Abstract
The transposition of Mu elements underlying Mutator activity in maize requires a transcriptionally active MuDR element. Despite variation in MuDR copy number and RNA levels in Mutator lines, transposition events are consistently late in plant development, and Mu excision frequencies are similar. Here, we report previously unsuspected and ubiquitous MuDR homologs that produce both RNA and protein. MuDR transcript levels are proportional to MuDR copy number, and homolog transcript levels increase in active Mutator lines. A subset of homologs exhibits constitutive transcription in MuDR− and epigenetically silenced MuDR lines, suggesting independent transcriptional regulation. Surprisingly, immunodetection demonstrated nearly invariant levels of MuDR and homolog protein products in all tested Mutator and non-Mutator stocks. These results suggest a strict control over protein production, which might explain the uniform excision frequency of Mu elements. Moreover, the nonfunctional proteins encoded by homologs may negatively regulate Mutator activity and represent part of the host defense against this transposon family.
INTRODUCTION
Optimized transmission and restricted transposition activity are characteristics of many transposable elements. These features combine to allow efficient transposon proliferation with a minimal cost to the host genome. A variety of regulatory mechanisms operating at both the transcriptional and post-transcriptional levels can result in specific temporal and spatial patterns of transposon activity. Some mechanisms are epigenetic and act through several generations. In plants, increased DNA methylation of promoter regions of autonomous elements such as Ac, En/Spm, and MuDR correlates with decreased production of transposase transcripts and loss of transposition reactions (Fedoroff and Chandler, 1994; Martienssen and Baron, 1994). In some cases, host methylation acts in concert with transposon autoregulation. For example, the TnpA transposase protein encoded by Spm binds to its own unmethylated promoter, an interaction that decreases transcription; in the absence of Spm-encoded products, the promoter becomes methylated, and both transcription and transposition cease (Schläppi et al., 1994). Introduction of a functional Spm element results in activation of the silenced promoter and resumption of transposon activity.
More dynamic regulation occurs during a single life cycle to limit the production or function of active transposase proteins. For example, tissue-specific RNA splicing restricts expression of the functional transposase of the Drosophila melanogaster P element to the germ line (Laski et al., 1986; Siebel et al., 1992). In bacteria, the inhibition of bacterial Tn10 transposition occurs through production of element-encoded antisense RNA (Simons and Kleckner, 1988). The maize transposable element Ac exhibits a negative dosage phenomenon: with increasing copy number and RNA levels, there is a lower excision frequency, and transposition events occur later in development (McClintock, 1951; Heinlein and Starlinger, 1991). Because protein levels correlate with Ac copy number (Fusswinkel et al., 1991), this behavior was hypothesized to require a reduction in the concentration of active transposase (Scofield et al., 1993). Experimentally, high concentrations of Ac transposase were shown to aggregate into inactive oligomers (Heinlein et al., 1994; Essers et al., 2000).
In contrast to the low copy number of Ac and Spm, active Mutator lines typically have five to 20 unmethylated, mobile copies of MuDR. The nonautonomous Mu elements also occur in multiple copies and have little or no homology with MuDR, except in the terminal inverted repeat (TIR) regions (Bennetzen, 1996). The nearly identical ∼215-bp TIRs of MuDR contain the promoters for the convergently transcribed mudrA and mudrB genes (Figure 1A). The transcripts for both genes are very abundant, particularly in contrast to the low levels of Ac and Spm transposase RNA (Walbot and Rudenko, 2001). The mudrA gene product, MURA protein, is likely the transposase, because it binds Mu TIRs in vitro (Benito and Walbot, 1997), it has a region of extensive homology with bacterial transposases (Eisen et al., 1994), and expression of mudrA (Lisch et al., 1999) or its fully spliced cDNA alone (Raizada and Walbot, 2000) is sufficient to program somatic excision. The function of MURB protein (mudrB gene product) is elusive, but it appears to be required for both somatic and germinal insertion (reviewed in Walbot and Rudenko, 2001).
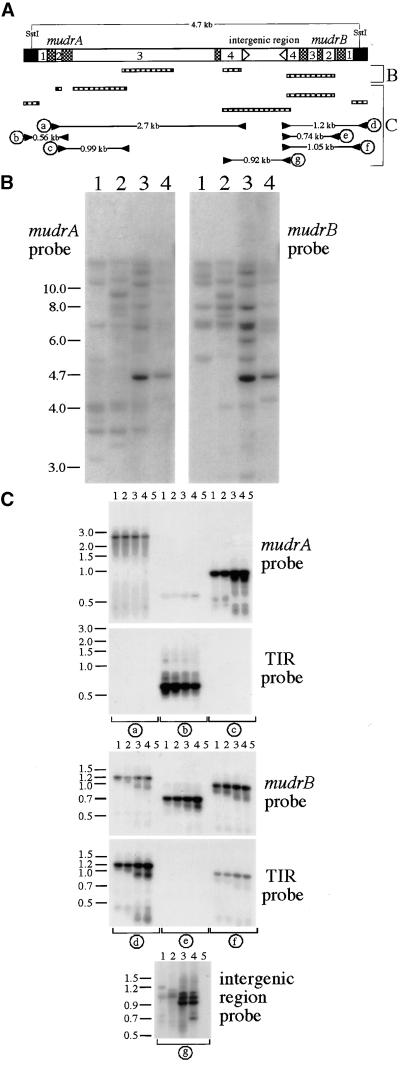
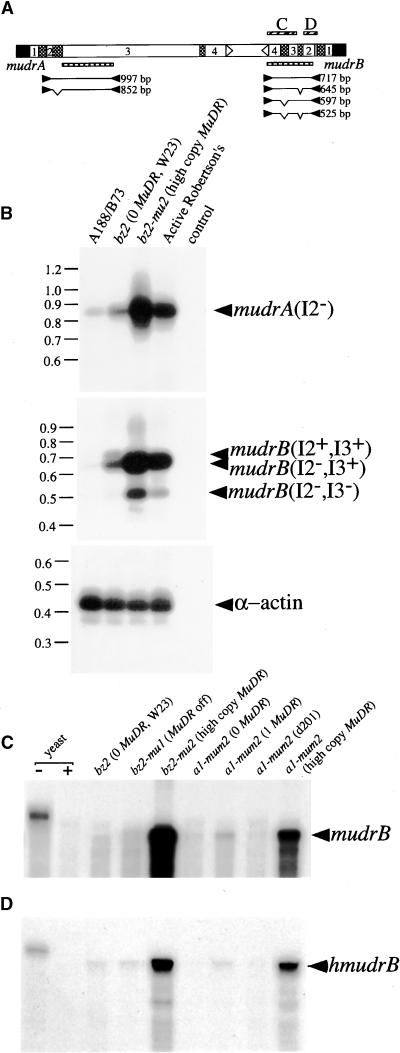
Figure 1.
Molecular Identification of mudrA and mudrB Homologous Sequences in Various Maize Lines.
(A) Diagram of the MuDR transposable element. MuDR terminates in nearly identical 215-bp TIRs (black boxes) and contains the convergently transcribed mudrA and mudrB genes, which are annotated with numbers indicating exons, cross-hatched boxes indicating introns, and arrowheads indicating poly(A) addition sites. Bracket B shows MuDR regions recognized by hybridization probes (hatched boxes) used to analyze the diagnostic 4.7-kb SstI fragment (B). Note that the mudrA probe was a mix of two mudrA-specific fragments. The hatched boxes in bracket C are probes (corresponding to components of MuDR) used in hybridization analysis; also shown are the PCR strategy, primer pairs, and expected sizes of successfully amplified products from the mudrA and mudrB genes analyzed in (C). The detailed description of the hybridization probes and PCR analysis can be found in Methods.
(B) DNA samples from mature embryos of two non-Mutator lines, A188/B73 hybrid (lanes 1) and bz2 inbred W23 (lanes 2), and two lines with transcriptionally active MuDR elements, bz2-mu2 (lanes 3) and Robertson's purple Mutator (lanes 4), were digested with SstI and processed by sequential DNA gel blot hybridization as described in (A).
(C) Ten nanograms of genomic DNA from the same maize lines in (B) were analyzed by PCR as described in (A). Lanes 5 are no DNA control samples.
Mutator activity is strictly regulated in plant development. Both somatic and germinal transposition events are restricted to the last few cell divisions during tissue development and do not correlate with transcript levels of MuDR-encoded genes (Walbot and Rudenko, 2001). Maize lines with a single copy of MuDR produce less transcript than do multiple-copy MuDR lines (Hershberger et al., 1991). Yet, a single copy of MuDR programs the same frequency and timing of somatic excision (Lisch et al., 1995) as those seen in multiple-copy lines (Walbot, 1991). Moreover, expression of a mudrA cDNA transgene from the cauliflower mosaic virus 35S promoter resulted in the late developmental pattern of Mu excisions observed in Mutator lines (Raizada and Walbot, 2000). The mudrA and mudrB transcripts are ubiquitously present in meristems, immature organs, and fully differentiated maize tissues (Hershberger et al., 1995; Joanin et al., 1997). The TIR next to mudrB directs high levels of reporter β-glucuronidase and luciferase protein expression in organ primordia, particularly floral primordia and developing pollen of transgenic plants (Raizada et al., 2001a). Polyclonal antibodies directed to MURB also demonstrated that the greatest accumulation was in immature organs (Donlin et al., 1995). Paradoxically, these studies demonstrate that MuDR transcripts and at least one of the predicted proteins are abundant in meristems and immature organs in which Mu transposition events are rarely observed. Consequently, MURA and MURB proteins, if produced in proportion to the MuDR copy number, must be either functionally or physically depleted until appropriate developmental stages. One mechanism might include the inhibition of MURA transposase by unknown host factors. Alternatively, specific features of post-transcriptional regulation or limited protein degradation might be responsible for the lack of functional MURA and MURB proteins.
To approach these questions experimentally, we surveyed diverse maize lines for the presence of unknown members of the MuDR/Mu family that might serve as host factors and thus contribute to the developmental regulation of Mutator activity. In this article, we report the identification of a large family of MuDR homologs. These heretofore unsuspected “host” elements are ubiquitously distributed in maize germplasm, and they produce transcripts and proteins in all Mutator and non-Mutator maize lines examined. Next, we demonstrate that in Mutator lines, mudrA and mudrB transcript levels are proportional to MuDR copy number, and that MuDR homologs yield substantially less transcript than do functional elements. By using antibodies directed to MURA and several antibodies to MURB, we found similar protein levels in all tested maize plants. This surprising result uncovers a translational or post-translational control mechanism affecting the accumulation of both MURA and MURB. A similar abundance of MURA transposase in various Mutator lines might explain the lack of correlation between MuDR copy number and excision frequencies. We also discuss the possible mechanisms by which the homolog proteins could modulate or interfere with MURA transposase to exert developmental host control of Mutator activities.
RESULTS
Identification of mudrA and mudrB Homologous Genes
In maize lines with a single, genetically active copy of MuDR, molecular-genetic analysis demonstrated cosegregation of Mutator activity with a unique 4.7-kb SstI DNA fragment (Chomet et al., 1991). In more typical Mutator lines, multiple copies of this 4.7-kb fragment are present, consistent with the failure of Mutator activity to segregate (Hershberger et al., 1991). As shown in Figure 1A, this diagnostic fragment is characteristic of an intact, unmethylated element, and it is absent in standard inbred lines (Chomet et al., 1991; Hershberger et al., 1991; Qin et al., 1991; Lisch et al., 1995). We confirmed these observations by DNA gel blot analysis of genomic DNA from two representative Mutator lines and two standard non-Mutator lines digested with SstI and hybridized separately to probes specific for mudrA and mudrB. As shown in Figure 1B, the expected 4.7-kb fragment is present in Mutator lines, as are various larger and smaller fragments that hybridize to one or both probes. The smaller fragments have been ascribed to deleted forms of MuDR that are generated in active Mutator lines (Hardeman and Chandler, 1993; Lisch and Freeling, 1994; Hershberger et al., 1995; Lisch et al., 1995). Larger fragments have been ascribed to methylation, which prevents digestion at the diagnostic SstI sites in the TIRs of MuDR, or to insertions into MuDR-like elements (i.e., 5.0-kb MuDRzc, Gutiérrez-Nava et al., 1998; 5.5-kb MuA, Qin and Ellingboe, 1990). Similarly, cross-hybridizing larger fragments could be detected in the two non-Mutator lines, but the 4.7-kb MuDR fragment was absent.
To assess the nature of the cross-hybridizing sequences, we subjected genomic DNA to polymerase chain reaction (PCR) analysis. We used primer pairs that would amplify the full-length coding regions of mudrA and mudrB genes as well as primer pairs that would address whether these genes are next to a TIR typical of a Mu element. In addition, we examined whether an intergenic region typical of a MuDR organization separates them. As shown in Figure 1C, this strategy yielded specific mudrA- and mudrB-related fragments in both non-Mutator and Mutator lines. Although the coding region–specific PCR products were of the expected length, multiple fragments resulted from amplification across the intergenic region in non-Mutator lines. These results establish that mudrA- and mudrB-related genes exist next to Mu TIRs in non-Mutator maize lines lacking a genetically active MuDR element, that at least some of these genes are organized in the same orientation as in MuDR, and that the intergenic regions vary in length.
Structural Characterization of mudrB and mudrA Homologous Genes
To verify that the PCR products represented genes, products from primer pairs b and d (TIR plus mudrA or mudrB) and e (mudrB only) were cloned and sequenced. Surprisingly, each product class was a heterogeneous family of MuDR-related molecules of similar size but defined by sequence divergence. Because all of the sequences appear to be homologs of wild-type MuDR-encoded genes, the letter h is added when referring to a particular product. Twenty-one different products resulted from the sequencing of the mudrB fragments. A graphic comparison of hmudrB sequences is shown in Figure 2A. The homologs share a suite of single-base substitutions but differ in the total number of mutations. This suggests that the diversity in the hmudrB gene family occurred predominantly through a stepwise incorporation of point mutations. A few products contain deletions and insertions of one or a few bases.
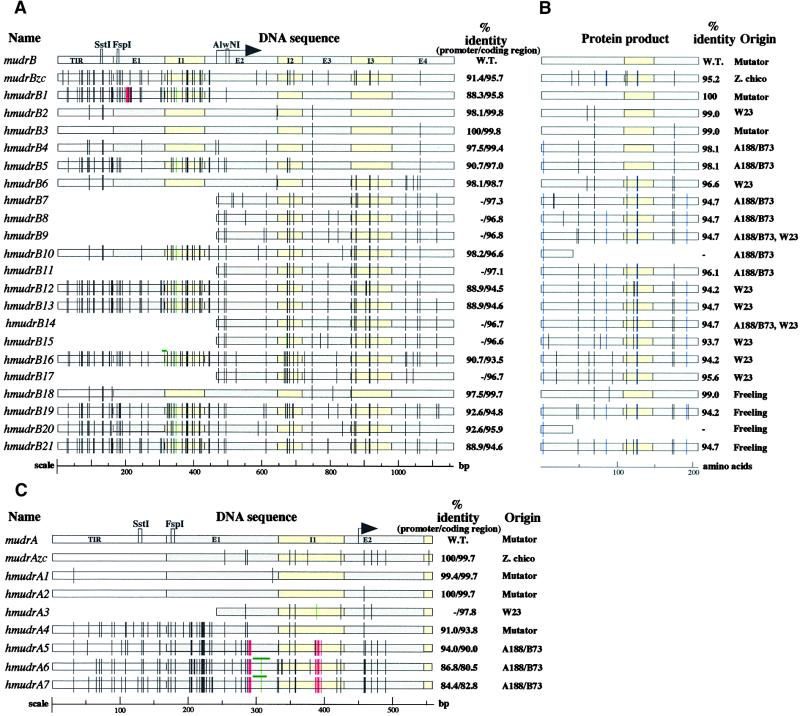
Figure 2.
Sequence Analysis of MuDR Homologs.
(A) Alignment and analysis of sequenced mudrB homologs. Wild-type (W.T.) mudrB is the top line, followed by the previously reported mudrB from the land race Zapalote chico, mudrBzc; the remaining sequences were recovered in this study, and their origins are listed in the column at right (B). Note that some homolog types were recovered from independent inbred lines. The TIR regions, introns, and exons are drawn to scale; the arrow indicates the translation initiation start site. Observed DNA mutations are indicated by vertical bars or boxes according to a color code: black, single base pair substitutions; green, insertions; red, deletions. As an internal control to account for potential point mutations that might have been derived as a result of clone propagation in Escherichia coli, six full-length wild-type mudrB genes were sequenced from an active Mutator line; these genes had the same sequence as the original MuDR isolate (Hershberger et al. 1995). In addition, single base DNA substitutions have been noted previously in three MuDR clones (Hershberger et al., 1995) and in the independently isolated MuA2 element (James et al., 1993).
(B) The MURB protein homologs encoded by each sequenced gene in (A) are shown as retaining intron 3 (207–amino acid form of MURB), with black bars corresponding to conservative substitutions and blue bars representing nonconservative substitutions. Truncated protein products are shown for hmudrB10 and hmudrB20, which share the same mutation creating a stop codon.
(C) Graphic alignment of sequenced regions corresponding to the mudrA gene. Annotations are the same as given in (A).
Sequence analysis demonstrated that the intron locations and intron/exon splice sites are conserved in hmudrB genes. Two of the sequenced products, hmudrB1 and hmudrB3, were obtained from a MuDR+ line, whereas the remaining 19 products were from inbred W23, hybrid A188/B73, and an a1 sh2 tester. Homologs are nearly identical to the mudrB-coding region: from 99.8% for hmudrB2 to 93.5% for hmudrB16. In contrast, the respective 5′ untranslated regions and the TIR B are more distant (98.1% identity for hmudrB2 and 90.7% for hmudrB16). There is no strict correlation, however, between mismatch levels in the coding and flanking noncoding regions. Translation of hmudrB open reading frames, illustrated in Figure 2B, predicts that all but two of the 21 hmudrB genes could encode MURB homologous proteins. With regard to the 207–amino acid form of MURB, which is encoded by the predominant transcript (Hershberger et al., 1995), the predicted proteins contain 0 to 13 amino acid substitutions.
To determine the type of selection acting on the mudrB gene family, we compared synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions in the protein-coding regions. In most cases in which no selection pressure is present, the frequency of synonymous and nonsynonymous substitutions is equal, a situation that has been observed in pseudogenes (Hughes, 1995). A Ka/Ks ratio >1 indicates that evolution of the gene family is under diversifying selection (Kreitman and Akashi, 1995). A Ka/Ks ratio <1 suggests selection for conservation of coding capacity (Kreitman and Akashi, 1995). Using the Genetics Computer Group (Madison, WI) Diverge program, we found that Ks exceeds Ka (Ka/Ks = 0.57), suggesting conservation of the gene. The same analysis applied to the protein encoded by the fully spliced transcript yields Ka/Ks = 0.47, suggesting the hypothesis of coding capacity conservation. These data are preliminary, however, because to distinguish between nucleotide substitutions derived through the action of natural selection and substitutions originating from spontaneous mutations, a larger number of hmudrB sequences is required.
Because the intact mudrA gene cannot be maintained in E. coli cells without undergoing mutations (Hershberger et al., 1991; Raizada and Walbot, 2000), only the region spanning the TIR, the 5′ untranslated region, and the first coding exon of the gene were characterized. These partial gene sequences are summarized in Figure 2C. As with mudrB, the mutation frequency is lower in the coding region than in the TIRs and untranslated regions. The homologs of mudrA fall into two classes: homologs from Mutator lines are 94 to 100% identical to MuDR in the coding regions sequenced, whereas homologs from non-Mutator lines are more diverse and share 80 to 98% identity. With the exception of Zapalote chico (Gutiérrez-Nava et al., 1998), the TIR A regions from inbred lines are 6 to 16% divergent from MuDR. Two additional mudrA homologs, hmudrA8 and hmudrA9, were isolated from active Mutator stocks (data not shown); they are more similar to the Mu5 transposable element. On the basis of internal sequence alignment, Mu5 was proposed to be a significantly diverged deletion derivative of MuDR (Chandler and Hardeman, 1992).
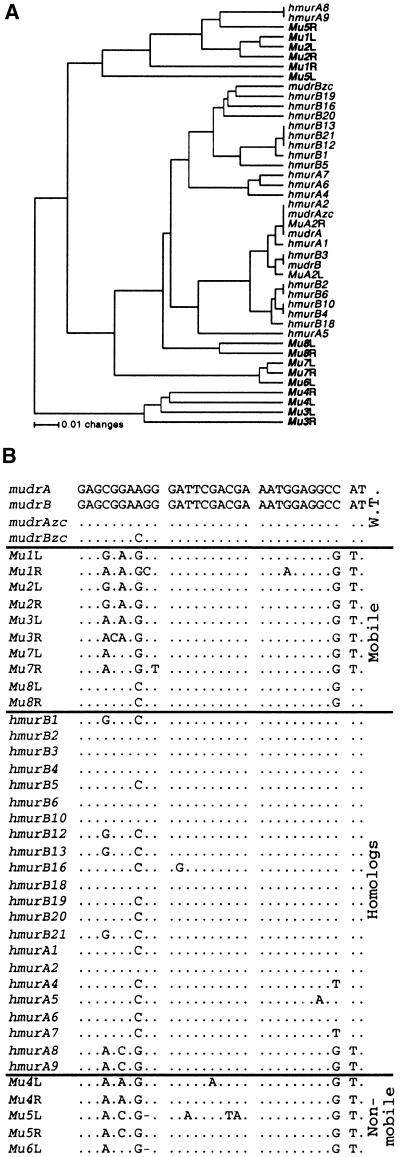
To assess evolutionary relationship between the MuDR homologs and other members of the MuDR/Mu family, we focused phylogenetic analysis on the terminal 189 bp, which is shared by all Mu elements. As shown in Figure 3A, all mudrA and the mudrB homologs except hmudrA8 and hmudrA9, the Mu5-like elements, cluster with MuDR and are distinct from other known Mu subfamilies (Bennetzen, 1996). This result is consistent with the hypothesis that the homologs are derived from MuDR by stepwise accumulation of mutations. Despite mutations, most of the TIRs characterized retain the transposase binding site (Benito and Walbot, 1997) and therefore should bind MURA (Figure 3B).
Figure 3.
Evolutionary Comparison between Members of MuDR/Mu Family.
(A) Unrooted relatedness tree of the TIRs of all sequenced Mu transposable elements. The left (L) and right (R) TIRs are shown; MuDR and related elements described in this study are shown in lightface, and the nonautonomous elements Mu1 to Mu8 are shown in boldface. The tree was generated by the UPGMA method using the Jukes-Cantor model for estimation of distances as implemented in the PAUP4.0b program (Sinauer, Sunderland, MA). Sequences were aligned manually with the aid of Sequencher 3.1.1 (GeneCodes, Ann Arbor, MI) software.
(B) Alignment of the 32-bp MURA transposase binding site. Elements were classified based on known genetic properties (Walbot and Rudenko, 2001). At the top (labeled W.T. for wild type) are the sequences for MuDR and MuDRzc. Next are the mobile Mu elements, then the hMuDR elements, and the nonmobile Mu elements. Dots represent matching bases, and dashes represent missing bases.
Organization of mudrB and mudrA Homologous Genes
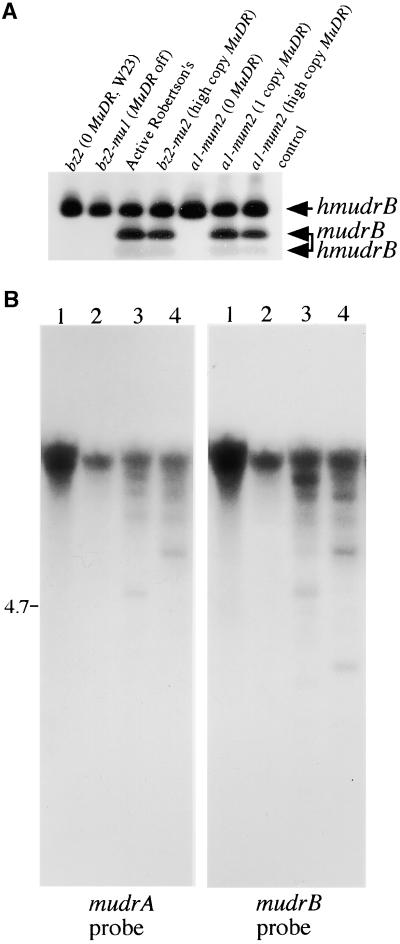
PCR analysis and sequencing results suggested that most of the larger bands that hybridized to both mudrB- and mudrA-specific probes (Figure 1B) could be hMuDR elements organized similarly to MuDR. They would have been overlooked in previous studies because point mutations have eliminated the diagnostic SstI sites in the TIRs of all mudrB and mudrA homologs from non-Mutator lines (Figure 2). Therefore, the number of bands >4.7 kb in SstI-digested DNA that are recognized by both mudrB and mudrA probes provides a minimum estimate of the putative hMuDR elements. As shown in Figure 1B, there are at least five such larger fragments in the A188/B73 hybrid and at least seven larger fragments in the W23 inbred line. Eight distinct mudrB homologs were sequenced from each of these lines, consistent with the minimal estimates from the DNA gel blot hybridization patterns.
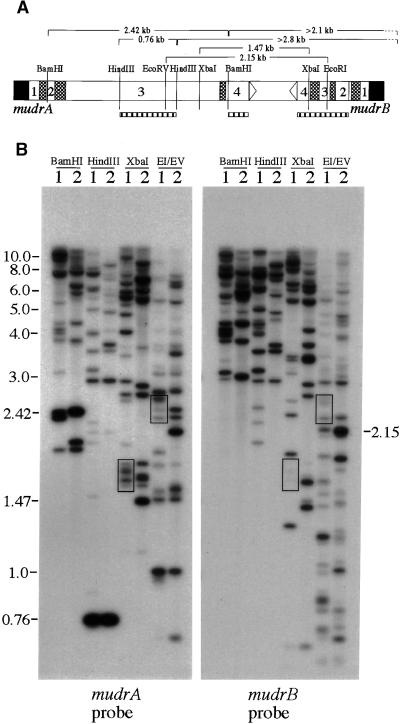
To provide additional evidence that most of the hmudrB and hmudrA genes are closely linked, samples from inbred W23 and a Mutator line were further assayed by restriction site mapping. As shown in Figure 4, BamHI and HindIII digests probed with a mudrA-specific probe detected the expected 2.42- and 0.76-kb DNA fragments in both lines, indicating conservation of these restriction sites in some of the putative hMuDR elements. The mudrA-specific fragments >2.1 kb in BamHI-digested W23 DNA and >2.8 kb in HindIII-digested W23 DNA would be expected to hybridize to a mudrB-specific probe if the hmudrA and hmudrB genes are closely linked. Indeed, the mudrB probe (Figure 4B) recognizes eight of 10 clearly visible bands in each of these digests.
Figure 4.
Molecular Organization of Homologous MuDR Genes.
(A) Diagram of MuDR and the sizes of expected restriction fragments analyzed in (B). The hatched boxes represent DNA hybridization probes. Note that the mudrA probe is composed of a mixture of exon 3– and exon 4–specific fragments, as described in Methods.
(B) DNA samples from the bz2 non-Mutator, W23 inbred line (lanes 1), and the bz2-mu2 active Mutator line (lanes 2) were digested with restriction enzymes as described in (A). Sequential DNA gel blot analysis with mudrA- and mudrB-specific probes demonstrates hybridization of both probes to fragments of the same size, as appropriate, based on the restriction map of MuDR. Some of the most prominent cohybridizing bands (boxes) that differ in size from the wild-type MuDR span the repeat-rich intergenic region, for which length variation has already been reported (Gutiérrez-Nava et al., 1998). Other features are discussed in the text. EI/EV, EcoRI/EcoRV.
PCR analysis suggested a convergent organization of the hmudrA and hmudrB genes, separated by an intergenic region of heterogeneous length in some hMuDR elements. The previously characterized MuDRzc element contains a 35-bp insertion of unknown origin and a 23-bp duplication of nearby sequences in its intergenic region (Gutiérrez-Nava et al., 1998). To confirm the polymorphic nature of the intergenic region in the hMuDR elements and to assess further the convergent organization of the hmudrA and hmudrB genes, we employed several digests. As shown in Figure 4B, an XbaI digest liberates a 1.47-kb fragment from the intergenic region of MuDR; this fragment hybridizes weakly with the mudrB probe, because there is only a 165-bp overlap with the expected fragment. As predicted, there is no 1.47-kb fragment present in XbaI-digested W23 DNA samples. Instead, we detected at least four fragments (∼1.65, 1.70, 1.77, and 1.80 kb) recognized by both gene probes (Figure 4B, bottom boxes). These fragment lengths are consistent with insertions in the intergenic regions of ∼180, 230, 300, and 330 bp, respectively. Similar results were obtained based on analysis of EcoRI/EcoRV double digests. Again, a group of bands slightly larger than the expected 2.15-kb product was detected (Figure 4B, top boxes). Collectively, these results are in accord with the length polymorphisms in the intergenic region observed by PCR analysis. On the basis of these results, we conclude that at least some of the hmudrB and hmudrA genes are separated by an intergenic region longer than that of MuDR. In subsequent analysis, we used double digests using one endonuclease with a known site in sequenced hmudrB elements and a series of other enzymes with sites in authentic MuDR. The resulting fragment patterns matched the map of MuDR, with heterogeneity in the length of the intergenic region (data not shown). Thus, even though we have not fully sequenced an hMuDR element, our results support the idea that most hmudrB and hmudrA genes have the same convergent orientation as do their wild-type counterparts in the MuDR element.
hMuDR Elements Are Transcriptionally Competent
In earlier studies, blot hybridization tests using RNA isolated from non-Mutator lines detected no MuDR transcripts (Chomet et al., 1991; Hershberger et al., 1991). These tests used total RNA, because MuDR transcripts are very abundant in Mutator lines, like actin and other highly expressed messages (Hershberger et al., 1995). More recently, Gutiérrez-Nava et al. (1998) detected mudrB-hybridizing transcripts in the poly(A)+ RNA fraction from the W23 inbred line. We suspected that these transcripts originated from hMuDR elements. To confirm these results and to estimate hMuDR relative expression levels, we examined poly(A)+ selected RNA from mature embryos of Mutator and non-Mutator lines. As shown in Figure 5B, reverse transcription (RT)–PCR was used to search for transcripts using primer pairs spanning the region across intron 2 of the mudrA gene or across the entire mudrB gene. An appropriately sized fragment (852 bp) characteristic of mudrA-related transcripts with properly joined exons 1 and 2 was amplified from representative MuDR+ and MuDR− lines (Figure 5B, top). Similarly, the major product with the mudrB-specific primers was 645 bp long, indicative of a transcript with intron 2 spliced out and intron 3 retained (Figure 5B, middle).
Figure 5.
RNA Expression Patterns.
(A) Diagram of primers (closed arrowheads), expected products (lines along with calculated sizes), and corresponding hybridization probes (hatched boxes) used in RT-PCR reported in (B) and the riboprobes (cross-hatched boxes) used in RPA analysis reported in (C) and (D).
(B) Semiquantitative RT-PCR demonstrates the presence of MuDR- and hMuDR-derived transcripts in mature embryos of two non-Mutator and two Mutator lines. Amplification of α-actin transcripts was included as an internal loading control. Arrowheads indicate the composition of individual products as retaining or lacking individual introns.
(C) RPA analysis to detect mudrB. Three active MuDR stocks show different levels of mudrB transcripts, reflecting MuDR copy numbers from 1 to >20. The position of the fully protected probe is indicated by the arrowhead at right; this corresponds to MuDR region 3782 to 4222 (see Table 2). Yeast RNA hybridized to the riboprobe and subsequently unprocessed (−) or processed (+) with the mixture of RNases served as an internal control.
(D) RPA analysis to detect hmudrB using probe 4298 to 4496, which is specific for a subset of homologs (see Tables 1 and 2). Other annotations are as in (C).
To confirm that RT-PCR–amplified products from non-Mutator lines are not derived from cryptic wild-type MuDR elements, representative RT-PCR products were verified by sequencing analysis to contain the expected polymorphisms (data not shown). In addition, total RNA samples from other lines were analyzed by ribonuclease protection assay (RPA), using a mudrB-specific riboprobe spanning MuDR regions 3782 to 4222. Consistent with the design of a riboprobe that had mismatches with each of the characterized mudrB homologs (Figure 2), none of the non-Mutator lines was expected to have full-length protected fragments. As shown in Figure 5C, only RNA derived from active MuDR stocks displayed full-length protection. Shorter protected fragments, as expected from hMuDR elements, were also detected. Furthermore, no full-length protected fragments were seen for Mutator active line d201, in which the mudrB gene of the single-copy MuDR element is deleted (Lisch et al., 1999). An epigenetically silenced high-copy MuDR line, with the now stable bz2-mu1 reporter allele, also contained no MuDR transcripts. Similar results were obtained using mudrA-specific RPA probes (data not shown). Collectively, the RPA analysis confirmed that MuDR transcripts are found only in active Mutator lines, whereas hMuDR transcripts are detected in both active and inactive Mutator lines.
By both RT-PCR and RPA, hMuDR transcripts from non-Mutator lines are at least 10- to 20-fold less abundant than are total hMuDR and MuDR transcripts in active Mutator lines. Therefore, on a conventional RNA gel blot using total RNA, hMuDR transcripts from non-Mutator lines could be below the level of detection.
Transcription Properties of hMuDR and MuDR Elements
To more precisely quantify hMuDR transcript abundance in the presence or absence of transcriptionally active MuDR elements, we performed extensive RPA analyses. Because hMuDR number and type vary between maize lines, expression from a single homolog cannot be monitored across all lines. As shown in Table 1, however, we designed three short riboprobes that specifically protect a subset of mudrB homolog classes in all lines examined. The RNase protection pattern generated by the probe spanning the 4332 to 4496 region is shown in Figure 5D. To allow relative quantification between hmudrB and mudrB RNA, the level of the mudrB transcripts in Mutator lines was examined using mudrB-specific probes that recognize regions of the same length as the listed hmudrB probes. These results are presented in Table 2.
Table 1.
Specificity of RNase Protection Probes Derived from hMuDR Elements
| Probe Source | Regiona | Protected hmudrB Types | No. of Unprotected Mismatches with mudrB |
|---|---|---|---|
| hmudrB21 | 4298–4496 | 5, 11, 12, 13, 14, 21 | 2 |
| hmudrB16 | 4332–4496 | 1, 5, 11, 12, 13, 14, 16, 17, 21 | 1 |
| hmudrB20 | 4087–4222 | 2, 3, 4, 5, 6, 7, 10, 18, 20 | 1 |
Because of variation in the length of the hMuDR elements, the nucleotide positions are taken from MuDR.
Table 2.
Quantification of hmudrB and mudrB Transcript Levels in Mutator and Non-Mutator Maize Lines
| Relative Expression in Probe Regions Indicated (%)b,c
|
||||||
|---|---|---|---|---|---|---|
| Line | Mutator | MuDR Copiesa | 4298–4496 | 4332–4496 | 4087–4222 | 3782–4222 |
| hmudrB | ||||||
| a1-mum2d | No | 0 | 1.0 (100) | 1.0 (100) | 1.0 (100) | |
| bz2 | No | 0 | 2.9 (100) | 3.9 (100) | 2.9 (100) | |
| bz2-mu1 | No | >20e | 3.3 (100) | 4.0 (100) | 2.8 (100) | |
| bz2-mu2 | Yes | 9–11 | 24.3 (26.6) | NDf | ND | |
| bz2-mu2 | Yes | 18–24 | 52.8 (28.8) | 59.4 (29.0) | 54.8 (31.7) | |
| a1-mum2 | Yes | 1 | 3.7 (20.9) | 3.7 (20.9) | 4.4 (24.7) | |
| a1-mum2 | Yes | 1(d201) | ND | 1.2 (100) | 1.4 (100) | |
| a1-mum2 | Yes | 6–8 | 16.7 (25.8) | 18.1 (25.9) | 21.5 (27.5) | |
| a1-mum2 | Yes | 10–12 | 26.7 (26.5) | ND | ND | |
| mudrB | ||||||
| a1-mum2 | No | 0 | 0 | 0 | 0 | 0 |
| bz2 | No | 0 | 0 | 0 | 0 | 0 |
| bz2-mu1 | No | >20e | 0 | 0 | 0 | 0 |
| bz2-mu2 | Yes | 9–11 | 9.6 | ND | ND | ND |
| bz2-mu2 | Yes | 18-24 | 18.6 | 21.1 | 19.5 | 22.3 |
| a1-mum2d | Yes | 1 | 1.0 | 1.0 | 1.0 | 1.0 |
| a1-mum2 | Yes | 1(d201) | 0 | 0 | 0 | 0 |
| a1-mum2 | Yes | 6–8 | 6.9 | 7.5 | 8.3 | 7.4 |
| a1-mum2 | Yes | 10–12 | 10.6 | ND | ND | ND |
Copy number was estimated from the intensity of hybridization to the 4.7-kb SstI band of MuDR by DNA blot hybridization.
Probe identity to various hMuDR elements is indicated in Table 1.
Percentage is calculated as the contribution of hmudrB transcripts in the total pool of hmudrB plus mudrB transcripts in the sample. Details are provided in Methods.
Hybridization to total RNA from this line was set as 1.0, and all other measurements are relative to it.
Copy number estimate from Walbot and Stapleton (1998).
ND, not determined.
The hmudrB transcript levels are very low in non-Mutator lines but increase dramatically in Mutator lines with transcriptionally active MuDR elements. In these lines, hmudrB transcripts constitute ∼25% of the total mudrB-related RNA (Table 2) and therefore reflect the increase in copy number–dependent MuDR expression. The positive response of at least some homologs to transcriptionally active MuDR elements could result from direct activation by MuDR-encoded products or through an indirect effect such as loss of methylation. The lack of abundant hmudrB transcripts in the epigenetically silenced bz2-mu1 Mutator line containing a high number of MuDR copies indicates that most hMuDR elements are also affected by silencing. Some hmudrB transcripts persist at the levels sustained in standard MuDR− inbred lines. Therefore, a subset of hMuDR elements is regulated in a manner distinct from MuDR.
In inbred lines, sequence analysis of RT-PCR products permits a rough estimate of the types of homologs with persistent transcription. For example, hmudrA4 transcripts, but not hmudrA1, -2, and -3 transcripts, were detected repeatedly in inbred samples. These homologs contain the modal number of mutations in protein-coding regions, but their promoters have diverged to a much greater extent. Similarly, mudrB homologs fall into two groups: hmudrB2, -3, -4, -6, and -18 possess three to four point mutations in their 162-bp promoter regions, whereas hmudrB1, -5, -12, -13, -16, -19, -20, and -21 contain 12 to 19 substitutions. Does the higher mutation density in the latter group account for novel promoter properties distinct from MuDR?
To answer this question, we exploited an AlwNI restriction site polymorphism at position 4448 (second exon of mudrB), which is lacking in all of the characterized hmudrB genes with more diverged promoters (Figure 2). Therefore, RT-PCR products digested with AlwNI should generate a fragment pattern that allows a general assessment of what group(s) of homologs is preferentially expressed. A 447-bp RT-PCR product corresponding to correctly joined exons 1, 2, and 3 was amplified in all tested lines. As shown in Figure 6A, after AlwNI digestion, this product would yield two smaller fragments of 287 and 160 bp if cDNAs were derived from the wild-type mudrB and/or hmudrB genes transcribed from less diverged promoters. Alternatively, this product would remain intact if the corresponding transcripts were from hmudrB genes with more diverged promoters. We found only the full-length RT-PCR product in standard inbred and epigenetically silenced Mutator lines. In contrast, both the full-length fragment and the two smaller fragments were found in active MuDR lines. Therefore, homologs with conserved and divergent promoters contribute to the transcript pools in active MuDR lines, but only hMuDR elements with more diverged promoters are represented in silenced and MuDR− lines.
Figure 6.
Independent Transcriptional Activity of hMuDR Elements Is Conferred by Evolutionarily Diverged Promoters.
(A) Gel-purified RT-PCR products corresponding to the MuDR region 4086 to 4722 were digested with AlwNI and analyzed by DNA gel blot analysis. The positions and compositions of three resulting products are indicated.
(B) The epigenetic status of hMuDR promoters was analyzed by DNA gel blot hybridization after FspI digestion of genomic DNA from two MuDR− lines (lanes 1 and 2) and two MuDR+ lines (lanes 3 and 4), as described in the legend to Figure 1.
The transcriptional independence of some hMuDR elements is a novel property. Epigenetic silencing of Mutator activity negatively affects MuDR transcription and is accompanied by the coordinate DNA methylation in TIRs of MuDR and all other Mu elements (Chandler and Walbot, 1986; Martienssen and Baron, 1994). Silencing initiates when a transcriptionally active MuDR element(s) is present, but maintenance of TIR methylation persists even if MuDR is segregated out of the stock (Lisch et al., 1995). Similar methylation patterns in Mu1 element ends have been found both in non-Mutator lines and in Mutator lines that are epigenetically silenced (Chandler et al., 1988). Therefore, we suspected that MuDR homologs would be methylated in non-Mutator lines. Symmetrical point mutations in most homologs create methylation-sensitive FspI recognition sites 7 and 13 bp downstream of the hmudrA and hmudrB transcription initiation sites, respectively. All sequenced hmudrB genes with diverged promoters contain these FspI sites (Figure 2). As shown in Figure 6B, hMuDR FspI sequences were heavily methylated in non-Mutator lines. Consistent with expectations from previous DNA methylation studies at the TIRs of Mu1 and MuDR elements (Bennetzen, 1996), partial demethylation of one or both FspI sites in some homologs was observed in active Mutator stocks. Therefore, a subset of hMuDR elements appears to be transcriptionally active despite epigenetic modification.
Characterization of MURA and MURB Proteins in Diverse Maize Lines
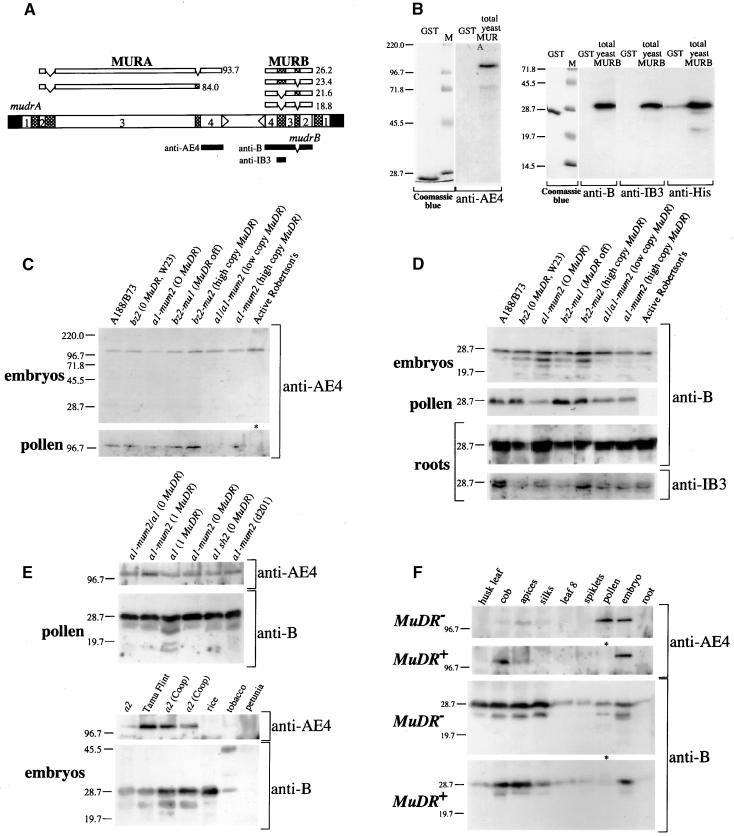
As shown in Figure 7A, active Mutator stocks express a complex set of MuDR transcripts. Two MURA proteins of 823 and 736 amino acids are predicted from fully spliced and intron 3–retained transcripts, respectively (Hershberger et al., 1995). For mudrB, the second (IB2) and the third (IB3) introns are retained in ∼10 and 90% of seedling RNA, respectively (Hershberger et al., 1995). Because both IB2 and IB3 are in frame with the major mudrB open reading frame (Figure 7A), it is possible that mudrB encodes four protein products ranging in size from 18.8 to 26.2 kD.
Figure 7.
Immunodetection of MURA and MURB Proteins.
(A) The alternatively spliced transcripts and calculated molecular masses of the respective protein products (in kD) are shown above the MuDR diagram. The polypeptide domains used for generation of antibodies are indicated below the diagram.
(B) Specificity of purified antibodies against the GST tag alone (5 μg) and yeast protein extracts (10 μg) expressing His-tagged full-length MURA and MURB proteins that were used for affinity purification of antibodies. M, molecular mass markers (in kD).
(C) Detection of MURA in mature embryos and pollen of various maize lines by immunoblot analysis. The sample array contains non-Mutator lines (hybrid A188/B73 and inbred W23), the a1-mum2 reporter line with no copies of MuDR, an epigenetically silenced bz2-mu1 high-copy MuDR line, and active Mutator lines, including the original purple Mutator line derived by Robertson (1978). As a result of pollen sterility, the pollen extract was prepared from a young tassel of the Robertson's line (asterisk).
(D) Immunodetection of MURB using different antibodies. The root samples were separated on higher percentage gels to demonstrate that the predominant MURB band consists of two distinct polypeptides. Other annotations are as in (C).
(E) Immunoanalysis to demonstrate the presence of MURB and MURA polypeptides in other maize lines, including those reported previously to have no MURB protein, and in other plant species.
(F) Accumulation of MURA and MURB proteins in developmentally staged tissues of non-Mutator inbred line W23 and the original Robertson's purple Mutator line.
To determine which of the predicted MuDR- and hMuDR-encoded proteins exist in vivo, we generated polyclonal antibodies against a bacterially expressed glutathione S-transferase (GST) fusion with the polypeptide sequence corresponding to the fourth exon of mudrA. In addition, antibodies were raised against GST fusions of the MURB (IB3 retained) protein and the 40–amino acid polypeptide segment specified by IB3 (Figure 7; see Methods). As shown in Figure 7B, the MURA-specific antibody detects a single ∼120-kD MURA protein in yeast expressing the fully spliced mudrA transcript; this protein was demonstrated previously to bind specifically to Mu TIRs (Benito and Walbot, 1997). The predicted molecular mass of this 823–amino acid protein is 93.9 kD; however, the anomalous electrophoretic mobility of the MURA protein has been noted (Benito and Walbot, 1997). Both MURB-specific antibodies detect a single polypeptide of 31.5 kD in yeast expressing MURB (IB3 retained) transcripts (Figure 7B).
As shown in Figure 7C, by immunodetection we found a single MURA polypeptide of 120 kD in both active Mutator and inbred non-Mutator lines of maize. Because our MURA-specific antibody is directed against the C-terminal part of the protein, it can detect only the full-length polypeptide (Hershberger et al., 1995). The major MURB/hMURB polypeptide is a doublet with an apparent molecular mass of ∼28.7 kD (Figure 7D). Because the majority of mudrB-related transcripts retain intron 3, we used the IB3 antibody to confirm that both major proteins contained this domain (Figure 7D). An antibody generated against IB2 did not recognize these proteins (data not shown). We conclude, therefore, that the major MURB-related polypeptides correspond to the most abundant mudrB transcript type that retains intron 3 but has intron 2 spliced out. A number of smaller polypeptides were also detected; a detailed analysis of alternative products will be presented elsewhere. Considering the disparity in MuDR-related transcript abundance among maize lines, we anticipated significant differences in protein content as well. Contrary to expectations, however, immunodetection experiments indicate that hMURA and hMURB proteins in non-Mutator maize inbred and epigenetically silenced MuDR lines attain nearly the same levels as do the presumed mixture of MURA/hMURA and MURB/hMURB proteins in active Mutator lines with multiple copies of MuDR. These results clearly demonstrate that significant post-transcriptional regulation of MuDR occurs.
Using polyclonal antibodies, Donlin et al. (1995) and Lisch et al. (1999) provided evidence that an ∼30-kD MURB protein was detectable only in active Mutator lines with a full-length MuDR element; they reported that the protein was differentially accumulated in a tissue-specific manner. In our initial experiments, hMURA and hMURB proteins were detected in inbred line W23, in the A188/B73 hybrid, and in a number of bz2 and a1 testers. Subsequently, we examined the same a1 sh2 tester, a non-Mutator line, that was used as a negative control by both Donlin et al. (1995) and Lisch et al. (1999). As shown in Figure 7E, both hMURB and hMURA proteins were readily detected in pollen protein extracts from this tester. Furthermore, the MURB-specific antibodies detected cross-reacting proteins of ∼28 kD in rice and tobacco, and MURA antibodies recognized MURA-like protein in tobacco (Figure 7E, bottom). This finding suggests the widespread distribution of MURA- and MURB-like proteins in angiosperms. Numerous mudrA-like genes have been sequenced in maize (Comelli et al., 1999), Arabidopsis (>53 hits), rice (GenBank accession numbers AP000366, AP000367, AB012392, and AB023047), and sorghum (GenBank accession number AF114171); a few mudrB-expressed sequence tags have been reported as well (GenBank accession number BE575286).
To determine how MURB/hMURB and MURA/hMURA protein levels change during plant development in Mutator and non-Mutator lines, we analyzed protein extracts prepared from developmentally staged tissues (Figure 7F). Both MURA and MURB proteins were detected ubiquitously in all tissues surveyed. The amount of MURA was considerably lower than that of MURB and was almost undetectable in tissues with low levels of MURB. Individual protein levels in green and nongreen tissues were similar in both immature and differentiated somatic and germinal stages (Figure 7F). The absence of the smaller forms of MURB in some lanes correlates with the somewhat lower abundance of MURB in these tissues and thus could represent a limitation of the detection method. Most strikingly, in tissues with detectable protein, the relative amounts of hMURA and hMURB in the standard W23 inbred line and MURA and MURB in the classic Robertson's Mutator line are very similar.
DISCUSSION
To gain insight into the unique developmental regulation of Mutator activities, we initiated experiments with two major purposes. First, we screened maize lines for the presence of MuDR/Mu-related elements as the “priority” candidates for host genes that could modulate Mutator activity. Second, we performed a detailed analysis of mudrA and mudrB expression at both the RNA and protein levels to search for any aspect of post-transcriptional regulation that might supply a developmental control function for the transposition of MuDR/Mu elements.
These approaches led to the discovery of a large family of hMuDR elements, evolutionary derivatives of MuDR (Figures 1 to 3). The hMuDR elements are organized like MuDR (Figures 1C and 2 to 4). At least some are transcribed in all Mutator and non-Mutator lines examined (Figure 5) and produce corresponding hMURA and hMURB proteins (Figure 7). Homologs yield substantially more transcript in the presence of an active MuDR element, and some appear to be subject to epigenetic silencing in parallel with MuDR elements (Figure 5 and Table 2). However, a subset of homologs with more divergent TIR promoters escapes transcriptional silencing of MuDR. As a consequence, these MuDR-like elements produce transcripts and proteins in all maize lines examined (Figures 6 and 7).
By examining MuDR expression, we demonstrated that mudrA and mudrB transcript levels are proportional to MuDR copy number in active Mutator lines (Figure 5 and Table 2). By immunodetection analysis, we found that there are similar levels and ubiquitous accumulation of the corresponding proteins in all maize lines tested (Figure 7). In non-Mutator lines and epigenetically silenced MuDR lines, the proteins are supplied by independently transcribed hMuDR elements, whereas in active MuDR stocks, protein pools are likely composed of MURA/hMURA and MURB/hMURB mixtures (Figures 5 to 7). Most importantly, in active Mutator stocks, protein levels are not proportional to MuDR copy number, perhaps explaining the similar somatic excision frequency observed in single-copy and multiple-copy lines. Furthermore, the similar protein levels observed in various maize lines could not be explained by protein degradation (Figure 7), suggesting the existence of a regulatory mechanism that controls protein production at the translational level. Finally, the likely presence of homolog MURA and MURB proteins plus authentic proteins may be important in establishing the late somatic timing of Mu element excision and insertion independent of MuDR copy number.
Ubiquitous MuDR Homologous Elements
Among plant DNA transposons, the most stringent regulatory mechanisms govern the activity of the MuDR/Mu transposons. This family of maize transposons was discovered relatively recently and may represent a case of rare reactivation of a normally silent element family. Nearly all currently known Mutator lines are derived by descent from a single active individual (Robertson, 1978). The independently discovered Cyclone (Cy/rCy) two-component transposable element system was later shown to be MuDR/Mu (Schnable and Peterson, 1989). Recently, a silent Mutator system that displays hybrid dysgenesis was discovered in an exotic maize line, Zapalote chico (Gutiérrez-Nava et al., 1998). Here, we established that multiple hMuDR elements exist in diverse maize lines (Figures 1 and 2). At the DNA sequence level, hMuDR elements are highly similar to MuDR (Figure 2), but the TIRs lack the SstI restriction digestion site that is routinely used to follow MuDR element copy number and methylation status. The absence of these sites explains why hMuDR elements were overlooked in previous studies. hMuDR elements apparently originated from MuDR by the stepwise incorporation of point mutations and a few insertion–deletion events. Diverse hMuDR elements are present in each maize line examined, and these define several groups of related elements (Figure 2).
Structurally, homologs have the same organization as MuDR: the hmudrA and hmudrB genes are convergently transcribed, with initiation within their respective TIRs (Figures 1C, 2, and 3). The two-gene organization of MuDR and hMuDR elements is unique in the Mutator superfamily of transposons, which includes bacterial (Eisen et al., 1994) and eukaryotic (Le et al., 2000) members. For example, the numerous Mu-related elements of Arabidopsis, termed MuLEs, have extended TIRs and a mudrA-related reading frame, but no mudrB (Le et al., 2000).
To date, we have not detected transposition of the hMuDR elements (data not shown), nor have any mutations that involve an hMuDR insertion been reported. The inability of hMuDR elements to transpose could reflect inefficient interactions with the MURA transposase, which binds in vitro to a 32-bp motif, the MURA transposase binding site (MBS), found from positions 25 to 56 in the TIRs (Benito and Walbot, 1997). The MBS is highly, but not perfectly, conserved among the mobile Mu elements. Subfamilies of nonmobile Mu elements contain single-base deletions or multiple-point mutations within the MBS at one or both ends of the element. Most hMuDR TIRs have MBS motifs nearly identical to those of the mobile MuDR element (Figure 3), but because we have not characterized intact elements, we do not know if the MBS at both element TIRs is conserved. Alternatively, it is possible that additional motifs within the 215-bp TIR are required for transposition in vivo (Benito and Walbot, 1997). The diversity of these natural TIR variants provides an invaluable tool for future analysis of the structural DNA features required in transposition reactions.
MuDR and hMuDR Transcripts
In active Mutator lines, hMuDR elements are regulated in parallel with MuDR and produce more stable transcripts in proportion to MuDR copy number. The total transcript level from the multiple hMuDR elements is always well below that of MuDR transcripts. Consequently, it appears that MuDR-derived products modulate either hMuDR transcription or transcript half-life. A similar conclusion was reached in a transgenic maize experiment in which the expression of β-gluc-uronidase and luciferase transgenes under the control of the mudrB TIR promoter was transiently and modestly increased by the presence of a transcriptionally active MuDR element (Raizada et al., 2001b). These authors proposed that MURA may protect TIRs from host methylation and thus indirectly increase transcription from elements with related promoters.
Demethylation is not a prerequisite for TIR promoter activity, however, because a subset of methylated hMuDR elements is transcriptionally active in standard inbred lines and in inactive Mutator stocks in the absence of MuDR. The relative levels of hMuDR transcripts in inbred MuDR− and epigenetically silenced MuDR+ lines are similar. The hMuDR elements with constitutive transcription have TIR promoters that are the most distinct from MuDR. Therefore, constitutive expression and insensitivity to MuDR are two experimental results that demonstrate that this subset of hMuDR elements is regulated independently. Further experiments are necessary to identify which promoter alterations might confer lower responsiveness to MuDR products and escape from epigenetic silencing.
Because homologous MuDR transcripts are produced in epigenetically silenced Mutator lines, MuDR silencing is unlikely to proceed via an RNA degradation pathway unless it is exquisitely targeted toward wild-type MuDR transcripts. RNA-based homology-dependent gene silencing requires significant similarity but not identity (Fagard and Vaucheret, 2000). Some of the constitutive hmudrB transcripts, defined by the lack of an AlwNI restriction site, have coding regions nearly identical to those of authentic mudrB (up to 97% identity for hmudrB5).
MuDR- and hMuDR-Encoded Proteins
Immunodetection using an antibody directed against MURA protein indicated that the full-length protein is produced by mudrA and hmudrA transcripts in somatic tissues and pollen of various maize lines. Similarly, two different antibodies, directed against MURB polypeptides, detected MURB proteins in all inbred and silenced Mutator lines examined. The ubiquitous expression of hMURA and hMURB in lines lacking MuDR contradicts previous reports that MURB antibodies detect cross-reactivity only in active Mutator lines (Donlin et al., 1995; Lisch et al., 1999). Using the same material (a1-mum2 MuDR and the a1 inbred lines) used in these previous studies, our MURB antibodies readily detected the homolog proteins (Figure 7). The observed discrepancy most likely was caused by differences in assay sensitivity.
In assessing whether hMuDR-derived transcripts are translated, we also discovered an unsuspected level of MuDR regulation: MURA and MURB protein levels are similar in all lines examined despite variation in RNA abundance. In contrast, the absolute amount of Ac transposase increases with Ac dosage (Fusswinkel et al., 1991), but the concentration of the effective protein is variable, depending on the level of protein self-aggregation (Heinlein et al., 1994). The lack of proportionality between MuDR transcript and protein levels likely explains a long-standing puzzle: the frequency of Mu element excision is similar in all lines (Walbot, 1991), whereas MuDR transcript abundance varies ∼50-fold among active Mutator lines (Chomet et al., 1991; Hershberger et al., 1991, 1995). Our results predict that there is a post-transcriptional step that determines protein production over a wide range of both mudrA and mudrB transcript levels. We consider it unlikely that protein degradation contributes, because the anti-MURA antibody detected no degradation products.
We favor translational control as the mechanism for achieving the protein levels observed, possibly mediated through the 5′ or 3′ UTRs (Walbot and Rudenko, 2001). Expression of the mudrA cDNA directed by the 35S promoter in transgenic maize results in Mu excision frequencies similar to those of a typical Mutator line (Raizada and Walbot, 2000). Preliminary antibody detection experiments with these transgenic lines found no increase in MURA levels compared with any other Mutator line (data not shown). This transgene and all mudrA and mudrB transcripts share some motifs in the 3′ UTR. Additional experiments are required to determine if the 3′ motifs are in fact responsible for the post-transcriptional regulation of translation.
A translational block that prevents MURA and MURB protein expression until late developmental stages is one plausible explanation for the stringent developmental timing. Mu somatic transpositions are restricted to terminal cell divisions (Levy and Walbot, 1990), and germinal events occur primarily just before and after meiosis (Walbot and Rudenko, 2001). Donlin et al. (1995) reported MURB in meristems and organ primordia, and we found that MURA and MURB levels are nearly invariant in pollen and certain somatic tissues, including embryos and floral primordia (Figure 7). Consequently, MURA and MURB functions rather than mere quantity must be important in developmental timing.
At least one function of MURA can be measured early in plant development, long before somatic excisions occur. In the absence of MuDR, Mu element TIRs are methylated (Bennetzen, 1996). Introduction of a transcriptionally active MuDR element or a MURA transgene results in the rapid demethylation of resident Mu elements early in development (Raizada and Walbot, 2000). These observations suggest that MURA is present and competent in DNA binding early in development. Furthermore, methylation accompanying the epigenetic silencing of Mu and MuDR elements (Chandler and Walbot, 1986; Martienssen and Baron, 1994) is maintained, despite the presence of hMURA proteins in both inbred and inactive Mutator lines. This evidence indicates that homolog-encoded MURA proteins are unable to interact with the TIRs in a manner that results in demethylation.
Because both mudrA and mudrB transcripts can undergo several alternative splicing events, multiple sizes of MURA and MURB proteins are predicted (Hershberger et al., 1995). A precise function has been assigned only for the 823–amino acid form of MURA, which is both necessary and sufficient to program somatic excision events (Raizada and Walbot, 2000). We detected several MURB size classes, but the pattern remained relatively constant (Figure 7). A precise role for MURB has not been established, but it is implicated as a requirement for Mu insertion. Spontaneous deletion derivatives of MuDR that lack mudrB do not support germinal insertion events (Lisch et al., 1999); in transgenic maize, expression of only MURA transposase leads to frequent somatic excision but no somatic insertions (Raizada and Walbot, 2000). Our results also indicate that none of the hMURB and hMURA proteins can substitute for the products of MuDR in the programming of transposition events.
Possible Contributions of the hMuDR Elements
It is possible that the diverse hMuDR elements are merely evolutionary relics that neither program transposition nor transpose. Indeed, it appears that these elements are now stable loci in maize (data not shown) and could be considered host genes that produce both RNA and protein products.
One striking feature of Mutator lines is that 10 to 100% of Mutator individuals are epigenetically silenced in each generation (Robertson, 1978, 1986; Walbot, 1986). Without active selection, Mutator activity is quickly lost. It is worthwhile to consider, therefore, whether the hMuDR elements are now part of the host defense against active MuDR/Mu elements. One example of the “poison protein” mechanism is seen when mutant transposase proteins encoded by numerous defective Mariner-like elements in Drosophila mauritiana and D. simulans directly impair wild-type transposase activity, leading to decreased excision frequency (Hartl et al., 1997). For P elements in D. melanogaster, a 66-kD protein resulting from alternative splicing represses transposition in both germinal and somatic cells (Misra and Rio, 1990). KP, a naturally occurring deletion derivative of P transposase, acts as a dominant negative regulator of the transposition reaction by forming nonfunctional oligomers with functional transposase (Andrews and Gloor, 1995). In maize, a defective En element derivative encoding an aberrant polypeptide causes developmentally delayed and less frequent excisions of En elements (Cuypers et al., 1988). If the hMuDR elements similarly encode defective proteins that repress Mutator functions, this form of regulation represents a case in which the host has in effect used the transposon against itself. A function in long-term host protection could explain the retention of hMuDR transcription and protein accumulation, despite that absence of any measurable transposon functions.
METHODS
Plant Material
Standard inbred and Mutator lines developed in this laboratory were used (Hershberger et al., 1995; Walbot and Stapleton, 1998). The a1-mum2 and a1 lines containing different numbers of MuDR copies were kindly provided by M. Freeling (University of California at Berkeley). Two a2 lines were obtained from the Maize Genetics Cooperation Stock Center (University of Illinois at Urbana/Champaign), and the HiII (A188/B73 hybrid) was a gift from Monsanto (St. Louis, MO). For molecular analysis, tissue samples were pooled from several sibling individuals of identical genotype and Mutator status.
Genomic DNA Isolation and Analysis
Plant genomic DNA was isolated using a modification of a published method (Mettler, 1987). Briefly, tissue was homogenized in liquid nitrogen and lysed in buffer containing 50 mM Tris-HCl, pH 8.0, 0.25 M sucrose, 50 mM NaCl, 20 mM EDTA, and 1% N-lauryl sarcosine. The mixture was incubated for 20 min at room temperature. Concentrations of NaCl and Tris in lysates were then adjusted to 1 M and 100 mM, respectively, and cetyl trimethylammonium bromide was added to 1% final concentration. After phenol/chloroform extraction, DNA was ethanol precipitated. Restriction endonuclease digestions and DNA gel blot hybridization used standard protocols. For polymerase chain reaction (PCR) amplification of genomic DNA, 10 or 100 ng of DNA, 1 μM of each forward and reverse primer, 0.2 mM deoxynucleotide triphosphates, and 0.75 units of Pfu DNA polymerase (Stratagene, La Jolla, CA) were used in a 50-μL reaction. PCR amplification shown in Figure 1 focused on the following MuDR regions: from 450 to 3142 (primer pair a), from 1 to 557 (primer pair b), from 450 to 1430 (primer pair c), from 3756 to 4942 (primer pair d), from 3756 to 4496 (primer pair e), from 3756 to 4795 (primer pair f), and from 2876 to 3788 (primer pair g). PCR-amplified products were cloned into pUC19 vector and sequenced from both strands using Dye Terminator Cycle Sequencing kits (Applied Biosystems, Foster City, CA). The hMuDR sequences generated in this study have been submitted to GenBank under accession numbers AF350331 through AF350360.
DNA Gel Blot Hybridization Probes
Transposon-specific probes were generated by PCR amplification from appropriate MuDR subclones described in Hershberger et al. (1995) and Raizada and Walbot (2000). mudrA probes were DNA fragments corresponding to MuDR positions from 1411 to 2167 and 2876 to 3130 (Figures 1A and 6B), from 450 to 557 and 691 to 1430 (Figure 1C), from 1411 to 2167 and 2876 to 3130 (Figure 4), and from 691 to 1430 (Figure 5). The mudrB probe corresponds to MuDR region from 3756 to 4496 in all experiments. The terminal inverted repeat (TIR) probe used in the experiment shown in Figure 1 was from 4727 to 4942, and the intergenic region–specific probe from 2866 to 3945. An actin probe was prepared as described in Raizada and Walbot (2000).
RNA Isolation and Analysis
Total plant RNA was prepared using RNeasy Plant Mini or RNeasy Maxi kits (Qiagen, Valencia, CA), according to the manufacturer's instructions. RNA was treated with RNase-free DNase I (Boehringer Mannheim, Indianapolis, IN) followed by phenol/chloroform extraction and ethanol precipitation. No DNA contamination was detected based on PCR amplification and inspection of RNA hybridization blots with mudrA- and mudrB-specific probes. The amount of RNA was determined by absorbance at 260 nm. Poly(A)+ fractions were recovered from purified total RNA using an Oligotex mRNA Midi Kit (Qiagen).
For reverse transcription (RT)–PCR analysis, cDNA was synthesized with Superscript II reverse transcriptase (Gibco BRL, Rockville, MD), according to the manufacturer's instructions using total RNA as the template and oligo(dT)12-18 as the primer. Reaction products were treated with RNase H, phenol/chloroform extracted, and subjected to 25 (Figure 5) or 40 (Figure 6) cycles of PCR amplification using gene-specific primers and Pfu DNA polymerase (Stratagene). cDNAs corresponding to MuDR regions from 473 to 1430 (mudrA) and from 3785 to 4477 (mudrB) were analyzed in Figure 5 and from 4087 to 4722 (mudrB) in Figure 6. RT-PCR products were separated on agarose gels and visualized by DNA gel blot hybridization using gene-specific probes.
Ribonuclease protection assays used a mixture of RNase A and RNase T1 as formulated in the RPA III kit (Ambion, Austin, TX) and were performed according to the manufacturer's recommendations. Ten micrograms of RNA was assayed using gene-specific single- stranded antisense riboprobes synthesized with T7 RNA polymerase (Ambion). Several different probes were used to assay the mudrA and mudrB genes. For the experiment described in Table 2, the relative levels of RNAs were compared by quantification of the fully protected fragments only. Signals were normalized to the intensity of protected α-actin transcripts.
Generation of Antibodies
DNA fragments corresponding to MuDR positions 2876 to 3130, 4477 to 3782, and 4084 to 3962 (Hershberger et al., 1995) were cloned as C-terminal additions into the glutathione S-transferase (GST) fusion protein expression vector pGEX 4T-1 (Amersham Pharmacia Biotech, Piscataway, NJ) to produce antigens specific for MURA, MURB (IB3 retained), and MURB IB3 polypeptides, respectively (Figure 7A). The purified fusion proteins were used to raise antibodies in rabbits. Polyclonal antisera were partially purified on columns with immobilized total Escherichia coli protein and with the bacterial GST used in fusion protein constructs. Subsequently, antibodies were affinity purified using immobilized MURA and MURB polypeptides. To ensure specificity, we used His-tagged full-length MURA and MURB (IB3 retained) proteins expressed in yeast cells (Benito and Walbot, 1997) that were purified under denaturing conditions on nickel–nitrilotriacetic acid agarose (Qiagen) and coupled to 3M Emphase Biosupport Medium AB1 resin (Pierce Chemical, Rockford, IL) to generate affinity matrices.
Plant Protein Preparations and Immunoblot Analysis
For preparation of total protein, plant tissues disrupted in liquid nitrogen were dissolved in grinding buffer containing 50 mM Tris-HCl, pH 8.0, 25 mM NaCl, 10 mM EDTA, 0.25 M sucrose, and 0.5 mM Pefabloc reagent (Boehringer Mannheim) at 4°C and passed through three layers of Miracloth (Calbiochem, San Diego, CA). The protein homogenates were immediately mixed with an equal volume of denaturation buffer containing 100 mM Tris-HCl, pH 6.8, 4% SDS, and 20% glycerol, and incubated at 75°C for 10 min. Finally, protein extracts were cleared by centrifugation at 14,000g for 10 min, and protein concentrations were estimated using DC protein assay kits (BioRad, Hercules, CA).
For protein gel blot analysis, 80 to 100 μg of total protein per sample was resolved on 10 to 12% polyacrylamide gels under denaturing conditions. Immunodetection of MURA and MURB proteins was performed using appropriate dilutions of primary antibodies, alkaline phosphatase–conjugated secondary antibodies (Sigma, St. Louis, MO), and a chemiluminescent substrate CSPD disodium 3-(4-methoxyspiro[1,2-dioxetane-3,2′-(5′-chloro)tricyclo(3.3.1.13,7)decan]-4-yl)phenyl phosphate (Tropix, Bedford, MA).
Acknowledgments
We thank D. Goodman, M. Fitzgerald, G. Nan, M. Raizada, and C. Yanofsky for critical reading of and comments on the manuscript. We are indebted to A. Ono and S. Kim for stimulating discussions, I. Zelinska for help with the figures, D. Petrov for phylogenetic analysis, and A. Bloom for administrative support. This research was supported by a grant from the National Institutes of Health (No. GM 49681).
References
- Andrews, J.D., and Gloor, G.B. (1995). A role for the KP leucine-zipper in regulating P-element transposition in Drosophila melanogaster. Genetics 141, 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito, M.-I., and Walbot, V. (1997). Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17, 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204, 195–229. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Hardeman, K.J. (1992). The Mu elements of Zea mays. Adv. Genet. 30, 77–122. [DOI] [PubMed] [Google Scholar]
- Chandler, V.L., and Walbot, V. (1986). DNA modification of a maize transposable element correlates with loss of activity. Proc. Natl. Acad. Sci. USA 83, 1767–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., Talbert, L.E., and Raymond, F. (1988). Sequence, genomic distribution and DNA modification of a Mu1 element from non-Mutator maize stocks. Genetics 119, 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet, P., Lisch, D., Hardeman, K.J., Chandler, V.L., and Freeling, M. (1991). Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics 129, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli, P., König, J., and Werr, W. (1999). Alternative splicing of two leading exons partitions promoter activity between coding regions of the maize homeobox gene Zmhox1a and Trap (transposon-associated protein). Plant Mol. Biol. 41, 615–625. [DOI] [PubMed] [Google Scholar]
- Cuypers, H., Dash, S., Peterson, P.A., Saedler, H., and Gierl, A. (1988). The defective En-I102 element encodes a product reducing the mutability of the En/Spm transposable element system of Zea mays. EMBO J. 7, 2953–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin, M.J., Lisch, D., and Freeling, M. (1995). Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell 7, 1989–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J.A., Benito, M.-I., and Walbot, V. (1994). Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22, 2634–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, L., Adolphs, R.H., and Kunze, R. (2000). A highly conserved domain of the maize Activator transposase is involved in dimerization. Plant Cell 12, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., and Vaucheret, H. (2000). (Trans)gene silencing in plants: How many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V., and Chandler, V. (1994). Inactivation of maize transposable elements. In Homologous Recombination and Gene Silencing in Plants, J. Paszkowski, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 349–385.
- Fusswinkel, H., Schein, S., Courage, U., Starlinger, P., and Kunze, R. (1991). Detection and abundance of mRNA and protein encoded by transposable element Activator (Ac) in maize. Mol. Gen. Genet. 225, 186–192. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Nava, M., Warren, C.A., Leon, P., and Walbot, V. (1998). Transcriptionally active MuDR, the regulatory element of the Mutator transposable element family of Zea mays, is present in some accessions of the Mexican land race Zapalote chico. Genetics 149, 329–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeman, K.J., and Chandler, V.L. (1993). Two maize genes are each targeted predominantly by distinct classes of Mu elements. Genetics 135, 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D.L., Lozovskaya, E.R., Nurminsky, D.I., and Lohe, A.R. (1997). What restricts the activity of mariner-like transposable elements? Trends Genet. 13, 197–201. [DOI] [PubMed] [Google Scholar]
- Heinlein, M., and Starlinger, P. (1991). Variegation patterns caused by transposable element Ac. Maydica 36, 309–316. [Google Scholar]
- Heinlein, M., Brattig, T., and Kunze, R. (1994). In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and petunia protoplasts. Plant J. 5, 705–714. [DOI] [PubMed] [Google Scholar]
- Hershberger, R.J., Warren, C.A., and Walbot, V. (1991). Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc. Natl. Acad. Sci. USA 88, 10198–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger, R.J., Benito, M.-I., Hardeman, K.J., Warren, C., Chandler, V.L., and Walbot, V. (1995). Characterization of the major transcripts encoded by the regulatory MuDR transposable element of maize. Genetics 140, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. (1995). Origin and evolution of HLA class-I pseudogenes. Mol. Biol. Evol. 12, 247–258. [DOI] [PubMed] [Google Scholar]
- James, M.G., Scanlon, M.J., Qin, M., Robertson, D.S., and Myers, A.M. (1993). DNA sequence and transcript analysis of transposon MuA2, a regulator of Mutator transposable element activity in maize. Plant Mol. Biol. 21, 1181–1185. [DOI] [PubMed] [Google Scholar]
- Joanin, P., Hershberger, R.J., Benito, M.I., and Walbot, V. (1997). Sense and antisense transcripts of the maize MuDR regulatory transposon localized by in situ hybridization. Plant Mol. Biol. 33, 23–36. [DOI] [PubMed] [Google Scholar]
- Kreitman, M., and Akashi, H. (1995). Molecular evidence for natural selection. Annu. Rev. Ecol. Syst. 26, 403–422. [Google Scholar]
- Laski, F.A., Rio, D.C., and Rubin, G.M. (1986). Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell 44, 7–19. [DOI] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A.A., and Walbot, V. (1990). Regulation of the timing of transposable element excision during maize development. Science 248, 1534–1537. [DOI] [PubMed] [Google Scholar]
- Lisch, D., and Freeling, M. (1994). Loss of Mutator activity in a minimal line. Maydica 39, 289–300. [Google Scholar]
- Lisch, D., Chomet, P., and Freeling, M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139, 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., Girard, L., Donlin, M., and Freeling, M. (1999). Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R., and Baron, A. (1994). Coordinate suppression of mutations caused by Robertson's Mutator transposons in maize. Genetics 136, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1951). Chromosome organization and gene expression. Cold Spring Harbor Symp. Quant. Biol. 16, 13–47. [DOI] [PubMed] [Google Scholar]
- Mettler, I. (1987). A simple and rapid method for minipreparation of DNA from tissue cultured plant cells. Plant Mol. Rep. 5, 346–349. [Google Scholar]
- Misra, S., and Rio, D.C. (1990). Cytotype control of Drosophila P element transposition: The 66 kd protein is a repressor of transposase activity. Cell 62, 269–284. [DOI] [PubMed] [Google Scholar]
- Qin, M., and Ellingboe, A.H. (1990). A transcript identified by MuA of maize is associated with Mutator activity. Mol. Gen. Genet. 224, 357–363. [DOI] [PubMed] [Google Scholar]
- Qin, M., Robertson, D.S., and Ellingboe, A.H. (1991). Cloning of the Mutator transposable element MuA2, a putative regulator of somatic mutability of the a1-Mum2 allele in maize. Genetics 129, 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada, M.N., and Walbot, V. (2000). The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell 12, 5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada, M.N., Benito, M.-I., and Walbot, V. (2001. a). MuDR transposon terminal inverted repeats contain multiple enhancer motifs and confer high expression in maize meristematic cells and pollen. Plant J. 25, 1–13. [DOI] [PubMed] [Google Scholar]
- Raizada, M.N., Brewer, K.V., and Walbot, V. (2001. b). A maize MuDR transposon promoter shows limited autoregulation. Mol. Gen. Genet., in press. [DOI] [PubMed]
- Robertson, D.S. (1978). Characterization of a mutator system in maize. Mutat. Res. 51, 21–28. [Google Scholar]
- Robertson, D.S. (1986). Genetics studies on the loss of Mu Mutator activity in maize. Genetics 113, 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schläppi, M., Raina, R., and Fedoroff, N. (1994). Epigenetic regulation of the maize Spm transposable element: Novel activation of a methylated promoter by TnpA. Cell 77, 427–437. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S., and Peterson, P.A. (1989). Genetic evidence of a relationship between two maize transposable element systems: Cy and Mutator. Mol. Gen. Genet. 215, 317–321. [Google Scholar]
- Scofield, S.R., English, J.J., and Jones, J.D.G. (1993). High-level expression of the Activator transposase gene inhibits the excision of Dissociation in tobacco cotyledons. Cell 75, 507–517. [DOI] [PubMed] [Google Scholar]
- Siebel, C.W., Fresco, L.D., and Rio, D.C. (1992). The mechanism of somatic inhibition of Drosophila P-element pre-messenger-RNA splicing: Multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 6, 1386–1401. [DOI] [PubMed] [Google Scholar]
- Simons, R.W., and Kleckner, N. (1988). Biological regulation by antisense RNA in prokaryotes. Annu. Rev. Genet. 22, 567–600. [DOI] [PubMed] [Google Scholar]
- Walbot, V. (1986). Inheritance of Mutator activity in Zea mays as assayed by somatic instability of the bz2-mu1 allele. Genetics 114, 1293–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V. (1991). The Mutator transposable element family of maize. Curr. Top. Genet. Eng. 13, 1–37. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and Rudenko, G.N. (2001). MuDR/Mu transposable elements of maize. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A. Lambowitz, eds (Washington, DC: American Society of Microbiology).
- Walbot, V., and Stapleton, A.E. (1998). Reactivation potential of epigenetically inactive Mu transposable elements of Zea mays L. decreases in successive generations. Maydica 43, 183–193. [Google Scholar]