Abstract
We used DNA sequencing and gel blot surveys to assess the integrity of the chloroplast gene infA, which codes for translation initiation factor 1, in >300 diverse angiosperms. Whereas most angiosperms appear to contain an intact chloroplast infA gene, the gene has repeatedly become defunct in ∼24 separate lineages of angiosperms, including almost all rosid species. In four species in which chloroplast infA is defunct, transferred and expressed copies of the gene were found in the nucleus, complete with putative chloroplast transit peptide sequences. The transit peptide sequences of the nuclear infA genes from soybean and Arabidopsis were shown to be functional by their ability to target green fluorescent protein to chloroplasts in vivo. Phylogenetic analysis of infA sequences and assessment of transit peptide homology indicate that the four nuclear infA genes are probably derived from four independent gene transfers from chloroplast to nuclear DNA during angiosperm evolution. Considering this and the many separate losses of infA from chloroplast DNA, the gene has probably been transferred many more times, making infA by far the most mobile chloroplast gene known in plants.
INTRODUCTION
Many genes have been lost from the chloroplast genome during plant and algal evolution. Most of these losses occurred in the murky interval between the original endosymbiosis of a cyanobacterium (with perhaps 2000 protein-coding genes) and the last common ancestor of all existing chloroplast genomes (with ∼210 protein-coding genes; Martin et al., 1998). Many other genes were lost during the early evolution of photosynthetic eukaryotes, often in parallel in different algal lineages, and some of these losses were the result of gene transfers to the nuclear genome (Martin et al., 1998). During the evolution of land plants, relatively few changes occurred to the set of genes found in chloroplast DNA (cpDNA) (Martin et al., 1998; Palmer and Delwiche, 1998). Nonetheless, the most recent changes are likely to provide the most information about the evolutionary mechanisms involved.
Among the six completely sequenced chloroplast genomes from angiosperms (excluding the nonphotosynthetic plant Epifagus virginiana; Wolfe et al., 1992a), 74 protein-coding genes are held in common and an additional five are present in only some species. These five genes are accD, ycf1, and ycf2 (pseudogenes in rice and maize; Hiratsuka et al., 1989; Maier et al., 1995), rpl23 (pseudogene in spinach; Thomas et al., 1988), and infA (pseudogene in tobacco, Arabidopsis, and Oenothera elata; Shinozaki et al., 1986; Wolfe et al., 1992b; Sato et al., 1999; Hupfer et al., 2000). Other chloroplast gene losses in angiosperms that have been confirmed by sequencing include rpl22, rps16, and ycf4 (open reading frame 184), all of which have been lost in some or all legumes (Gantt et al., 1991; Nagano et al., 1991; Doyle et al., 1995; K.H. Wolfe, unpublished data), and ycf2 and ndhF, both of which have been lost repeatedly in a variety of angiosperms (Downie et al., 1994; Neyland and Urbatsch, 1996; Smith, 2000; M. Ireland, H. Deiderick, Y.-L. Qiu, and J.D. Palmer, unpublished data). Additional gene losses have been suggested by DNA gel blot hybridization surveys (Downie and Palmer, 1992) but have not been confirmed by sequencing.
For several of these chloroplast gene losses (i.e., infA, rps16, ycf1, ycf2, and ycf4), it is unknown whether they reflect successful gene transfer to the nucleus or loss of the gene from the cell entirely. Among the better understood cases, only rpl22 in legumes constitutes a classic transfer of a chloroplast gene to the nucleus, with the protein being imported back into the chloroplast by means of a transit peptide (Gantt et al., 1991). The spinach chloroplast rpl23 locus is a pseudogene and has been functionally replaced by a nuclear gene similar to the homologous cytosolic ribosomal protein gene—a case of gene substitution (Bubunenko et al., 1994; see also Yamaguchi and Subramanian, 2000). A similar cytosolic gene–for–plastid gene substitution led to the chloroplast accD locus becoming a pseudogene in grasses (Konishi et al., 1996), and a mitochondrial gene–for–chloroplast gene substitution appears to have caused the loss of rpl21 from cpDNA in an ancestor of angiosperms and gymnosperms (Martin et al., 1990; additional analyses not shown). Finally, repeated loss of chloroplast ndhF probably reflects repeated biochemical loss of the chloroplast NADH dehydrogenase complex, the product of 11 chloroplast genes and an undetermined number of nuclear genes (Wakasugi et al., 1994; M. Ireland, H. Deiderick, Y.-L. Qiu, and J.D. Palmer, unpublished data).
Here, we have analyzed the loss of infA, which codes for translation initiation factor 1, and show that it is a second example of chloroplast-to-nucleus gene transfer in land plants. We suspected that the chloroplast infA pseudogenes in tobacco and other species indicated transfer(s) of the gene to the nucleus rather than simple gene losses because infA is an essential gene in Escherichia coli (Cummings and Hershey, 1994). In E. coli, three initiation factors (IFs) mediate the coming together of the mRNA, ribosome, and initiator tRNA-Met to begin translation. IF1 is the smallest of these factors (only 72 residues long) and consists of an RNA binding domain whose precise molecular role is unclear (Sette et al., 1997; Battiste et al., 2000). The initiation of translation in organelles is thought to be similar to that in E. coli, although much less is known about the organelle systems (Pel and Grivell, 1994; Yu and Spremulli, 1998). The gene for chloroplast IF1 was first reported by Sijben-Müller et al. (1986) from spinach cpDNA. Chloroplast IF2 and IF3 proteins, encoded by nuclear genes, have been characterized in Euglena gracilis (Ma and Spremulli, 1992; Lin et al., 1994), and candidate nuclear genes for chloroplast IF2 and IF3 are present in the Arabidopsis nuclear genome sequence.
To study the evolution of infA further, we characterized the chloroplast locus from >300 diverse angiosperms and investigated infA-like genes in the nucleus. We show that chloroplast infA has been lost repeatedly in angiosperm evolution and that four nuclear infA genes characterized thus far probably result from independent events of chloroplast-to-nucleus gene transfer.
RESULTS
Eleven Independent Losses of Chloroplast infA Revealed by DNA Sequencing
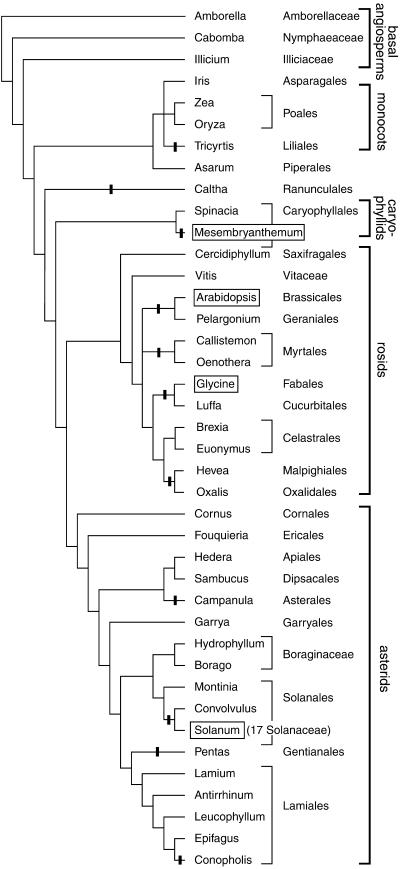
Combining new sequence data with seven sequences available from GenBank, we were able to study the structure of the chloroplast infA locus in 56 angiosperms, as summarized in Figure 1. We focused initially on the family Solanaceae and other asterids to determine the extent to which the infA loss in tobacco is shared by related species. infA is a pseudogene in all 17 Solanaceae species examined (representing 16 genera), suggesting a single loss of the gene in an ancestor of this family. There is no start codon in any species, three species have frameshifts, and in tomato the first 124 bp have been deleted. Analysis of nucleotide substitutions in infA within the Solanaceae indicates nearly equal numbers of substitutions at first, second, and third codon positions, consistent with it being a pseudogene. infA is also a pseudogene in Convolvulus (Convolvulaceae), representing the sister family to the Solanaceae. The gene may have become nonfunctional in an ancestor of these lineages, although all of the inactivating mutations in the Convolvulus gene are at different sites from those in the Solanaceae and so seem to have been acquired independently.
Figure 1.
Losses of Chloroplast infA during Angiosperm Evolution Based on Analysis of Sequenced infA Loci.
Shown are all angiosperms whose chloroplast infA loci have been sequenced, except that only one representative of the 17 sequenced Solanaceae species is shown. Black bars indicate lineages with chloroplast infA pseudogenes or complete loss. The branching order and taxonomic classifications are based on Soltis et al. (1999) and Olmstead et al. (2000). Boxed names indicate species from which a nuclear infA gene was identified.
To survey the extent of chloroplast infA loss more widely among asterids, the locus was sequenced from representatives of 10 of the 11 major lineages of asterids identified by molecular analyses (Olmstead et al., 2000). Three more infA pseudogenes were found, in Pentas (Rubiaceae), Campanula (Campanulaceae), and Conopholis (Orobanchaceae) (Figure 1). The loss of infA in the nonphotosynthetic plant Conopholis is in contrast to the presence of an intact gene in its close relative Epifagus (Wolfe et al., 1992b). On the basis of an interpretation of the distribution of infA losses and retentions in terms of the species' phylogeny, we conclude that there were at least four independent losses within the asterid clade (Figure 1).
The infA locus was sequenced (or analyzed from GenBank) for 12 rosids and found to be a pseudogene or entirely missing in eight of them (Figure 1). The defining characteristics of the pseudogenes range from multiple frameshifts in Luffa and Hevea to only half of the gene being present with a highly divergent sequence in Pelargonium. The intact chloroplast infA genes in Cercidiphyllum and Vitis represent what are clearly the two basal lineages of rosids (Soltis et al., 1999). The other 10 rosids represent a very large, monophyletic group that accounts for 39% of total eudicot species diversity (Magallón et al., 1999; Soltis et al., 1999). Only two apparently intact infA genes were discovered within this group, from Brexia and Euonymus (Celastraceae; Euonymus infA is truncated by three amino acids but is otherwise intact and differs from the intact Brexia gene by only three nucleotide substitutions). We infer four separate losses of infA among the sequenced rosids (Figure 1).
In more limited sequencing surveys of other angiosperm lineages, we found two other unambiguous examples of infA loss from cpDNA and one potential case. Chloroplast pseudogenes were sequenced from Caltha (Ranunculaceae) and the monocot Tricyrtis (Liliaceae). Because the sister lineages to both of these two groups contain a functional gene (Figure 1), the losses most likely are independent. The functional status of chloroplast infA from ice plant (Mesembryanthemum crystallinum; Aizoaceae) is unclear. A single frameshift (a 5-bp insert) near the 3′ end of the gene removes the last five codons and replaces them with a 41-codon extension. Because an intact and expressed infA gene is present in the nucleus of ice plant (see below), we have taken the ice plant chloroplast locus to be a pseudogene. Including ice plant, we estimate that infA has been lost from cpDNA 11 times among the 56 angiosperms examined by DNA sequencing (Figure 1).
Thirteen Additional Losses of Chloroplast infA Inferred from DNA Gel Blots
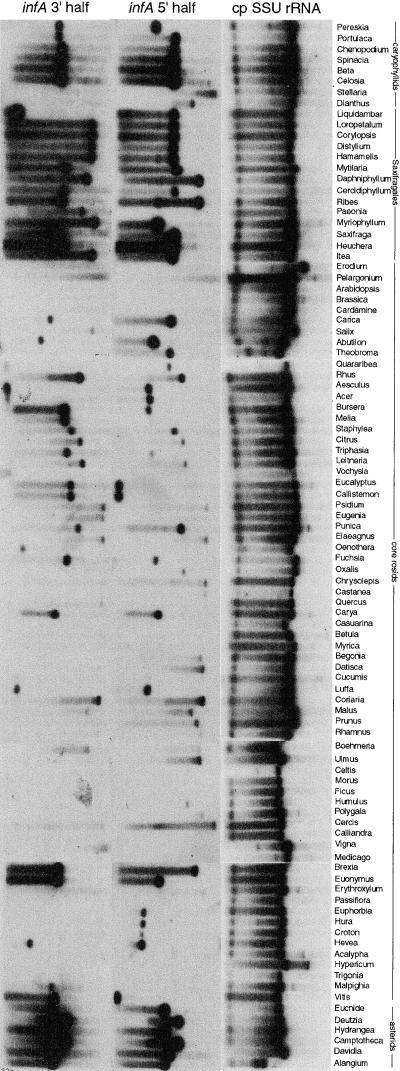
DNA gel blot hybridizations were used to survey rapidly a large number of angiosperms for cases of probable loss of chloroplast infA. This approach, performed on a much smaller scale (17 angiosperms), was used previously to successfully identify the only known case of chloroplast-to-nucleus transfer among land plants (Gantt et al., 1991). Probes for the 5′ and 3′ halves of chloroplast infA from Antirrhinum were hybridized to filters containing total DNAs from 280 species of angiosperms, representing 276 genera and 169 families (Figure 2). Chloroplast gene “loss” was inferred if there was no detectable hybridization on an overexposed autoradiograph against two layers of controls: good hybridization to the DNA in question using other chloroplast probes (such as the chloroplast 16S rRNA probe used in Figure 2) and good hybridization to other DNAs with the probe in question. This inference assumes that chloroplast genes are in high-copy number relative to (potentially transferred) nuclear genes of single-copy or low-copy number. This assumption seems safe, because the total DNAs were all made from green leaves, in which cpDNA copy number is invariably at least hundreds, and usually thousands, per cell (Bendich, 1987).
Figure 2.
Evidence for Loss of Chloroplast infA from DNA Gel Blot Hybridization.
The three probes named at top were hybridized sequentially to a filter containing BamHI digests of total DNA from 280 angiosperms, 96 of which are shown here. Note that some infA genes have a BamHI site near their middle. SSU, small subunit.
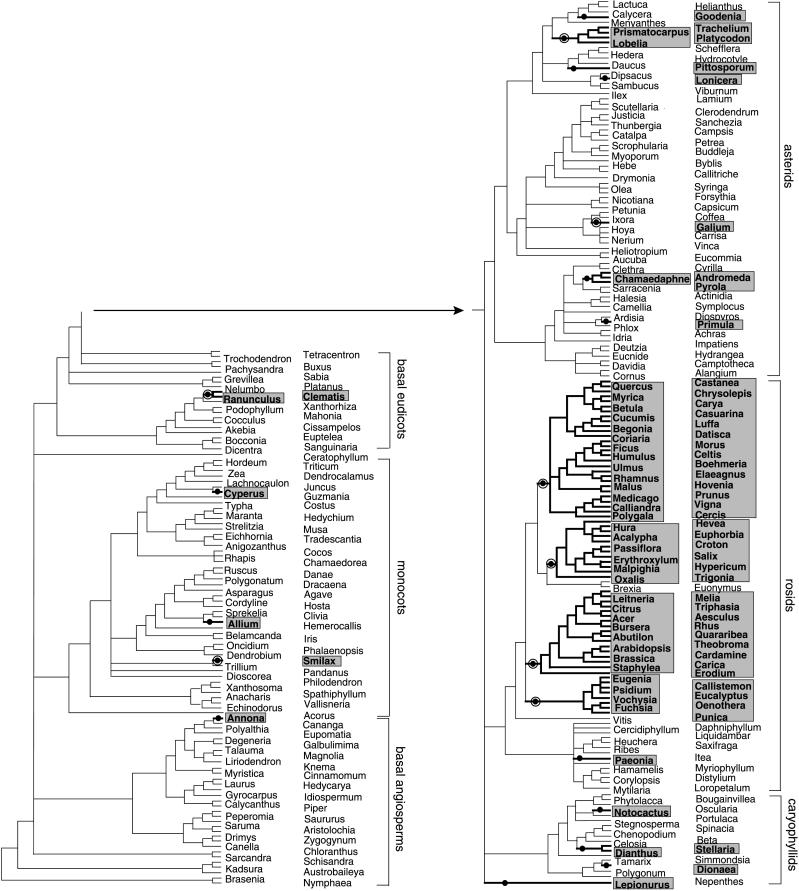
All taxa showing either weak (i.e., markedly diminished relative to the controls described above) or no hybridization with one or both infA probes were scored as having a defunct chloroplast infA gene. These taxa are indicated in Figure 3, which depicts the current best hypothesis of phylogenetic relationships among the 280 angiosperms (see www.bio.indiana.edu/~palmerlab for details on tree construction). Apart from the rosids, there are 16 scattered and small groups that showed weak or no hybridization (Figure 3; results for two of these are depicted in the topmost 18 species in Figure 2). The gel blot inference of infA loss has been confirmed by sequencing for four of these 16 groups (see bull's-eyes on Figure 3 and legend to Figure 3; the other 12 groups were not sequenced). We conclude from these results that chloroplast infA has probably been lost, in all cases recently, 16 times among the examined angiosperms, excluding rosids.
Figure 3.
Losses of Chloroplast infA during Angiosperm Evolution Based on DNA Gel Blot Hybridization Results.
The infA probes used were both from Antirrhinum. Plants with significantly diminished or no hybridization are shaded (see Figure 2 and text). Plant names beside the tree alternate in two columns. Loss lineages are shown in boldface on the tree and are marked with bullets. Bull's-eye bullets indicate the eight loss lineages for which loss was confirmed by sequencing of chloroplast infA from at least one member of the lineage. Within rosids, the sequenced member(s) was included on the blots; for the other four sequence-validated loss lineages, a different member of the loss lineage was sequenced than was on the blots. In particular, Tricyrtis (Figure 2) is the sister taxon to Smilax relative to all other taxa on the blots, Caltha is likewise sister to the Clematis-Ranunculus clade, Pentas is sister to Galium, and Campanula belongs within the Lobelia-Trachelium clade.
The situation among the rosids, in particular the “core” rosids, is quite different and less certain. Of the 14 examined members of the most basal rosid clade, only Paeonia has a defunct chloroplast infA gene as determined by hybridization (Figures 2 and 3), and Vitis, the only examined member of the next most basal group, has an intact chloroplast infA gene (Figures 1 to 3). However, 65 of the 67 remaining core rosids showed diminished to no hybridization with one or both infA probes (Figures 2 and 3). As described in the preceding section and depicted in Figure 1, all eight sequenced species from this group of 65 core rosids have defunct chloroplast infA genes (and three of these eight—Callistemon, Luffa, and Hevea—were deliberately chosen for sequencing because they have some of the strongest hybridization signals; Figure 2). These sequencing confirmations, combined with those for nonrosids reported in the preceding paragraph, lend considerable weight to our inference of probable infA loss for all 65 core rosids with diminished or no hybridization. We also noted a good correlation between the degree of diminished infA hybridization and the degree of infA sequence divergence. For example, the intact and strongly hybridizing chloroplast infA of Cercidiphyllum is 93% identical to the probe infA sequence from Antirrhinum, the moderately hybridizing infA pseudogene from Hevea is 84% identical, and the very weakly hybridizing Oxalis pseudogene is only 73% identical.
Brexia and Euonymus, the only two of the 67 core rosids with normal (i.e., strong) hybridization to infA (Figure 2), were both confirmed by sequencing to contain intact chloroplast infA genes (see preceding section and Figure 1). These two species form a clade within the core rosids whose position, if correct (and it receives moderately strong support in the analysis of Soltis et al. [1999]), implies the four separate but phylogenetically clustered (i.e., all similarly deep) losses of chloroplast infA within core rosids shown in Figures 1 and 3. Assuming these four losses within core rosids, the hybridization results indicate a total of five infA losses among all examined rosids and a total of 21 losses among all 280 angiosperms surveyed by hybridization (Figure 3). As already described, eight of these losses have been sequence validated, leaving three additional losses (in Conopholis, Solanaceae, and Mesembryanthemum) inferred solely from DNA sequencing. In total, we infer 24 separate losses of chloroplast infA among the 309 angiosperms investigated by DNA sequencing and/or hybridization.
Nuclear Genes with Chloroplast Transit Sequences
Candidate nuclear genes encoding chloroplast IF1 were identified by TBLASTN searches (Altschul et al., 1997) against GenBank using the spinach cpDNA-encoded IF1 protein as a query. Matching expressed sequence tag (EST) sequences were found from four species: Arabidopsis, soybean, tomato, and ice plant (Figure 4). The chloroplast loci in these four species are pseudogenes (Figure 1). The Arabidopsis ESTs correspond to a nuclear gene on chromosome 4 that was sequenced by the genome project (Mayer et al., 1999) but whose structure was predicted incorrectly (see Methods). The ESTs from the other three species are inferred to be transcripts of genes located in the nucleus because they have poly(A) tails and because the chloroplast loci from the same species are not intact.
Figure 4.
Multiple Alignment of IF1 Protein Sequences from the Four Nuclear infA Genes and Two Representative Chloroplast Genes.
The alignment was made using ClustalW 1.8 (Thompson et al., 1994), with the most common residue at each position highlighted. Predicted chloroplast transit peptides are shown by lowercase letters, and their predicted cleavage sites (Emanuelsson et al., 1999) are marked by large X symbols. The alignment of the transit peptides is not meant to indicate that they share a common ancestor (see text). cp, chloroplast; nuc, nuclear.
The nuclear genes encode proteins of 138 to 156 residues, compared with 77 residues for the spinach chloroplast gene, due to long N-terminal extensions (Figure 4). The subcellular locations of the proteins were predicted by using three computer programs, all of which use neural network methods (Table 1). All three programs predicted the N-terminal extensions of the soybean, tomato, and ice plant proteins to be transit peptides directing protein import into the chloroplast stroma. For the Arabidopsis protein, the ChloroP program (Emanuelsson et al., 1999) predicted that it is also imported into chloroplasts, but rather surprisingly, the two newer programs predicted it to be mitochondrial (Table 1). These two programs, TargetP (Emanuelsson et al., 2000) and Predotar (N.M. Peeters, A. Chapron, D. Lancelin, O. Grandjean, A. Giritch, H. Wintz, and I. Small, unpublished data), both aim to achieve accurate discrimination between chloroplast and mitochondrial transit peptides, whereas the older ChloroP does not attempt to identify mitochondrial transit peptides.
Table 1.
Transit Peptide Prediction Scores by Different Computer Methodsa
| ChloroPcTP Score |
TargetP
|
Predotar
|
||||
|---|---|---|---|---|---|---|
| Sequence | cTP Score |
mTP Score |
RCb | cTP Score |
mTP Score |
|
| Tomato | 0.556 | 0.863 | 0.059 | 1 | 0.620 | 0.001 |
| Soybean | 0.536 | 0.590 | 0.237 | 4 | 0.853 | 0.122 |
| Ice plant | 0.539 | 0.963 | 0.114 | 1 | 0.791 | 0.032 |
| Arabidopsis | 0.515 | 0.061 | 0.671 | 2 | 0.100 | 0.636 |
Boldface numbers indicate scores exceeding the programs' thresholds for prediction of chloroplast (cTP) or mitochondrial (mTP) transit peptides. Scores are not comparable among different programs, but the maximum score possible is 1.0 in all cases.
Reliability class (RC) of TargetP prediction on a scale of 1 (confident prediction) to 5 (weak prediction).
The subcellular locations of the soybean and Arabidopsis InfA proteins were examined by fusing the full-length InfA precursor proteins to the green fluorescent protein (GFP). Gene constructs encoding GFP fusion proteins were introduced by microprojectile bombardment of tobacco or Arabidopsis leaves, resulting in individual transformed cells expressing the GFP constructs. In both species, GFP fluorescence colocalized with chlorophyll fluorescence in chloroplasts of epidermal cells (Figure 5). GFP fused to soybean InfA was clearly localized to chloroplasts in a tobacco guard cell (Figures 5A to 5C), whereas GFP fused to Arabidopsis InfA can be seen in the smaller chloroplasts of an epidermal pavement cell (Figures 5D to 5F). The Arabidopsis InfA precursor directed GFP exclusively to the chloroplast, with no evidence of colocalization to mitochondria.
Figure 5.
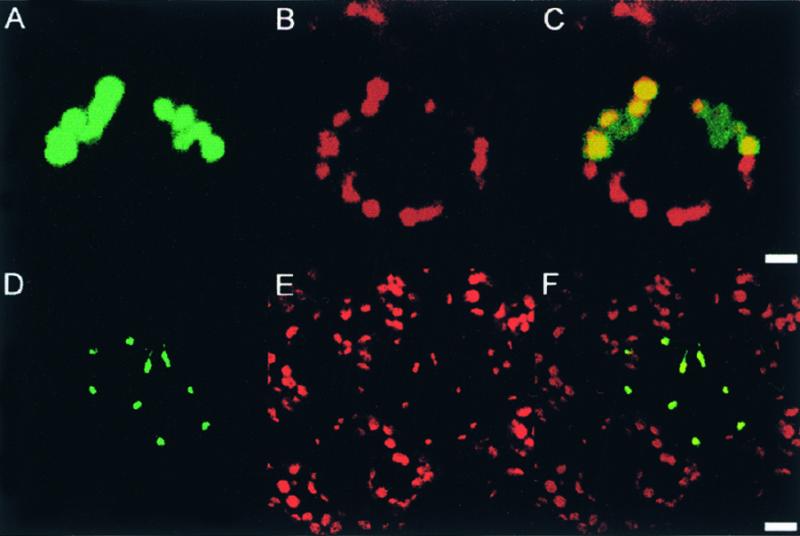
Localization of Fusion Proteins Encoded by Nuclear InfA-gfp Constructs to Chloroplasts.
The soybean InfA-gfp and Arabidopsis InfA-gfp were introduced into epidermal cells of tobacco and Arabidopsis using microprojectile bombardment. The localization of GFP in individual transformed cells was detected by confocal laser scanning microscopy 3 days after bombardment.
(A) to (C) pGmInfA-gfp in a tobacco guard cell. The images show two guard cells; the upper cell shows expression of pGmInfA-gfp. GFP fluorescence (A). Chlorophyll fluorescence (B). Merged images (C). Bar in (C) = 5 μm.
(D) to (F) Arabidopsis epidermal pavement cells with one cell expressing pAtInfA-gfp. The chloroplasts in epidermal pavement cells are smaller than those in guard cells. GFP fluorescence (D). Chlorophyll fluorescence (E). Merged images (F). Bar in (F) = 10 μm.
The Arabidopsis nuclear infA gene contains one intron (Figure 4). This occurs within the transit peptide coding region, unlike pea rpl22, whose intron is at the transit/mature peptide junction (Gantt et al., 1991). Sizing of polymerase chain reaction (PCR) products indicated that there are no introns in the nuclear genes from soybean, tomato, and ice plant (data not shown). BLAST searches (Altschul et al., 1997) did not reveal significant similarities between the four transit peptide sequences and any other sequence database entries.
Phylogenetic Analyses
Phylogenetic analyses were used to help infer the number and timing of infA transfer(s) to the nucleus. At one extreme, if there was a single transfer of infA in a common ancestor of rosids, asterids, and caryophyllids, then all four nuclear infA sequences would be expected to cluster together in the tree at a position corresponding to this ancestor (i.e., at the base of the dicots or earlier). At the other extreme, if the nuclear infA sequences were derived from four separate transfers, then each nuclear sequence would be expected to group together with a chloroplast infA sequence from a closely related plant.
Estimating the phylogenetic placement of the nuclear infA sequences is challenging because the short gene carries a limited amount of phylogenetic information. Also, the rapid evolution of nuclear genes as compared to chloroplast genes (Wolfe et al., 1987) means that phylogenetic trees drawn from a mixture of chloroplast and multiple nuclear sequences are likely to be affected by long-branch attraction. This artifact, in which the long branches tend to clump together even if they are not each other's closest relatives, can occur if some branches in a tree are much longer than others (Felsenstein, 1978). We took several precautionary steps to minimize the risk of long-branch attraction (see Methods). Most importantly, we analyzed the nuclear infA sequences one at a time, finding the optimal position for each nuclear sequence on a tree that did not have any other long branches.
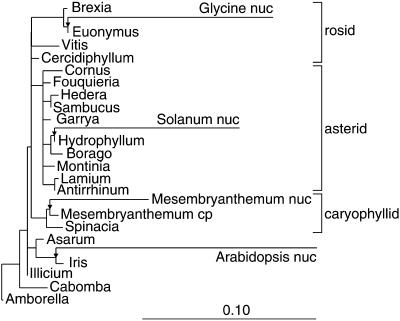
Our starting point for estimating the origins of the nuclear genes was the angiosperm phylogenetic tree recently proposed by Soltis et al. (1999) based on combined rbcL, atpB, and nuclear small subunit rRNA sequences from 560 species. We chose to use this “reference phylogeny” for angiosperms, in preference to estimating angiosperm phylogeny from the infA sequences themselves, because the data of Soltis et al. (1999) are much more likely to yield a correct tree (their study used 20 times more nucleotide sites and 10 times more species than are available for infA). We then took a set of 21 chloroplast infA sequences (20 intact genes and the ice plant chloroplast pseudogene), constrained the phylogenetic relationship among these 21 sequences to be the same as that in Soltis et al. (1999), and then found the optimal placement of each of the four nuclear infA sequences onto this constrained tree using maximum likelihood methods (Figure 6).
Figure 6.
Phylogenetic Analysis of Nuclear and Chloroplast infA Sequences.
The diagram summarizes the maximum likelihood trees found when each of the four nuclear infA sequences was added individually to a constrained topology (the species phylogeny from Figure 1) for the 21 chloroplast sequences. The arrows indicate the most likely placement of the tomato (Solanum), soybean (Glycine), ice plant (Mesembryanthemum), and Arabidopsis nuclear sequences, and all branch lengths are drawn to scale horizontally. Mesembryanthemum cp denotes the ice plant chloroplast pseudogene. The branch lengths for the chloroplast sequences were identical in the four trees, allowing them to be merged. cp, chloroplast; nuc, nuclear.
The nuclear sequence from ice plant clusters with its chloroplast pseudogene, suggesting a recent gene transfer in this lineage. The tomato nuclear sequence falls within the chloroplast sequences from asterids, which also suggests a recent transfer. The soybean nuclear sequence clusters with Euonymus and Brexia, the only core rosids with intact chloroplast genes (Figures 1 to 3). This is the expected phylogenetic position for a rosid nuclear sequence if a gene transfer occurred in the core rosid lineage after it separated from the Vitis lineage. The Arabidopsis nuclear gene also would be expected to cluster with Brexia and Euonymus, regardless of whether it and the soybean nuclear gene were formed by one or two separate gene transfers in rosids, but instead it clusters with the monocot Iris, which is entirely implausible (see Discussion).
DISCUSSION
How Many Independent infA Transfers to the Nucleus?
Our results indicate that the chloroplast infA gene has been lost (either entirely or has become a pseudogene) ∼24 separate times in the 309 angiosperms examined in this study (Figures 1 and 3). Intact and expressed nuclear infA genes were characterized from four diverse angiosperms that have lost the chloroplast gene. Are these nuclear genes orthologs derived from a single ancient transfer or paralogs derived from independent transfers?
Ice Plant
We are confident that the ice plant nuclear sequence represents a gene transfer independent of the other three, one that took place relatively recently in the caryophyllid lineage. Phylogenetic analysis implies that this nuclear gene was formed after the ice plant lineage diverged from the spinach lineage (Figure 6). It seems unlikely that the nuclear and chloroplast pseudogene sequences from the same species would cluster together on the tree if this were anything other than a recent transfer. In addition, the transit peptide of ice plant InfA shows little similarity to InfA transit peptides in the other species (Figure 4). Consistent with this being a recent transfer, the ice plant gene is the least divergent of the four nuclear infA genes (Figure 6).
Tomato
The tomato nuclear gene is probably also the product of an independent transfer event. The sporadic pattern of chloroplast infA losses among asterids, with most asterids containing intact chloroplast genes (Figures 1 and 3), is most consistent with a within-asterid loss, and transfer of infA, probably in the common ancestor of the Solanaceae/Convolvulaceae. Also consistent with an asterid-specific transfer is the placement of the tomato nuclear gene within the clade of asterid chloroplast sequences in phylogenetic analyses (Figure 6). Although there is some sequence similarity between the tomato and soybean transit peptides (Figure 4), the statistical significance of this similarity was estimated at only P ≈ 0.05, using a test based on sequence shuffling (PRSS algorithm; Pearson and Lipman, 1988). In summary, there is little evidence for a shared, ancient transfer of the tomato and any other nuclear infA genes; instead, both types of phylogenetic evidence (sequence-based phylogeny and distribution of infA losses) favor a separate transfer within asterid evolution.
Arabidopsis and Soybean
Whether the nuclear sequences from the rosids Arabidopsis and soybean represent two independent transfers is less clear. The interpretation shown in Figures 1 and 3 of four separate infA losses within core rosids, with Arabidopsis and soybean belonging to two different loss lineages, certainly implies independent transfers of their nuclear infA genes. Furthermore, there is a notable lack of similarity, in two respects, between the transit peptides of these two rosid nuclear infA genes, also suggesting that they arose by separate transfers. First, the Arabidopsis and soybean transit peptides show absolutely no evidence of homology at the level of sequence similarity (Figure 4). Only 12% of chloroplast transit peptides from orthologous Arabidopsis and legume genes are this different in sequence (see Methods), and the infA situation is more consistent with separate transfers than a single common transfer. Second, there is an intron in the transit peptide region of the Arabidopsis gene but not the soybean gene. This difference also is more consistent with separate origins for the two transit peptides, because it is uncommon for orthologous nuclear genes from these two rosids to be different in intron/exon organization (94% of introns were conserved in a sample of 16 genes that we examined; see Methods). The congruence of these three lines of evidence leads us to favor two separate transfers of the Arabidopsis and soybean genes. However, we cannot rule out a single common transfer followed by intron loss/gain and either unusually rapid transit peptide evolution or, as in rice mitochondrial rps11 (Kadowaki et al., 1996), gene duplication in the nucleus followed by independent gain of different transit peptides by the duplicate copies.
If there was a single transfer of infA in the rosids, then the intact infA genes of Brexia and Euonymus would reflect either the misplacement of this clade in current phylogenies (i.e., these two belong at the base of core rosids) or the clade's singular retention of intact chloroplast infA sequences after gene transfer at the base of the core rosids. Note that the infA phylogeny fundamentally does not bear on the number of rosid transfers because the single and double transfer scenarios predict the same phylogenetic result: placement of both the Arabidopsis and soybean nuclear infA sequences with the intact chloroplast infA sequences from Brexia and Euonymus. The soybean sequence attaches essentially where expected (Figure 6), but the Arabidopsis placement with the monocot Iris is biologically impossible (barring horizontal gene transfer) because the inferred transfer postdates the divergence of monocots from the lineage leading to Arabidopsis.
Overall, we conclude that the four nuclear infA genes probably arose via four independent chloroplast-to-nucleus transfer events, although we cannot rule out a smaller number (three or even two) of more ancient transfers. We regard the evidence in favor of independent transfer as strongest for ice plant and weakest for the two rosids. If our preferred hypothesis of four separate transfers is correct, then each of the other scattered losses of chloroplast infA (Figures 1 and 3) quite possibly reflects yet another independent case of infA transfer, although obviously this remains conjectural in the absence of nuclear gene isolation and characterization. The lack of similarity of any of the four characterized infA transit peptides to any sequences in the databases raises the possibility that these arose de novo from sequences not used previously for chloroplast targeting. This possibility is strongest for Arabidopsis, given that its entire genome has been sequenced (Arabidopsis Genome Initiative, 2000). In contrast, a number of recently transferred plant genes of mitochondrial origin have recruited preexisting transit peptides from long-established nuclear genes encoding mitochondrial proteins (Kadowaki et al., 1996; Figueroa et al., 1999; Kubo et al., 1999; Adams et al., 2000).
What Factors Drive Organelle Gene Transfer to the Nucleus?
Several hypotheses for why organelle genes are transferred to the nucleus have been discussed in recent reviews (Doolittle, 1998; Martin and Herrmann, 1998; Berg and Kurland, 2000; Blanchard and Lynch, 2000; Palmer et al. 2000). Briefly, there are three major hypotheses: (1) the relatively high frequency of organelle DNA escape to the nucleus provides numerous opportunities for successful functional gene transfers and is essentially a one-way process; (2) the progressive accumulation of detrimental mutations in asexual organelle genomes by Muller's ratchet favors transfer; and (3) smaller, streamlined organelle genomes are favored selectively. All of these factors could be operating in some eukaryotes in which gene transfer is ongoing, but only the first seems likely to be significant in flowering plants. The rate of nucleotide substitution in plant chloroplast and mitochondrial genomes is very low, much lower than in the nucleus (Wolfe et al. 1987; Laroche et al. 1997), and thus the overall effects of Muller's ratchet are likely to be minimal. As for streamlining, the “losses” of infA from the chloroplast genome mostly represent highly diverged pseudogenes, and there does not appear to be any genomic streamlining accompanying its transfer.
Why Are Some Organelle Genes Transferred More Often Than Others?
Most of the chloroplast genes (e.g., rbcL, atpB, matK, rps2, and rps4) that have been sequenced widely for molecular phylogenetic purposes are intact in all of the hundreds to thousands of photosynthetic angiosperms in which they have been examined. The only genes that have been lost repeatedly are infA, described here, and ndhF, whose many losses more likely reflect repeated loss of the entire multisubunit chloroplast NADH dehydrogenase complex rather than repeated gene transfer (Wakasugi et al., 1994; Neyland and Urbatsch, 1996; M. Ireland, H. Deiderick, Y.-L. Qiu, and J.D. Palmer, unpublished data). A few other chloroplast genes are thought to have been lost repeatedly during angiosperm evolution (Downie and Palmer, 1992), but in only one case (ycf2) has this been confirmed by sequencing (Downie et al., 1994), and no nuclear ycf2 genes have been reported.
At present, therefore, infA stands out as an unusually unstable angiosperm chloroplast gene, having been lost from cpDNA on many separate occasions and transferred to the nucleus multiple times, perhaps as often as it has been lost. One might wonder whether the chloroplast copy is functional in any angiosperms, but estimation of the ratio of nonsynonymous to synonymous nucleotide substitutions among the intact genes (the Ka/Ks ratio of Li [1993]; data not shown) shows that the intact genes are still subject to purifying selection (or have been until very recently) and therefore probably are still functional. No chloroplast genes other than infA have been reported to have undergone multiple evolutionary transfers to the nucleus, and none is an obvious candidate for similar multiple transfers within the context of angiosperm evolution. Among all photosynthetic eukaryotes, however, there are many candidate genes for multiple transfers. These are the genes—each with one documented nuclear transfer—that have been lost twice (12 genes) or three times (14 genes) among the five major lineages of completely sequenced plastid genomes analyzed by Martin et al. (1998).
The evolution of infA in angiosperm cpDNA is in some ways similar to that of rps10 in angiosperm mitochondrial DNA (Adams et al., 2000), with multiple losses and parallel transfers to the nucleus in both cases. It is striking that many of the recent gene losses from both chloroplast and mitochondrial genomes in plants involve genes for ribosomal proteins or other translation components. These include gene transfers of chloroplast infA and rpl22, gene substitutions of chloroplast rpl21 and rpl23 (see Introduction), and uncharacterized losses of several mitochondrial ribosomal protein genes in addition to the transfers of rps10 (Adams et al., 2000; Palmer et al., 2000).
Why have some genes been transferred to the nucleus more often than others? Conversely, why do a few genes remain in the organelle genomes of almost all eukaryotes? Several hypotheses as to why some genes but not others remain in organelles, discussed in detail elsewhere (Doolittle, 1998; Martin and Herrmann, 1998; Lang et al., 1999; Race et al., 1999; Palmer et al., 2000; Pérez-Martínez et al., 2000), can be mentioned: (1) some highly hydrophobic membrane proteins may be difficult to import into organelles and to insert into the appropriate membrane (Popot and de Vitry, 1990; Claros et al., 1995); (2) hydrophobic mitochondrial proteins might be misrouted to the endoplasmic reticulum if synthesized in the cytosol (von Heijne, 1987); (3) the presence of certain organellar proteins in the cytosol might have adverse effects (Herrmann, 1997; Martin and Schnarrenberger, 1997); and (4) expression of some genes is directly and rapidly regulated by the redox state of the organelle and therefore selection favors retention of these genes in the organelle (Allen, 1993; reviewed in Race et al., 1999). These hypotheses seek to explain why some types of genes are transferred infrequently, if ever. There is direct experimental evidence for hypothesis 1 in mitochondria (Claros et al., 1995) and hypothesis 4 in chloroplasts (Pfannschmidt et al., 1999). Gene size may also play a role, because it affects both the likelihood of intact gene transfer to the nucleus and the likelihood of the transferred gene being damaged by mutation before it acquires a transit peptide. Other factors, such as the possible presence of cryptic organelle targeting sequences internal to some proteins, may also be important (Adams et al., 2000; Kubo et al., 2000).
In the case of infA, most of these hypotheses fail to explain why the gene was not lost and transferred to the nucleus early in chloroplast evolution. InfA is a small hydrophilic protein, so it should have been easy to transfer long ago. Its expression is presumably not regulated by redox potential because it has been lost from the chloroplast genome in many angiosperms as well as in three algal lineages (Martin et al., 1998). Therefore, it is perhaps more surprising that infA survived in the chloroplast genome during the millions of years of evolution leading to angiosperms than that it has been transferred to the nucleus many times in different angiosperms. The question then becomes whether any other chloroplast genes have been lost and transferred in angiosperms at rates approaching that of infA.
METHODS
Chloroplast DNA Sequencing
The species sequenced are listed at www.bio.indiana.edu/~palmerlab. Chloroplast sequences were determined by direct sequencing of single-stranded DNA templates produced by asymmetrical polymerase chain reaction (PCR) (Gyllensten, 1989; Kaltenboek et al., 1992) using either chloroplast DNA (cpDNA) or total cellular DNA. PCR primers were located at the proximal ends of the flanking genes rps8 and rpl36. The single-stranded PCR product was cleaned of excess primers and deoxynucleotide triphosphates by spin dialysis (Centricon 100 microconcentrators; Amicon, Inc., Beverly, MA). Purified template was sequenced by standard dideoxy methods using 32P or 35S with Sequenase (Amersham Life Science, Tallaght, Ireland) or by automated sequencing (PE Biosystems, Norwalk, CT). PCR primers and internal primers were used for sequencing. Both strands within the genic region were sequenced to resolve ambiguities. All new sequences reported here have been submitted to GenBank (accession numbers AF347615 to AF347666).
DNA Gel Blot Hybridizations
All species surveyed are listed at www.bio.indiana.edu/~palmerlab. Blots were prepared and hybridizations were performed as described (Qiu et al., 1998), except that the temperature for hybridization and washes was 55°C.
Nuclear Sequences
All nuclear sequences were identified initially by BLAST searches against GenBank and dbEST. The soybean infA cDNA clone corresponding to accession number AI440898 was obtained from Incyte Genomics (St. Louis, MO), and the sequence was completed at the 3′ end. The ice plant and tomato sequences analyzed here are assemblies of expressed sequence tags (ESTs) from dbEST. Accession numbers are AI823024, BE130300, and AA762038 (ice plant) and AI782841, AW624662, and AW223254 (tomato).
The Arabidopsis genomic region on chromosome 4 was sequenced independently by American and European groups participating in the genome project (Mayer et al., 1999). The GenBank accession numbers for these sequences are AF080120 (clone F2P3) and AL049876 (clone T22B4). These genomic sequences are identical, but the two groups' intron/exon predictions (named F2P3.7 and T22B4.150 or AT4g11170) for the infA-like region disagree with each other and with our interpretation. We reanalyzed the region using MZEF (Zhang, 1998) and predicted a different, two-exon structure for infA (bases 89,932 to 90,011 and 90,231 to 90,576 of AL049876), whose splice sites were subsequently confirmed by the ESTs AI995770 and BE039009.
Gene Fusions between infA and Green Fluorescent Protein
Gene fusions between the soybean and Arabidopsis infA cDNAs and the gene encoding the jellyfish green fluorescent protein (GFP) were constructed to investigate the subcellular targeting of InfA–GFP fusion proteins. The construct pGmInfA–GFP was generated by amplifying the entire soybean infA open reading frame (but without the stop codon) by using Pfu polymerase and the oligonucleotide primers InfX (5′-GCTCTAGATGTTCACCTCACTCC-3′) and InfFS (5′-CGCGTCGACGAGGGGGTGCTGCTG-3′). In pAtInfA–GFP, the entire Arabidopsis infA open reading frame except the stop codon was amplified from an EST clone (GenBank accession number AI995770; purchased from Incyte Genomics) by using the primers AtInfF (5′-GCTCTAGAAAATGCTTCAACTCTGCTCCAC-3′) and AtInfR (5′-CGCGTCGACGAGTCTCTACTACTCATTCTGAA-3′). To generate the various gene fusions, the PCR-amplified infA fragments were digested with XbaI and SalI and cloned upstream of the GFP gene (CD3-326; Davis and Vierstra, 1998). In each case, transcriptional control was provided by the cauliflower mosaic virus 35S promoter, and transcription termination and polyadenylation signals were provided by the nopaline synthase (nos) terminator (Davis and Vierstra, 1998). Leaves of tobacco or Arabidopsis were bombarded with tungsten particles (0.7 μm) coated with pGmInfA–GFP or pAtInfA–GFP by using a Bio-Rad PDS-1000/He particle delivery system, as described elsewhere (Hibberd et al., 1998). After 3 days, samples were cut into 1 × 1-cm sections, placed on a glass slide, and viewed by confocal scanning laser microscopy (TCS-NT, DM1RB light microscope stand; Leica, Wetzlar, Germany). Images were captured as described elsewhere (Hibberd et al., 1998).
Phylogenetic Analysis
Phylogenetic analyses were conducted at the University of Sydney Bioinformatics Supercomputing Facility. The input data were a DNA sequence alignment corresponding to a ClustalW alignment (Thompson et al., 1994) of the deduced amino acid sequences that had been edited by eye. The 5′ extensions and the extreme 3′ ends of the sequences were excluded from analysis (the alignment is available at www.bio.indiana.edu/~palmerlab). To minimize long-branch attraction, we omitted several chloroplast sequences with relatively long branches (Zea, Oryza, Leucophyllum, Epifagus, and all pseudogenes except ice plant), and the gene from Iris was sequenced instead to represent monocots. To avoid long branches leading to nonangiosperm outgroups, we determined sequences from Amborella, Cabomba, and Illicium for use as outgroups. Recent multigene phylogenetic studies have found that these represent the earliest lineages of angiosperms (Mathews and Donoghue, 1999; Parkinson et al., 1999; Qiu et al., 1999; Soltis et al., 1999).
The variable codon sites in the remaining set of 20 intact chloroplast infA sequences (Figure 6) were surveyed using a method similar to that of Andrews et al. (1998) (L.S. Jermiin, S.R. Wilson, and S. Easteal, unpublished data). There was substantial base composition heterogeneity at codon position 3, so nucleotides at these sites were recoded as purine/pyrimidine. The alignments were analyzed using the maximum likelihood program TrExML (Wolf et al., 2000), which implements the F84 model of nucleotide substitutions. Using an iterative approach, we estimated a transition/transversion ratio of 1.86 and relative rates of change of 2.26, 1.00, and 7.32 at codon positions 1, 2, and 3, respectively, for the species tree. Searches of the 20-species data set using these parameters resulted in almost 10,000 trees that did not differ significantly from the most likely tree by the test of Kishino and Hasegawa (1989). The species tree of Soltis et al. (1999) was among these good trees, implying that the infA data were compatible with this tree, so it was used as the framework for subsequent analyses. The same transition/transversion and rate parameters were used in the estimation of the most likely placement for the ice plant chloroplast sequence (which grouped with spinach, as expected) and then to place each of the four nuclear genes separately onto this 21-species tree (Figure 6). If all four nuclear sequences were added simultaneously, then the maximum likelihood tree grouped them together with the ice plant chloroplast pseudogene as their closest relative. This is biologically impossible and is almost certainly due to a long-branch attraction artifact.
Transit Peptide and Intron Comparisons
Transit peptide predictions were made using ChloroP version 1.1 (Emanuelsson et al., 1999; www.cbs.dtu.dk/services/ChloroP), TargetP version 1.01 (Emanuelsson et al., 2000; www.cbs.dtu.dk/services/ TargetP), and Predotar version 0.5 (N.M. Peeters, A. Chapron, D. Lancelin, O. Grandjean, A. Giritch, H. Wintz, and I. Small, unpublished data; www.inra.fr/Internet/Produits/Predotar).
To compare the level of similarity seen in the soybean and Arabidopsis InfA transit peptides with that in other genes, we identified all legume (Fabaceae) proteins in Swissprot having an annotated chloroplast transit peptide with a specified cleavage site. These were compared with the Arabidopsis genome sequence (www.arabidopsis.org) by using BLASTP, and putative orthologs were identified on a mutual-best-hits basis (Makalowski and Boguski, 1998). The transit peptides were then compared using the PRSS shuffling algorithm as implemented in the FASTA package version 33 (Pearson and Lipman, 1988). Of the 58 transit peptides compared, seven (12%) had less significant scores (i.e., were more dissimilar) than the soybean/Arabidopsis InfA comparison had.
To estimate the extent to which intron locations are conserved between soybean and Arabidopsis genes, we examined all soybean genomic sequences in GenBank, and 16 apparently orthologous soybean/Arabidopsis sequence pairs were identified. Among these, 83 introns were conserved (i.e., they had coincident locations in the two species relative to an alignment of the amino acid sequences), and five introns (6%) were unique to one species or the other.
Acknowledgments
We thank Tim Littlejohn for hospitality, Hans Bohnert for ice plant DNA, and Ian Small and Olof Emanuelsson for preprints. This work was supported by Royal Irish Academy and Enterprise Ireland travel awards to K.H.W.; National Science Foundation Grant No. BSR-9107827 to R.G.O.; National Institutes of Health Grant No. GM-35087 to J.D.P.; U.S. Department of Agriculture Fellowship No. 95-38420-2214 to K.L.A.; a University of Sydney U2000 Fellowship to L.S.J.; a Biotechnology and Biological Science Research Council Sir David Phillips Research Fellowship to J.M.H.; and European Commission Contract No. BIO4 CT972245 to N.T.L., T.A.K., J.M.H., and J.C.G.
References
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. [DOI] [PubMed] [Google Scholar]
- Allen, J.F. (1993). Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, T.D., Jermiin, L.S., and Easteal, S. (1998). Accelerated evolution of cytochrome b in simian primates: Adaptive evolution in concert with other mitochondrial proteins? J. Mol. Evol. 47, 249–257. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Battiste, J.L., Pestova, T.V., Hellen, C.U., and Wagner, G. (2000). The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5, 109–119. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J. (1987). Why do chloroplasts and mitochondria contain so many copies of their genome? Bioessays 6, 279–282. [DOI] [PubMed] [Google Scholar]
- Berg, O.G., and Kurland, C.G. (2000). Why mitochondrial genes are most often found in nuclei. Mol. Biol. Evol. 17, 951–961. [DOI] [PubMed] [Google Scholar]
- Blanchard, J.L., and Lynch, M. (2000). Organellar genes: Why do they end up in the nucleus? Trends Genet. 16, 315–320. [DOI] [PubMed] [Google Scholar]
- Bubunenko, M.G., Schmidt, J., and Subramanian, A.R. (1994). Protein substitution in chloroplast ribosome evolution: A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240, 28–41. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., Perea, J., Shu, Y., Samatey, F.A., Popot, J.-L., and Jacq, C. (1995). Limitations to in vitro import of hydrophobic proteins into yeast mitochondria: The case of a cytoplasmically synthesized apocytochrome b. Eur. J. Biochem. 228, 762–771. [PubMed] [Google Scholar]
- Cummings, H.S., and Hershey, J.W. (1994). Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F. (1998). You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311. [DOI] [PubMed] [Google Scholar]
- Downie, S.R., and Palmer, J.D. (1992). Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In Molecular Systematics of Plants, P.S. Soltis, D.E. Soltis, and J.J. Doyle, eds (New York: Chapman and Hall), pp. 14–35.
- Downie, S.R., Katz-Downie, D.S., Wolfe, K.H., Calie, P.J., and Palmer, J.D. (1994). Structure and evolution of the largest chloroplast gene (ORF2280): Internal plasticity and multiple gene loss during angiosperm evolution. Curr. Genet. 25, 367–378. [DOI] [PubMed] [Google Scholar]
- Doyle, J.J., Doyle, J.L., and Palmer, J.D. (1995). Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst. Bot. 20, 272–294. [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network–based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1978). Cases in which parsimony and compatibility methods will be positively misleading. Syst. Zool. 27, 401–410. [Google Scholar]
- Figueroa, P., Gomez, I., Holuigue, L., Araya, A., and Jordana, X. (1999). Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron-sulphur subunit of succinate dehydrogenase and expression by alternative splicing. Plant J. 18, 601–609. [DOI] [PubMed] [Google Scholar]
- Gantt, J.S., Baldauf, S.L., Calie, P.J., Weeden, N.F., and Palmer, J.D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyllensten, U. (1989). Direct sequencing of in vitro amplified DNA. In PCR Technology, H.A. Erlich, ed (New York: Stockton Press), pp. 45–60.
- Herrmann, R.G. (1997). Eukaryotism, towards a new interpretation. In Eukaryotism and Symbiosis, H.E.A. Schenk, R.G. Herrmann, K.W. Jeon, N.E. Muller, and W. Schwemmler, eds (Vienna: Springer), pp. 73–118.
- Hibberd, J.M., Linley, P.J., Khan, M.S., and Gray, J.C. (1998). Transient expression of green fluorescent protein in various plastid types following microprojectile bombardment. Plant J. 16, 627–632. [Google Scholar]
- Hiratsuka, J., et al. (1989). The complete sequence of the rice (Oryza sativa) chloroplast genome: Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217, 185–194. [DOI] [PubMed] [Google Scholar]
- Hupfer, H., Swiatek, M., Hornung, S., Herrmann, R.G., Maier, R.M., Chiu, W.L., and Sears, B. (2000). Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Gen. Genet. 263, 581–585. [DOI] [PubMed] [Google Scholar]
- Kadowaki, K., Kubo, N., Ozawa, K., and Hirai, A. (1996). Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15, 6652–6661. [PMC free article] [PubMed] [Google Scholar]
- Kaltenboek, B., Spatafora, B.J.W., Zhang, X., Kousoulas, K.G., Blackwell, M., and Storz, J. (1992). Efficient production of single-stranded DNA as long as 2 kb for sequencing of PCR-amplified DNA. BioTechniques 12, 164–171. [PubMed] [Google Scholar]
- Kishino, H., and Hasegawa, M. (1989). Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29, 170–179. [DOI] [PubMed] [Google Scholar]
- Konishi, T., Shinohara, K., Yamada, K., and Sasaki, Y. (1996). Acetyl-CoA carboxylase in higher plants: Most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 117–122. [DOI] [PubMed] [Google Scholar]
- Kubo, N., Harada, K., Hirai, A., and Kadowaki, K. (1999). A single nuclear transcript encoding mitochondrial RPS14 and SDHB of rice is processed by alternative splicing: Common use of the same mitochondrial targeting signal for different proteins. Proc. Natl. Acad. Sci. USA 96, 9207–9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, N., Jordana, X., Ozawa, K., Zanlungo, S., Harada, K., Sasaki, T., and Kadowaki, K. (2000). Transfer of the mitochondrial rps10 gene to the nucleus in rice: Acquisition of the 5′ untranslated region followed by gene duplication. Mol. Gen. Genet. 263, 733–739. [DOI] [PubMed] [Google Scholar]
- Lang, B.F., Gray, M.W., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33, 351–397. [DOI] [PubMed] [Google Scholar]
- Laroche, J., Li, P., Maggia, L., and Bousquet, J. (1997). Molecular evolution of angiosperm mitochondrial introns and exons. Proc. Natl. Acad. Sci. USA 94, 5722–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H. (1993). Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36, 96–99. [DOI] [PubMed] [Google Scholar]
- Lin, Q., Ma, L., Burkhart, W., and Spremulli, L.L. (1994). Isolation and characterization of cDNA clones for chloroplast translational initiation factor-3 from Euglena gracilis. J. Biol. Chem. 269, 9436–9444. [PubMed] [Google Scholar]
- Ma, L., and Spremulli, L.L. (1992). Immunological characterization of the complex forms of chloroplast translational initiation factor 2 from Euglena gracilis. J. Biol. Chem. 267, 18356–18360. [PubMed] [Google Scholar]
- Magallón, S., Crane, P.R., and Herendeen, P.S. (1999). Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Mo. Bot. Gard. 86, 297–372. [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., and Kössel, H. (1995). Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Makalowski, W., and Boguski, M.S. (1998). Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. USA 95, 9407–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Schnarrenberger, C. (1997). The evolution of the Calvin cycle from prokarytic to eukaryotic chromosomes: A case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 32, 1–18. [DOI] [PubMed] [Google Scholar]
- Martin, W., Lagrange, T., Li, Y.F., Bisanz-Seyer, C., and Mache, R. (1990). Hypothesis for the evolutionary origin of the chloroplast ribosomal protein L21 of spinach. Curr. Genet. 18, 553–556. [DOI] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and Donoghue, M.J. (1999). The root of angiosperm phylogeny inferred from duplicate phytochrome genes. Science 286, 947–950. [DOI] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777. [DOI] [PubMed] [Google Scholar]
- Nagano, Y., Matsuno, R., and Sasaki, Y. (1991). Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr. Genet. 20, 431–436. [DOI] [PubMed] [Google Scholar]
- Neyland, R., and Urbatsch, L. (1996). Phylogeny of subfamily Epidendroideae (Orchidaceae) inferred from ndhF chloroplast gene sequences. Am. J. Bot. 83, 1195–1206. [Google Scholar]
- Olmstead, R.G., Kim, K.-J., Jansen, R.K., and Wagstaff, S.J. (2000). The phylogeny of the Asteridae sensu lato based on chloroplast ndhF gene sequences. Mol. Phylogenet. Evol. 16, 96–112. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D., and Delwiche, C.F. (1998). The origin and evolution of plastids and their genomes. In Molecular Systematics of Plants. II. DNA Sequencing, D.E. Soltis, P.S. Soltis, and J.J. Doyle, eds (Boston: Kluwer Academic Publishing), pp. 375–409.
- Palmer, J.D., Adams, K.L., Cho, Y., Parkinson, C.L., Qiu, Y.L., and Song, K. (2000). Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson, C.L., Adams, K.L., and Palmer, J.D. (1999). Multigene analyses identify the three earliest lineages of extant flowering plants. Curr. Biol. 9, 1485–1488. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., and Lipman, D.J. (1988). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel, H.J., and Grivell, L.A. (1994). Protein synthesis in mitochondria. Mol. Biol. Rep. 19, 183–194. [DOI] [PubMed] [Google Scholar]
- Pérez-Martínez, X., Vázquez-Acevedo, M., Tolkunova, E., Funes, S., Claros, M.G., Davidson, E., King, M.P., and González-Halphen, D. (2000). Unusual location of a mitochondrial gene: Subunit III of cytochrome c oxidase is encoded in the nucleus of Chlamydomonad algae. J. Biol. Chem. 275, 30144–30152. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt, T., Nilsson, A., and Allen, J.F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397, 625–628. [Google Scholar]
- Popot, J.-L., and de Vitry, C. (1990). On the microassembly of integral membrane proteins. Annu. Res. Biophys. Chem. 19, 369–403. [DOI] [PubMed] [Google Scholar]
- Qiu, Y.L., Cho, Y., Cox, J.C., and Palmer, J.D. (1998). The gain of three mitochondrial introns identifies liverworts as the earliest land plants. Nature 394, 671–674. [DOI] [PubMed] [Google Scholar]
- Qiu, Y.L., Lee, J., Bernasconi-Quadroni, F., Soltis, D.E., Soltis, P.S., Zanis, M., Zimmer, E.A., Chen, Z., Savolainen, V., and Chase, M.W. (1999). The earliest angiosperms: Evidence from mitochondrial, plastid and nuclear genomes. Nature 402, 404–407. [DOI] [PubMed] [Google Scholar]
- Race, H.L., Herrmann, R.G., and Martin, W. (1999). Why have organelles retained genomes? Trends Genet. 15, 364–370. [DOI] [PubMed] [Google Scholar]
- Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., and Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. [DOI] [PubMed] [Google Scholar]
- Sette, M., van Tilborg, P., Spurio, R., Kaptein, R., Paci, M., Gualerzi, C.O., and Boelens, R. (1997). The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijben-Müller, G., Hallick, R.B., Alt, J., Westhoff, P., and Herrmann, R.G. (1986). Spinach plastid genes coding for initiation factor IF-1, ribosomal protein S11 and RNA polymerase alpha-subunit. Nucleic Acids Res. 14, 1029–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.F. (2000). Phylogenetic resolution within the tribe Episcieae (Gesneriaceae): Congruence of ITS and ndhF sequences from parsimony and maximum-likelihood analyses. Am. J. Bot. 87, 883–897. [PubMed] [Google Scholar]
- Soltis, P.S., Soltis, D.E., and Chase, M.W. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- Thomas, F., Massenet, O., Dorne, A.M., Briat, J.F., and Mache, R. (1988). Expression of the rpl23, rpl2 and rps19 genes in spinach chloroplasts. Nucleic Acids Res. 16, 2461–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1987). Why mitochondria need a genome. FEBS Lett. 198, 1–4. [DOI] [PubMed] [Google Scholar]
- Wakasugi, T., Tsudzuki, J., Ito, S., Nakashima, K., Tsudzuki, T., and Sugiura, M. (1994). Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 91, 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, M.J., Easteal, S., Kahn, M., McKay, B.D., and Jermiin, L.S. (2000). TrExML: A maximum-likelihood approach for extensive tree-space exploration. Bioinformatics 16, 383–394. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Li, W.H., and Sharp, P.M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84, 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Morden, C.W., and Palmer, J.D. (1992. a). Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H., Morden, C.W., Ems, S.C., and Palmer, J.D. (1992. b). Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J. Mol. Evol. 35, 304–317. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., and Subramanian, A.R. (2000). The plastid ribosomal proteins: Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- Yu, N.J., and Spremulli, L.L. (1998). Regulation of the activity of chloroplast translational initiation factor 3 by NH2- and COOH-terminal extensions. J. Biol. Chem. 273, 3871–3877. [DOI] [PubMed] [Google Scholar]
- Zhang, M.Q. (1998). Identification of protein-coding regions in Arabidopsis thaliana genome based on quadratic discriminant analysis. Plant Mol. Biol. 37, 803–806. [DOI] [PubMed] [Google Scholar]