A chromatin-targeting module? Nat. Struc. Biol. 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Wittschieben, B., Otero, G., de Bizemont, T., Fellows, J., Erdjument-Bromage, H., Ohba, R., Li, Y., Allis, C.D., Tempst, P., and Svejstrup, J.Q. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P., and Hayes, J.J. (1999). Chromatin disruption and modification. Nucleic Acids Res. 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000. a). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000. b). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Wyrick, J.J., Holstege, F.C.P., Jennings, E.G., Causton, H.C., Shore, D., Grunstein, M., Lander, E.S., and Young, R.A. (1999). Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402, 418–421. [DOI] [PubMed] [Google Scholar]