Abstract
Post-transcriptional gene silencing (PTGS) is a sequence-specific RNA degradation mechanism that is widespread in eukaryotic organisms. It is often associated with methylation of the transcribed region of the silenced gene and with accumulation of small RNAs (21 to 25 nucleotides) homologous to the silenced gene. In plants, PTGS can be triggered locally and then spread throughout the organism via a mobile signal that can cross a graft junction. Previously, we showed that the helper component–proteinase (HC-Pro) of plant potyviruses suppresses PTGS. Here, we report that plants in which PTGS has been suppressed by HC-Pro fail to accumulate the small RNAs associated with silencing. However, the transgene locus of these plants remains methylated. Grafting experiments indicate that HC-Pro prevents the plant from responding to the mobile silencing signal but does not eliminate its ability to produce or send the signal. These results demonstrate that HC-Pro functions downstream of transgene methylation and the mobile signal at a step preceding accumulation of the small RNAs.
INTRODUCTION
Post-transcriptional gene silencing (PTGS) is a sequence-specific RNA degradation mechanism first discovered in transgenic plants (Napoli et al., 1990; Smith et al., 1990; van der Krol et al., 1990). Related processes have been found in diverse eukaryotic organisms including Neurospora, in which it is called quelling, and a variety of animal systems, in which it is referred to as RNA interference or RNAi (Vaucheret et al., 1998; Fire, 1999; Grant, 1999; Kooter et al., 1999; Ding, 2000; Matzke et al., 2001). Sequence-specific RNA degradation is triggered by double stranded RNA (dsRNA) in a variety of organisms (Montgomery and Fire, 1998; Waterhouse et al., 1998; Sharp, 1999; Bass, 2000; Matzke et al., 2001). In plants, PTGS can be induced by RNA viruses, many of which replicate via dsRNA intermediates. Finally, in both plants and Caenorhabditis elegans, the process can be triggered locally and then spread to distant parts of the organism (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Fire et al., 1998; Jorgensen et al., 1998; Palauqui and Vaucheret, 1998; Voinnet et al., 1998). The relatedness of these sequence-specific RNA degradation processes in different organisms is evidenced by their requirement for a conserved set of gene products (Sharp and Zamore, 2000; Matzke et al., 2001), including a protein with homology to translation factor eIF2C (Tabara et al., 1999; Catalanotto et al., 2000; Fagard et al., 2000), an RNA-dependent RNA polymerase (RdRp) (Cogoni and Macino, 1999a; Dalmay et al., 2000; Mourrain et al., 2000; Smardon et al., 2000), and proteins with homology to DNA helicases and RNase D (Cogoni and Macino, 1999b; Ketting et al., 1999). However, at this point, neither the roles of these various gene products nor the mechanisms for induction, maintenance, and spread of sequence-specific RNA degradation are clearly understood.
Several molecular features characterize the sequence-specific RNA degradation processes found in diverse organisms. Studies in both plants and Drosophila have shown that silencing is accompanied by the accumulation of small RNAs (21 to 25 nucleotides) of both sense and antisense orientation that are homologous to the silenced locus (or input dsRNA) (Hamilton and Baulcombe, 1999; Bass, 2000; Hammond et al., 2000; Zamore et al., 2000). In plants, transgene-induced PTGS is associated with methylation of the transcribed region of the affected transgene (Wassenegger and Pélissier, 1998; Kooter et al., 1999; Matzke et al., 2001), and it is possible that some sort of chromatin modification is also a feature of silencing in other organisms (Wolffe and Matzke, 1999). Methylation recurs in subsequent generations of a silenced organism and may be involved in the maintenance of the silenced state. It has been suggested that the small RNAs associated with PTGS may comprise the mobile silencing signal that spreads silencing throughout the organism and/or the signal between cytoplasm and nucleus that leads to perpetuation of transgene methylation.
Certain plant viruses encode proteins that can suppress PTGS (Anandalakshmi et al., 1998; Beclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Voinnet et al., 1999, 2000). The identification of such proteins provides a novel approach to understanding silencing in plants. Here, we used one such suppressor of PTGS, the helper component–proteinase (HC-Pro) protein encoded by a plant potyvirus, to help dissect the steps in the plant silencing pathway. We determined where HC-Pro acts to suppress transgene-induced gene silencing relative to known hallmarks of the process: the small RNAs, transgene methylation, and the mobile silencing signal. The results suggest that the accumulation of high levels of the small RNAs is not required for either transgene methylation or the production or transmission of the mobile silencing signal.
RESULTS
The β-Glucuronidase–Silenced Transgenic Line 6b5: A Model System
To determine where HC-Pro acts with respect to the known hallmarks of PTGS, we needed a well-characterized system of PTGS. The transgenic tobacco line 6b5 (Elmayan and Vaucheret, 1996) fulfills this requirement on the basis of several criteria: (1) it is silenced for the uidA transgene encoding the easily assayed reporter enzyme β-glucuronidase (GUS); (2) silencing of GUS in line 6b5 is at the post-transcriptional level and occurs whether the plant is homozygous or hemizygous for the transgene (Elmayan and Vaucheret, 1996); (3) this line was used in the original grafting experiments that demonstrated the mobile silencing signal in plants (Palauqui et al., 1997); and (4) high levels of the small RNAs associated with PTGS accumulate in this line (Hamilton and Baulcombe, 1999).
The tobacco etch virus (TEV) P1/HC-Pro sequence has been previously demonstrated to suppress transgene-induced PTGS in several other silenced lines (Anandalakshmi et al., 1998; Kasschau and Carrington, 1998). To determine the effect of HC-Pro on the silencing of GUS in line 6b5, we crossed a homozygous 6b5 transgenic plant with a homozygous transgenic tobacco plant (line X-27-8) expressing high levels of the TEV P1/HC-Pro sequence. The offspring of this cross (termed 6b5 × HC-Pro) have one copy of the silenced GUS locus from the 6b5 transgenic line and one copy of the P1/HC-Pro locus from the X-27-8 transgenic line. As a control for these experiments, the 6b5 line was crossed with a nontransformed (NT) plant of the same genetic background as the X-27-8 line (Nicotiana tabacum cv xanthi NC). The offspring of this control cross (termed 6b5 × NT) have one copy of the silenced GUS locus from the 6b5 transgenic line but no copies of the P1/HC-Pro transgene. Plants from the F1 generation of each cross were then characterized for the expression of GUS using both quantitative GUS assays and RNA gel blot analyses. Because the onset of PTGS in each generation of the 6b5 line occurs as the plants mature, we performed this characterization as a function of time after germination to follow the progression of silencing in these lines.
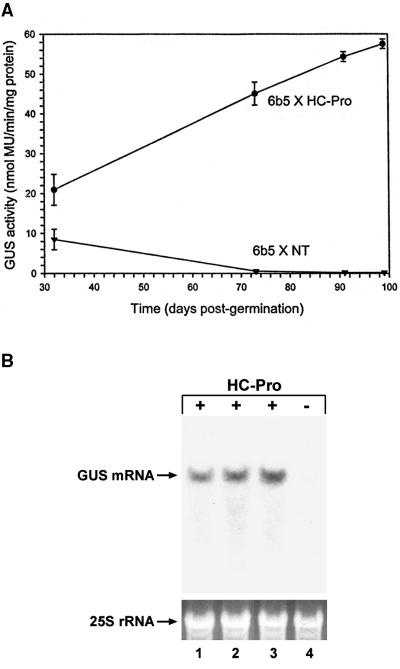
6b5 × NT plants displayed a moderate level of GUS activity at 32 days after germination, and the activity declined 30-fold over the next 2 months to a nearly undetectable level (Figure 1A, 6b5 × NT; each time point is an average of activities from eight individual plants). In contrast, 6b5 × HC-Pro plants displayed significantly higher GUS activity at the initial time point, and the activity continued to increase over the next 2 months to a level ∼200-fold higher than in 6b5 × NT plants at ∼3 months after germination (Figure 1A, 6b5 × HC-Pro; each time point is an average of activities from three individual plants). As expected, the GUS mRNA failed to accumulate in 6b5 × NT plants (Figure 1B, lane 4) but accumulated to high levels in the 6b5 × HC-Pro plants (Figure 1B, lane 3). Suppression of silencing was also obtained when a P1/HC-Pro–expressing locus was introduced into line 6b5 by crossing with either of two other independent transgenic lines, TEV-B and TEV-I (Figure 1B, lanes 1 and 2, respectively). Both the TEV-B and the TEV-I transgenic lines have been previously shown to suppress PTGS (Anandalakshmi et al., 1998). Thus, introduction of a P1/HC-Pro–expressing locus into line 6b5 is accompanied by dramatic increases in both GUS enzyme activity and the accumulation of the GUS mRNA, indicating that introduction of the P1/HC-Pro locus interferes with the post-transcriptional silencing of the GUS transgene in the subsequent generation.
Figure 1.
Expression of HC-Pro Suppresses Post-Transcriptional Gene Silencing of the GUS Transgene in Line 6b5.
(A) Time-course analysis of GUS enzyme activity in transgenic tobacco in the presence (6b5 × HC-Pro) or absence (6b5 × NT) of an HC-Pro–expressing locus. Each point represents the average of activity in three (6b5 × HC-Pro) or seven (6b5 × NT) individual plants. The standard deviation is indicated for all points, but it is too small to see at the later times for the 6b5 × NT line. MU, 4-methylumbelliferone.
(B) RNA gel blot showing the level of GUS mRNA in transgenic plants in the presence or absence of an HC-Pro–expressing locus. Total RNA was isolated from offspring of crosses between the 6b5 GUS-silenced line and three independent transgenic lines expressing high levels of HC-Pro (lane 1, 6b5 × TEV-B; lane 2, 6b5 × TEV-I; lane 3, 6b5 × X-27-8) or a nontransformed control plant (lane 4, 6b5 × NT). Ethidium bromide staining of 25S rRNA is shown as a loading control; 10 μg total RNA was loaded per lane.
HC-Pro Suppression of Silencing Interferes with Accumulation of the 21- to 25-Nucleotide RNAs Associated with PTGS
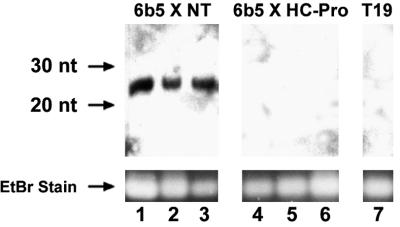
A species of small RNAs (21 to 25 nucleotides) is associated with PTGS in a variety of silencing systems (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Zamore et al., 2000), and these RNAs have been shown to accumulate to high levels in the 6b5 transgenic line (Hamilton and Baulcombe, 1999). To determine if these small RNAs accumulate when PTGS has been suppressed by introduction of an HC-Pro–expressing locus, we examined the 6b5 × NT and 6b5 × HC-Pro plants described above for the presence of the GUS small RNAs. The GUS-expressing transgenic line T19 was used as a negative control for these experiments because it has been shown to lack the small RNAs (Hamilton and Baulcombe, 1999). Nucleic acids were isolated from the plants (seven individual plants of each kind) and enriched for low molecular weight species, and the samples were examined for accumulation of the small GUS RNAs using RNA gel blot analysis. As expected, no small RNAs were detectable in the T19 control plants (Figure 2, lane 7), whereas high levels accumulated in the 6b5 × NT plants (Figure 2, lanes 1 to 3). However, in the 6b5 × HC-Pro plants, in which silencing of GUS was suppressed by expression of HC-Pro, no small GUS RNAs were detectable (Figure 2, lanes 4 to 6). These results were highly reproducible. All seven individual 6b5 × NT plants accumulated high levels of small RNAs, whereas none of the seven individual 6b5 × HC-Pro plants accumulated detectable levels of these RNAs. Densitometric analyses suggest that accumulation of the small RNAs in the 6b5 × HC-Pro plants would have been detected if present at a level two orders of magnitude lower than that in the 6b5 × NT plants. Thus, HC-Pro suppression of transgene-induced gene silencing is correlated with a dramatic decrease in the accumulation of the small RNAs that are associated with PTGS. This result suggests that HC-Pro suppresses silencing at a step upstream of the accumulation of the small RNAs.
Figure 2.
Suppression of PTGS by HC-Pro Interferes with the Accumulation of the Small RNAs Associated with Silencing.
RNA gel blot showing the accumulation of small GUS RNAs in GUS-silenced plants (6b5 × NT; lanes 1 to 3) but not in plants in which silencing has been suppressed by HC-Pro (6b5 × HC-Pro; lanes 4 to 6). RNA isolated from three individual plants of each line is shown. Lane 7 shows that the small GUS RNAs are not detected in the GUS-expressing control line T19. The migration of 20- and 30-nucleotide (nt) DNA oligomers is indicated on the left. Ethidium bromide (EtBr) staining of the predominant RNA species found in samples enriched for small RNAs (see Methods) is shown as a loading control.
Transgene Methylation Is Reset in Line 6b5 and Recurs within Days after Germination in the Presence or Absence of HC-Pro
PTGS is often correlated with methylation of the transcribed region of the silenced transgene (Wassenegger and Pélissier, 1998; Kooter et al., 1999). Because PTGS is reset at every generation in line 6b5 and recurs in a developmental fashion (Figure 1A), it is possible that transgene methylation is also reset and then restored in the next generation. However, reinitiation of PTGS begins very soon after germination, and transgene methylation in line 6b5 has not previously been examined at early time points. To determine if methylation of the GUS transgene in line 6b5 is reset during reproduction and if HC-Pro interferes with this process, we developed a polymerase chain reaction (PCR)–based technique to detect methylation using the small amounts of genomic DNA that can be obtained from very small individual tobacco plants at early times after germination.
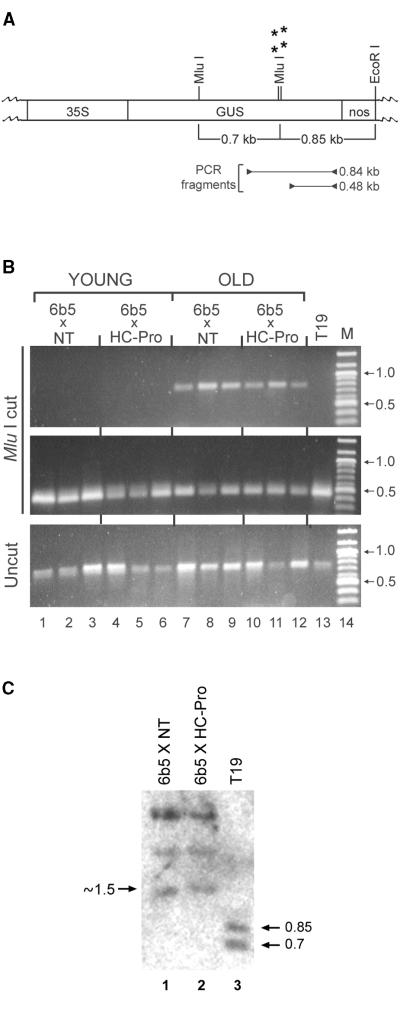
Genomic DNA was isolated from individual 6b5 × NT and 6b5 × HC-Pro tobacco plants at 1, 5, or 45 days after germination and digested with the methylation-sensitive enzyme MluI. Three primers were then used to amplify two partially overlapping portions of the GUS transgene. To ascertain the presence of methylation at specific MluI sites, a reverse primer near the 3′ end of the GUS coding region was used in conjunction with a forward primer located 0.84 kilobase pair (kbp) away at a site upstream of two closely spaced MluI sites that were shown to be methylated in the older 6b5 plants (Figure 3A, PCR fragment of 0.84 kb; MluI sites that are methylated in older 6b5 plants are indicated by asterisks). The same reverse primer was also used in conjunction with a second forward primer located 0.48 kbp upstream of the reverse primer but downstream of the MluI sites that were shown to be methylated in older 6b5 plants (Figure 3A, PCR fragment of 0.48 kb). If the MluI sites in a particular plant were methylated, then the enzyme would be unable to cut the genomic DNA, and both a 0.84- and a 0.48-kbp fragment would be amplified. However, if the DNA were not methylated at those sites, then the enzyme would cut and only the small fragment would be amplified. All undigested DNAs should also produce the large fragment because the selective disappearance of this band is expected to depend on digestion of the DNA at the MluI sites located between the two forward primers.
Figure 3.
Transgene Methylation Occurs Even When Silencing Is Suppressed by HC-Pro.
(A) A partial restriction map of the GUS gene is indicated. 35S indicates the location of the cauliflower mosaic virus 35S promoter, GUS indicates the coding region for the GUS transgene, and nos indicates the nopaline synthase terminator. Asterisks mark the location of the MluI sites that are methylated in both the 6b5 × NT plants (silenced for the GUS transgene) and the 6b5 × HC-Pro plants (silencing of the GUS transgene suppressed by HC-Pro). The location of PCR primers used to generate DNA fragments of 0.84 and 0.48 kbp in (B) are indicated.
(B) Agarose gel electrophoresis of PCR fragments amplified from MluI-cut genomic DNA samples (top and middle panels) or the same DNAs before digestion (bottom panel) isolated from 6b5 × NT and 6b5 × HC-Pro plants at 1 day after germination (lanes 1 to 6) or 45 days after germination (lanes 7 to 12). DNA isolated from the GUS-expressing control line T19 at 45 days after germination is shown in lane 13. The top panel shows amplification of the 0.84-kbp fragment shown in (A) and is indicative of methylation at the MluI sites marked by asterisks in (A). The middle panel shows amplification of the 0.48-kbp fragment from the same MluI-cut samples shown in the top panel. The bottom panel shows amplification of the 0.84-kbp band from the same DNAs shown in the top and middle panels before digestion. Lane 14 in each panel shows the migration of molecular weight standards in kbp, two of which are identified in each panel for comparison.
(C) DNA gel blot of EcoRI-MluI–cut DNA from GUS-silenced 6b5 × NT plants (lane 1), 6b5 × HC-Pro plants, in which silencing has been suppressed by HC-Pro (lane 2), or T19, a GUS-expressing control line (lane 3). The position of the 0.85- and 0.7-kbp DNA fragments resulting from complete digestion with the methylation-sensitive MluI restriction enzyme are indicated at right. The position of the ∼1.5-kbp band predicted if MluI digestion is incomplete is indicated at left.
Using this method, we found that MluI-digested DNA from all of the 45-day-old 6b5 plants produced the pattern predicted for methylated DNA: amplification of both the long (Figure 3B, top panel, lanes 7 to 12) and the short (Figure 3B, middle panel, lanes 7 to 12) fragments. Furthermore, the methylation occurred regardless of the presence of HC-Pro (cf. lanes 7 to 9 showing three independent 6b5 × NT samples with lanes 10 to 12 showing three independent 6b5 × HC-Pro samples). As expected, MluI-cut DNA from the GUS-expressing T19 control plants produced the pattern predicted for unmethylated DNA: the large fragment was not amplified (Figure 3B, top panel, lane 13) but the small fragment was (Figure 3B, middle panel, lane 13). The loss of large fragment amplification in T19 DNA depended on MluI digestion because the band was amplified from equivalent amounts of the same samples before digestion (Figure 3B, bottom panel, lane 13).
In contrast to the results obtained at 45 days after germination, none of the 6b5 MluI-cut DNA samples were methylated at 1 day after germination, regardless of the presence or absence of HC-Pro, as assayed by failure to amplify detectable levels of the large fragment (Figure 3B, top panel, lanes 1 to 6). As with the DNA from the unmethylated T19 control plants, the small fragment was amplified in these samples (Figure 3B, middle panel, lanes 1 to 6), and the large fragment was amplified from equivalent amounts of the samples before MluI digestion (Figure 3B, bottom panel, lanes 1 to 6). At 5 days after germination, ∼80 to 90% of the individual 6b5 × NT plants (eight of nine individuals tested) and 6b5 × HC-Pro plants (eight of 10 individuals tested) were methylated as assayed by this method (data not shown). These results suggest that these particular MluI sites within the 3′ region of the GUS trangene are not methylated at early times after germination and that de novo methylation occurs at these sites within a few days after germination. This resetting of methylation occurs even when PTGS of the transgene is suppressed by the presence of HC-Pro.
Transgene Methylation Is Maintained when Silencing Is Suppressed by HC-Pro
To confirm that HC-Pro does not interfere with transgene methylation, DNA gel blot analysis was used to determine the methylation status of the GUS transgene in the 6b5 × NT and 6b5 × HC-Pro plants. The GUS-expressing transgenic line T19 was again used as a negative control because the GUS locus is not methylated in this line (English et al., 1996; Figure 3B, lane 13). Genomic DNA extracted from these plants was digested with EcoRI and the methylation-sensitive restriction enzyme MluI and then examined by DNA gel blot analysis. Complete digestion of the MluI and EcoRI sites within the GUS locus results in bands of 0.7 and 0.85 kbp (Figure 3A; English et al., 1996), and we detected these two bands in control line T19 (Figure 3C, lane 3). This result confirms that the MluI sites within the T19 GUS transgene are not methylated. In contrast, MluI-EcoRI digestion of the GUS-silenced 6b5 × NT plants failed to produce the 0.7- and 0.85-kbp bands, giving instead three larger bands of ∼1.5, 2.1, and 3.5 kbp (Figure 3C, lane 1). The two larger of these three new bands result from cleavage upstream of the GUS coding region within the tobacco genomic DNA, and we are therefore unable to predict their expected size. The 1.5-kbp band, however, is approximately the size expected if the two MluI sites marked by asterisks in Figure 3A are both methylated and the upstream MluI site is unmethylated. A band of this size has been reported for the GUS-silenced transgenic line T4 and attributed to methylation at the two downstream MluI sites (English et al., 1996). This result, together with the PCR analyses, suggests that the GUS locus in the 6b5 × NT plants, like that in the T4 GUS-silenced line, is methylated at specific MluI sites within the 3′ coding region of the GUS transgene. As predicted by the previous PCR analysis, a banding pattern identical to that in the GUS-silenced 6b5 × NT plants was seen in the 6b5 × HC-Pro plants (Figure 3C, lane 2). Reprobing the blot with a nitrite reductase probe showed that all the DNA samples had been digested to the same extent (data not shown). Thus, the two 3′ proximal MluI sites that were methylated in the GUS coding region of 6b5 × NT plants were also methylated in the 6b5 × HC-Pro plants. The failure of HC-Pro to interfere with the methylation status of the GUS transgene, even though it suppressed silencing of that transgene, suggests that HC-Pro does not suppress PTGS by actively demethylating the transgene or by preventing the initial onset of methylation. Thus, HC-Pro is predicted to suppress PTGS at a step downstream of the onset and maintenance of transgene methylation.
Suppression of PTGS by HC-Pro Does Not Eliminate the Ability to Produce and Send the Mobile Silencing Signal
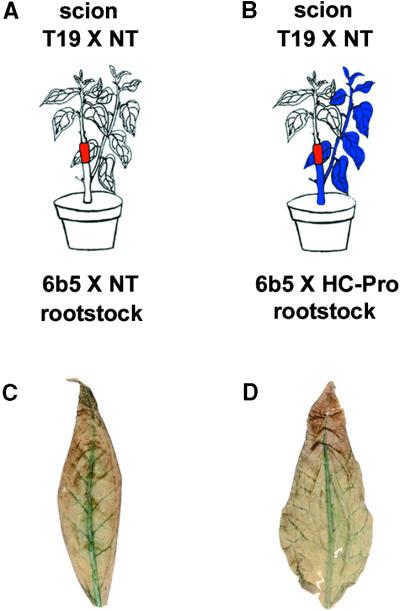
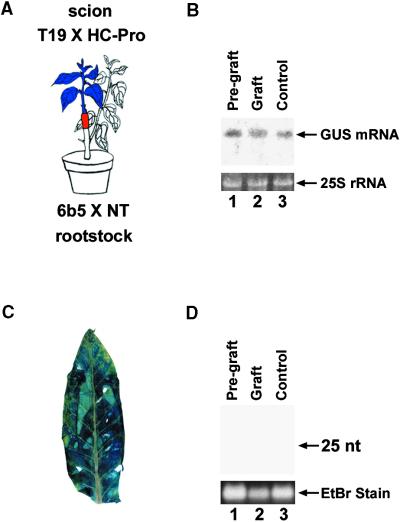
It is known that PTGS involves production of a mobile signal molecule that can cross a graft junction and induce silencing of a homologous transgene in a graft (Palauqui et al., 1997; Palauqui and Vaucheret, 1998; Voinnet et al., 1998). To determine if HC-Pro suppression of PTGS interferes with the production or transmission of this mobile silencing signal, we devised a set of grafting experiments (Figures 4A and 4B). The scion in both experiments was the GUS-expressing transgenic line T19 × NT. The rootstocks were either the 6b5 × NT control plants in which GUS is silenced (Figure 4A) or the 6b5 × HC-Pro plants in which the GUS silencing is suppressed by HC-Pro (Figure 4B).
Figure 4.
Suppression of PTGS by HC-Pro Does Not Interfere with the Production or Transmission of the Mobile Silencing Signal.
(A) Diagram of control experiment showing the effect of grafting a GUS-expressing plant (T19 × NT) onto a GUS-silenced rootstock (6b5 × NT): the GUS transgene in the scion becomes silenced.
(B) Diagram of experiment showing the effect of grafting a GUS-expressing plant (T19 × NT) onto a rootstock (6b5 × HC-Pro) in which silencing of GUS has been suppressed by HC-Pro: the GUS transgene in the scion becomes silenced even though silencing in the rootstock itself has been suppressed.
(C) GUS-histochemical staining of a leaf from the scion in the grafting experiment shown in (A).
(D) GUS-histochemical staining of a leaf from the scion in the grafting experiment shown in (B).
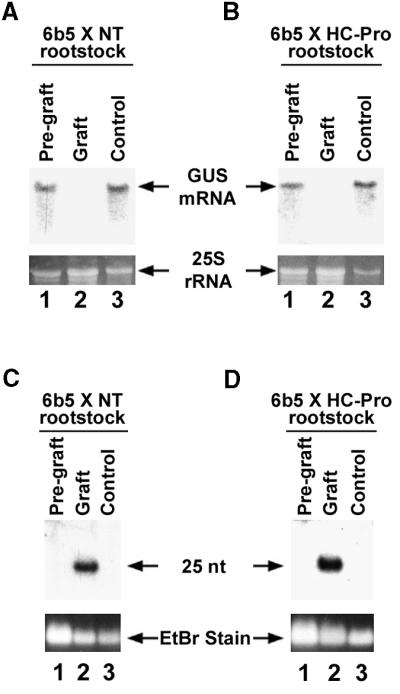
As expected, when the GUS-silenced control plant was used as rootstock, it was consistently able to produce and send the mobile silencing signal and induce silencing of the GUS transgene in the scion. Silencing in the scion was evidenced by the absence of histochemical staining for GUS activity in leaves of the scion (Figure 4C), by the failure to detect GUS mRNA in these leaves (Figure 5A, lane 2, Graft), and by the presence of the small RNAs in those same leaves (Figure 5C, lane 2, Graft). These results were reproducibly found in two independent experiments involving seven grafted plants. As a control, we showed that just before being grafted, each GUS-expressing T19 × NT plant used as a scion was accumulating GUS mRNA (Figure 5A, lane 1, Pre-graft) but not the small RNAs associated with PTGS (Figure 5C, lane 1, Pre-graft). Furthermore, the plants used as the source of scions in each grafting experiment were allowed to grow back and were sampled again at the same time as the grafted scions. In all cases, these nongrafted T19 × NT controls continued to accumulate GUS mRNA (Figure 5A, lane 3, Control) and failed to accumulate the small RNAs (Figure 5C, lane 3, Control).
Figure 5.
Suppression of PTGS by HC-Pro Does Not Interfere with the Production or Transmission of the Mobile Silencing Signal: Molecular Analyses.
(A) RNA gel blot showing accumulation of GUS mRNA in T19 × NT plants before and after being grafted onto GUS-silenced 6b5 × NT rootstocks. Total RNA was isolated from T19 × NT plants just before grafting (lane 1, Pre-graft). Four weeks after grafting, RNA was isolated from the T19 × NT scion (lane 2, Graft) and from the plant used as the source of the scion (lane 3, Control).
(B) RNA gel blot showing accumulation of GUS mRNA in T19 × NT plants before and after being grafted onto rootstocks (6b5 × HC-Pro) in which GUS silencing was suppressed by HC-Pro. Total RNA was isolated from T19 × NT plants just before grafting (lane 1, Pre-graft). Four weeks after grafting, RNA was isolated from the scion (lane 2, Graft) and from the plant used as the source of the scion (lane 3, Control).
(C) RNA gel blot showing the accumulation of small RNAs in T19 × NT plants before and after being grafted onto silenced rootstocks (6b5 × NT). The T19 × NT pre-graft, graft, and control RNAs described in (A) were fractionated as described in Methods to enrich for small RNA and examined for 21- to 25-nucleotide (nt) RNAs derived from the GUS gene.
(D) RNA gel blot analyses showing the level of small RNAs in T19 × NT plants before and after being grafted onto rootstocks (6b5 × HC-Pro) in which GUS silencing was suppressed by HC-Pro. The T19 × NT pre-graft, graft, and control samples described in (B) were fractionated as described in Methods to enrich for small RNA and examined for 21- to 25-nucleotide RNAs derived from the GUS gene.
Ethidium bromide (EtBr) staining of 25S rRNA was used as loading control for the RNA gel blots shown in (A) and (B). Ethidium bromide staining of the predominant band in samples enriched for small RNAs was used as loading control in the RNA gel blots shown in (C) and (D).
Interestingly, the 6b5 × HC-Pro rootstock was also consistently able to silence its scion (Figures 4B, 4D, 5B, and 5D) even though silencing in the rootstock itself was suppressed by HC-Pro. A T19 × NT scion grafted onto a 6b5 × HC-Pro rootstock behaved exactly like one grafted onto a GUS-silenced 6b5 × NT rootstock (Figure 4, compare panels C and D; Figure 5, compare panels A and B; Figure 5, compare panels C and D). Again, this result was highly reproducible, occurring in two independent experiments involving seven grafted plants. Thus, even though HC-Pro suppresses silencing, it does not eliminate the ability to produce and send the mobile silencing signal.
Similar results were obtained using different transgenic lines. One set of experiments used T19 × NT scions as in the above-described experiments, but the rootstocks were derived from a different GUS-silenced line (line 106; Ulker et al., 1999). The results of these grafts were nearly identical to those reported above: the 106 × HC-Pro rootstock, in which PTGS was suppressed by HC-Pro, was as capable of silencing the T19 × NT scion as the 106 × NT rootstock (data not shown). A third series of experiments used 6b5 × HC-Pro and 6b5 × NT rootstocks but had different GUS-expressing lines as scions (lines 23b9 and 23b10; Elmayan and Vaucheret, 1996). Again, we found that silencing of GUS was induced in the scion. In this case, however, the silencing induced by the 6b5 × HC-Pro rootstocks was not as great as that induced by the silenced 6b5 × NT rootstocks and was confined primarily to the veins of the leaf (data not shown). This result suggests that HC-Pro may, in some circumstances, either reduce the amount or delay the transmission of the mobile silencing signal. All together, these results indicate that suppression of PTGS by HC-Pro does not eliminate production of the mobile silencing signal or prevent its systemic movement.
HC-Pro Works Downstream of the Mobile Silencing Signal
Our result that HC-Pro does not eliminate the mobile silencing signal suggests that suppression of silencing by HC-Pro reflects an inability to respond to the signal. To determine if a transgenic line that expresses HC-Pro can still respond to the silencing signal, we again used grafting experiments, but in this case, the HC-Pro transgene was expressed in the scion instead of the rootstock (Figure 6A). The rootstock in these experiments was 6b5 × NT, a line that is silenced for GUS and known to produce and send the silencing signal. The plants used for the scion (T19 × HC-Pro) contained an expressing GUS locus from line T19 and an HC-Pro–expressing locus from line U-6B. Four weeks after grafting, the T19 × HC-Pro scions displayed GUS enzyme activity (Figure 6C), accumulated GUS mRNA at approximately the same level as before grafting (Figure 6B, compare lanes 1 and 2), and failed to accumulate small RNAs (Figure 6D, lane 2). These results were consistently found in two independent experiments involving seven grafted plants. Thus, a scion expressing HC-Pro fails to respond to the mobile silencing signal and continues to express the targeted transgene. This result suggests that HC-Pro interferes either with the perception of the mobile silencing signal by the grafted scion or with the ability of the plant to respond to that signal. Thus, HC-Pro suppression of PTGS occurs at a step downstream of the mobile silencing signal.
Figure 6.
Expression of HC-Pro Prevents Silencing in Response to the Mobile Silencing Signal from a Silenced Rootstock.
(A) Diagram of experiment showing the effect of grafting a plant expressing both GUS and HC-Pro (T19 × HC-Pro) onto a GUS-silenced rootstock (6b5 × NT): the scion continues to express GUS.
(B) RNA gel blot showing the accumulation of GUS mRNA in T19 × HC-Pro plants before and after being grafted onto GUS-silenced rootstocks (6b5 × NT). RNA was isolated from the T19 × HC-Pro plants just before grafting (lane 1, Pre-graft). Four weeks after grafting, RNA was isolated from the T19 × HC-Pro scions (lane 2, Graft) and from the plants used as the source of the scions (lane 3, Control). Ethidium bromide staining of 25S rRNA is shown as a loading control.
(C) GUS-histochemical staining of a leaf from the scion in the grafting experiment shown in (A).
(D) RNA gel blot showing the accumulation of GUS small RNAs in T19 × HC-Pro scions before and after being grafted onto GUS-silenced rootstocks (6b5 × NT). The pre-graft, graft, and control RNAs described in (B) were fractionated to enrich for small RNA, as described in Methods, and examined for 21- to 25-nucleotide (nt) RNAs derived from the GUS gene. Ethidium bromide (EtBr) staining of the predominant band of RNA found in samples enriched for small RNA is shown as a loading control.
DISCUSSION
In the work reported here, we identify where HC-Pro suppression of silencing occurs relative to three features of PTGS: the accumulation of the 21- to 25-nucleotide RNAs, the methylation of the silenced transgene, and the mobile silencing signal. The results allow us to draw inferences about the connection between these features of PTGS and how they are ordered with respect to one another in the silencing pathway. In combination with previously reported work, our data suggest a working model for transgene-induced gene silencing, which is presented in Figure 7 and discussed below.
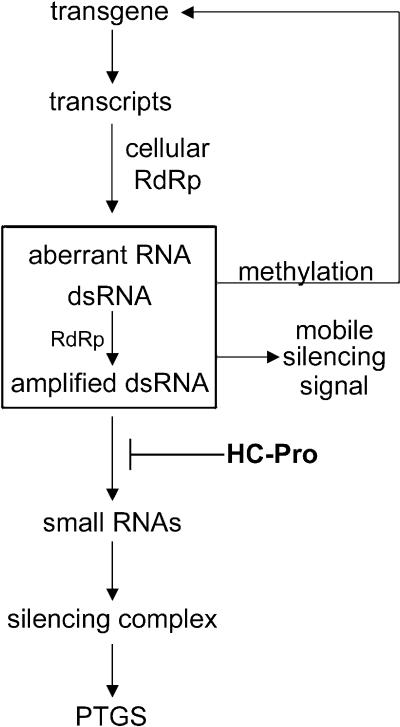
Figure 7.
A Working Model for Transgene-Induced PTGS.
Transcription of a transgene results in production of dsRNA and perhaps other aberrant RNAs either directly or via a cellular RdRp. The dsRNA may be amplified by action of the same RdRp and also is a precursor to the 21- to 25-nucleotide RNAs (small RNAs), which are incorporated into a silencing complex that directs sequence-specific degradation of RNA. Molecules that signal systemic spread of silencing and methylation of the transgene are produced at a step preceding the HC-Pro suppression of PTGS. HC-Pro suppression of silencing occurs at a step before accumulation of the small RNAs either by preventing their production or by rendering them unstable.
We hypothesize that HC-Pro suppresses PTGS via an interaction with one or more cellular proteins that are either components of the silencing machinery or regulators of the pathway. Our results show that HC-Pro interferes with the accumulation of the small RNAs associated with silencing. Because the small RNAs associated with PTGS derive from cleavage of dsRNA by a putative RNase III–like enzyme (Bass, 2000; Zamore et al., 2000), HC-Pro may target the process at this step. There are several possible mechanisms by which HC-Pro might block this enzyme. HC-Pro might prevent the enzyme from binding to the dsRNA template or block the cleavage step so that the small RNAs are never produced. Alternatively, it could block at a step downstream of cleavage, somehow interfering with incorporation of the small RNAs into the silencing complex and thereby rendering them unstable. Thus, our model shows HC-Pro suppression of PTGS occurring upstream of accumulation of the small RNAs (Figure 7).
Although methylation within the transcribed regions of transgenes has been detected in many cases of PTGS in plants, the relevance of this methylation and whether it is a cause or a consequence of silencing remains unclear (Wassenegger and Pélissier, 1998; Kooter et al., 1999; Matzke et al., 2001). Our work shows that the silenced GUS locus in the 6b5 transgenic line is methylated at specific MluI sites within the 3′ region of the coding sequence. This particular pattern of methylation appears to be associated with PTGS because it has been reported for a different GUS-silenced line and is absent from a control GUS-expressing line (English et al., 1996; Figure 3). PTGS is reset at every generation in line 6b5 and recurs in a developmental fashion. If HC-Pro is present, then the reestablishment of PTGS is interrupted and silencing is suppressed (Figure 1). Here, we show that methylation of the GUS transgene is also reset and restored in a developmental fashion in line 6b5. However, in contrast to PTGS, the restoration of methylation is not suppressed by HC-Pro—it occurs with the same kinetics in the presence or the absence of HC-Pro (Figure 3 and text). The trigger(s) for this de novo methylation are uncertain and are conceivably different in different PTGS systems, though many models propose an RNA-directed DNA methylation signal (Wassenegger and Pélissier, 1998; Kooter et al., 1999; Pélissier et al., 1999; Matzke et al., 2001). The fact that HC-Pro can suppress PTGS in the 6b5 line without a dramatic change in the methylation of the transgene suggests that the molecule(s) that signals DNA methylation during PTGS is produced at a step that occurs before HC-Pro interference. Thus, our model shows HC-Pro functioning downstream from production of the signal that directs transgene methylation (Figure 7).
In plants, the systemic movement of silencing can be followed using grafting experiments, and this characteristic gave us a unique opportunity to determine where HC-Pro functions relative to the mobile silencing signal. We found that HC-Pro suppression of PTGS did not eliminate the ability of the plant to produce or send the signal (Figures 4 and 5), though these experiments cannot rule out the possibility that HC-Pro partially interferes with systemic signaling. However, the most dramatic finding from our grafting experiments was that the presence of HC-Pro prevented the plant from responding to the mobile silencing signal (Figure 6). This result shows clearly that HC-Pro works downstream of the mobile signal, as indicated in our working model (Figure 7).
A number of other plant virus proteins have been shown to suppress PTGS (Brigneti et al., 1998; Voinnet et al., 1999, 2000), and these proteins may act at different places than HC-Pro, perhaps defining different steps in the silencing pathway. This result is, in fact, predicted by the behavior of various viral suppressors in the reversal of silencing assay developed in the Baulcombe lab (Brigneti et al., 1998; Voinnet et al., 1999). More recently, it has been shown that a newly identified viral suppressor of PTGS, the p25 cell-to-cell movement protein encoded by potato virus X (PVX), appears to suppress PTGS by targeting the mobile silencing signal (Voinnet et al., 2000). Thus, the mechanism of action of the PVX p25 suppressor stands in contrast to that of HC-Pro, which acts at a step downstream of the mobile signal (Figure 6). In light of that result, it may not be surprising that mixed infections with PVX and any of a variety of potyviruses (which encode HC-Pro) produce a synergistic disease (Vance et al., 1995). It may be that such synergistic diseases, which are common in plants, often result from the interaction of viruses that suppress gene silencing at different points in the pathway (Marathe et al., 2000; Matzke et al., 2001).
Do the small RNAs signal either systemic PTGS or transgene methylation? One of the more intriguing aspects of PTGS is the ability to spread throughout the plant via movement of a signaling molecule presumed to be an RNA. In addition, the sequence-specific RNA degradation occurs in the cytoplasm yet signals the methylation of homologous DNA sequences in the nucleus. The small RNAs have been proposed as the signals for both these events. However, two lines of evidence that derive from experiments using viral suppressors of PTGS contradict the idea that the small RNAs are the mobile silencing signal. First, HC-Pro suppression of silencing interferes with accumulation of the small RNAs in transgene-induced PTGS (Figure 2; Llave et al., 2000) but does not eliminate the production or movement of the silencing signal (Figure 4). Second, the PVX p25 protein interferes with the mobile silencing signal but does not affect the accumulation of small RNAs produced in the viral RdRp-dependent branch of PTGS (Voinnet et al., 2000). The fact that HC-Pro can suppress RNA degradation and small RNA accumulation without a dramatic suppression of either the systemic spread of PTGS or transgene methylation raises the possibility that these latter two events are initiated in the same manner and that neither is signaled by the small RNAs. Although the identity of the signaling molecule(s) in PTGS remains unknown, these viral suppressor studies suggest that this signaling molecule(s) is a precursor of the small RNAs (Figure 7).
The presence of small RNAs has been associated with both transgene-induced gene silencing and virus-induced gene silencing (VIGS) (Hamilton and Baulcombe, 1999). It has recently been reported that these two branches of the silencing pathway differ in that transgene-induced gene silencing requires the cellular RdRp whereas VIGS does not (Dalmay et al., 2000). This result, together with our earlier finding that HC-Pro interferes with both VIGS and transgene-induced gene silencing (Anandalakshmi et al., 1998), suggests that HC-Pro works downstream of the cellular RdRp. However, it remains to be seen how the two branches of gene silencing are interrelated and if HC-Pro affects VIGS in the same way that it affects transgene-induced PTGS.
Our methylation results are consistent with previously reported work in which HC-Pro expressed from a virus could reverse silencing of a transgene encoding green fluorescent protein (GFP) but did not interfere with the methylation of that transgene (Jones et al., 1999). However, they are in apparent conflict with a recent report that suppression of PTGS by HC-Pro causes partial loss of methylation of a GUS transgene with a premature stop codon (Llave et al., 2000). Several differences in the two experimental systems could be responsible for this conflict. In particular, Llave et al. (2000) used a transgene with a nonsense codon, leading to the possibility that nonsense-mediated decay (NMD) is involved in their system in addition to PTGS. NMD is an evolutionarily conserved pathway in which mRNAs that contain a premature stop codon are selectively degraded. There appears to be some overlap between NMD and PTGS because three out of six different genes required for NMD in C. elegans were also found to be required for persistance of RNAi (Domeier et al., 2000). If there is a partial overlap between NMD and PTGS, but the two pathways differ in certain regulatory features and mechanisms, it could explain the difference between the results reported here using a transgene encoding a translatable GUS mRNA (pure PTGS) and those of Llave and colleagues using a transgene encoding a GUS mRNA with a premature stop codon (PTGS/NMD overlap). In addition, the system of Llave et al. (2000) shows only small and variable differences in methylation as a function of silencing or its suppression by HC-Pro. Methylation can be highly variable (Patterson et al., 1993) and depends on the age of the plant (this report); therefore, the small differences in methylation observed by Llave et al. (2000) may simply reflect variation between individual plants at different stages in development and not be an effect of HC-Pro. Our current results point to the value of a time-course analysis to correlate gene silencing and transgene methylation.
A number of cellular proteins required for gene silencing have been identified via mutant screens, and these can be divided into two general classes based on whether they affect development. For example, Arabidopsis plants that are defective in RdRp (sgs/sde mutants; Dalmay et al., 2000; Mourrain et al., 2000) are unable to carry out transgene-induced gene silencing but appear otherwise normal. In contrast, Arabidopsis ago1 silencing mutants, which are impaired in PTGS because of a mutation in an eIF2C-like molecule, exhibit marked developmental abnormalities and are infertile (Fagard et al., 2000). Similarly, tobacco plants expressing HC-Pro show developmental aberrations and, when introduced into Arabidopsis, the HC-Pro phenotype is very similar to that of an ago-1 mutation (T.H. Smith and V.B. Vance, unpublished data). This result raises the possibility that HC-Pro may function via a direct or indirect interaction with AGO-1. It has recently been reported that HC-Pro interacts in the yeast two-hybrid system with a cellular calmodulin-related protein (termed rgs-CaM) that also suppresses PTGS and results in developmental aberrations when overexpressed in tobacco (Anandalakshmi et al., 2000). This HC-Pro–interacting protein is a good candidate for a cellular intermediate in HC-Pro suppression of PTGS.
METHODS
Transgenic Tobacco Lines
The β-glucuronidase (GUS)–silenced lines 6b5 (Elmayan and Vaucheret, 1996) and 106 (Ulker et al., 1999) and GUS-expressing transgenic lines 23b9, 23b10 (Elmayan and Vaucheret, 1996), and T19 (English et al., 1996) have been previously described. Line U-6B (Carrington et al., 1990), carrying wild-type P1/helper component–proteinase (HC-Pro) sequence from tobacco etch virus (TEV), and control line Nicotiana tabacum cv Havana 425, carrying only the binary vector sequence, were provided by J. Carrington (Washington State University, Pullman, WA). Transgenic lines TEV-I and TEV-B, which express mutant versions of the TEV P1/HC-Pro sequence, were used in crosses with the GUS–silenced 6b5 transgenic line. The TEV-I and TEV-B lines have been previously shown to reverse silencing in the T4 GUS–silenced transgenic line (Anandalakshmi et al., 1998). TEV-I contains a three–amino acid insertion near the amino terminus of HC-Pro (Shi et al., 1997), and TEV-B has a three–amino acid insertion near the carboxy terminus of P1 (Anandalakshmi et al., 1998). Tobacco line X-27-8 is N. tabacum cv Xanthi NC transformed with the wild-type P1/HC-Pro sequence. Line X-27-8 was constructed for the experiments reported here by transformation with Agrobacterium carrying the binary plasmid used to make transgenic line U-6B (provided by J. Carrington; Carrington et al., 1990).
GUS Histochemical Staining and Quantitative Assay
GUS histochemical staining was performed as previously described (Anandalakshmi et al., 1998). Briefly, leaves were abraded lightly with carborundum, fixed in 90% acetone for 20 min, vacuum infiltrated with a solution of 50 mM sodium phosphate, pH 7.2, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 2 mg/mL X-Gluc (cyclohexylammonium salt), and incubated overnight at 37°C in a humid oven. GUS fluorometric assays were performed as previously described (Elmayan and Vaucheret, 1996), except that incubation was at room temperature instead of 37°C.
RNA Gel Blot Analyses
Total nucleic acid was isolated from leaf tissues by phenol/chloroform extraction of samples ground to a fine powder in liquid nitrogen and allowed to thaw into 5 volumes of a buffer containing 0.1 M NaCl, 2% SDS, 50 mM Tris/HCl, pH 9.0, 10 mM EDTA, and 20 mM β-mercaptoethanol. RNA gel blot analyses were performed as described by Vance (1991) using 10 μg total nucleic acid per lane. The probe for GUS mRNA was 32P-labeled randomly primed cDNA from polymerase chain reaction (PCR)–amplified fragments representing the entire GUS coding region. Ethidium bromide–stained 25S rRNA is shown in each figure as a loading control.
Low molecular weight RNAs were isolated by polyethylene glycol precipitation of total RNA, separated by denaturing polyacrylamide gel electrophoresis (15 μg/lane), and blotted to nylon membrane exactly as previously described (Hamilton and Baulcombe, 1999). The probe for GUS small RNA was 32P-labeled RNA transcribed in the sense direction from the 3′ 700 nucleotides of the GUS coding region. The small RNAs (5 μg/lane) were also separated by agarose gel electrophoresis and stained with ethidium bromide. The predominant stainable species of RNA on these gels is a band that runs at ∼400 bp and is shown as a loading control. At least seven plants each of the 6b5 × NT and 6b5 × HC-Pro lines were analyzed individually for the small RNAs.
Grafting Experiments
Plants used for the grafting experiments were ∼9 weeks old. A wedge-grafting procedure was performed as previously described (Palauqui et al., 1997). Rootstocks were prepared by removing and discarding the top 5 to 6 cm of the plant. A vertical cut ∼1 to 2 cm long was made in the center of the stem. Scions were prepared by cutting the top 3 to 4 cm of the plant and trimming the bottom of the stem into a wedge. The graft junctions were secured using cotton string and parafilm. For the first week, the graft junction and scion were covered with plastic wrap and misted lightly with water to increase the humidity and prevent dehydration. RNA was isolated from both the rootstock and the scion plants immediately before grafting (pregraft samples). Approximately 4 weeks later, midsize leaves were removed from the scions for GUS histochemical staining and isolation of RNA (graft samples). At the same time, equivalent midsize leaves from the plants used as the source of the scions were also removed for RNA isolation (control samples).
Methylation Analyses—PCR Technique
Total genomic DNA was extracted at 1, 5, and 45 days after germination from individual plants of the experimental lines 6b5 × NT and 6b5 × HC-Pro and T19 control line plants at 45 days after germination using a DNeasy plant mini kit (Qiagen, Valencia, CA). Equal amounts of DNA (∼200 ng) were digested for 8.5 hr at 37°C with 30 units of MluI restriction endonuclease. A primer pair flanking the two closely spaced MluI sites marked by asterisks in Figure 3A was designed. The sequence of the 5′ forward primer was 5′-CATTACCCTTACGCTGAAGAGATGCT-3′ and that of the 3′ primer was 5′-GTTTTTCACCGAAGTTCATGCCAGT-3′. This primer pair amplified a 0.84-kbp band indicative of methylation at the MluI sites when amplified from MluI-cut genomic DNA samples. A second set of primers amplified a 0.48-kbp fragment using the same reverse 3′ primer and a forward 5′ primer with the sequence 5′-AATGTAATGTTCTGCGAC-GCTCAC-3′. Genomic DNA samples (either uncut or cut with MluI) were subjected to PCR amplification using each primer pair and 40 PCR amplification cycles. Genomic DNAs were analyzed from at least three individual plants at each time point. The first cycle consisted of a 10-min 94°C denaturation step, a 1-min 60°C annealing step, and a 2-min 72°C extension step. The remaining cycles used a 1-min 94°C denaturation step, a 1-min 60°C annealing step, and a 2-min 72°C extension step. All PCR products were separated by size on a 1.5% agarose gel and stained with ethidium bromide for analysis. Size markers (100-bp ladder; New England Biolabs, Inc., Beverly, MA) were run on each gel to facilitate comparison between gels.
DNA Gel Blot Analysis
Total genomic DNA was extracted from leaf tissue using the Qiagen DNeasy plant mini kit and digested overnight at 37°C with EcoRI and MluI restriction endonucleases. DNA gel blot analysis was performed as previously described (Vance and Huang, 1988) using ∼10 μg of digested DNA per lane, and blots were probed with 32P-labeled randomly primed cDNA from a PCR-amplified fragment representing the entire GUS coding region. The experiments were performed at least three times, in each case using DNA samples isolated from leaves of several individual plants. A pBluescript plasmid containing the entire coding region of GUS was cut with EcoRI and MluI and run on the gel to provide size markers for unmethylated EcoRI-MluI–cut GUS DNA. Lambda DNA cut with the restriction enzyme HindIII was used as a size standard. The blots were stripped and rehybridized with a 32P-labeled randomly primed cDNA to N. tabacum nitrite reductase as a control to verify that the restriction digests had gone to completion.
Acknowledgments
We thank W. Thompson, S. Spiker, and A. Weissenger for the GUS-silenced transgenic line 106 and O. Voinnet for helpful discussions of the work. This work was supported by grants to V.B.V. from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program, Plant Pathology and Genetic Mechanisms Panels, and by a grant from Akkadix Corporation, La Jolla, CA. A.C.M. was supported by a National Science Foundation Industry/Graduate Research Traineeship.
References
- Anandalakshmi, R., Pruss, G.J., Xin, G., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandalakshmi, R., Marathe, R., Xin, G., Herr, J.M., Jr., Mau, C., Mallory, A.C., Pruss, G., Bowman, L., and Vance, V.B. (2000). A calmodulin-related protein suppresses post-transcriptional gene silencing in plants. Science 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Bass, B. (2000). Double-stranded RNA as a template for gene silencing. Cell 101, 235–238. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.-C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of non-viral (trans) genes. Virology 252, 313–317. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.-X., Ji, L.-H., Ding, S.-W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Carrington, J.C., Freed, D.D., and Oh, C.-S. (1990). Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J. 9, 1347–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., Assalin, G., Macino, G., and Cogoni, C. (2000). Gene silencing in worms and fungi. Nature 404, 245. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. a). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. b). Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Ding, S.W. (2000). RNA silencing. Curr. Opin. Biotechnol. 11, 152–156. [DOI] [PubMed] [Google Scholar]
- Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L., and Mango, S.E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289, 1928–1930. [DOI] [PubMed] [Google Scholar]
- Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. [Google Scholar]
- English, J.J., Mueller, E., and Baulcombe, D.C. (1996). Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO-1, QDE-2 and RDE-1 are related proteins required for PTGS in plants, quelling in fungi and RNAi in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 39, 806–811. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. (1999). Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell 96, 303–306. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279, 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RnaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Kooter, J.M., Matzke, M.A., and Meyer, P. (1999). Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4, 340–347. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., and Carrington, J.C. (2000). Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97, 13401–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe, R., Anandalakshmi, R., Smith, T.H., Pruss, G.J., and Vance, V.B. (2000). RNA viruses as inducers, suppressors and targets of post-transcriptional gene silencing. Plant Mol. Biol. 43, 295–306. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Matzke, A.J.M., Pruss, G., and Vance, V.B. (2001). RNA-based silencing strategies in plants. Curr. Opin. Genet., in press. [DOI] [PubMed]
- Montgomery, M.K., and Fire, A. (1998). Double stranded RNA as a mediator in sequence specific genetic silencing and co-suppression. Trends Genet. 14, 255–258. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.-C., and Vaucheret, H. (1998). Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95, 9675–9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.-C., Elmayan, T., Pollien, J.-M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific posttranscriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.I., Thorpe, C.J., and Chandler, V.L. (1993). Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135, 881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélissier, T., Thalmair, S., Kempe, D., Sänger, H.L., and Wassenegger, M. (1999). Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 27, 1625–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.A. (1999). RNAi and double-strand RNA. Genes Dev. 13, 139–141. [PubMed] [Google Scholar]
- Sharp, P.A., and Zamore, P.D. (2000). RNA interference. Science 287, 2431–2433. [DOI] [PubMed] [Google Scholar]
- Shi, X.M., Miller, H., Verchot, J., Carrington, J., and Vance, V.B. (1997). Mutations in the region encoding the central domain of helper component–proteinase (HC-Pro) eliminate potato virus X/potyviral synergism. Virology 231, 35–42. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith, C.J., Watson, C.F., Bird, C.P., Ray, J., Schuch, W., and Grierson, D. (1990). Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol. Gen. Genet. 224, 477–481. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Ulker, B., Allen, G.C., Thompson, W.F., Spiker, S., and Weissenger, A.K. (1999). A tobacco matrix attachment region reduces the loss of transgene expression in the progeny of transgenic tobacco plants. Plant J. 18, 253–263. [Google Scholar]
- Vance, V.B. (1991). Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 182, 486–494. [DOI] [PubMed] [Google Scholar]
- Vance, V.B., and Huang, A.H.C. (1988). Expression of a lipid body protein gene during maize seed development. J. Biol. Chem. 263, 1476–1481. [PubMed] [Google Scholar]
- Vance, V.B., Berger, P.H., Carrington, J.C., Hunt, A.G., and Shi, X.M. (1995). 5′ proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206, 583–590. [DOI] [PubMed] [Google Scholar]
- Van der Krol, A.R., Mur, L.A., Beld, M., Mol, J.N.M., and Stuitje, A.R. (1990). Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to suppression of gene expression. Plant Cell 2, 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Mourrain, P., Palauqui, J.-C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signaling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Pinto, Y.M., and Baulcombe, D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., and Baulcombe, D.C.. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wassenegger, M., and Pélissier, T. (1998). A model for RNA-mediated gene silencing in higher plants. Plant Mol. Biol. 37, 349–362. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe, A.P., and Matzke, M.A. (1999). Epigenetics: Regulation through repression. Science 286, 481–486. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]