Abstract
During early seed development, nuclear divisions in the endosperm are not followed by cell division, leading to the development of a syncytium. The simple organization of the Arabidopsis endosperm provides a model in which to study the regulation of the cell cycle in relation to development. To monitor nuclear divisions, we constructed a HISTONE 2B::YELLOW FLUORESCENT PROTEIN gene fusion (H2B::YFP). To validate its use as a vital marker for chromatin in plants, H2B::YFP was expressed constitutively in Arabidopsis. This enabled the observation of mitoses in living root meristems. H2B::YFP was expressed specifically in Arabidopsis syncytial endosperm by using GAL4 transactivation. Monitoring mitotic activity in living syncytial endosperm showed that the syncytium was organized into three domains in which nuclei divide simultaneously with a specific time course. Each mitotic domain has a distinct spatiotemporal pattern of mitotic CYCLIN B1;1 accumulation. The polar spatial organization of the three mitotic domains suggests interactions between developmental mechanisms and the regulation of the cell cycle.
INTRODUCTION
Regulation of the cell cycle in plants and animals involves a remarkable number of conserved genes and mechanisms (Huntley and Murray, 1999; Mironov et al., 1999). In animals, regulation of the entry into G1 has proven to be critical for some developmental steps, such as Drosophila wing patterning (Johnston and Edgar, 1998) and the integration between trophic factors and proliferation (Conlon and Raff, 1999; Galloni and Edgar, 1999). Recently, an essential block of entry into mitosis has been characterized for the coordination between proliferation and gastrulation in Drosophila (Grosshans and Wieschaus, 2000; Mata et al., 2000; Seher and Leptin, 2000). It has been shown in plants that endogenous cell cycle regulation is very precise in meristems, where cells are produced in roots (Berger et al., 1998a), and in the shoot apex (Meyerowitz, 1997; Laufs et al., 1998). However, little is known about the general mechanisms that govern cell proliferation in relation to patterning in plants. To undertake such an analysis, we propose to use a simple developmental system, the syncytial endosperm in Arabidopsis.
Several developmental programs in multicellular organisms are characterized by multiple nucleate structures called syncytia or coenocytes. These structures originate either from the fusion of multiple cells, as in osteoclasts (Jotereau and Le Douarin, 1978), or from a single cell in which nuclear divisions proceed without cytokinesis. The best-studied examples of the latter type are the Drosophila oocyte (Foe et al., 1993) and the hyphae of the fungus Aspergillus nidulans (Doonan, 1992). In these systems, nuclei divide synchronously and thus provide simple models in which to study the developmental control of the cell cycle (Edgar and Lehner, 1996). In plants, the development of syncytia is typical of the endosperm (Vijayaraghavan and Prabhakar, 1984; Lopes and Larkins, 1993; Berger, 1999). Endosperm is included in angiosperm seed and is considered an accessory embryo (Friedman, 1992). It develops from one of the two products of the double fertilization process typical of flowering plants. In the embryo sac, the diploid central cell fuses with one of the two male gametes delivered by the pollen tube and produces a triploid zygotic product that develops as the endosperm (Vijayaraghavan and Prabhakar, 1984; Lopes and Larkins, 1993).
Extensive cytological studies on the development of endosperm have been performed with Arabidopsis and closely related species (Vandendries, 1909; Schultz and Jensen, 1974; Vijayaraghavan and Prabhakar, 1984; Mansfield and Briarty, 1990a, 1990b; Schneitz et al., 1995; Scott et al., 1998; Brown et al., 1999). In these species, the endosperm develops as a syncytium within a few days after fertilization and comprises a few hundred nuclei. This syncytium is a large cell that ultimately divides into individual cells by a process called cellularization (Olsen et al., 1995; Brown et al., 1999; Otegui and Staehelin, 2000). Endosperm cells can adopt a limited number of fates and are organized along a main polar axis. The Arabidopsis endosperm thus constitutes a simple developmental model for cell cycle control and cell fate determination.
We have obtained plant lines that express the fusion of an endogenous Arabidopsis HISTONE 2B (H2B) with the green fluorescent protein (GFP) yellow variant (YFP). Chromosome condensation and segregation were observed in vivo, which enabled measurement of the dynamic parameters for mitotic phases in root meristem cells. Endosperm-specific expression of the H2B::YFP fusion protein was achieved using a GAL4–upstream activating sequences (UAS) enhancer trap strategy (Haseloff, 1999). Time lapse confocal observations showed that successive rounds of mitosis punctuate syncytial endosperm development in Arabidopsis. After the initial three synchronous cycles of nuclear division, independent mitotic domains are established in which nuclei either experience coordinated divisions or increase in size, presumably after endoreduplication. Furthermore, such mitotic domains are functionally delineated by the expression of the Arabidopsis mitotic CYCLIN B1;1. Mitotic domains form distinct compartments organized along the apical–basal polarity axis. These compartments correspond to three areas identified previously by a characteristic cytological structure (Mansfield and Briarty, 1990b; Scott et al., 1998; Brown et al., 1999), suggesting that precise spatial regulation of the expression of cell cycle control genes may play a role in the functional organization of the developing syncytial endosperm.
RESULTS
Stable Expression of H2B::YFP in Arabidopsis
To visualize the expression of H2B, we fused the YFP variant of GFP to the C terminus of the Arabidopsis H2B gene (Figure 1). The chimeric gene was placed under transcriptional control of the 35S cauliflower mosaic virus promoter and transformed in Arabidopsis by using Agrobacterium-mediated root transformation. Fluorescence due to YFP expression was observed in roots 7 days after transformation. Three stable lines expressing H2B::YFP were obtained, and homozygous progeny were further characterized. In these plants, fluorescence with emission characteristics of YFP was detected in nuclei in roots, hypocotyl, stems, and flowers. Morphogenesis and fertility were identical to those of wild-type plants. Moreover, transgenic lines for H2B::YFP and wild-type plants possessed similar root growth kinetics (2 ± 0.2 mm/day [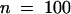 ] and 2 ± 0.15 mm/day [
] and 2 ± 0.15 mm/day [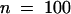 ], respectively).
], respectively).
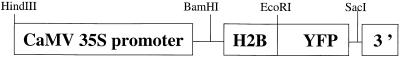
Figure 1.
H2B::YFP Fusion Constructs.
Scheme of the T-DNA left border region of transfer vector pBI121 in which the EcoRI is deleted. Fusion was created by the insertion of the H2B cDNA between the BamHI and EcoRI sites of either pBI121 or pBIB. CaMV 35S promoter, 35S promoter from the cauliflower mosaic virus; H2B, histone 2B from Arabidopsis amplified by reverse transcription–polymerase chain reaction and homolog to ATH2B (GenBank accession number Y07745); YFP, modified sequence of yellow fluorescent protein (Haseloff, 1999); 3′, nopaline synthase terminator.
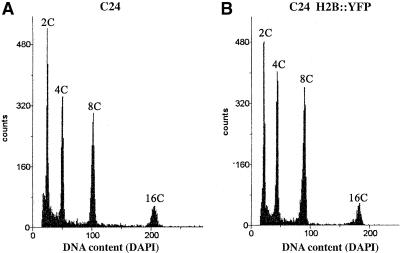
It was conceivable that the expression of H2B::YFP could affect chromatin structure and perturb the regulation of the cell cycle. We performed analyses of the proportion of seedling cells in the different phases of the cell cycle by fluorescence-activated cell sorting (FACS). Nuclear DNA content was determined by measuring 4′,6-diamidino-2-phenylindole (DAPI) fluorescence (Figure 2). In control plants, nuclei were distributed in four groups corresponding to diploid cells in G1 or G2 phase and to cells in which endoreduplication had taken place (4C and 8C values). A similar distribution of nuclear DNA was observed in plants expressing H2B::YFP. This finding strongly suggested that, overall, the regulation of the cell cycle was not affected significantly by the expression of H2B::YFP. In conclusion, the expression of H2B::YFP does not appear to interfere with the function of the plant cell, and the fluorescent marker can be considered vital.
Figure 2.
Analysis of the Repartition of Cell Cycle Phases in Plants Expressing H2B::YFP.
Nuclei of root cells were purified and labeled with DAPI, and fluorescence was analyzed by FACS. The phases of the cell cycle were identified as 2C (G1) or 4C (G2) of nuclear DNA content. Nuclei with higher DNA content (8C and 16C) have undergone endoreduplication. Endoreduplication and the repartition of cells between the phases of the cell cycle were not significantly different between plants that did not (A) or did (B) express the H2B::YFP fusion protein.
Localization of H2B::YFP in Nuclei of Plant Cells
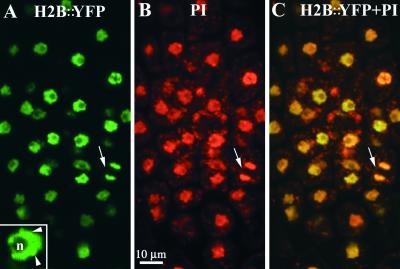
H2B::YFP fluorescence observed in fixed tissues of seedlings was limited to structures that corresponded to nuclei (Figure 3A). No fluorescence was observed in the cytoplasm, and the YFP signal was much greater than the low levels of autofluorescence observed in roots and young leaves of nontransgenic plants. Detection of H2B::YFP fluorescence by using confocal microscopy enabled a more precise determination of the nuclear morphology. The nucleolus showed characteristic circular cross-sections with low levels of fluorescence compared with the surrounding nucleoplasm. Speckled structures (<0.5 μm in diameter) with high levels of fluorescence were present at the nuclear envelope as well as at the periphery of the nucleolus. H2B::YFP fluorescence in nuclei colocalized with structures labeled by propidium iodide (Figures 3B and 3C), as expected for colocalization of chromatin and DNA. Propidium iodide also labeled nucleic acids present in organelles that were not labeled by H2B::YFP. Colocalization of YFP and propidium iodide signals was observed in condensed chromosomes during mitosis (Figure 3, arrows). The H2B::YFP fusion protein thus appeared to be a valid marker for chromatin organization in plant nuclei.
Figure 3.
Colocalization of H2B::YFP Fluorescence and DNA Stained with Propidium Iodide in Fixed Root Meristems.
(A) Confocal section of a root meristem. H2B::YFP fluorescence is shown in green. A more detailed image of one nucleus (inset) shows the nucleolus (n) and dense chromatin foci (arrowheads).
(B) DNA stained with propidium iodide (PI) is shown in red. Condensed chromosomes during late anaphase are visible (arrow). PI fluorescence corresponding to organelle DNA is visible as spots around each nucleus.
(C) The H2B::YFP signal colocalizes with the PI signal in nuclei during interphase and mitosis.
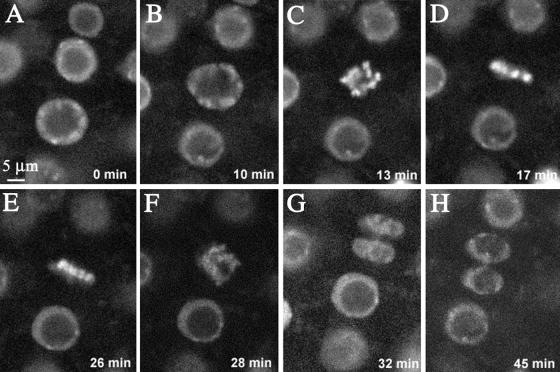
Confocal microscopy applied to live seedlings enabled us to study in vivo the dynamics of chromatin distribution in nuclei during interphase and mitosis. Dynamic views are available at http://www.ens-lyon.fr/~ifobis/RDP/pagesweb/Francais/groupes/embryogenese/F.Berger/mitose%20animee.html. Nuclei observed in root meristems were characterized by a structure similar to that observed in fixed material (Figure 4). After cell division, pairs of daughter cells were recognizable (Figure 4H). These cells had small nuclei in G1 phase that could be distinguished from the larger nuclei of cells that did not belong to a pair. These cells were characterized by a large nucleolus surrounded by a thin band of nucleoplasm and were probably in S phase or G2 phase of the cell cycle (Figure 4A). We performed in vivo observation of mitosis in the root meristem and were able to determine its kinetic parameters by using time lapse confocal microscopy (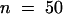 dividing cells). The entry into mitosis was characterized by chromatin condensation. During prophase (8 to 15 min), the nucleolus disappeared and the nucleus lost its spherical shape (Figure 4B). Chromatin gradually condensed into chromosomes that fluoresced brightly (Figure 4C). Chromosomes migrated and aligned in a band of high intensity, the metaphase plate (Figure 4D) (5 to 12 min). The initiation of anaphase was marked by separation of sister chromatids (Figure 4E); chromosomes appeared as individual small rods that rapidly migrated away from the metaphase plate (Figure 4F) (3 to 4 min). Telophase (15 to 20 min) was initiated when chromosomes reached poles opposite the initial position of the metaphase plate (Figure 4G). The bright fluorescence associated with chromosomes gradually became more diffuse. In each daughter cell, the nucleus reacquired its spherical shape and the nucleolus reappeared slowly (Figure 4H).
dividing cells). The entry into mitosis was characterized by chromatin condensation. During prophase (8 to 15 min), the nucleolus disappeared and the nucleus lost its spherical shape (Figure 4B). Chromatin gradually condensed into chromosomes that fluoresced brightly (Figure 4C). Chromosomes migrated and aligned in a band of high intensity, the metaphase plate (Figure 4D) (5 to 12 min). The initiation of anaphase was marked by separation of sister chromatids (Figure 4E); chromosomes appeared as individual small rods that rapidly migrated away from the metaphase plate (Figure 4F) (3 to 4 min). Telophase (15 to 20 min) was initiated when chromosomes reached poles opposite the initial position of the metaphase plate (Figure 4G). The bright fluorescence associated with chromosomes gradually became more diffuse. In each daughter cell, the nucleus reacquired its spherical shape and the nucleolus reappeared slowly (Figure 4H).
Figure 4.
Dynamics of the Nuclear Structure and of the Chromatin from G2 Phase to G1 Phase in Living Root Epidermal Cells.
(A) The cell at the bottom of the optical section was observed by using time lapse confocal microscopy during mitosis. The time elapsed from the first recognizable sign of nuclear envelope breakdown is indicated. Chromatin condensation foci are visible at the nuclear envelope and at the surface of the nucleolus.
(B) In prophase, the nucleus loses its spherical shape and shows condensed chromatin aggregates.
(C) These aggregates further condense into recognizable chromosomes.
(D) Chromosomes align along the metaphase plate.
(E) and (F) During anaphase, chromosomes separate (E) and migrate to opposite poles of the cell (F).
(G) During telophase, chromosomes decondense.
(H) A pair of cells results from the division of the G2 cell shown in (A).
Syncytial Development of Arabidopsis Endosperm Is Divided into Successive Stages Characterized by Total Number of Nuclei
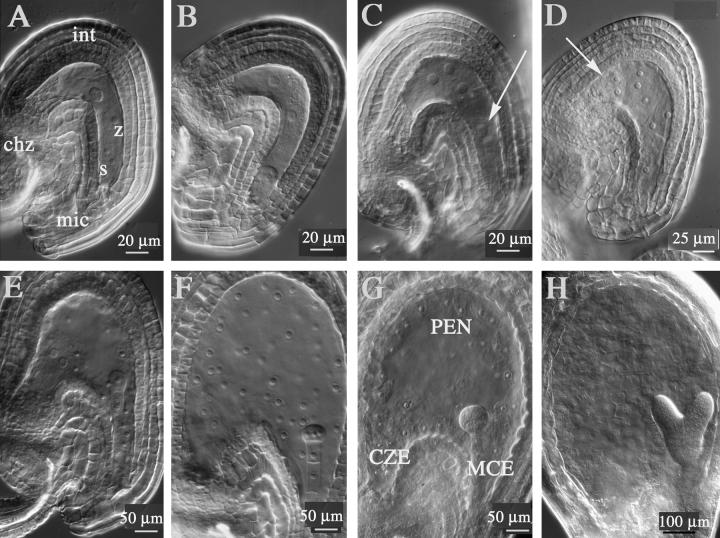
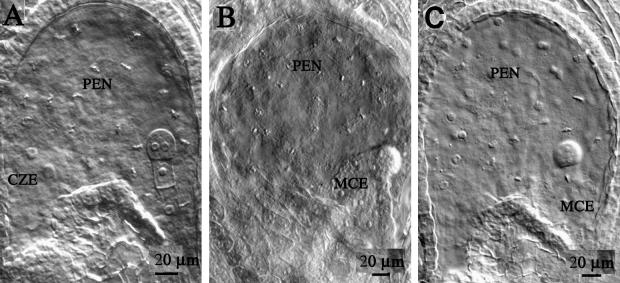
We characterized the main features of early endosperm development by using whole mounts of isolated developing seed. The endosperm develops from the central cell fertilized by one of the two male gametes that are delivered by the pollen tube at the micropylar pole of the ovule (Figure 5A). The zygotic endosperm is occupied by a large vacuole, and its large nucleus is located initially close to the zygote. It divides into two nuclei (Figure 5B). This division is not followed by the division of the cell, which becomes binucleated. Further nuclear divisions take place without cytokinesis, and a syncytial structure develops (Figures 5C to 5G). Nuclei gradually occupy the whole periphery of the endosperm (Figures 5E to 5G), with the exception of nuclei located at the chalazal pole (Figures 5E and 5F) and nuclei surrounding the embryo (Figure 5G). Each endosperm nucleus is surrounded by a denser cytoplasm, and this unit has been termed the nuclear cytoplasmic domain (Brown et al., 1999). Nuclei in the chalazal endosperm are larger, with large nucleoli (Figure 5E). After the embryo globular stage, cellularization of the syncytium is initiated. The growth of cell walls progresses from the area surrounding the embryo throughout the peripheral endosperm (Figure 5H). This represents the end of the syncytial phase of endosperm development. Thus, this phase is characterized by a large number of nuclear divisions, by massive growth of the endosperm cell wall, and by morphological and cytological polarization along the micropyle–chalazal axis. This polarity axis is divided into three domains: the micropylar endosperm (MCE), the peripheral endosperm (PEN), and the chalazal endosperm (CZE) (Mansfield and Briarty, 1990a, 1990b; Scott et al., 1998).
Figure 5.
Main Developmental Stages of the Syncytial Endosperm.
Whole mounts of cleared seed were observed by using differential interference contrast (DIC) optics.
(A) The zygotic endosperm is tube shaped and surrounded by four to five cell layers of integuments (int). It is separated from the micropyle (mic) by the zygote (z) and the degenerating synergids (s). At the opposite pole, the chalazal proliferating tissue (chz) separates the endosperm from the vascular elements of the funiculus.
(B) The first nuclear division is not followed by the division of the central cell, which becomes binucleate. At this stage, one nucleus is located closer to the zygote and the other is closer to the chalazal pole. Both nuclei occupy a central position in the endosperm tube.
(C) The four-nuclei–stage endosperm is the result of the second round of syncytial nuclei division. One nucleus migrates to the vicinity of the zygote (arrow).
(D) The third round of nuclear division takes place perpendicular to the polar axis of the now enlarged endosperm tube. A pair of nuclei is located close to the zygote, and one or two nuclei are located at the chalazal pole (arrow).
(E) At the two-cell embryo stage, one to four nuclei occupy the cleft formed above the chalaza. Those nuclei are usually larger than the nuclei located in the periphery and are characterized by large nucleoli.
(F) Quadrant-stage embryo with stage VIII endosperm.
(G) The globular embryo protrudes inside the endosperm cavity and is surrounded by a layer of endosperm nuclei. Three morphologically distinct zones can be identified: the micropylar pole occupied by a dense cytoplasmic embryo-surrounding region (MCE); the peripheral endosperm (PEN), which is the largest domain in terms of total number of nuclei and total surface area; and, at the chalazal pole, a pocket that contains few nuclei and constitutes the chalazal endosperm (CZE). The PEN is composed of one layer of nuclear cytoplasmic domains evenly spaced along the entire surface of the endosperm.
(H) The endosperm becomes cellularized at the embryo torpedo stage. A section of the surface of the endosperm shows hexagonal and pentagonal endosperm cell sections.
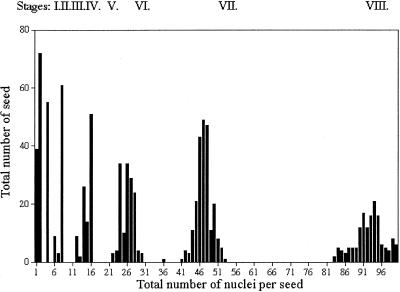
The observation of early stages of development in which two, four, and eight nuclei were present suggested that nuclear divisions were synchronous and that the total number of nuclei increased exponentially. To test this hypothesis, we counted nuclei for each developing seed in a population of 1000 seed at different developmental stages taken at random from 120 siliques of 40 different plants. For each seed, the embryo developmental stage was scored. It appeared that seed were divided into subpopulations according to the total number of nuclei in the endosperm (Figure 6). Those main subpopulations were represented by developing seed with two, four, eight, 12 to 16, 24 to 27, 46 to 48, and 90 to 96 nuclei. After the embryo dermatogen stage, the total number of nuclei per developing seed was difficult to determine accurately, but we estimated that before cellularization the endosperm contained ∼200 nuclei (196.5, 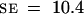 ,
, 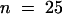 ). Thus, we defined developmental stages I to IX (Table 1). The correlation between endosperm development and embryo development was clear but not absolute. For example, a two-cell–stage embryo could be surrounded by an endosperm at stage VI or VII. Similarly, the growth of the ovule integument did not strictly follow endosperm development, and seed size was not strictly uniform for a given endosperm developmental stage (data not shown).
). Thus, we defined developmental stages I to IX (Table 1). The correlation between endosperm development and embryo development was clear but not absolute. For example, a two-cell–stage embryo could be surrounded by an endosperm at stage VI or VII. Similarly, the growth of the ovule integument did not strictly follow endosperm development, and seed size was not strictly uniform for a given endosperm developmental stage (data not shown).
Figure 6.
Developmental Stages of the Syncytial Endosperm as Defined by the Total Number of Nuclei.
A large population of developing seed was divided into classes defined by the total number of endosperm nuclei. It appears that seed are divided into subpopulations that correspond to distinct stages of development.
Table 1.
Developmental Stages of the Arabidopsis Endosperm
| Endosperm Developmental Stage | No. of Endosperm Nuclei | Embryo Developmental Stage |
Time after Fertilization (hr) | Cytological Events |
|---|---|---|---|---|
| I | 1 | Zygote | 0 | One large nucleus close to the zygote. |
| II | 2 | Zygote | 2–4 | First syncytial mitosis. The two nuclei migrate to opposite poles of the endosperm. |
| III | 4 | Zygote | NDa | Second syncytial mitosis. Nuclei division planes are parallel to the main polar axis. |
| IV | (6)–8 | Elongated zygote | ND | Third syncytial mitosis. Nuclei division planes are perpendicular to the main polar axis. One or two nuclei migrate to the chalazal pole, and the six or seven other nuclei become tightly linked to the endosperm peripheral cell wall. Nuclei of the CZE do not enter mitosis together with nuclei of the PEN. |
| V | 12–16 | Elongated zygote, one cell |
ND | Fourth syncytial mitosis. Nuclei divisions are not synchronous, and nuclei of the CZE are larger than nuclei in the PEN. |
| VI | 24–28 | One cell, two cell | 12–18 | Fifth syncytial mitosis. The CZE contains one to four large nuclei. Nuclei in the MCE undergo synchronous divisions earlier than nuclei in the PEN. |
| VII | 44–48 | Two cell, quadrant, octant |
24 | Sixth syncytial mitosis. The delay between nuclei division in the MCE and the PEN becomes more pronounced. In the PEN, coordinated nuclei divisions take place as a wave. The PEN and the MCE become independent domains of cyclin B1;1 expression. |
| VIII | 90 | Octant | 30 | Seventh syncytial mitosis. A layer of nuclear cytoplasmic domains surrounds the embryo, and the MCE is the only part of the endosperm with two layers of nuclear cytoplasmic domains. |
| IX | 200 | Dermatogen-globular | 36–60 | Eighth syncytial mitosis restricted to the PEN. Divisions in the MCE are completely independent of divisions in the PEN. The CZE contains large and small nuclei. |
ND, not determined.
GAL4-Mediated Transactivation of the H2B::YFP Fusion Protein in the Endosperm Shows Higher Ploidy of CZE Nuclei and Allows Direct Visualization of Distinct Mitotic Domains in the Syncytial Endosperm
If mitoses were strictly synchronous, the nine successive stages of development defined in terms of total nuclei number should be one, two, four, eight, 16, 32, 64, 128, and 256. This was not observed, and after stage III, each major stage was preceded by substages (Figure 6). For example, in addition to the large pool of seed with eight nuclei, there were seed with six and seven nuclei. Similarly, seed with 12, 14, and 15 nuclei were found in addition to the stage 16 nuclei. These findings suggested that nuclear divisions were not strictly synchronous in the syncytium after stage III. This hypothesis was supported by the observation of larger nuclei in the CZE (Figure 5E). It was possible that those large nuclei did not divide like the other endosperm nuclei. To test this hypothesis, we directly monitored nuclei division in endosperm by using the H2B::YFP fusion protein.
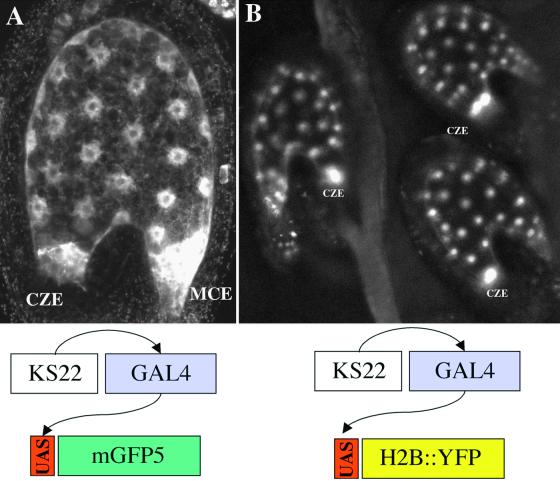
The cauliflower mosaic virus 35S promoter was not active during embryogenesis before the torpedo stage and did not present any activity in the syncytial endosperm. To transactivate the expression of the H2B::YFP fusion protein, we used the enhancer trap line KS22, in which the modified yeast transcription activator GAL4::VP16 is expressed in the endosperm (Haseloff, 1999). In this line, GAL4::VP16 drives the expression of the GFP variant mGFP5, which is targeted to the endoplasmic reticulum (ER) (Haseloff et al., 1997). Thus, seed of the KS22 line contain fluorescent endosperm that can be imaged using confocal microscopy (Figure 7A). Plants of line KS22 were transformed with a construct in which the gene coding for the fusion protein H2B::YFP was placed under the control of the GAL4-responsive element UAS. Living seed of transformed plants were characterized by fluorescent endosperm nuclei as a consequence of H2B::YFP expression (Figure 7B). The level of mGFP5 fluorescence in the ER was decreased compared with that in line KS22. Seed development did not appear to be affected, and progeny of double homozygotes for the KS22 enhancer and the promUAS-H2B::YFP construct were obtained.
Figure 7.
Transactivation of H2B::YFP in the Endosperm.
(A) The GAL4::VP16 enhancer trap line KS22 expresses the ER-targeted variant mGFP5 in the endosperm in the plant seed. This outlines the ER-rich MCE and CZE and the nuclear cytoplasmic domain in the PEN. Nuclei appear as black discs surrounded by brighter nuclear envelopes.
(B) The KS22 line was transformed with promUAS-H2B::YFP. This causes specific labeling of chromatin in endosperm nuclei. All nuclei appear similar in diameter and intensity of labeling, with the exception of larger and brighter nuclei in the CZE.
In the KS22 line, the enhancer-dependent expression of H2B::YFP was detectable from the eight-nuclei stage. In all seed in which the endosperm contained >16 nuclei, larger and brighter nuclei were present in the CZE (Figure 7B). The number of large nuclei in the CZE varied from one to four in developing seed until stage VII. Although quantification of GFP fluorescence in living seed was made difficult by differential attenuation of the signal as a consequence of the thickness of the seed integument, the larger size and greater fluorescence observed in CZE nuclei strongly suggested that they contained a much greater quantity of chromatin and that their ploidy was higher than that of nuclei present in the other domains of the endosperm.
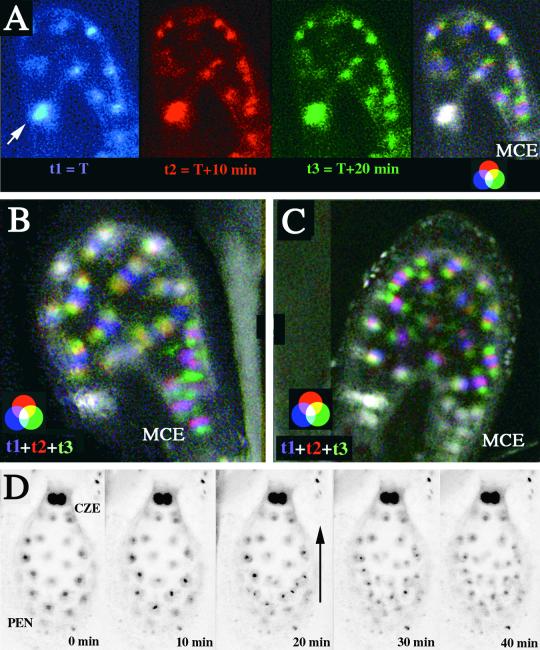
This asynchrony of divisions between nuclei present in the CZE and nuclei present in the other part of the endosperm was confirmed by direct observation of mitoses in syncytial endosperm (Figure 8). Observations were conducted for 12 hr using time lapse confocal microscopy on living seed of the KS22 line transformed with the construct promUAS-H2B::YFP. Typically, each experiment corresponded to one transition between two developmental steps. Time series corresponding to the figures are available at http://www.ens-lyon.fr/~ifobis/RDP/pagesweb/Francais/groupes/embryogenese/F.Berger/albumen2%20animee.html. With the exception of the large CZE nuclei, all nuclei present in the PEN and the MCE divided synchronously, and nuclear division was complete within 20 to 30 min (Figure 8A) (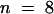 ). Coordinated nuclear divisions in the PEN and the MCE were no longer simultaneous in seed older than stage V (
). Coordinated nuclear divisions in the PEN and the MCE were no longer simultaneous in seed older than stage V (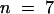 ). After coordinated nuclear divisions in the PEN, nuclei in the MCE divided synchronously with a 10-min delay (Figure 8B). The delay between the divisions in the two domains was longer in older developmental stages, leading to the establishment of two independent mitotic domains (Figure 8C). In endosperm older than stage VI, time lapse observations showed a wave of nuclear divisions crossing the peripheral endosperm from the MCE pole toward the CZE (Figure 8D). Thus, nuclear divisions were no longer synchronous but were still coordinated. The wave of nuclear divisions did not affect nuclei present in the CZE. At these stages, the CZE contained both large nuclei and a few nuclei similar in size to PEN nuclei.
). After coordinated nuclear divisions in the PEN, nuclei in the MCE divided synchronously with a 10-min delay (Figure 8B). The delay between the divisions in the two domains was longer in older developmental stages, leading to the establishment of two independent mitotic domains (Figure 8C). In endosperm older than stage VI, time lapse observations showed a wave of nuclear divisions crossing the peripheral endosperm from the MCE pole toward the CZE (Figure 8D). Thus, nuclear divisions were no longer synchronous but were still coordinated. The wave of nuclear divisions did not affect nuclei present in the CZE. At these stages, the CZE contained both large nuclei and a few nuclei similar in size to PEN nuclei.
Figure 8.
Time Lapse Observation of Nuclear Division in Mitotic Domains of the Syncytial Endosperm.
Time lapse imaging of nuclei in KS22 seed expressing H2B::YFP enabled us to monitor nuclear division during endosperm development. Optical sections were recorded every 10 min for 12 hr. Only three consecutive sections are displayed, corresponding to the time at which nuclear divisions were observed. Each section is color coded with the fundamental colors red, blue, and green. The superposition of the three images clearly shows nuclei that do not divide (white) and nuclei that undergo mitosis (color).
(A) Transition from stage V to stage VI. Two large nuclei in the CZE do not participate in the synchronous division that affects all other nuclei in the endosperm (arrow).
(B) Transition from stage VI to stage VII. Superposition of three color-coded consecutive sections obtained every 10 min during a 12-hr time lapse recording. CZE nuclei do not divide and appear white. White nuclei in the PEN divide perpendicular to the plane of observation. PEN synchronous nuclear division precedes by 10 min MCE nuclear division, resulting in different color sequences.
(C) A similar recording of nuclear division in a stage VII endosperm shows that MCE nuclear division takes place independently from coordinated PEN mitoses. MCE nuclei thus appear white, as do CZE nuclei. A few PEN nuclei located close to the chalazal pole do not divide until the next period (data not shown). This results from the fact that nuclear division takes place in the PEN as a wave from the MCE toward the CZE.
(D) A series of consecutive optical sections from a 14-hr time lapse recording of the transition from stage VII to stage VIII. Negative images of optical sections are displayed. The endosperm is observed from the chalazal pole. CZE nuclei are larger and brighter than all other nuclei and do not experience mitosis when it is observed in the PEN. A wave of nuclear division crosses the PEN, as indicated by the arrow. The four nuclei closest to the CZE large nuclei do not divide and instead become part of the CZE.
The organization of the endosperm into three mitotic domains was observed in fixed seed as well (Table 2). When mitoses were observed in the PEN of seed older than stage IV, the CZE contained one to four large endosperm nuclei that never divided (Figure 9A). Typical mitosis was not observed in the CZE in seed at any stage of syncytial development (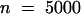 ). In seed older than stage VIII, synchronous mitoses in the PEN and the MCE were never observed simultaneously (Figures 9B and 9C).
). In seed older than stage VIII, synchronous mitoses in the PEN and the MCE were never observed simultaneously (Figures 9B and 9C).
Table 2.
Localization of Mitotic Domains in the Syncytial Endosperm of Arabidopsis
| Total No. of Divisions | All Nuclei Except CZE | All Nuclei Except MCE and CZE | Division of Nuclei in MCE Only |
|---|---|---|---|
| 169 | 92 (stages IV to VII) | 68 (stages VIII and IX) | 9 (stages VIII and IX) |
Figure 9.
Mitotic Domains in Arabidopsis Endosperm Observed in Fixed Seed.
Whole mounts of cleared seed are observed with DIC optics.
(A) Endosperm at the transition between stages VI and VII with a two-cell embryo. Metaphase plates are detected in all nuclei with the exception of the two large nuclei of the CZE.
(B) Transition from stage VIII to stage IX. All nuclei in the PEN are in anaphase, whereas nuclei in the MCE are in interphase.
(C) Endosperm at stage VIII with a dermatogen embryo. Nuclei in the MCE are in metaphase, whereas nuclei in the PEN are in interphase.
In conclusion, the syncytial endosperm appeared to be divided into three mitotic domains in which mitosis occurs synchronously or in a coordinated manner. Those domains corresponded to the MCE, PEN, and CZE, as defined according to their cytological characteristics and positions.
Mitotic Cyclin Activity Parallels the Organization of Mitotic Domains in the Endosperm Syncytium
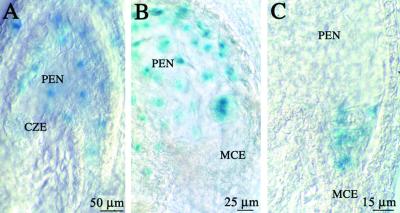
We used the marker line FA4C to visualize the expression of the mitotic cyclin CYCLIN B1;1 (Colon-Carmona et al., 1999). In this transgenic line, the expression of the reporter gene uidA was under the control of the promoter of the gene encoding the mitotic CYCLIN B1;1. The β-glucuronidase (GUS) protein was fused to the destruction box responsible for cyclin degradation after anaphase. Hence, the GUS protein was produced at the G2/M transition and subsequently degraded. The differential control of the cell cycle in the domains MCE, PEN, and CZE was supported by the pattern of expression of the cyclin B1;1. In the reporter line FA4C, GUS activity was observed in the endosperm in a low percentage of the seed (0.6%, 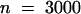 ). This probably reflects the small temporal window of expression of the cyclin B1;1. We never observed a uniform GUS activity in endosperm older than stage V (
). This probably reflects the small temporal window of expression of the cyclin B1;1. We never observed a uniform GUS activity in endosperm older than stage V (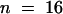 ). In some seed older than stage V, the GUS activity was present in PEN nuclei but not in the large nuclei of the CZE (Figure 10A). We never observed any GUS activity in CZE nuclei, but some low activity might be present in the cytoplasm. As early as stage VII, the mitotic domains PEN and MCE could be distinguished in terms of CYCLIN B1;1 activity. In the FA4C line, GUS activity was detected exclusively in the PEN (Figure 10B) or in the MCE (Figure 10C). This showed that the mitotic domains are functionally defined by the characteristic regulation of the expression of cyclin B1;1.
). In some seed older than stage V, the GUS activity was present in PEN nuclei but not in the large nuclei of the CZE (Figure 10A). We never observed any GUS activity in CZE nuclei, but some low activity might be present in the cytoplasm. As early as stage VII, the mitotic domains PEN and MCE could be distinguished in terms of CYCLIN B1;1 activity. In the FA4C line, GUS activity was detected exclusively in the PEN (Figure 10B) or in the MCE (Figure 10C). This showed that the mitotic domains are functionally defined by the characteristic regulation of the expression of cyclin B1;1.
Figure 10.
CYCLIN B1;1 Is Differentially Expressed in the Syncytial Endosperm Mitotic Domains MCE, PEN, and CZE.
Whole mounts of cleared FA4C seed are observed after GUS staining. This reflects the expression of the CyclinB1;1::GUS fusion protein.
(A) Endosperm at stage VI with a two-cell embryo. GUS stain is present in all nuclei of the PEN but not in the CZE occupied by four nuclei (only three nuclei are visible).
(B) A stage VIII endosperm with GUS expression restricted to the PEN. The octant embryo shows a strong GUS signal on two cells of the embryo proper.
(C) Endosperm at stage VII. The GUS stain is localized in the MCE of a two-cell embryo.
DISCUSSION
H2B::YFP Is a Vital Marker for Chromatin Structure and for Monitoring the Cell Cycle in Plants
Although transient expression of GFP fusion proteins in onion epidermal cells is useful and informative to show protein localization (Scott et al., 1999), it remains essential to develop viable plant lines expressing GFP-based cytological markers. Such lines have been obtained for the microtubule (Marc et al., 1998) and actin (Kost et al., 1998) cytoskeletons using proteins with affinity for the target structures. GFP has been targeted to the nucleus in transgenic plants (Grebenok et al., 1997; Chytilova et al., 1999) using nuclear localization signal sequences. This enabled the visualization of nuclei and the determination of the time of nuclear envelope breakdown at the onset of mitosis. However, this marker did not withstand fixation and did not report any structural element of the nucleus with the exception of the nucleolus. Therefore, we designed a fusion of a major structural component of the chromatin with YFP, a brighter spectrum-shifted variant of the GFP (Haseloff, 1999). Histones are major structural constituents of the chromosomes, and viable expression of the human H2B::GFP has been obtained in cell lines (Kanda et al., 1998). It was reported recently that the expression of a GFP fused to a fragment of a Histone 2A in Drosophila does not perturb development (Clarkson and Saint, 1999). We showed that the expression of the fusion H2B::YFP in Arabidopsis plants perturbed neither plant growth nor morphogenesis and reproduction. FACS analyses further demonstrated that the regulation of the cell cycle is not affected by the expression of H2B::YFP. We showed that H2B::YFP fluorescence is located exclusively in the nucleus and not in the cytoplasm or organelles. Moreover, H2B::YFP fluorescence colocalizes with the DNA stain propidium iodide in the nucleus. Chromatin condensation into chromosomes during mitosis provided further evidence for specific vital labeling of chromatin by the H2B::YFP fusion protein.
Investigation of the dynamic parameters of mitosis has been performed in unusual cell types such as Haemanthus endosperm cells (Bajer, 1958; Bajer and Molè-Bajer, 1972) and Tradescantia stamen hair cells (Wolniak, 1987). The dynamics of meiosis in maize have been reported using a vital chromosome stain (Yu et al., 1997). Using transgenic plants that express the H2B::YFP fusion protein, we report the dynamics of mitosis in cells within whole organs. Our experiments were performed in root meristems by using time lapse confocal microscopy. On average, mitosis takes 45 min in root meristem cells, and the longest phases are the metaphase and the telophase. The time scale observed is compatible with previous observations in animal cells (Enger et al., 1968) but is much more rapid than that reported for endosperm cells in Haemanthus, in which the entire mitosis lasts 2.5 hr and anaphase lasts 26 min (Bajer, 1958). Moreover, we determined the phase of the cell cycle in individual cells in the division zone of the root meristem. During G1 phase, nuclei are smaller and have less developed nucleoli than during the S and G2 phases. In conclusion, H2B::YFP is a vital marker useful for monitoring the phases of the cell cycle in living cells.
GAL4 Transactivation Strategy Combined with Specific Markers Allows Specific Developmental Studies in Plants
Transactivation of gene expression using the modified yeast transcriptional activator GAL4 was performed originally in Drosophila (Brand and Perrimon, 1993). This approach has provided a powerful method to investigate interactions between cell cycle regulation and developmental mechanisms in Drosophila (Neufeld and Edgar, 1998; Mata et al., 2000). Transactivation strategies using modified GAL4 (Guyer et al., 1998; Haseloff, 1999) and the yeast transcription factor GH4 have been tested in plants (Moore et al., 1998). In Arabidopsis, a library of enhancer trap lines with cell type–specific GAL4 expression has been constructed (Haseloff, 1999). To be able to label endosperm nuclei, we used the enhancer trap line KS22, which is characterized by endosperm-specific expression of GAL4::VP16 and mGFP5. Plants of the KS22 line, transformed with the H2B::YFP gene placed under the control of the UAS promoter, were characterized by fluorescent nuclei in the endosperm. Surprisingly, the expression of the mGFP5 reporter was selectively silenced after transformation with H2B::YFP, suggesting that caution is warranted when multiple transactivation events are required. This could result from epigenetic control of cytosine methylation, which was shown to affect GAL4 binding to DNA (Gälwailer et al., 2000). The use of GAL4 enhancer trap lines in plants should result in efficient and modular ways to drive tissue-specific expression. This technique should provide new insights that global overexpression cannot provide. Moreover, flow cytometry could be used to sort nuclei of specific cell types in transgenic plants that express the H2B::YFP fusion protein under the control of cell identity–specific promoters.
Control of the Cell Cycle Is Linked to the Establishment of Mitotic Domains in the Endosperm
The cytological characteristics of syncytial endosperm development have been studied in Arabidopsis (Mansfield and Briarty, 1990a, 1990b; Schneitz et al., 1995; Scott et al., 1998; Brown et al., 1999) and in closely related crucifers (Vandendries, 1909; Schultz and Jensen, 1974; Vijayaraghavan and Prabhakar, 1984). Those studies provide a detailed account of endosperm organization at the cytological level and are in agreement with our own cytological observations. Here, we report that endosperm syncytial development can be divided into distinct developmental stages based on the total number of nuclei. Eight successive rounds of mitoses take place during the syncytial stage and define developmental stages I to IX. We have characterized each stage by cytological events based on our own observations and descriptions from the various sources cited above (Table 1). Such stages provide a basis for studies of cell cycle regulation and the characterization of mutants in endosperm development.
Time lapse observations demonstrated that the Arabidopsis syncytial endosperm is divided into mitotic domains. In the MCE and the PEN, all nuclei divide synchronously, and the mitotic activity of each domain appears to be independent of mitotic activity in the other domains. The organization of mitotic domains in Arabidopsis endosperm appears to be different from that observed in Drosophila embryos. Drosophila syncytial embryogenesis is characterized by 13 synchronous divisions (Foe et al., 1993), and mitotic domains are established after cellularization of the syncytium (O'Farrell et al., 1989). An exception to this rule is observed during the last cycle before cellularization, when posterior nuclei that later develop as progenitors of germ cells divide later than the other nuclei (Su et al., 1998a). Two other exceptions are seen in polar nuclei that do not participate to mitosis and yolk nuclei that remain at the center of the blastoderm and endoreduplicate (Su et al., 1998b). In contrast to Drosophila embryos, Arabidopsis endosperm mitotic domains are defined much earlier, as shown by the expression of cyclin B1;1 demonstrated by the GUS activity in line FA4C (Colon-Carmona et al., 1999). With the exception of the initial stages of syncytial development, CYCLIN B1;1 was never distributed uniformly but was restricted alternatively to nuclei of the mitotic domains PEN and MCE. This finding suggests that precise control of cyclin accumulation constitutes part of the overall program responsible for syncytial development. The PEN appears to be organized as a network of cytoplasmic bridges that connect nuclei rather than as a continuous layer. Due to the extreme thinness of its cytoplasmic strands, the PEN may not be functionally connected to the MCE cytoplasm. PEN and MCE therefore may function as independent units.
Distinctive regulation of the cell cycle was observed in a third domain of the Arabidopsis endosperm, the CZE. During the syncytial stage, nuclear division was never observed directly in the CZE. Nuclei labeled by the H2B::YFP fusion protein were characterized by much greater fluorescence than nuclei in the PEN. Moreover, mitotic division was never observed in the CZE. This strongly suggests that ploidy in the CZE is greater than in the other mitotic domains. Thus, we hypothesize that endoreduplication takes place in the CZE. This idea is supported by the absence of any observation of the mitotic CYCLIN B1;1 in CZE nuclei. Whether the mitotic CYCLIN B1;1 is present in the CZE cytoplasm remains to be determined using immunolocalization and in situ hybridization. Thus, M phase is likely to be bypassed in the CZE during each cell cycle, leading to endoreduplication after stage IV. Endoreduplication is a widespread phenomenon in plants (Traas et al., 1998). It is characteristic of maize endosperm and takes place during reserve storage (Sun et al., 1999). Arabidopsis endosperm does not accumulate reserves, and recent analyses of the degree of ploidy in mature Arabidopsis seed have shown that endoreduplication is very limited (Matzk et al., 2000). This is in agreement with our observation of endoreduplication limited to a few nuclei in the CZE. Recently, new key regulators for endoreduplication were isolated from maize (Sun et al., 1999), Medicago sativa (Cebolla et al., 1999), and Arabidopsis (Jacqmard et al., 1999). The identities of all putative regulators of endoreduplication suggest that M phase is bypassed as a consequence of the lack of activation of the cyclin/cyclin kinase complex. This would prevent the translocation of the complex to the nucleus. If the presence of CYCLIN B1;1 activity in the cytoplasm of the CZE were confirmed, this would suggest that such a mechanism would be involved in CZE endoreduplication.
The distinction between the three mitotic domains is established progressively during development. The mitotic domains are organized along the main polar axis of the endosperm. In cereals, endosperm differentiation is characterized by the definition of distinct territories along the micropylar–chalazal axis (Berger, 1999; Olsen et al., 1999), where specific genes are expressed (Doan et al., 1996; Opsahl-Ferstad et al., 1997; Bonello et al., 2000). The domains MCE, PEN, and CZE defined in Arabidopsis probably are functionally relevant to other species, and genes involved in endosperm polarization remain to be characterized. The central cell is polarized along the micropylar–chalazal axis (Webb and Gunning, 1990; Christensen et al., 1997). As in various animals with embryos that develop from large eggs (Gavis, 1997), the polarization of Arabidopsis endosperm might originate from maternal determinants present in the central cell. Gametophytic mutants in which endosperm proliferation is affected have been isolated (Chaudhury et al., 1997; Grossniklaus et al., 1998; Ohad et al., 1999). The potential effects of imprinting on the expression of the male genome have been shown to affect the extension of the CZE (Scott et al., 1998). Thus, it is likely that gametophytic components regulate endosperm polarity, which in turn would affect locally the regulation of the cell cycle.
METHODS
Plant Material
The Arabidopsis thaliana wild-type ecotype Landsberg erecta was obtained from the Nottingham Arabidopsis Centre (Nottingham, UK). The ecotype C24 was obtained from H. Goodman (University of Cambridge, UK). The marker line FA4C used to report mitosis was constructed as reported by Colon-Carmona et al. (1999). Line KS22 came from a library of enhancer trap lines (Haseloff, 1999).
Plant Growth Conditions
Plants were grown at 20°C in a growth chamber with a 12-hr-day/12-hr-night cycle until they formed rosettes. Flowering was then induced at 22°C with a 16-hr-day/8-hr-night cycle in a greenhouse. Seed grown in vitro were initially sterilized in 5% sodium hypochlorite and stratified on growth medium at 4°C in the dark for 2 to 3 days. The growth medium was 0.8% agarose and 1% sucrose in Murashige and Skoog (1962) medium, pH 5.7. Plants used for confocal microscopic root observation were grown in growth chambers, as described previously (Berger et al., 1998b). Root length measurements were made 10 days after germination.
Construction of a Histone Reporter Gene
Routine DNA manipulations were performed according to Sambrook et al. (1989). An Arabidopsis cDNA coding for HISTONE 2B (H2B) was obtained by reverse transcription–polymerase chain reaction (PCR) amplification of an Arabidopsis C24 cDNA template using the following primers: primer 1, 5′-GGCGGATCCAACAATGGCGAAGGCAGATAAGAAACCAG-3′; and primer 2, 5′-GGCGAATTCTCCAGCTCCAGCAGAACTCGTAAACTTCGTAACCG-3′. The forward primer introduced a BamHI restriction site at the 5′ end of the H2B sequence, and the reverse primer introduced an EcoRI site at the 3′ end. PCR reactions were performed using Vent DNA polymerase (New England Biolabs, Beverly, MA), and the parameters were as follows: 35 cycles at 94°C for 30 sec, 55°C for 1 min, and 72°C for 1 min. One of the clones obtained, which was identical to the gene coding for H2B present in the database (GenBank accession number Y07745), was used to construct the H2B::Yellow Fluorescent Protein (H2B::YFP) vector. For studies involving transient and stable expression in Arabidopsis roots, the PCR product was digested with BamHI and EcoRI and ligated into either BamHI-EcoRI-cut binary vector pBI121 (Bevan, 1984) or the modified vector pBIB (Becker, 1990), both of which contain the gene encoding YFP (Haseloff, 1999) (Figure 1). This resulted in an in-frame fusion of H2B and YFP. In the pBI121 vector, the chimeric sequence was placed under the transcriptional regulation of the cauliflower mosaic virus 35S promoter and the nopaline synthase terminator, and in the pBIB vector, it was placed under the control of the yeast transactivator GAL4 (with upstream activating sequences) and the nopaline synthase terminator. The resultant plasmids were mobilized into Agrobacterium tumefaciens strain LBA 4044 (Clontech, Palo Alto, CA) through electroporation. Kanamycin at 50 mg L−1 was used as a selective agent for pBI121, and hygromycin at 50 mg L−1 was used as a selective agent for pBIB. Arabidopsis root transformation, line selection, and regeneration of transgenic lines were performed according to published procedures (Bechtold et al., 1993).
FACS Analyses
Experiments were performed on wild-type seedlings and seedlings expressing the construct prom35S-H2B::YFP at 7 days after germination. Seedlings were fixed and cut into pieces. Nuclei were extracted and purified through a 50-μm sieve according to instructions from Partec (Jena, Germany). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) according to the instructions from Partec. DAPI fluorescence was measured with a FACScan (PASIII; Partec) using standard optics. Data analyses were performed using Peak and Cluster Analyses and Statistics software (Partec).
Cytological Preparations
Developing seed were fixed in ethanol:acetic acid (3:1) for 1 hr followed by a partial rehydration series in ethanol. For stages younger than the globular embryo stage, fixation was performed in ethanol:acetic acid (8:1) for 30 min. Clearing was obtained after 5 to 30 min in a derivative of Hoyer's medium (chloral hydrate:distilled water:glycerol [80:30:10 g]). The preparations can be used for 1 hr at room temperature and can be conserved at 4°C for 24 hr. Observations were performed with a Nikon (Tokyo, Japan) microscope using differential interference contrast (DIC) optics. Nuclei were counted from images seen on a video monitor attached to the microscope. A glass plate allowed us to mark each nucleus observed throughout the endosperm while shifting from one focal plane to the other. This procedure was used to avoid missing a nucleus or counting one nucleus twice.
β-Glucuronidase (GUS) staining was performed as follows. Siliques were fixed in acetone (90%) for 1 hr at 20°C. Siliques were then infiltrated for 15 min with 2 mM ferricyanide and 2 mM ferrocyanide in Na phosphate buffer. The samples were further assayed for GUS activity according to established procedures. Whole seed were cleared using Hoyer's medium before observation with DIC optics.
To investigate the colocalization of the H2B::YFP fusion protein and DNA, we used propidium iodide. Eight-day-old seedlings were fixed overnight in formaldehyde. Fixed seedlings were incubated for 30 min in 20 mg L−1 RNase A at 37°C. Seedlings were incubated overnight with 5 μg L−1 propidium iodide in 0.1 M arginine, pH 12.4, and further rinsed in 0.1 M arginine, pH 8.0. Seedlings were observed after mounting in glycerol.
Confocal Microscopy
Optical sections were obtained on fixed and fresh roots by using an inverted confocal microscope (model LSM510; Zeiss, Jena, Germany). Fresh roots were observed with a ×40 water immersion apochromat objective (Zeiss), and fixed tissues were observed using a ×63 plan neofluar objective (Zeiss).
Time lapse observation of nuclear division in seed expressing H2B::YFP in the endosperm was obtained using seed prepared as follows. We carefully prepared the material such that seed remained attached to the funiculus and to the septum. Open siliques were kept in small culture chambers and remained alive for at least 12 hr. In certain cases, we managed to keep seed alive for 24 hr. Only seed at younger stages (younger than endosperm stage V) were fragile and difficult to keep alive in our culture conditions. Seed death was easily recognized by arrest of the small movement of nuclei, by loss of green fluorescent protein fluorescence, and eventually by shrinkage of the seed. Fresh seed were observed with a ×20 long working distance objective. Optical sections were obtained after a 15.6-sec scan every 10 min. Time series were obtained overnight, and a nuclear division event was observed once per time series. This lasted ∼30 min, and the results were displayed as a succession of three consecutive optical sections. YFP fluorescence was monitored with a 505- to 550-nm band pass emission filter (488-nm excitation line). Propidium iodide was visualized using the 543-nm excitation line with a long pass 585-nm emission filter.
Image Processing
Transparencies and film negatives were scanned (Nikon Coolscan; Nikon, Tokyo, Japan), and confocal optical sections were imported as psd files and processed using Adobe Photoshop 3.0 (Mountain View, CA).
Acknowledgments
We thank Jim Murray and Margit Menges for help with FACS analyses. We thank Jean-Emmanuel Faure for critical reading of the manuscript. This work was supported by the Institut National de la Recherche Agronomique (INRA), the Gatsby Charitable Foundation, the Biotechnology and Biological Science Research Council (BBSRC), and the ALLIANCE program. F.B. is supported by INRA. This work was supported in part by U.S. Department of Agriculture Grant No. 95-37304-2228 to P.D. The ALLIANCE program and BBSRC and INRA supported collaborations between C.B.-L., F.B., and J.H.
References
- Bajer, A.S. (1958). Cine-micrographic studies on mitosis in endosperm. IV. Exp. Cell Res. 14, 245–256. [DOI] [PubMed] [Google Scholar]
- Bajer, A.S., and Molè-Bajer, J. (1972). Spindle dynamics and chromosome movements. Int. Rev. Cytol. 3 (suppl.), 34.–65. [Google Scholar]
- Bechtold, C., Ellis, P., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, F. (1999). Endosperm development. Curr. Opin. Plant Biol. 2, 28–32. [DOI] [PubMed] [Google Scholar]
- Berger, F., Hung, C.-Y., Dolan, L., and Schiefelbein, J. (1998. a). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194, 235–245. [DOI] [PubMed] [Google Scholar]
- Berger, F., Haseloff, J., Schiefelbein, J., and Dolan, L. (1998. b). Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr. Biol. 8, 421–430. [DOI] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12, 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonello, J.-F., Opsahl-Ferstad, H.-G., Perez, P., Dumas, C., and Rogowsky, P.M. (2000). ESR genes show different levels of expression in the same region of maize endosperm. Gene 246, 219–227. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brown, R.C., Lemmon, B.E., Nguyen, H., and Olsen, O.-A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12, 32–42. [Google Scholar]
- Cebolla, A., Vinardell, J.M., Kiss, E., Oláh, B., Roudier, F., Kondorosi, A., and Kondorosi, E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18, 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64. [Google Scholar]
- Chytilova, E., Macas, J., and Galbraith, D.W. (1999). Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann. Bot. 83, 645–654. [Google Scholar]
- Clarkson, M., and Saint, R.A. (1999). His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18, 457–462. [DOI] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin–GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Conlon, I., and Raff, M. (1999). Size control in animal development. Cell 96, 235–244. [DOI] [PubMed] [Google Scholar]
- Doan, D.N., Linnestad, C., and Olsen, O.A. (1996). Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Mol. Biol. 31, 877–886. [DOI] [PubMed] [Google Scholar]
- Doonan, J.H. (1992). Cell division in Aspergillus. J. Cell Sci. 103, 599–611. [DOI] [PubMed] [Google Scholar]
- Edgar, B.A., and Lehner, C.F. (1996). Developmental control of cell cycle regulators: A fly's perspective. Science 274, 1646–1651. [DOI] [PubMed] [Google Scholar]
- Enger, M.D., Tobey, R.A., and Saponara, A.G. (1968). RNA synthesis in Chinese hamster cells. I. Differential synthetic rate for ribosomal RNA in early and late interphase. J. Cell Biol. 36, 583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V.E., Odell, G.M., and Edgar, B.A. (1993). Mitosis and morphogenesis in the Drosophila embryo. In The Development of Drosophila melanogaster, M. Bate and A. Martinez Arias, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 149–300.
- Friedman, W.E. (1992). Evidence of a pre-angiosperm origin of endosperm: Implications for the evolution of flowering plants. Science 255, 336–339. [DOI] [PubMed] [Google Scholar]
- Galloni, M., and Edgar, B. (1999). Cell autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126, 2365–2375. [DOI] [PubMed] [Google Scholar]
- Gälwailer, L., Conlan, R.S., Mader, P., Palme, K., and Moore, I. (2000). The DNA-binding activity of GAL4 is inhibited by methylation of the GAL4 binding site in plant chromatin. Plant J. 23, 143–157. [DOI] [PubMed] [Google Scholar]
- Gavis, E.R. (1997). Expeditions to the pole: RNA localization in Xenopus and Drosophila. Trends Cell Biol. 7, 485–492. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J., Lambert, G.M., and Galbraith, D.W. (1997). Characterization of the targeted nuclear accumulation of GFP within the cells of transgenic plants. Plant J. 12, 685–696. [Google Scholar]
- Grosshans, J., and Wieschaus, E. (2000). A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101, 523–531. [DOI] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Guyer, D., Tuttle, A., Rouse, S., Volrath, S., Johnson, M., Potter, S., Görlach, J., Goff, S., Crossland, L., and Ward, E. (1998). Activation of latent transgenes in Arabidopsis using a hybrid transcription factor. Genetics 149, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J. (1999). GFP variants for multispectral imaging of living cells. Methods Cell Biol. 58, 139–151. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley, R.P., and Murray, J.A.H. (1999). The plant cell cycle. Curr. Opin. Plant Biol. 2, 440–446. [DOI] [PubMed] [Google Scholar]
- Jacqmard, A., De Veylder, L., Segers, G., de Almeida Engler, J., Bernier, G., Van Montagu, M., and Inze, D. (1999). Expression of CKS1At in Arabidopsis thaliana indicates a role for the protein in both the mitotic and the endoreduplication cycle. Planta 207, 496–504. [DOI] [PubMed] [Google Scholar]
- Johnston, L.A., and Edgar, B.A. (1998). Wingless and Notch regulate cell cycle arrest in the developing Drosophila wing. Nature 394, 82–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jotereau, F.V., and Le Douarin, N.M. (1978). The developmental relationship between osteocytes and osteoclasts: A study using the quailchick nuclear markers in endochondral ossification. Dev. Biol. 63, 253–265. [DOI] [PubMed] [Google Scholar]
- Kanda, T., Sullivan, K.F., and Wahl, G.M. (1998). Histone–GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377–385. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.-H. (1998). A GFP–mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Grandjean, O., Jonal, C., Kien, K., and Traas, J. (1998). Cellular parameters of the shoot apical meristem in Arabidopsis. Plant Cell 10, 1375–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M.A., and Larkins, B.A. (1993). Endosperm origin, development, and function. Plant Cell 5, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990. a). Development of the free-nuclear endosperm in Arabidopsis thaliana. Arabidopsis Inf. Serv. 27, 53–64. [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990. b). Endosperm cellularization in Arabidopsis thaliana. Arabidopsis Inf. Serv. 27, 65–72. [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T.-H., McCubbin, A.G., and Cyr, R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., Curado, S., Ephrussi, A., and Rorth, P. (2000). Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating String/Cdc25 proteolysis. Cell 101, 511–522. [DOI] [PubMed] [Google Scholar]
- Matzk, F., Meister, A., and Schubert, I. (2000). An efficient screen for reproductive pathways using mature seeds in monocots and dicots. Plant J. 21, 97–108. [DOI] [PubMed] [Google Scholar]
- Meyerowitz, E.M. (1997). Controls of cell division patterns in developing shoots and flowers of Arabidopsis thaliana. Cold Spring Harbor Symp. Quant. Biol. 62, 369–375. [PubMed] [Google Scholar]
- Mironov, V., De Veylder, L., Van Montagu, M., and Inzé, D. (1999). Cyclin-dependent kinases and cell division in plants: The nexus. Plant Cell 11, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, I., Gälweiler, L., Grosskopf, D., Schell, J., and Palme, K. (1998). A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. USA 95, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neufeld, T.P., and Edgar, B.A. (1998). Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 6, 784–790. [DOI] [PubMed] [Google Scholar]
- O'Farrell, P.H., Edgar, B.A., Lakich, D., and Lehner, C.F. (1989). Directing cell division during development. Science 246, 635–640. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Micheali, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, O.-A., Brown, R.C., and Lemmon, B.E. (1995). Pattern and process of wall formation in developing endosperm. Bioessays 17, 812–830. [Google Scholar]
- Olsen, O.-A., Linnestad, C., and Nichols, S.E. (1999). Developmental biology of the cereal endosperm. Trends Plant Sci. 4, 253–257. [DOI] [PubMed] [Google Scholar]
- Opsahl-Ferstad, H.-G., Le Deunff, E., Dumas, C., and Rogowsky, P.M. (1997). ZmMCE, a novel endosperm-specific gene ex-pressed in a restricted region around the maize embryo. Plant J. 12, 235–246. [DOI] [PubMed] [Google Scholar]
- Otegui, M., and Staehelin, L.A. (2000). Syncytial-type cell plates: A novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12, 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schneitz, K., Hülskamp, M., and Pruitt, R.E. (1995). Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 7, 731–743. [Google Scholar]
- Schultz, P., and Jensen, W.A. (1974). Capsella embryogenesis: The development of the free nuclear endosperm. Protoplasma 80, 183–205. [Google Scholar]
- Scott, A., Wyatt, S., Tsou, P.-L., Robertson, D., and Strömgren, A. (1999). Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques 26, 1127–1132. [DOI] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341. [DOI] [PubMed] [Google Scholar]
- Seher, T., and Leptin, M. (2000). Tribbles, a cell cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr. Biol. 10, 623–629. [DOI] [PubMed] [Google Scholar]
- Su, T.T., Campbell, S.D., and O'Farrell, P.H. (1998. a). Cell cycle program in Drosophila germ cells. Dev. Biol. 196, 160–170. [DOI] [PubMed] [Google Scholar]
- Su, T.T., Sprenger, F., DiGregorio, P.J., Campbell, S.D., and O'Farrell, P.H. (1998. b). Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation and is associated with localized dephosphorylation. Genes Dev. 12, 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traas, J., Hülskamp, M., Gendreau, E., and Höfte, H. (1998). Endoreduplication and development: Rule without dividing? Curr. Opin. Plant Biol. 1, 498–503. [DOI] [PubMed] [Google Scholar]
- Vandendries, R. (1909). Contribution à l'histoire du développement des crucifères. Cellule 25, 414–458. [Google Scholar]
- Vijayaraghavan, M.R., and Prabhakar, K. (1984). The endosperm. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 319–376.
- Webb, M.C., and Gunning, B.E.S. (1990). Embryo sac development in Arabidopsis thaliana. I. Megasporogenesis including the microtubular cytoskeleton. Sex. Plant Reprod. 3, 244–256. [Google Scholar]
- Wolniak, S.M. (1987). Lithium alters mitotic progression in stamen hair cells of Tradescantia in a time-dependent and reversible fashion. Eur. J. Cell Biol. 44, 286–293. [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., Chan, A., Sweeney, M., and Dawe, K. (1997). Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]