Abstract
The plastid division apparatus (called the plastid-dividing ring) has been detected in several plant and algal species at the constricted region of plastids by transmission electron microscopy. The apparatus is composed of two or three rings: an outer ring in the cytosol, an inner ring in the stroma, and a middle ring in the intermembrane space. The components of these rings are not clear. FtsZ, which forms the bacterial cytokinetic ring, has been proposed as a component of both the inner and outer rings. Here, we present the ultrastructure of the outer ring at high resolution. To visualize the outer ring by negative staining, we isolated dividing chloroplasts from a synchronized culture of a red alga, Cyanidioschyzon merolae, and lysed them with nonionic detergent Nonidet P-40. Nonidet P-40 extracted primarily stroma, thylakoids, and the inner and middle rings, leaving the envelope and outer ring largely intact. Negative staining revealed that the outer ring consists of a bundle of 5-nm filaments in which globular proteins are spaced 4.8 nm apart. Immunoblotting using an FtsZ-specific antibody failed to show immunoreactivity in the fraction containing the filament. Moreover, the filament structure and properties are unlike those of known cytoskeletal filaments. The bundle of filaments forms a very rigid structure and does not disassemble in 2 M urea. We also identified a dividing phase–specific 56-kD protein of chloroplasts as a candidate component of the ring. Our results suggest that the main architecture of the outer ring did not descend from cyanobacteria during the course of endosymbiosis but was added by the host cell early in plant evolution.
INTRODUCTION
Eukaryotic cells contain organelles that are duplicated and inherited by daughter cells during cell division. Among these organelles, mitochondria and plastids are unique in that they contain DNA–protein complexes (nucleoids) and machinery sufficient for protein synthesis. It is generally believed that mitochondria and plastids arose from prokaryotic endosymbionts during eukaryotic evolution. The endosymbiotic theory states that the progenitor of mitochondria was an α-proteobacterium and that the plastid ancestor was a cyanobacterium (Gray, 1992). Mitochondria and plastids multiply by the division of preexisting organelles, as do bacteria (Leech et al., 1981; Kuroiwa, 1982, 1991; Possingham and Lawrence, 1983; Boffey and Lloyd, 1988). Of the characterized mitochondrial and plastid genomes, however, none has a complete set of genes sufficient for self-replication. Mitochondria that cannot synthesize proteins as a result of large genome deletions (petite mutant) (Attardi and Schatz, 1988) and plastids that lack ribosomes (Hashimoto and Possingham, 1989) still can proliferate. Therefore, it is believed that products from the host (nuclear) genomes perform mitochondrial and plastid division. Nuclear regulation of organelle division remains poorly understood, but in plastid division both the plastid-dividing ring (PD ring) (summarized in Kuroiwa et al., 1998; Kuroiwa, 2000) and FtsZ (summarized in Osteryoung and Pyke, 1998; Pyke, 1999) play important roles.
The PD ring was identified first in red alga (Mita et al., 1986). It has been detected in several species of Chlorophyta (green algae and terrestrial plants) (Hashimoto, 1986; Tewinkel and Volkmann, 1987; Hashimoto and Possingham, 1989; Oross and Possingham, 1989; Chida and Ueda, 1991; Duckett and Ligrone, 1993a, 1993b; Ogawa et al., 1995; Robertson et al., 1996), Rhodophyta (Mita et al., 1986; Suzuki et al., 1994), and Chromophyta (Hashimoto, 1997; Beech and Gilson, 2000) and is thought to be ubiquitous in the plant kingdom (summarized in Kuroiwa, 1998; Kuroiwa et al., 1998). In most organisms, the PD ring consists of a double ring structure, with one ring on the cytosolic face of the outer envelope (outer ring) and one on the stromal face of the inner envelope (inner ring). In the unicellular red alga Cyanidioschyzon merolae, a middle ring also was identified in the intermembrane space (Miyagishima et al., 1998a).
The PD rings of most plants are small and have been detected only during the late stage of plastid division. However, in some algal species, the PD ring can be detected clearly just before the onset of constriction. Of these species, C. merolae is an ideal organism for studying the PD ring because it contains a single chloroplast and mitochondrion that divide in a specific order before cytokinesis. In addition, the division of each organelle can be synchronized by subjecting the cells to a light/dark cycle (Suzuki et al., 1994). These features allow us to observe the PD ring readily, although it appears only briefly during plastid division. In a series of studies using C. merolae, we determined the mode of formation, contraction, and disassembly of the PD ring. The inner ring forms before the middle and outer rings (Miyagishima et al., 1998b). As the outer rings contract, they grow thicker and maintain a constant volume. The middle and inner rings do not change in thickness, and their volumes decrease at a constant rate in proportion with contraction (Miyagishima et al., 1999a). The inner and middle rings disassemble in parallel as they contract and disappear just before the completion of chloroplast division. In contrast, the outer rings remain in the cytosol after chloroplast division (Miyagishima et al., 2001). Some of these morphological changes have been reported in Chlorophyta (Chida and Ueda, 1991; Ogawa et al., 1995). The differences among the three rings suggest that the protein compositions of the rings are different (Miyagishima et al., 2001).
In addition to morphological studies, molecular genetic studies show that plastid division requires nucleus-encoded homologs of FtsZ (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998), a protein that forms a cytokinetic ring in bacteria beneath the plasma membrane (Bi and Lutkenhaus, 1991; reviewed in Bramhill, 1997). Disruption of ftsZ in the moss Physcomitrella patens (Strepp et al., 1998) and antisense repression of FtsZ in Arabidopsis (Osteryoung et al., 1998) produced giant chloroplasts in each cell. Plant nuclear genes encoding FtsZ also have been identified in Chlorophyta (Osteryoung and Vierling, 1995; Osteryoung et al., 1998; Strepp et al., 1998; Gaikwad et al., 2000; Mori and Tanaka, 2000), Rhodophyta (Takahara et al., 1999, 2000a, 2000b), and Chromophyta (Fraunholz et al., 1998; Beech et al., 2000), and most of them are highly conserved compared with their cyanobacterial counterparts. Arabidopsis FtsZ (AtFtsZ1-1) (Osteryoung et al., 1998) and pea FtsZ (Gaikwad et al., 2000) were imported into chloroplasts in vitro. Recently, a plant nuclear homolog of MinD, which determines the division site in bacteria, was demonstrated to determine the division site of chloroplasts in Arabidopsis (Colletti et al., 2000). These results show that significant components of bacterial cell division have descended to chloroplasts. Thus, it is reasonable to predict that FtsZ localizes to the inner surface of the plastid division site in the same manner as does bacterial FtsZ.
On the basis of these and morphological results, it is suggested that the inner PD ring contains FtsZ but that the outer ring is composed of proteins other than FtsZ (Kuroiwa et al., 1998; Miyagishima et al., 1998a). On the other hand, another type of FtsZ (AtFtsZ2-1 and AtFtsZ2-2) that lacks obvious transit peptides was identified in Arabidopsis. AtFtsZ2-1 was shown not to be imported into chloroplasts in vitro (Osteryoung et al., 1998). Antisense repression of AtFtsZ2-1 also generates giant chloroplasts (Osteryoung et al., 1998). On the basis of these results, it is hypothesized that chloroplast-targeted and nontargeted forms of FtsZ are components of the inner and outer PD rings, respectively (Osteryoung and Pyke, 1998; Osteryoung et al., 1998; Erickson, 2000; Margolin, 2000; Osteryoung, 2000). Although double or triple PD rings have been observed to be electron dense in several plants by use of transmission electron microscopy, the FtsZ ring has never been observed directly in thin sections in several bacteria. Some preliminary experiments suggest that FtsZ is not localized to the PD ring (Fraunholz et al., 1998; Kuroiwa et al., 1999). Whether or not this hypothesis is correct, proteins other than FtsZ are likely major components of the two rings. These other proteins may account for the differences between the two rings and likewise may cause the rings to be visible directly by transmission electron microscopy, unlike bacterial FtsZ rings. Nevertheless, the localization of FtsZ or other proteins to the PD ring has not been proved, and morphological and molecular genetic studies are incomplete.
We investigated the ultrastructure of the PD ring at high resolution to determine the main components of the outer ring, which has been observed only as electron-dense deposits in thin sections. For this purpose, we isolated dividing chloroplasts with PD rings (Miyagishima et al., 1999c) and observed the outer ring by negative staining with the aid of a detergent. We found that the main constituent of the outer PD ring is a bundle of novel filaments other than FtsZ that is 5 nm in diameter. Our results highlight the significance of a system newly created by host cells that regulates the bacterial symbiont and is the primary constituent of the plastid division apparatus.
RESULTS
Visualization of the Outer PD Ring by Negative Staining with Nonidet P-40
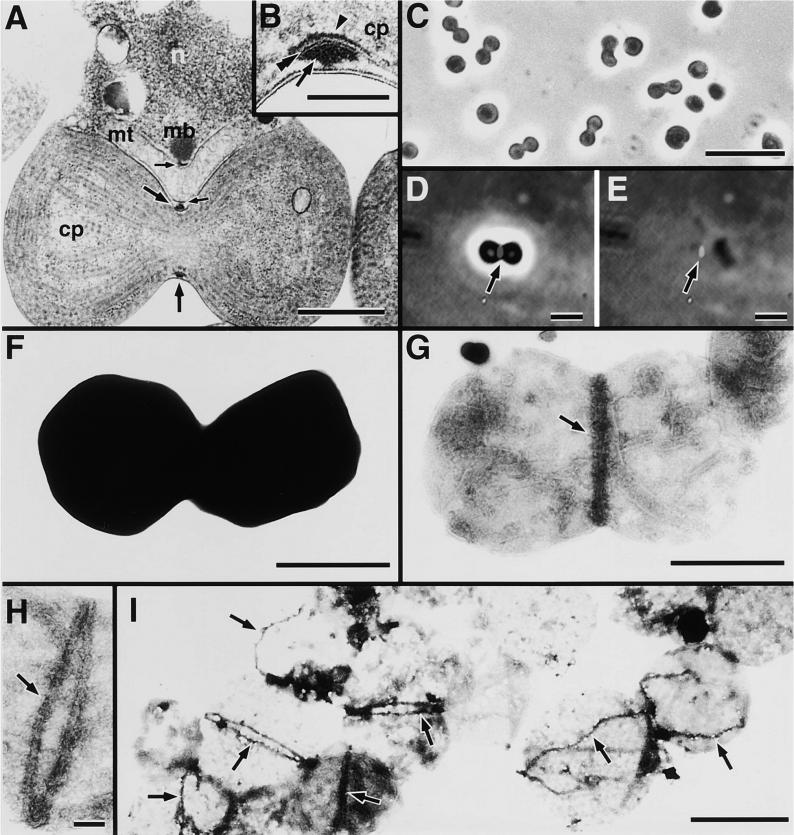
Thin sections of C. merolae clearly show the triple ring structure of the PD ring; one ring rests on the cytosolic face of the outer envelope (outer ring), one on the stromal face of the inner envelope (inner ring), and one in the intermembrane space (middle ring) (Miyagishima et al., 1998a) (Figures 1A and 1B). In C. merolae, the mitochondrion-dividing ring also is observed at the division site of a mitochondrion (Kuroiwa et al., 1993) (Figure 1A). In Figure 1B, the outer, middle, and inner PD rings are measured at 80 × 25 nm, 80 × 2 nm, and 100 × 5 nm (width × thickness), respectively. In thin sections, ring structures are observed only as electron-dense lumps; further ultrastructural information on the rings is not available. We thought that to reveal the ultrastructure at still higher resolution, negative staining would be appropriate, as with other cellular structures. Because negative staining can visualize only the surface of material, to use negative staining here we isolated dividing chloroplasts with the PD ring from synchronized C. merolae culture, according to the method described previously (Miyagishima et al., 1999c) (Figure 1C). However, isolated dividing chloroplasts were too electron dense (Figure 1F). To obtain negative images of the PD ring, it was necessary to remove some of the chloroplast structure other than the PD ring. Chloroplasts were treated with several detergents, and chloroplasts and outer PD rings were viewed by fluorescence microscopy. The outer PD ring was visualized by labeling surface proteins of chloroplasts with N-hydroxy-sulfo-succinimidyl (NHS)–biotin and detected by fluorescein isothiocyanate (FITC)–avidin, as described previously (Miyagishima et al., 1999c) (Figure 1D).
Figure 1.
Visualization of the Outer PD Ring by Negative Staining after Treatment with Nonidet P-40.
(A) Thin section of a C. merolae cell containing a dividing chloroplast and mitochondrion showing the PD ring and the mitochondrion-dividing ring. The section cut the cell at the center perpendicular to the equator.
(B) Magnified cross-sectional view of the PD ring showing that the PD ring is composed of three rings.
(C) Phase-contrast image of isolated dividing chloroplasts from a synchronized culture.
(D) The outer PD ring of an isolated dividing chloroplast was visualized as green fluorescence by labeling surface proteins of the chloroplast with NHS-biotin and detecting them with FITC-avidin.
(E) Chloroplasts shown in (D) were lysed by the addition of solution containing 0.5% Nonidet P-40 from the edge of the cover glass. The outer PD ring remained insoluble.
(F) and (G) Negative staining images of isolated chloroplasts before (F) and after (G) 0.1% Nonidet P-40 treatment.
(H) The outer PD ring was clearly observed as a closed ring by negative staining after Nonidet P-40 treatment in oblique samples.
(I) Low magnification images frequently showed that the outer PD rings are attached to the outer envelopes of chloroplasts.
Small arrows indicate the mitochondrion-dividing ring. Large arrows, arrowhead, and double arrowhead indicate the outer, inner, and middle PD rings, respectively. cp, chloroplast; mb, microbody; mt, mitochondrion; n, nucleus. Bars in (A), (F), and (G) = 500 nm; bars in (B) and (H) = 100 nm; bar in (C) = 5 μm; bars in (D) and (E) = 2 μm; bar in (I) = 1 μm.
In conclusion, we found that the nonionic detergent Nonidet P-40 at concentrations >0.1% solubilized a portion of the chloroplast structure but spared the outer PD ring (Figures 1D and 1E; 0.5% Nonidet P-40 solution was used). Other detergents listed in Methods could not solubilize chloroplasts of C. merolae as efficiently as Nonidet P-40 could, even at high concentrations. Insoluble material from Nonidet P-40 lysates expected to contain the outer PD rings was collected by centrifugation at 10,000g and negatively stained with phosphotungstic acid (Figures 1G to 1I). Nonidet P-40 removed most of the stroma and thylakoids, and negative images of the envelope and outer PD ring were obtained (Figures 1F and 1G). Some of the outer PD rings, which dried obliquely, were clearly observed as closed rings (Figure 1H) and were uniform in width (40 nm in Figure 1H). At low magnification, outer PD rings clinging to outer envelopes were evident at high frequency (Figure 1I) as dumbbell-shaped chloroplasts before Nonidet P-40 treatment (Figure 1C). Some of these rings were closed, whereas others were cut at one point (Figure 1I). When the supernatant from a centrifugation at 10,000g was further centrifuged at 20,000g and the pellet was negatively stained, many small particles were observed but the rings were absent (data not shown). This indicated that most of the outer PD ring remained insoluble in the presence of ⩾0.1% (not shown) Nonidet P-40 and pelleted by centrifugation at 10,000g.
The Outer PD Ring Remains Insoluble but the Inner and Middle Rings Disassemble in Nonidet P-40
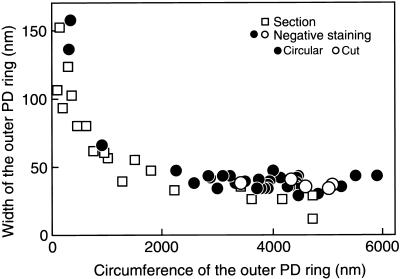
We also compared the morphology of the outer rings after Nonidet P-40 treatment with the in vivo morphology and further examined the effect of Nonidet P-40 on the inner part of the plastid division apparatus (the middle and inner PD rings), which was not visible by negative staining. First, the width versus circumference (or length in the case of cut rings) after Nonidet P-40 treatment was measured and compared with that in sections of whole cells measured previously (Miyagishima et al., 1999a). The results of negative staining after Nonidet P-40 were identical to those in sections of whole cells in vivo (Figure 2). Although it was possible that Nonidet P-40 extracted some minor accessory proteins, these results suggest that the main architecture of the outer PD ring was preserved.
Figure 2.
Comparison of the Width of the Outer PD Rings after Nonidet P-40 Treatment with the in Vivo Width.
Width versus circumference of the outer PD ring (open squares) measured previously on thin sections of cells (Miyagishima et al., 1999a) and by negative staining after 0.1% Nonidet P-40 treatment (closed and open circles). Closed rings (closed circles) and rings cut at one point (open circles) were plotted. In cell sections, the two parameters could not be measured in the same section (e.g., in Figure 1A, only the width could be measured). The width was plotted versus the circumference as follows. The width of the outer PD ring was plotted versus the diameter of the chloroplast at the equator. The chloroplast diameters were converted to circumferences by using the graph of circumference versus diameter (Miyagishima et al., 1999a).
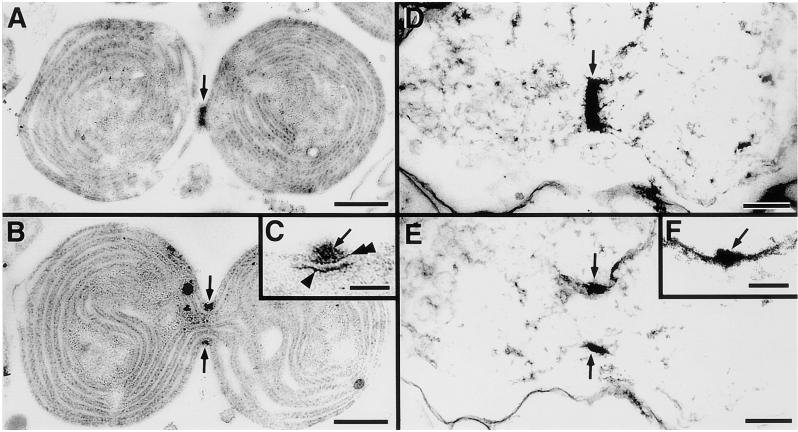
We further examined the structural integrity of the outer PD ring after Nonidet P-40 treatment and the effect of Nonidet P-40 on inner parts of the plastid division apparatus (middle and inner PD rings). Isolated chloroplasts (Figures 3A to 3C) and insoluble fractions from Nonidet P-40 treatment (Figures 3D to 3F) were embedded in resin, and serial thin sections were analyzed by transmission electron microscopy. Figures 3A and 3B and Figures 3D and 3E show serial thin sections of identical samples. Nonidet P-40 clearly extracted most of the thylakoids except for the envelope membrane (Figures 3D and 3E). The diameters at the ring equators in Figures 3A and 3B and Figures 3D and 3E are similar at 250 nm (Figures 3B and 3E), and the widths and thicknesses of the outer PD rings are nearly identical, measuring ∼50 × 20 nm (Figures 3A and 3B). The electron density of the outer PD ring was not diminished by Nonidet P-40 (Figure 3). These data support the results shown in Figure 2 and confirm the fact that in addition to the width, the thickness and density of the outer PD ring were conserved after Nonidet P-40 treatment. In contrast to the outer PD ring, higher magnification clearly showed that the inner and middle PD rings disappeared completely after Nonidet P-40 treatment (Figures 3C and 3F). These results suggest that Nonidet P-40 solubilized the main architecture of the two rings and that the outer and two PD rings inside the plastid have different biochemical properties.
Figure 3.
Comparison of the Morphology of the Outer, Middle, and Inner PD Rings after Nonidet P-40 Treatment with That in Isolated Chloroplasts in Thin Sections.
(A) to (C) Serial thin sections of an isolated chloroplast showing a tangential section (A) and a cross-section (B) of the PD ring. The cross-sectional image of the PD ring is magnified in (C).
(D) to (F) Serial thin sections of the insoluble portion after 0.1% Nonidet P-40 treatment showing a tangential section (D) and a cross-section (E) of the PD ring. The cross-sectional image of the PD ring is magnified in (F).
Arrows, arrowhead, and double-arrowhead indicate the outer, inner, and middle PD rings, respectively. Bars in (A) and (B) = 500 nm; bars in (C) and (F) = 50 nm; bars in (D) and (E) = 200 nm.
The Outer PD Ring Is a Bundle of 5-nm Filaments
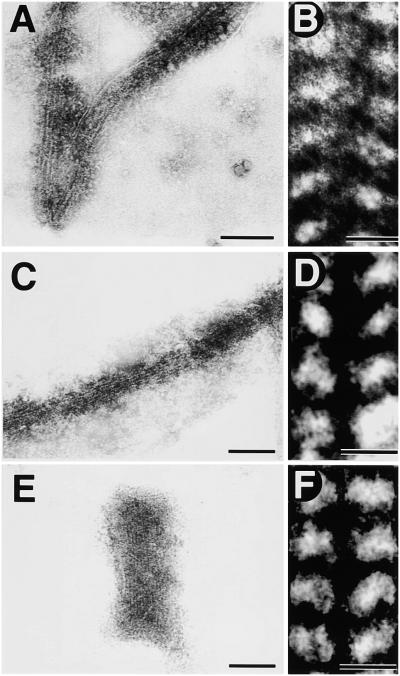
The results described above demonstrate that at least the main architecture of the outer PD ring was preserved structurally after treatment with Nonidet P-40. We further examined the ultrastructure of the outer PD ring and its changes during contraction (Figure 4). A magnified view of part of the outer PD ring revealed that it was composed of a number of filaments ∼5 nm in diameter (in negative images, the filament was observed as an electron-lucent structure) arranged uniformly in a line (Figure 4A; the belt of the ring was bent at the bottom). Two edges of the bundle were sharp, the filaments were arranged uniformly from edge to edge, and neither a higher density in the center nor a lower density at either edge was observed. At still higher magnification and contrast between two adjacent filaments, the filament was observed as a uniform line of globular proteins ∼5 nm in diameter (Figure 4B).
Figure 4.
Magnified Images of the Outer PD Ring Negatively Stained.
(A) The outer ring is composed of a number of filaments ∼5 nm in diameter.
(B) Higher magnification of two adjacent filaments showing straight arrays of globular proteins along the filaments.
(C) and (D) Negative images of the ring at early stages of contraction.
(E) and (F) Negative images of the ring at late stages of contraction.
Bars in (A), (C), and (E) = 100 nm; bars in (B), (D), and (F) = 5 nm.
Figures 4C and 4D and Figures 4E and 4F show negative images of the outer PD ring at the early and late stages of contraction, when it was 50 and 140 nm wide, respectively. Figure 4C shows part of a ring, whereas Figure 4D shows an entire ring. These images show that the 5-nm filaments were arrayed in the same manner at every stage of contraction (Figures 4C and 4E) and demonstrate that the intervals of filaments and proteins did not change during contraction (Figures 4D and 4F). The spacing between filaments was measured as 6.4 ± 0.6 nm (average ±sd, n = 49). The proteins were spaced 4.8 ± 0.4 nm apart along the filament (n = 43). In a previous study, we demonstrated that the outer ring contracts without changing its volume and density by widening and thickening (Miyagishima et al., 1999a). These data indicate that compaction of filaments does not occur during contraction, but the filaments may coil around the constricted region while maintaining the 6.4-nm interval toward the exterior to widen and thicken the bundle of filaments. Some amorphous structures in addition to the ring often were observed at an earlier stage of contraction (Figures 4A and 4B), but details of these structures could not be obtained.
FtsZ Is Not a Component of the Filaments
Although the components of the PD ring are not known, it is hypothesized that chloroplast-targeted and nontargeted forms of FtsZs are components of the inner and outer PD rings, respectively (Osteryoung et al., 1998). The in vivo structure of FtsZ polymers in bacteria or plants has not been visualized, but much has been learned from polymers of bacterial FtsZ assembled in vitro. Although these in vitro polymers have shown a range of structures depending on the experimental conditions, including sheets (Erickson et al., 1996), tubules (Bramhill and Thompson, 1994; Mukherjee and Lutkenhaus, 1994), and mini rings (Erickson et al., 1996), all are composed of the same protofilaments (summarized in Lu et al., 2000). In Escherichia coli FtsZ, adjacent protofilaments are 5.3 to 5.4 nm apart (Erickson et al., 1996; Lu et al., 2000), and the FtsZ is arranged at 4.3-nm intervals (Erickson et al., 1996). The 6.4-nm spacing of filaments and 4.8-nm spacing of proteins of the outer PD ring (Figure 4) resemble or are slightly larger than those of the protofilament of bacterial FtsZ.
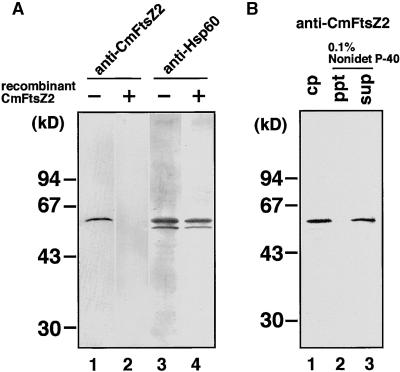
To determine whether FtsZ is a component of the 5-nm filament organizing the outer PD ring, we performed immunoblot analysis. We identified two kinds of FtsZ proteins in C. merolae, one (CmFtsZ2) that resembles a cyanobacterial counterpart and the other (CmFtsZ1) that resembles an α-proteobacterial counterpart (Takahara et al., 2000b). CmFtsZ1 probably functions in mitochondria, and CmFtsZ2 is localized in chloroplast fractions by immunoblot analysis (Takahara et al., 2000b). Genomic DNA gel blot analyses show that CmftsZ2 (cyanobacteria type) and CmftsZ1 (α-proteobacteria type) are single-copy genes in the C. merolae genome (Takahara et al., 2000b). At present, it is not clear whether FtsZ2 is imported into chloroplasts through the envelope or localizes on the cytosolic surface of the outer envelope. The anti-CmFtsZ2 antiserum was confirmed to specifically react with recombinant CmFtsZ2 protein in E. coli lysate (Takahara et al., 2000b). To further examine whether the antiserum specifically reacts with FtsZ in total protein of C. merolae, we performed competition experiments (Figure 5A). The antiserum detected a 60-kD band in total protein of C. merolae, and no other bands cross-reacted with the antiserum (Figure 5A, lane 1). In contrast, when the antiserum was preincubated with recombinant CmFtsZ2, the 60-kD band was not detected (Figure 5A, lane 2). Preimmune serum at the same dilution did not detect any bands (not shown). The 60- and 58-kD bands detected by anti-Hsp60 antibody were not diminished by preincubation with recombinant CmFtsZ2 (Figure 5A, lanes 3 and 4). These results indicated that the 60-kD band was recognized by the same subset of antibodies in the antiserum that recognized the recombinant CmFtsZ2 protein and confirmed that the 60-kD band is identical to CmFtsZ2. The specificity of the antiserum was confirmed above; then, using this antiserum, we examined whether or not CmFtsZ2 is involved in the insoluble fraction from Nonidet P-40 treatment that contains the 5-nm filament. The 60-kD protein corresponding to CmFtsZ2 was clearly detected in isolated chloroplasts by an anti-CmFtsZ2 antibody (Figure 5B, lane 1). However, when the same amount of chloroplast was treated with 0.1% Nonidet P-40, the antibody did not react with the pellet (Figure 5B, lane 2) but reacted with the supernatant (Figure 5B, lane 3). The densities of the two bands were approximately the same. These results show that FtsZ2 was solubilized by Nonidet P-40, unlike the 5-nm filament, and suggest that FtsZ2 is not the 4.8-nm protein component of the filament.
Figure 5.
Immunoblot of FtsZ by Using Anti-CmFtsZ2 Antiserum.
(A) Specificity of anti-CmFtsZ2 antiserum. Total protein (100 μg) of C. merolae were separated by SDS-PAGE and reacted with the antiserum raised against recombinant CmFtsZ2 protein (lanes 1 and 2) or anti-Hsp60 antibody (lanes 3 and 4), respectively. Antibodies were preincubated with purified recombinant CmFtsZ2 protein (lanes 2 and 4).
(B) Localization of FtsZ. Isolated dividing chloroplasts containing 100 μg of protein (cp), pellet (ppt), and the supernatant (sup) obtained from the same amount of chloroplasts by 0.1% Nonidet P-40 treatment were separated by SDS-PAGE. FtsZ (60-kD band) was detected in the chloroplasts and the supernatant but not in the pellet.
The Bundle of the Filament Is a Relatively Insoluble Structure
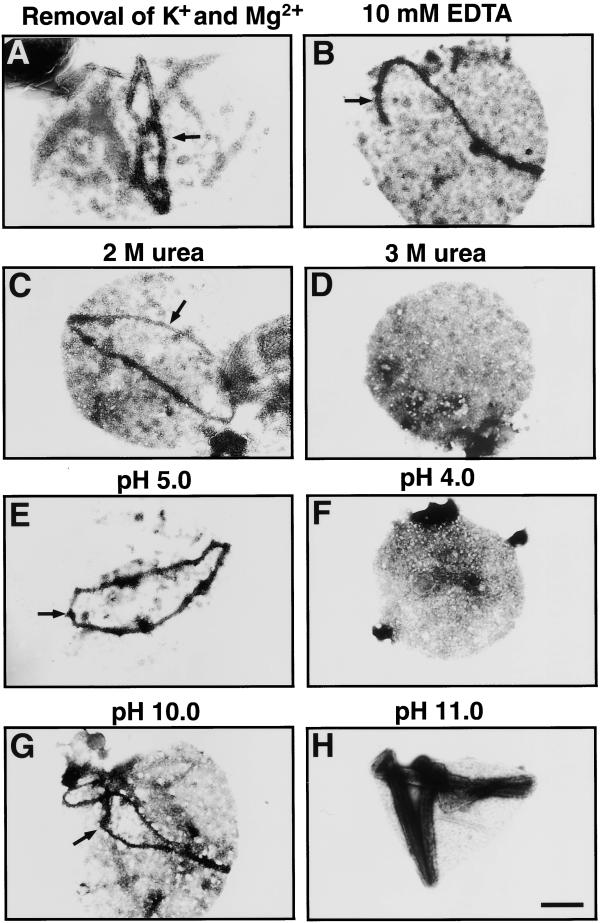
Compared with established cytoskeletal filaments, the filament composing the outer PD ring, with its 5-nm diameter and straight arrangement of proteins within, resembles only FtsZ (described above) and tubulin protofilaments. However, most of the steps in the experiments described above were performed at low temperature (on ice or at 4°C), which is known to depolymerize the tubulin protofilament. The immunoblot analyses shown in Figure 5 indicated that the 5-nm filament is different from FtsZ-based structure in sensitivity to Nonidet P-40. On the basis of these properties, it is suggested that the filament is a novel cytoskeletal structure. To further examine the biochemical properties of the 5-nm filament, we treated insoluble fractions of Nonidet P-40 lysates with several reagents and then negatively stained the treated fractions. Some examples of negative staining are shown in Figure 6; the results are summarized in Table 1. The bundle of filaments remained insoluble without KCl and MgCl2 or by chelating divalent cations with EDTA or in the presence of high salt (Figures 6A and 6B, and Table 1). However, the filaments were disassembled by agents that dissociate hydrophobic bonds (>3 M urea or >1 M NaClO4) or at low or high pH (pH < 4.0 or pH > 11.0; Figures 6C to 6H and Table 1). We verified that protease treatment or denaturing Laemmli sample buffer breaks the filaments (Table 1). In this experimental condition, nucleotide triphosphate, nucleotide diphosphate, nucleotide thiotriphosphate, and Ca2+ produced no detectable changes in the outer PD rings (Table 1). In summary, the 5-nm filament was relatively insoluble, and the solubility properties resemble those of intermediate filaments such as desmin and vimentin (Hubbard and Lazarides, 1979; Lazarides and Granger, 1982), although the dimension is different (intermediate filaments are 10 nm in diameter). In addition, the bundle of filaments was so rigid that it could not be broken by freezing and thawing. Furthermore, the envelope membrane could not be stripped from the outer PD ring by any treatment without disassembly of the ring, suggesting that the outer ring is anchored firmly to the outer envelope.
Figure 6.
The Stability of the Outer PD Ring in Several Media.
(A) K+- and Mg2+-depleted medium.
(B) 10 mM EDTA.
(C) and (D) 2 M (C) or 3 M (D) urea.
(E) and (F) 0.1 M acetate/sodium acetate, pH 5.0 (E) or 4.0 (F).
(G) and (H) 0.1 M sodium carbonate/sodium hydrogen carbonate, pH 10.0 (G) or 11.0 (H).
Arrows indicate the outer PD ring. Bar in (H) = 500 nm for (A) to (H).
Table 1.
Effects of Chemical or Physical Factors on Dissociation of the Outer PD Ring
| Treatment | Concentration/pH | Efficiencya |
|---|---|---|
| Reagents reducing hydrophobic bonds |
||
| Urea | 2 M | − |
| 3 M | + | |
| NaClO4 | 0.5 M | − |
| 1 M | + | |
| Removal of salts | ||
| Removal of K+ and Mg2+ | − | |
| High ionic solution | ||
| NaCl | 3 M | − |
| Chelating reagent | ||
| EDTA | 10 mM | − |
| Reducing reagent | ||
| DTT | 5 mM | − |
| Bases | ||
| 0.1 M Sodium carbonate/ | pH 11.0 | + |
| Sodium hydrogen carbonate | pH 10.0 | − |
| pH 9.0 | − | |
| Acids | ||
| 0.1 M Acetate/Sodium acetate | pH 5.0 | − |
| pH 4.0 | + | |
| pH 3.0 | + | |
| Nucleotidesb | ||
| NTP | 5 mM | − |
| NDP | 5 mM | − |
| NTPγS | 5 mM | − |
| Protease | ||
| Thermolysin | 1 mg/mL | + |
| Others | ||
| CaCl2 | 5 mM | − |
| Laemmli sample buffer | + | |
| Freezing and thawing | − |
(−) and (+), poor and high efficiency on dissociation of the outer PD ring, respectively.
Adenine or guanine nucleotides were used respectively.
Identification of a Candidate for the Protein Composing the Filament
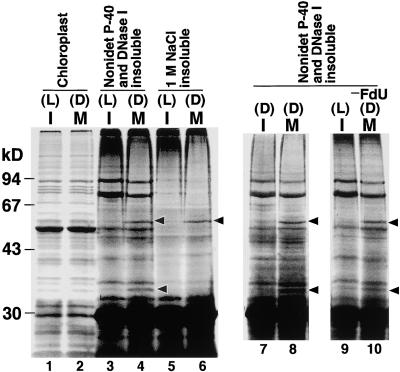
Because the filament of the outer PD ring appears different from established cytoskeletal filaments, we attempted to obtain candidates for the component. Our current morphological study of the PD ring in C. merolae suggested that the proteins composing the PD rings exist on the envelopes only in the dividing phase (Miyagishima et al., 2001). Therefore, candidate proteins should be bound to the envelopes only during the dividing phase and enriched in the insoluble fraction of Nonidet P-40 treatment. Although the bundle of filaments could not be separated from the envelope membrane, to remove as much other protein as possible, we used osmotic shock to burst isolated chloroplasts and then lysed them with 0.5% Nonidet P-40 and DNase I. DNase I was added to remove the chloroplast nucleoids, which remain insoluble even in the presence of Nonidet P-40. Negative staining confirmed that the outer PD rings were enriched in the insoluble fraction, as seen in Figure 1I (data not shown). For comparison, nondividing chloroplasts also were isolated and treated in the same manner. The results of SDS-PAGE and silver staining are shown in Figure 7.
Figure 7.
SDS-PAGE Analysis of Isolated Chloroplasts and Fractions in Which the Outer PD Rings Are Enriched.
In the synchronous culture, cells were in interphase during light periods (L) and in M phase during dark periods (D). Nondividing and dividing chloroplasts were isolated from interphase (I) and M phase (M) synchronous cultures, respectively, and chloroplasts containing 5 μg of protein were analyzed (lanes 1 and 2). The chloroplasts were then lysed in hypotonic medium containing 0.5% Nonidet P-40 and 100 μg/mL DNase I. Pellets obtained from chloroplasts containing 1 mg of protein were analyzed (lanes 3 and 4). Proteins of the pellet were further extracted with 1 M NaCl, and the insoluble pellets were analyzed (lanes 5 and 6). To exclude differences between light and dark conditions, nondividing chloroplasts were isolated from a concentrated culture during the dark period, in which cells were in interphase. Then, the chloroplasts were lysed using the same treatment as was used for lanes 3 and 4 (lanes 7 and 8). Dividing chloroplasts were isolated from M phase culture synchronized without 5-fluorodeoxyuridine (−FdU; lanes 9 and 10). Arrowheads indicate specific bands in the dividing phase.
Although no difference was detected in band patterns between nondividing chloroplasts and dividing chloroplasts at the level of total proteins (Figure 7, lanes 1 and 2), in the insoluble fraction of Nonidet P-40 and DNase I treatment, 56- and 33-kD dividing phase–specific bands were enriched (Figure 7, lanes 3 and 4). When the insoluble fraction was further treated with 1 M NaCl, which did not disassemble the bundle of filaments (Table 1), the 33-kD protein was extracted, and only the 56-kD protein remained insoluble as a candidate (Figure 7, lanes 5 and 6). However, it is possible that the phase-specific localization of the protein relies on the light and dark rather than on the division cycle of the chloroplast, because cells were synchronized by light and dark cycles (interphase during the light period and M phase during the dark period). To test this idea, we isolated nondividing chloroplasts from a concentrated culture during the dark period (in a concentrated culture, cells do not divide even during the dark periods). In this case, the 56- and 33-kD bands were identified specifically in the dividing phase (Figure 7, lanes 7 and 8). Finally, we tested the possibility that these proteins were synthesized in response to the addition of 5-fluorodeoxyuridine, which was added to prepare highly synchronized M phase culture in a reproducible manner. Without 5-fluorodeoxyuridine, the two proteins were detected only in the dividing phase (Figure 7, lanes 9 and 10). From these results, we conclude that the 56- and 33-kD proteins were located in/on the chloroplast membrane specifically in the dividing phase and that the 56-kD protein is a strong candidate for the component of the 5-nm filament.
DISCUSSION
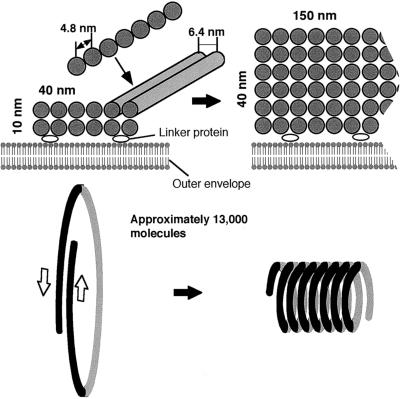
By negative staining and with the aid of a detergent, we demonstrated that the main architecture of the outer PD ring is a bundle of novel 5-nm filaments. Although it is possible that Nonidet P-40 extracted some minor accessory proteins, it was confirmed that at least the main architecture of the outer PD ring was preserved. Observations by negative staining in this study and previous observations of thin sections allow us to present a model of the ultrastructure of the outer PD ring and its dynamics during contraction (Figure 8). Negative staining in this (Figures 2 and 4) and previous (Miyagishima et al., 1998b, 1999a) studies show that when formed, the outer PD ring is initially ∼40 nm wide and 10 nm thick. The outer PD ring widens and thickens during contraction, maintaining a constant volume and density, and finally becomes >150 nm wide and >40 nm thick (Miyagishima et al., 1998b, 1999a; Figures 2 and 4). The 6.4-nm spacing of filaments and 4.8-nm spacing of proteins remained constant in all stages of contraction (Figure 4). Thus, the ring was initially formed of 6 × 2 filaments (width × thickness), and once contraction started, the filaments coiled around the constricted region toward the exterior to widen and thicken the bundle of filaments to 24 × 6 without loss of components (Figure 8).
Figure 8.
Scheme of the Main Architecture of the Outer PD Ring and Its Change during Contraction.
The outer PD ring is composed mainly of filaments arranged in rows at 6.4-nm intervals. Along each filament, proteins are spaced 4.8 nm apart. The bundle of filaments probably adheres to the outer envelope via some linker proteins. The ring is a bundle of 12 filaments, two filaments thick and six filaments wide, which contain 13,000 components. By the final stage of contraction, the bundle changes so that it is six filaments thick and 24 filaments wide, without loss of components. Adjacent filaments slide side by side in the opposite direction to widen the bundle, as shown. A filament sometimes coils upon itself and thickens the bundle.
One possible mechanism for this process involves adjacent filaments that slide in opposite directions to widen the bundle as the filaments coil around each other and thicken the bundle (Figure 8). Because the tips of the filaments remained undetectable, the number of individual continuous filaments composing the bundle is not clear. Taking into account the circumference of the ring, which is initially ∼5000 nm, one outer ring is composed of ∼13,000 molecules of 4.8-nm protein. This is similar to the 15,000 bacterial FtsZ molecules estimated to exist in an average E. coli cell (Dai and Lutkenhaus, 1992; Lu et al., 1998). Therefore, the number of components is not the reason that the PD ring has been observed directly as an electron-dense ring and the bacterial FtsZ ring has not. At present, it is not known whether the filament moves by itself or if some motor proteins are involved. In this regard, no structures that look like motor proteins, which are generally very large (>10 nm), were detected in this study. It is notable that only filaments in the lowest layer face the outer envelope and that the filaments are not plugged into the envelope membrane. This finding suggests that the filament itself does not have the ability to adhere directly to the outer envelope and that some linker proteins probably bind the bundle of filaments to the outer envelope specifically at the division site. In the green algae Nannochloris bacillaris (Ogawa et al., 1995) and Trebouxia potteri (Chida and Ueda, 1991), the outer PD ring appeared as a bundle of filaments in tangential sections, suggesting that our finding in Rhodophyta is probably applicable to Chlorophyta as well.
Although no protein has been localized to the PD ring, FtsZ has been proposed as a candidate. On the basis of molecular genetic studies in Arabidopsis (Osteryoung et al., 1998), it is hypothesized that chloroplast-targeted and nontargeted forms of FtsZs are components of the inner and outer PD rings, respectively (Osteryoung and Pyke, 1998; Osteryoung et al., 1998; Erickson, 2000; Margolin, 2000; Osteryoung, 2000). This hypothesis suggests that plastid division is performed by almost the same machinery as bacterial division, which descended from cyanobacteria and forms inside and outside the site of plastid division. On the basis of this hypothesis, the outer FtsZ ring was added after the endosymbiosis of cyanobacterium.
Contrary to this hypothesis, our results suggest that the main architecture of the outer PD ring is a bundle of novel 5-nm filaments that is more rigid than structures based on FtsZ. FtsZ was extracted by Nonidet P-40, whereas the electron density and dimensions of the outer PD ring were preserved after Nonidet P-40 treatment (Figures 3 and 5). Nevertheless, at present, we cannot rule out the possibility that another unidentified FtsZ, which does not cross-react with anti-CmFtsZ2 polyclonal antibody, remains insoluble in the PD ring subfraction after Nonidet P-40 treatment, unlike CmFtsZ2. In this regard, no additional plastid-type ftsZ genes have been detected by genomic DNA gel blot analysis under low-stringency conditions (Takahara et al., 2000b). The outer PD ring is also resistant to still higher concentration (1.0%) of Nonidet P-40. An insoluble property such as that reported here for the 5-nm filament has not been reported for FtsZ. The isoelectric point of the 56-kD candidate component was calibrated around 6.5 by two-dimensional PAGE (data not shown), whereas isoelectric points of FtsZs (as in the case of plant FtsZs, the N-terminal extensions were excepted) were calculated between 4.5 and 5.5 by DNASIS software (Hitachi Software Engineering). Even if another FtsZ is involved in the outer PD ring subfraction or FtsZ beside the outer PD ring was extracted by Nonidet P-40, this study suggests that the 5-nm filament is not identical to FtsZ filament and that the primary constituent of the outer ring that visualizes the ring as an electron-dense structure is not FtsZ but the novel 4.8-nm protein. Our results are consistent with the fact that the bacterial division apparatus based on FtsZ has not been observed directly, but the division apparatus of plastids has been observed directly as an electron-dense ring by transmission electron microscopy.
In primitive algae, FtsZs that function in mitochondria have been identified (Beech et al., 2000; Takahara et al., 2000b). However, because budding yeast and Caenorhabditis elegans do not have sequences homologous with ftsZ anywhere in their nuclear genomes, most eukaryotic mitochondria divide via a newly created system that includes dynamin GTPase instead of via a system of bacterial origin (Bleazard et al., 1999; Labrousse et al., 1999; reviewed in Erickson, 2000; Margolin, 2000; Osteryoung, 2000). The host cell, which also was once a bacterium, does not divide using an FtsZ ring but uses a contractile ring based on actin and type II myosin. This study suggests that although the bacterial division system is partially preserved in plastids, a new main system was added to plastid division early in plant evolution, probably to regulate the plastid division.
Previously, it was suggested that actin could be involved in the outer PD ring (summarized in Kuroiwa et al., 1998). However, in this study, any helical structure similar to F-actin could not be detected around the outer PD ring. Isolated dividing chloroplasts did not stain with FITC-phalloidin (Miyagishima et al., 1999c). In red alga Cyanidium caldarium, the contractile ring was labeled by anti-actin antibody by using immunoelectron microscopy (Takahashi et al., 1998), but the PD ring was not (H. Takahashi, S. Miyagishima, T. Kuroiwa, unpublished data). From these results, it is probable that the plastid division apparatus does not contain actin. Previous work indicating that phalloidin staining of the PD ring is restricted in some algal species may in fact have detected the contractile ring for cytokinesis rather than the PD ring. Those algae have a single chloroplast per cell, and the cell and the chloroplast divide in the same plane at almost the same time. Moreover, the cells are too small to distinguish the contractile and PD rings by fluorescence microscopy.
The ultrastructure of the inner and middle PD rings could not be visualized in this study because the two rings were extracted by Nonidet P-40. Our morphological studies of the PD ring have shown that throughout the entire organelle division cycle, the behavior of the outer PD ring is quite distinct from that of the inner and middle rings (Miyagishima et al., 1998b, 1999a, 2001). The differences in sensitivity to Nonidet P-40 between these two rings and the outer PD ring could be due to differences in protein composition. The inner PD ring and FtsZ are extracted by Nonidet P-40. Therefore, it is possible that FtsZ interacts with the inner ring. Nevertheless, because the inner PD ring is observed as an electron-dense ring, unlike the bacterial division apparatus, the electron-dense inner ring is likely to be a structure distinct from the FtsZ ring.
Several treatments of the outer PD ring suggest that the ring is relatively insoluble. At the final stage of contraction, the outer ring becomes a large condensed structure >150 nm wide and >40 nm thick. Our observations suggest that the outer PD ring disassembles and disappears from the surface inward over 1 to 2 hr (Miyagishima et al., 2001). It will be interesting to determine how the cytosol disassembles the ring after chloroplast division and how such a rigid bundle of filaments contracts. Identification of the components of the filament and further study at the molecular level are needed. We identified a 56-kD protein as a candidate component of the 5-nm filament (Figure 7). This protein was detected only in the dividing phase of chloroplasts and was enriched in the pellet after Nonidet P-40 treatment. As shown in Figure 7, the insoluble fractions from chloroplasts containing 1 mg of protein were separated in each lane by SDS-PAGE. On the assumption that one outer ring contains 13,000 molecules of 4.8-nm protein, 1 mg of chloroplast protein should contain ∼100 fmol of the 4.8-nm protein, an amount that is clearly detectable by silver staining. We are currently investigating the primary structure of the protein. Further study should illuminate what proteins and mechanisms the host eukaryotic cell created to control the division of the cyanobacterial endosymbionts and then convert them to plastids.
METHODS
Isolation of Dividing and Nondividing Chloroplasts from Synchronous Culture
Cyanidioschyzon merolae were synchronized according to Suzuki et al. (1994). Cells were cultured in Allen's medium at pH 2.5 (Allen, 1959). Flasks were shaken under continuous light (40 W/m2) at 40°C. The cells were subcultured to <107 cells/mL and then synchronized by subjecting them to a 12-hr-light/12-hr-dark cycle at 45°C while the medium was aerated. After the second cycle, cells multiply synchronously. During the light period, cells are in interphase, whereas during the dark period, the chloroplast, mitochondrion, and nucleus divide in that order before cytokinesis. To isolate dividing and nondividing chloroplasts, we harvested cells at the middle of the second dark and third light periods, respectively. In some experiments, to obtain highly synchronized M phase culture, we treated the cells with 5-fluorodeoxyuridine (Sigma) at a concentration of 10 μg/mL at the onset of the second dark period (Miyagishima et al., 1999c). The nondividing chloroplasts also were isolated from concentrated culture (∼3 × 107 cells/mL), in which cells did not divide even under the light/dark cycles. The chloroplasts were isolated from synchronized culture, as described previously (Miyagishima et al., 1999c). Isolated chloroplasts suspended in isolation medium (20 mM Tris, pH 7.6, 5 mM MgCl2, 5 mM KCl, 5 mM EGTA, and 300 mM sucrose) were kept on ice until use. Total protein concentrations from chloroplast fractions were measured with a protein assay kit (Bio-Rad).
Treatment of Chloroplasts with Detergents under Fluorescence Microscopy
To visualize the outer plastid-dividing ring (PD ring) by fluorescence microscopy, we labeled surface proteins of isolated dividing chloroplasts with N-hydroxy-sulfo-succinimidyl biotin (sulfo-NHS-biotin; Pierce Chemical Co.) and detected them using fluorescein isothiocyanate (FITC)–avidin (Amersham Pharmacia) using an epifluorescence microscope (BHS-RFC; Olympus, Tokyo, Japan), as described by Miyagishima et al. (1999c). Isolated dividing chloroplasts labeled with FITC-avidin (∼3 μL; 1 to 5 mg protein/mL) were gently sandwiched between a glass slide (76 × 26 mm) and a cover glass (24 × 24 mm) to avoid crushing the chloroplasts. To view phase–contrast images of chloroplasts, we poured 3 μL of isolation medium containing detergents at different concentrations into the sandwich from the edges of the cover glass at room temperature. In this experiment, we used Triton X-100, Nonidet P-40, sodium cholate, digitonin, and 3-([3-cholamidopropyl]dimethylammonio)propanesulfonic acid. The flow of added medium was monitored by phase–contrast imaging. The effects of the detergents on chloroplasts and the outer PD ring were determined by changes in the phase–contrast and fluorescence images, respectively, under blue light excitation.
Nonidet P-40 Treatment and Negative Staining
Isolated dividing chloroplasts were lysed in isolation medium and 0.1% Nonidet P-40 at a concentration of 1 mg total protein/mL for 1 hr on ice. A portion of untreated chloroplasts was removed by centrifugation at 800g for 5 min, and the insoluble fraction was collected by centrifugation at 10,000g for 15 min at 4°C. Pellets were suspended and fixed in a mixture containing 1% glutaraldehyde (distilled grade; TAAB, Calleva Park, Aldermaston, UK), 300 mM sucrose, and 20 mM sodium cacodylate, pH 7.2, on ice for 30 min. The fixed sample was washed twice with distilled water, absorbed on carbon-coated copper grids, and negatively stained with 0.5% phosphotungstic acid, pH 7.0. The samples were examined with an electron microscope (model JEM-1200EX; JEOL, Tokyo, Japan), and electron micrographs were taken at magnifications of ×10,000 to ×100,000.
Transmission Electron Microscopy for Thin Sections
Transmission electron microscopy of whole cells was performed as described previously (Miyagishima et al., 1999b). For isolated chloroplasts and the insoluble fraction of Nonidet P-40 extraction, samples were fixed in a mixture containing 1% glutaraldehyde, 300 mM sucrose, and 20 mM sodium cacodylate, pH 7.2, on ice for 1 hr. Samples were pelleted, washed twice with 20 mM sodium cacodylate, and postfixed in 1% osmium tetroxide diluted in 20 mM sodium cacodylate for 2 hr on ice. Fixed samples were collected by centrifugation and resuspended in a small volume of 2% agarose (type IX, ultra-low gelling temperature; Sigma). Samples embedded in agarose were cut into 1-mm cubes, dehydrated in a graded ethanol series at room temperature, and embedded in Spurr's resin (Spurr, 1969). Serial thin sections (each 70 nm thick) were stained with uranyl acetate and lead citrate and examined by electron microscopy. Electron micrographs were taken at magnifications of ×10,000 to ×30,000.
SDS-PAGE and Immunoblot Analysis Using Anti-FtsZ Antiserum
Pellets of isolated chloroplasts and Nonidet P-40–insoluble fraction of chloroplasts were lysed in 1 × Laemmli sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.01% bromphenol blue). Supernatants from Nonidet P-40 treatment (10,000g for 15 min) were enriched 20-fold by ultrafiltration with Centricon-10 (10-kD cutoff; Millipore, Bedford, MA) and mixed with one-third volume of 4 × Laemmli sample buffer. All samples were incubated at 65°C for 20 min, separated on 10% SDS–polyacrylamide gels, and transferred onto polyvinylidene difluoride membranes (Immobilon; Millipore) at 100 V for 1.5 hr. The membrane was blocked for 1 hr at room temperature with 3% gelatin in Tris-buffered saline (TBS) and incubated at room temperature for 1 hr with anti-CmFtsZ2 mouse antiserum (Takahara et al., 2000b) diluted at 1:1000. The membrane was washed four times with TBS plus 0.1% Tween 20, and the primary antibody was detected by alkaline phosphatase–conjugated goat anti–mouse IgG (Bio-Rad) diluted at 1:1000. After a 1-hr incubation with secondary antibody at room temperature, the blots were washed three times with TBS plus 0.1% Tween 20 and finally with TBS before the color reaction was developed, according to the manufacturer's instructions (Bio-Rad).
Antibody specificity was examined by a competition experiment. Recombinant CmFtsZ2 eluted from Ni-NTA resin (Takahara et al., 2000b) was dialyzed to TBS at 4°C for 12 hr. The final concentration of the recombinant protein was 20 mg/mL. Pellets of C. merolae cells were lysed in 1 × Laemmli sample buffer and incubated at 65°C for 20 min. Immunoblotting was performed as described above, except that before probing membranes, 1 mL of the diluted antiserum was preincubated for 2 hr in 3% gelatin in TBS with 1 mg of recombinant CmFtsZ2 protein. As a control, rabbit anti-Hsp60 polyclonal antibody (SPA-804; Stressgen, Victoria, British Columbia, Canada) diluted at 1:1000 was used in the same manner as anti-CmFtsZ2 antiserum and detected by alkaline phosphatase–conjugated goat anti–rabbit IgG (Bio-Rad) diluted at 1:1000.
Treatment of the Outer PD Ring with Several Reagents
Isolated dividing chloroplasts containing 1 mg of protein were lysed with Nonidet P-40, and the insoluble fraction was pelleted as described above. The pellet was resuspended in 200 μL of different media (listed below) and held at 37°C (reagents 10 to 15) or on ice (other reagents) for 1 hr: reagents 1 to 7, 20 mM Tris, pH 7.6, 5 mM MgCl2, and 5 mM KCl containing 2 M urea, 3 M urea, 0.5 M NaClO4, 1 M NaClO4, 3 M NaCl, 10 mM EDTA, or 5 mM DTT; reagent 8, 20 mM Tris, pH 7.6; reagent 9, 20 mM Tris, pH 7.6, 5 mM CaCl2; reagents 10 to 15, 20 mM Tris, pH 7.6, 5 mM MgCl2, and 5 mM KCl containing 5 mM ATP, ADP, GTP, GDP, ATPγS, or GTPγS; reagents 16 to 18, 100 mM sodium carbonate/sodium hydrogen carbonate, pH 11.0, 10.0, or 9.0; reagents 19 to 21, 100 mM acetate/sodium acetate, pH 5.0, 4.0, or 3.0; reagent 22, 20 mM Tris, pH 7.6, 5 mM KCl, 5 mM MgCl2, and 5 mM CaCl2 containing thermolysin (type X protease; Sigma) at a concentration of 1 mg/mL; and reagent 23, Laemmli sample buffer. Effects of freezing and thawing on the outer PD ring were examined by freezing at −20°C and thawing on ice gradually. Samples were pelleted by centrifugation at 10,000g for 15 min at 4°C and examined by negative staining, as described above.
Fractionation of the Outer PD Ring
Dividing and nondividing chloroplasts were lysed, and protein concentrations were normalized (1 mg/mL) in sucrose-free isolation medium containing 0.5% Nonidet P-40 and 100 μg/mL DNase I (DN-25; Sigma) for 1 hr on ice. Insoluble fractions were pelleted as described above and then suspended in 20 mM Tris, pH 7.6, and 1 M NaCl on ice for 1 hr. High-salt-washed samples were pelleted by centrifugation at 10,000g for 15 min and washed with sucrose-free isolation medium. Pellets were lysed in 1 × Laemmli sample buffer and separated by SDS-PAGE. Polypeptides were detected by silver staining using a commercial kit (Bio-Rad).
Acknowledgments
This work was supported by a research fellowship to S.M. (No. 10148) from the Japanese Society for the Promotion of Science for Young Scientists and by grants to T.K. (Nos. 12446222 and 12874111) from the Ministry of Education, Science, and Culture of Japan and from the Program for the Promotion of Basic Research Activities for Innovative Biosciences.
References
- Allen, M.B. (1959). Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Microbiol. 32, 270–277. [DOI] [PubMed] [Google Scholar]
- Attardi, G., and Schatz, G. (1988). Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4, 289–333. [DOI] [PubMed] [Google Scholar]
- Beech, P.L., and Gilson, P.R. (2000). FtsZ and organelle division in protists. Protist 151, 11–16. [DOI] [PubMed] [Google Scholar]
- Beech, P.L., Nheu, T., Schultz, T., Herbert, S., Lithgow, T., Gilson, P.R., and McFadden, G.I. (2000). Mitochondrial FtsZ in a chromophyte alga. Science 287, 1276–1279. [DOI] [PubMed] [Google Scholar]
- Bi, E., and Lutkenhaus, J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffey, S.A., and Lloyd, D. (1988). Division and Segregation of Organelles. (Cambridge, UK: Cambridge University Press).
- Bramhill, D. (1997). Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13, 395–424. [DOI] [PubMed] [Google Scholar]
- Bramhill, D., and Thompson, C.M. (1994). GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91, 5813–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida, Y., and Ueda, K. (1991). Division of chloroplasts in a green alga, Trebouxia potteri. Ann. Bot. 67, 435–442. [Google Scholar]
- Colletti, K.S., Tattersall, E.A., Pyke, K.A., Froelich, J.E., Stokes, K.D., and Osteryoung, K.W. (2000). A homologue of the bacterial division site–determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10, 507–516. [DOI] [PubMed] [Google Scholar]
- Dai, K., and Lutkenhaus, J. (1992). The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174, 6145–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett, J.G., and Ligrone, R. (1993. a). Plastid-dividing rings in the liverwort Odontoschisma denudatum (Mart) Dum. (Jungermanniales, Hepaticae). G. Bot. Ital. 127, 318–319. [Google Scholar]
- Duckett, J.G., and Ligrone, R. (1993. b). Plastid-dividing rings in ferns. Ann. Bot. 72, 619–627. [Google Scholar]
- Erickson, H.P. (2000). Dynamin and FtsZ: Missing links in mitochondrial and bacterial division. J. Cell Biol. 148, 1103–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, H.P., Taylor, D.W., Taylor, K.A., and Bramhill, D. (1996). Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraunholz, M.J., Moerschel, E., and Maier, U.G. (1998). The chloroplast division protein FtsZ is encoded by a nucleomorph gene in cryptomonads. Mol. Gen. Genet. 260, 207–211. [DOI] [PubMed] [Google Scholar]
- Gaikwad, A., Babbarwal, V., Pant, V., and Mukherjee, S.K. (2000). Pea chloroplast FtsZ can form multimers and correct the thermosensitive defect of an Escherichia coli ftsZ mutant. Mol. Gen. Genet. 263, 213–221. [DOI] [PubMed] [Google Scholar]
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357. [DOI] [PubMed] [Google Scholar]
- Hashimoto, H. (1986). Double-ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 135, 166–172. [Google Scholar]
- Hashimoto, H. (1997). Electron-opaque annular structure girdling the constricting isthmus of the dividing chloroplasts of Heterosigma akashiwo (Raphydophyceae, Chromophyta). Protoplasma 197, 210–216. [Google Scholar]
- Hashimoto, H., and Possingham, J.V. (1989). Division and DNA distribution in ribosome-deficient plastids of the barley mutant “albostrians.” Protoplasma 149, 20–23. [Google Scholar]
- Hubbard, B.D., and Lazarides, E. (1979). Copurification of actin and desmin from chicken smooth muscle and their copolymerization in vitro to intermediate filaments. J. Cell Biol. 80, 166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa, T. (1982). Mitochondrial nuclei. Int. Rev. Cytol. 75, 1–59. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128, 1–62. [Google Scholar]
- Kuroiwa, T. (1998). The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20, 344–354. [Google Scholar]
- Kuroiwa, T. (2000). The discovery of the division apparatus of plastids and mitochondria. J. Electron Microsc. 49, 123–134. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Suzuki, K., and Kuroiwa, H. (1993). Mitochondrial division by an electron-dense ring in Cyanidioschyzon merolae. Protoplasma 175, 173–177. [Google Scholar]
- Kuroiwa, T., Kuroiwa, H., Sakai, A., Takahashi, H., Toda, K., and Itoh, R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181, 1–41. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Takahara, M., Miyagishima, S., Ohashi, Y., Kawamura, F., and Kuroiwa, H. (1999). The FtsZ protein is not located on outer plastid dividing rings. Cytologia 64, 333–342. [Google Scholar]
- Labrousse, A.M., Zappaterra, M.D., Rude, D.A., and van der Bliek, A.M. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826. [DOI] [PubMed] [Google Scholar]
- Lazarides, E., and Granger, B.L. (1982). Preparation and assay of the intermediate filament proteins desmin and vimentin. Methods Enzymol. 85, 488–508. [DOI] [PubMed] [Google Scholar]
- Leech, R.M., Thomson, W.W., and Platt-Aloika, K.A. (1981). Observations on the mechanism of chloroplast division in higher plants. New Phytol. 87, 1–9. [Google Scholar]
- Lu, C., Stricker, J., and Erickson, H.P. (1998). FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima: Quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskeleton 40, 71–86. [DOI] [PubMed] [Google Scholar]
- Lu, C., Reedy, M., and Erickson, H.P. (2000). Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182, 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, W. (2000). Organelle division: Self-assembling GTPases caught in the middle. Curr. Biol. 10, 328–330. [DOI] [PubMed] [Google Scholar]
- Mita, T., Kanbe, T., Tanaka, K., and Kuroiwa, T. (1986). A ring structure around the dividing plane of the Cyanidium caldarium chloroplast. Protoplasma 130, 211–213. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Takahashi, H., Kuroiwa, H., and Kuroiwa, T. (1998. a). Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J. Electron Microsc. 47, 269–272. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Takahashi, H., Kuroiwa, H., and Kuroiwa, T. (1998. b). Orderly formation of the double ring structures for plastid and mitochondrial division in the unicellular red alga Cyanidioschyzon merolae. Planta 206, 551–560. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Kuroiwa, H., and Kuroiwa, T. (1999. a). Real-time analyses of chloroplast and mitochondrial division and differences in the behavior of their dividing rings during contraction. Planta 207, 343–353. [Google Scholar]
- Miyagishima, S., Itoh, R., Toda, K., Kuroiwa, H., Nishimura, M., and Kuroiwa, T. (1999. b). Microbody proliferation and segregation cycle in the single-microbody alga Cyanidioschyzon merolae. Planta 208, 326–336. [Google Scholar]
- Miyagishima, S., Itoh, R., Aita, S., Kuroiwa, H., and Kuroiwa, T. (1999. c). Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga Cyanidioschyzon merolae. Planta 209, 371–375. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). The timing and manner of disassembly of the apparatuses for chloroplast and mitochondrial division in the red alga Cyanidioschyzon merolae. Planta, in press. [DOI] [PubMed]
- Mori, T., and Tanaka, I. (2000). Isolation of the ftsZ gene from plastid-deficient generative cells of Lillium longiflorum. Protoplasma 214, 57–64. [Google Scholar]
- Mukherjee, A., and Lutkenhaus, J. (1994). Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176, 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, S., Ueda, K., and Noguchi, T. (1995). Division apparatus of chloroplast in Nannochloris bacillaris (Chlorophyta). J. Phycol. 31, 132–137. [Google Scholar]
- Oross, J.W., and Possingham, J.V. (1989). Ultrastructural features of the constricted region of dividing plastids. Protoplasma 150, 131–138. [Google Scholar]
- Osteryoung, K.W. (2000). Organelle fission: Crossing the evolutionary divide. Plant Physiol. 123, 1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Pyke, K.A. (1998). Plastid division: Evidence for a prokaryotically derived mechanism. Curr. Opin. Plant Biol. 1, 475–479. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Vierling, E. (1995). Conserved cell and organelle division. Nature 376, 473–474. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., Stokes, K.D., Rutherford, S.M., Percival, A.L., and Lee, W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possingham, J.V., and Lawrence, M.E. (1983). Controls to plastid division. Int. Rev. Cytol. 84, 1–56. [Google Scholar]
- Pyke, K.A. (1999). Plastid division and development. Plant Cell 11, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, E.J., Rutherford, S.M., and Leech, R.M. (1996). Characterization of chloroplast division using the Arabidopsis mutant arc5. Plant Physiol. 112, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr, A.R. (1969). A low-viscosity resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–34. [DOI] [PubMed] [Google Scholar]
- Strepp, R., Scholz, S., Kruse, S., Speth, V., and Reski, R. (1998). Plant molecular gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95, 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Ehara, T., Osafune, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (1994). Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 63, 280–288. [PubMed] [Google Scholar]
- Takahara, M., Takahashi, H., Matsunaga, S., Sakai, A., Kawano, S., and Kuroiwa, T. (1999). Two types of ftsZ genes isolated from the unicellular primitive red alga Galdieria sulphuraria. Plant Cell Physiol. 40, 784–791. [DOI] [PubMed] [Google Scholar]
- Takahara, M., Takahashi, H., Matsunaga, S., Sakai, A., Kawano, S., and Kuroiwa, T. (2000. a). Isolation, characterization, and chromosomal mapping of an ftsZ gene from the unicellular primitive red alga Cyanidium caldarium RK-1. Curr. Genet. 37, 143–151. [DOI] [PubMed] [Google Scholar]
- Takahara, M., Takahashi, H., Matsunaga, S., Miyagishima, S., Sakai, A., Kawano, S., and Kuroiwa, T. (2000. b). A putative mitochondrial ftsZ gene is encoded in the unicellular primitive red alga Cyanidioschyzon merolae. Mol. Gen. Genet. 264, 452–460. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Takano, H., Kuroiwa, H., Itoh, R., Toda, K., Kawano, S., and Kuroiwa, T. (1998). A possible role for actin dots in the formation of the contractile ring in the ultra-micro alga Cyanidium caldarium RK-1. Protoplasma 202, 91–104. [Google Scholar]
- Tewinkel, M., and Volkmann, D. (1987). Observations on dividing plastids in the protonema of the moss Funaria hygrometrica Sibth: Arrangement of microtubules and filaments. Planta 172, 309–320. [DOI] [PubMed] [Google Scholar]