Abstract
Syntaxins are a large group of proteins found in all eukaryotes involved in the fusion of transport vesicles to target membranes. Twenty-four syntaxins grouped into 10 gene families are found in the model plant Arabidopsis thaliana, each group containing one to five paralogous members. The Arabidopsis SYP2 and SYP4 gene families contain three members each that share 60 to 80% protein sequence identity. Gene disruptions of the yeast (Saccharomyces cerevisiae) orthologs of the SYP2 and SYP4 gene families (Pep12p and Tlg2p, respectively) indicate that these syntaxins are not essential for growth in yeast. However, we have isolated and characterized gene disruptions in two genes from each family, finding that disruption of individual syntaxins from these families is lethal in the male gametophyte of Arabidopsis. Complementation of the syp21-1 gene disruption with its cognate transgene indicated that the lethality is linked to the loss of the single syntaxin gene. Thus, it is clear that each syntaxin in the SYP2 and SYP4 families serves an essential nonredundant function.
INTRODUCTION
The syntaxins are a large evolutionarily conserved family of proteins required for fusion of transport vesicles in eukaryotic cells. Individual syntaxins reside on the organelles of the endomembrane system in which it is believed that they assemble with other proteins of the SNARE (for soluble N-ethylmaleimide sensitive factor attachment protein receptor) family to form t-SNARE complexes. The t-SNARE complex serves as a binding site for v-SNAREs on the transport vesicle, and association of the v-SNARE with the t-SNARE complex is thought to drive membrane fusion (McNew et al., 2000).
The Saccharomyces cerevisiae genome encodes eight syntaxins (Pelham, 1999), two of which, Sso1p and Sso2p, are functionally redundant (Aalto et al., 1993). Gene disruptions of all yeast syntaxins have been characterized, with only the ER syntaxin Ufe1p, the Golgi syntaxin Sed5p, and one of either Sso1p or Sso2p (plasma membrane syntaxins) being essential for viability, although mutations of the other syntaxins result in large defects in vesicle trafficking (reviewed in Pelham, 1999). The effect of the loss of a syntaxin gene in a multicellular eukaryote has been examined in only a few cases. The Caenorhabditis elegans unc-64 mutant, which corresponds to a plasma membrane syntaxin, has pleiotrophic defects in coordination and neurotransmitter release (Ogawa et al., 1998). Mutants in Drosophila melanogaster syntaxin 1 can be examined in flies that are mosaics of mutant and wild-type cells; these flies show multiple defects in many organs, including a failure to complete cellularization in the eye (Schulze and Bellen, 1996; Burgess et al., 1997). In the plant Arabidopsis thaliana, mutations in the KNOLLE gene, which encodes a syntaxin (now called SYP111; see Sanderfoot et al., 2000) involved in cytokinesis, lead to defects in the embryo, eventually resulting in seedling lethality (Lukowitz et al., 1996).
The genome of Arabidopsis encodes 24 syntaxins that we have reclassified recently using the name syntaxin of plants or SYP (Sanderfoot et al., 2000). We have described eight main groups (SYP1 to SYP8), most of which are orthologous to syntaxins of yeast or mammals, although some are novel to plants. In general, each of the SYP groups is encoded by small gene families of one to five paralogous members. Some research has suggested that individual members of particular gene families have distinct localizations and thus may have distinct functions (Sato et al., 1997; Bassham et al., 2000); however, to clearly indicate the function of individual syntaxins would require analysis of mutants in the syntaxin genes.
Methods have been developed for the identification of target-specific gene disruptions in large insertion-mutagenized collections. “Transferred-” or T-DNA of Agrobacterium tumefaciens or transposons, such as the dSpm from Zea mays, are used as a random insertion mutagen to disrupt genes in Arabidopsis (Krysan et al., 1999; Tissier et al., 1999). From such collections, we isolated Arabidopsis plants containing T-DNA or transposon insertions in syntaxins of the SYP2 and SYP4 gene families and examined the effect of the gene disruptions. Disruptions of the yeast orthologs of the SYP2 and SYP4 gene families (Pep12p and Tlg2p, respectively) are viable (Becherer et al., 1996; Holthuis et al., 1998). However, we have found that disruption of single members of each gene family is lethal to the male gametophyte of Arabidopsis, and thus plants cannot be recovered that lack both copies of a syntaxin gene. Complementation of the syp21-1 gene disruption by its cognate transgene restores pollen viability and allows the isolation of plants lacking both copies of SYP21, indicating the lethality is linked directly to the syp21-1 gene disruption. These results indicate that the SYP2 and SYP4 gene families are essential in plants. Furthermore, the fact that disruptions of single genes in each family are lethal indicates that members of each gene family have unique essential functions and, despite the high degree of sequence identity between the gene family members, are not redundant.
RESULTS
Isolation of Syntaxin Mutants
Analysis of the recently completed Arabidopsis genome (Arabidopsis Genome Initiative, 2000) has indicated that 24 syntaxins are found in this model plant (Sanderfoot et al., 2000). We have subdivided these syntaxins into eight groups that contain 10 gene families of one to five paralogous members (Sanderfoot et al., 2000). Our initial focus has been on the genes of the SYP2 and SYP4 families.
The SYP2 family is represented by three genes (SYP21, SYP22, and SYP23) that encode syntaxins that reside on the prevacuolar compartment of Arabidopsis cells (Sanderfoot et al., 1999), although SYP22 also has been reported to localize to the vacuole in some cell types (Sato et al., 1997). The members of the SYP2 family are most similar to the yeast prevacuolar syntaxin Pep12p (Becherer et al., 1996) and to mammalian syntaxins 7 and 13 (which reside on various endosomal compartments; Prekeris et al., 1999). SYP21 (formerly called AtPEP12; Bassham et al., 1995) shares ∼60% amino acid identity with either SYP22 (AtVAM3; Sato et al., 1997) or SYP23 (AtPLP; Zheng et al., 1999), whereas SYP22 and SYP23 share 80% identity. Similarly, the SYP4 family is represented by three members (SYP41, SYP42, and SYP43) that encode syntaxins that localize to the trans-Golgi network (Bassham et al., 2000). The members of the SYP4 family are most similar to the yeast late-Golgi/endosomal-localized syntaxin Tlg2p (Abeliovich et al., 1998; Holthuis et al., 1998; Séron et al., 1998) and mammalian trans-Golgi network-localized syntaxin 16 (Simonsen et al., 1998). SYP41 (AtTLG2a) and SYP43 proteins are 84% identical, and each is 58 or 61% identical (respectively) to SYP42 (AtTLG2b).
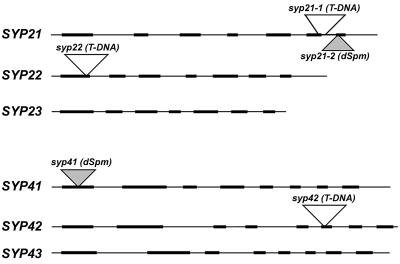
Gene-specific primers were synthesized for each member. Using these reagents, we screened the pools of seeds mutagenized with T-DNA or dSpm transposon insertions (see Methods). In each gene family, insertions were found in two of the three members, with SYP21 having two alleles (Figure 1). In all cases, insertion of the T-DNA or dSpm into a particular gene was confirmed by cloning and sequencing of the genomic DNA bordering the insertions. We discuss in detail the characterization of plants carrying a T-DNA insertion in SYP21 (called syp21-1). Mutations in the other syntaxins shown in Figure 1 have similar phenotypes as syp21-1.
Figure 1.
Arabidopsis Syntaxin Mutants.
Schematic representation of the loci of the SYP2 and SYP4 gene family members, with exons indicated by thick lines and noncoding regions by thin lines. The insertion sites are indicated by triangles (open triangle, T-DNA; filled triangle, dSpm). The insertion in syp21-1 results in a small deletion in the SYP21 locus, which is indicated by the extension of the sides of the triangle to indicate approximately the extent of the deletion.
Characterization of syp21-1
The SYP21 locus is found on chromosome V of Arabidopsis and is contained within the bacterial artificial chromosome F5E19 (GenBank accession number AL391147) encoded in the opposite orientation between nucleotides 66,967 and 69,302. The T-DNA insertion in syp21-1 results in the deletion of 65 nucleotides of SYP21 sequence (nucleotides 67,274 to 67,339), removing the end of exon 6 and part of the following intron. The T-DNA was rearranged during integration into the genome, because both ends of the insertion correspond to the left border; however, at least one complete copy of the T-DNA is present, because the glufosinate resistance gene encoded within the T-DNA is functional. Such rearrangements during integration of the T-DNA are not uncommon (e.g., De Buck et al., 1997). No significant rearrangements or deletions are found with the other T-DNA insertions (syp22 and syp42), although both of the dSpm insertions (syp21-2 and syp41) appear to contain multiple rearranged copies of the transposon within the locus.
Initial examination of the pool containing the syp21-1 insertion revealed several sibling plants carrying the same T-DNA insertion. Further examination showed that all of the recovered plants were heterozygous for the syp21-1 insertion (i.e., SYP21/syp21-1). No obvious growth or developmental defects were seen in the plants heterozygous for syp21-1. Representative plants were allowed to self-fertilize, and the resulting progeny were examined for the state of the SYP21 locus. Using the glufosinate resistance gene contained within the T-DNA to initially score the presence of the syp21-1 insertion, we examined the progeny. Although we expected 3:1 segregation of glufosinate resistance in the progeny, instead we observed ∼1:1 segregation (66 resistant:71 sensitive; 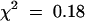 ; P > 0.2). Polymerase chain reaction (PCR) analysis of the genomic DNA of 24 representative glufosinate resistant plants revealed that all contained the syp21-1 insertion in a heterozygous state (SYP21/syp21-1). Subsequently, we examined a similar number of seedlings germinated in the absence of selection. PCR analysis of the genomic DNA showed 13 SYP21/syp21-1 plants, 11 plants lacking the syp21-1 insertion (SYP21/SYP21), and no plants that were homozygous for the insertion (syp21-1/syp21-1). Analysis of the selfed progeny of the SYP21/syp21-1 plants in subsequent generations revealed similar 1:1 segregation of the syp21-1 insertion (47 resistant:53 sensitive;
; P > 0.2). Polymerase chain reaction (PCR) analysis of the genomic DNA of 24 representative glufosinate resistant plants revealed that all contained the syp21-1 insertion in a heterozygous state (SYP21/syp21-1). Subsequently, we examined a similar number of seedlings germinated in the absence of selection. PCR analysis of the genomic DNA showed 13 SYP21/syp21-1 plants, 11 plants lacking the syp21-1 insertion (SYP21/SYP21), and no plants that were homozygous for the insertion (syp21-1/syp21-1). Analysis of the selfed progeny of the SYP21/syp21-1 plants in subsequent generations revealed similar 1:1 segregation of the syp21-1 insertion (47 resistant:53 sensitive; 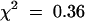 , P > 0.2) and the lack of homozygous plants. Figure 2 shows a representative analysis of 14 selfed progeny (the F3 of an original SYP21/syp21-1 plant isolated from the pool) germinated in the absence of selection. As judged by PCR of genomic DNA from these plants, eight of the 14 contain the syp21-1 insertion (Figure 2, top). PCR with primers that span the insertion site indicate that one copy of the SYP21 locus is still intact (Figure 2, bottom); thus, these plants are heterozygous (SYP21/syp21-1). The segregation patterns of these plants was not due to a second unlinked T-DNA disruption elsewhere in the genome, because all glufosinate-resistant plants we examined carried the syp21-1 insertion, indicating that these plants contained a single T-DNA. The lack of any progeny homozygous for syp21-1 suggested that the loss of SYP21 function is lethal. Furthermore, the pattern of segregation suggested a syp21-1– associated defect in one of the gametes.
, P > 0.2) and the lack of homozygous plants. Figure 2 shows a representative analysis of 14 selfed progeny (the F3 of an original SYP21/syp21-1 plant isolated from the pool) germinated in the absence of selection. As judged by PCR of genomic DNA from these plants, eight of the 14 contain the syp21-1 insertion (Figure 2, top). PCR with primers that span the insertion site indicate that one copy of the SYP21 locus is still intact (Figure 2, bottom); thus, these plants are heterozygous (SYP21/syp21-1). The segregation patterns of these plants was not due to a second unlinked T-DNA disruption elsewhere in the genome, because all glufosinate-resistant plants we examined carried the syp21-1 insertion, indicating that these plants contained a single T-DNA. The lack of any progeny homozygous for syp21-1 suggested that the loss of SYP21 function is lethal. Furthermore, the pattern of segregation suggested a syp21-1– associated defect in one of the gametes.
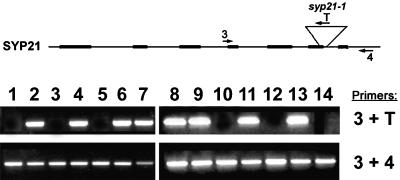
Figure 2.
PCR Analysis of the syp21-1 Insertion.
Genomic DNA was extracted from 14 selfed progeny of a SYP21/syp21-1 plant sowed in the absence of glufosinate. At the top is a schematic diagram of the SYP21 locus with the primer binding sites indicated. PCR was performed on the genomic DNA by using primers 3 and T to indicate the presence of the T-DNA (top). Eight plants were found to carry the syp21-1 insertion, whereas six lacked the T-DNA. Of the plants that carried the syp21-1 insertion, primers spanning the insertion site (primers 3 and 4, bottom) could still amplify the correct size band, indicating that all these plants were heterozygous (SYP21/syp21-1).
Similar analysis of plants carrying the syp21-2, syp22, syp41, and syp42 disruptions indicates that only heterozygous progeny (with similar segregation patterns to syp21-1) can be recovered from these mutants as well.
syp21-1 Defect Occurs First in Pollen
To examine in which of the gametes the syp21-1–associated defect occurs, we crossed heterozygous plants as either a pollen or egg donors to wild-type plants from the Columbia accession (the background of the syp21-1 mutant), and the progeny of the crosses were examined. Use of wild-type pollen to fertilize SYP21/syp21-1 pistils results in many progeny that carry the syp21-1 insertion, suggesting that a syp21-1 egg is viable. However, examination of >100 progeny from a cross in which SYP21/syp21-1 pollen was used to fertilize Columbia pistils revealed no glufosinate-resistant progeny, thus indicating that none carried the syp21-1 insertion. Crosses of the syp22 and syp41 gene disruptions to their wild-type accessions (Wassilewskija and Columbia, respectively) showed a similar inability to pass the mutation through the pollen. As with syp21-1, progeny carrying the insertions could only be recovered when wild-type pollen was used to fertilize the syp22 or syp41 pistils. These results indicate that gene disruptions in other syntaxins show similar pollen defects.
Because these results indicated that the syp21-1 defect was associated with pollen, we examined the ability of pollen from SYP21/syp21-1 plants to germinate in vitro (Li et al., 1999). Although the in vitro germination rate of pollen was variable, pollen from SYP21/syp21-1 plants consistently germinated at ∼50 to 60% of the rate of wild-type pollen (Table 1). Similar results of decreased in vitro pollen germination were seen with heterozygous plants carrying an insertion in SYP42 (SYP42/syp42; Table 1). Examination of the germinated pollen from the SYP21/syp21-1 plants revealed no obvious defects with respect to nuclei number, organelle morphology, or overall appearance at the level of light microscopy. Unfortunately, the variability in the germination rate of even wild-type pollen makes further analysis of pollen phenotype problematic. The low in vitro germination rate of SYP21/syp21-1 plants together with the inability to pass the syp21-1 insertion through the pollen indicates that syp21-1 pollen is most likely inviable.
Table 1.
In Vitro Germination of Pollen
| Experiment
|
|||||
|---|---|---|---|---|---|
| Flower Genotypea | 1 | 2 | 3 | 4 | 5 |
| Wild type | 34% | 26% | 51% | 48% | 56% |
| SYP21/syp21-1 | 21% | 17% | 28% | 26% | 21% |
| syp21-1::T7-SYP21 | 28% | 32% | 37% | 39% | 44% |
| SYP42/syp42 | NDb | ND | 38% | 30% | 29% |
Flowers from independent plants of the indicated genotype were applied to agar plates and allowed to germinate. At least 300 pollen grains were counted for each plant.
ND, not determined.
Complementation of the syp21-1–Associated Lethality
To prove that the observed phenotype is directly associated with loss of SYP21 function, we attempted to complement the mutation with a SYP21 transgene. Plants constitutively expressing a transgene encoding an epitope-tagged version of SYP21 (T7-SYP21) have been described (Sanderfoot et al., 1999), and pollen from these homozygous plants was used to fertilize SYP21/syp21-1 heterozygotes, resulting in several F1 progeny that carried a T7-SYP21 transgene and the syp21-1 insertion, both in a heterozygous state. Several representative plants were allowed to self, and their progeny were examined. When selected on glufosinate and hygromycin (to select for T7-SYP21), 55 plants were resistant to both agents, whereas 40 were sensitive. This is a weak fit ( , 0.1 < P < 0.05) to a 8:4 segregation (expected if pollen lacking both markers is not viable). Twenty-one resistant plants were examined by PCR. As expected from the resistance to both agents, all carried the T7-SYP21 transgene as well as the syp21-1 insertion. Thirteen were of the SYP21/syp21-1 genotype, and eight were now homozygous for syp21-1 (consistent with a predicted 5:3 segregation;
, 0.1 < P < 0.05) to a 8:4 segregation (expected if pollen lacking both markers is not viable). Twenty-one resistant plants were examined by PCR. As expected from the resistance to both agents, all carried the T7-SYP21 transgene as well as the syp21-1 insertion. Thirteen were of the SYP21/syp21-1 genotype, and eight were now homozygous for syp21-1 (consistent with a predicted 5:3 segregation;  , P > 0.2). Figure 3 shows PCR analysis of genomic DNA extracted from representative plants of each genotype. The F2 syp21-1::T7-SYP21 plants were indistinguishable from the wild type and were fertile. DNA was extracted from representative F3 plants, and all progeny carried the T7-SYP21 transgene and were homozygous for the syp21-1 disruption. The expression of T7-SYP21 also increased the in vitro germination of the pollen from the F3 plants almost to the wild-type levels (Table 1). The fact that the expression of T7-SYP21 does not always completely restore the in vitro pollen germination rate (Table 1) may explain the low fit of the segregation to the predicted 8:4, because some pollen expressing both markers may still fail to germinate. These results indicated that the presence of the T7-SYP21 transgene can complement the syp21-1 mutation. This further indicates that the observed lethality is due to loss of SYP21 function, suggesting that despite the high degree of homology between the members of the SYP2 gene family, each member has a unique essential function.
, P > 0.2). Figure 3 shows PCR analysis of genomic DNA extracted from representative plants of each genotype. The F2 syp21-1::T7-SYP21 plants were indistinguishable from the wild type and were fertile. DNA was extracted from representative F3 plants, and all progeny carried the T7-SYP21 transgene and were homozygous for the syp21-1 disruption. The expression of T7-SYP21 also increased the in vitro germination of the pollen from the F3 plants almost to the wild-type levels (Table 1). The fact that the expression of T7-SYP21 does not always completely restore the in vitro pollen germination rate (Table 1) may explain the low fit of the segregation to the predicted 8:4, because some pollen expressing both markers may still fail to germinate. These results indicated that the presence of the T7-SYP21 transgene can complement the syp21-1 mutation. This further indicates that the observed lethality is due to loss of SYP21 function, suggesting that despite the high degree of homology between the members of the SYP2 gene family, each member has a unique essential function.
Figure 3.
Complementation of the syp21-1 by T7-SYP21.
(A) Schematic of the SYP21 locus and of the transgene encoding the T7-SYP21. Binding sites for primers 1, 2, 3, 4, and T are indicated by the arrows. Note that the T7-SYP21 transgene lacks the binding site for primer 4.
(B) PCR analysis of genomic DNA extracted from representative plants of the SYP21/SYP21 (lane 1), SYP21/SYP21::T7-SYP21 (lane 2), SYP21/syp21-1 (lane 3), SYP21/syp21-1::T7-SYP21 (lane 4), and syp21-1::T7-SYP21 (lane 5) genotypes. PCR with primers 1 and 2 amplify a band from the 5′ region of the SYP21 locus as well as from the T7-SYP21 transgene (top). Primers 3 and 4, which overlap the T-DNA insertion (middle), show that both copies of the SYP21 locus can be disrupted only in the presence of the T7-SYP21 transgene. The rearranged T-DNA insertion of syp21-1 is >5 kb and is too large to be efficiently amplified under the conditions used in this experiment. Using a T-DNA specific primer (T) together with primer 3, the presence of the T-DNA in syp21-1 plants can be observed (bottom).
Expression of syp21-1 Protein
To examine whether the syp21-1 protein is produced in plants carrying insertions, we extracted protein from SYP21/syp21-1 and syp21-1::T7-SYP21 seedlings. If expressed, the syp21-1 message would code for a truncated protein that would lack the C-terminal membrane anchor and may no longer be membrane associated. We examined both total protein (Figure 4) and soluble and microsomal extracts of the seedlings by using specific antisera to SYP21. We could never detect any additional anti-SYP21–reactive proteins in the extracts other than the endogenous SYP21 (in SYP21/syp21-1 seedlings) or the epitope tagged T7-SYP21 (in syp21-1::T7-SYP21 seedlings), even upon prolonged exposure (data not shown). These results suggest that the syp21-1 protein (or mRNA) is unstable or is not produced. The fact that we could not detect the endogenous SYP21 in the syp21-1::T7-SYP21 plants confirms our PCR-based definition of these plants as homozygous for the syp21-1 insertion.
Figure 4.
Protein Gel Blot Analysis of syp21-1 Plants.
Leaf tissue was homogenized, and equivalent amounts of total protein from wild-type (lane 1), SYP21/syp21-1 plants (lane 2), and syp21-1::T7-SYP21 plants (lane 3) were separated by SDS-PAGE and blotted with SYP21 antiserum. Note the absence of the endogenous SYP21 in the syp21-1::T7-SYP21 plants; instead, only the epitope-tagged T7-SYP21 (indicated by an arrowhead) is found. The T7-SYP21 runs as a smaller band than the endogeous protein because the N terminus was truncated during addition of the epitope tag (see Sanderfoot et al., 1999).
DISCUSSION
We have attempted to address the function of the syntaxins of the SYP2 and SYP4 gene families of Arabidopsis. Because each of these gene families contains three paralogous members that show high protein sequence identity, it was possible that the individual members of each family would perform redundant functions. However, prior studies have suggested that this was not the case, because individual members within each group were found to have distinct localizations (Sato et al., 1997; Sanderfoot et al., 1999; Bassham et al., 2000). Consistent with these results, we have found that gene disruptions in single syntaxin genes from these families are lethal to the male gametophyte, indicating that each member of these gene families has a unique essential function.
The inability to acquire homozygous syntaxin mutants is somewhat surprising. In the case of both SYP22 and SYP41, a second expressed protein of ∼80% sequence identity also is found (SYP23 and SYP42, respectively) yet cannot complement the loss of its paralog. It is likely that each member of the gene family has specialized into a unique essential function. Consistent with this idea, we have found that SYP41 and SYP42 are localized to distinct domains of the TGN (Bassham et al., 2000). Unfortunately, because the lethality of the syntaxin mutants occurs at such an early time during development (i.e., during development of the male gametophyte), we were unable to precisely determine for which function these proteins are essential. Because it is clear that syntaxins play a major role in the fusion of transport vesicles (McNew et al., 2000), it is most likely that the absence of a syntaxin creates a block in vesicle traffic within the endomembrane system. The lethality induced by loss of a syntaxin would occur due to an inability to deliver essential cargo to the organelles on which these syntaxins reside. The nature of this cargo remains unclear, but considering the many potential roles for the late endosomal/vacuolar system in growth and development (De, 2000), this cargo could be involved in a great many essential processes.
The loss of a single syntaxin appears to be lethal to the male gametophyte. Each of the syntaxins we have examined at the protein level appears to be produced ubiquitously in all tissues examined, including pollen (Conceição et al., 1997; Sato et al., 1997; data not shown), which is consistent with a possible function for these proteins in pollen. Yet, because of the near ubiquitous expression of these syntaxins and important roles proposed for syntaxins in vesicular traffic, we do not believe that the only essential role for these syntaxins is in pollen. Although it is possible, it is unlikely that all of the syntaxins mutated thus far will have unique essential functions in these particular cells. Rather, we think that the male gametophyte is the first opportunity to “notice” the loss of the particular syntaxin, and because viable pollen cannot be isolated that carries the mutated syntaxin gene, homozygous plants cannot be produced. Why a syp21-1 egg is viable is unclear. One speculation is that because the large egg cell contains much more cytoplasm than a pollen grain, sufficient SYP21 mRNA or protein (derived from the heterozygous megasporocyte before meiosis) remains to allow development until the pollen delivers a functional SYP21 gene.
Note that the pollen-associated lethality of syp21-1 can be rescued by expression of a cognate transgene (T7-SYP21). This clearly indicated that the lethality was due to the loss of SYP21 function in the pollen. Note also that the T7-SYP21 is being expressed at low levels in these lines, yet the small amount of protein (see Figure 4, lane 3) is sufficient to allow pollen to survive. This suggests that the complementation is not due to overexpression of the transgene; in fact, it appears that the amount of T7-SYP21 protein is much less than the endogenous SYP21 protein. The amount of T7-SYP21 being produced is also less than the amount of SYP22 in these plants (see Sanderfoot et al., 1999). This is further evidence that the members of the SYP2 family have distinct functions, because SYP22 is present at much higher levels than T7-SYP21 yet cannot complement the defect in the syp21-1 pollen.
Because a homozygous gene disruption of a syntaxin can be isolated in the presence of a complementing transgene, we now are attempting to complement syntaxin mutants by using conditional expression of the transgene (i.e., the transgene is under the control of an inducible or repressible promoter). Although such conditional expression systems are somewhat unpredictable in plants, we hope to be able to produce plants whose only functional syntaxin gene can be transcriptionally controlled, thus allowing us to examine the function of a particular syntaxin in an intact plant. Such experiments are currently in progress.
In conclusion, we have been able to show that the presence of gene families encoding the Arabidopsis syntaxins of the SYP2 and SYP4 types does not indicate genetic redundancy. Instead, each syntaxin gene appears to have a unique essential function in the Arabidopsis cell. Furthermore, unlike their yeast orthologs, members of these gene families are essential to the viability of plant cells.
METHODS
Seed Pools of T-DNA or dSpm Mutants
Seed pools of T-DNA–mutagenized Arabidopsis thaliana were acquired from the Arabidopsis Biological Resource Center (Columbus, OH) or from the Arabidopsis Functional Genomics Center (http://afgc.stanford.edu). Seeds carrying dSpm insertions were acquired from the Sainsbury Laboratory at the John Innes Centre (Norwich, UK).
Screening of the insertion mutagenized collections for null mutants in each Arabidopsis syntaxin was performed at Mendel Biotechnology Inc. by using a polymerase chain reaction (PCR)–based methodology similar to that of Krysan et al. (1999). Nested gene-specific primers were designed from the 5′ and 3′ untranslated region of each syntaxin. Nested sets of primers also were created specific to each of the T-DNA or transposon ends (called right and left borders). All possible combinations of gene-specific and T-DNA/transposon primers were used in PCR to detect rare insertion events within the target gene. Amplified DNA fragments were sequenced to determine the insertion point relative to the target gene. Insertion events were deconvoluted from the pools to single plants. Further characterizations of the single plants were conducted at Michigan State University. Sequences of the gene-specific and “border” primers are found in Table 2.
Table 2.
Primers Described In This Study
| Gene | Mutant Allelea | Primer Name | Sequence |
|---|---|---|---|
| SYP21 | Forward | CATCATCTTCGTCTTCGTTTGTTCCC | |
| Nested forward | GAGAGATTCGAGAGAAGAGACGAG | ||
| Reverse | CAATTTGGTTGAATGGGGAAGATGGAGT | ||
| Nested reverse | GTTTCCATAGATTCGCTTGATGCTTGAAG | ||
| Primer 1b | GACAAGGTTACAGATATCCGAATTGGTG | ||
| Primer 2b | ACCTGAACTGGATACTGAATCTTTAAG | ||
| Primer 3b | TAGAAGAAGAGCTTGCTGCTGGGAGAT | ||
| Primer 4b | Same as reverse from above | ||
| syp21-1c | T-DNA left (primer Tb) | CGCACTCAAGTCTTTCATCTACGGCAATGACC | |
| T-DNA left nested | AGATAGCTGGGCAATGGAATCCGAGGAGG | ||
| syp21-2 | dSpm 5′ | GGTGCAGCAAAACCCACACTTTTACTTC | |
| dSpm 5′ nested | CCGACACTCTTTAATTAACTGACATC | ||
| dSpm 3′ | CTTATTTCAGTAAGAGTGTGGGGTTTTGG | ||
| dSpm 3′ nested | GACACTCCTTACCTTTTTTCTTGTA | ||
| SYP22 | Forward | GAGATTAGAGAAAACTCCGATAACCAA | |
| Forward nested | ATCAATACCGGAGTCTCCACGTTTCAG | ||
| Reverse | ACAACAAAAAGCCACACACGCACACA | ||
| Reverse nested | CAATAGAACCTCAAACCGAACTGAACCA | ||
| syp22d | T-DNA left | CTCATCTAAGCCCCCATTTGGACGTGAATG | |
| T-DNA left nested | TTGCTTTCGCCTATAAATACGACGGATCG | ||
| T-DNA right | TGGGAAAACCTGGCGTTACCCAACTTAAT | ||
| SYP41 | Forward | CGATTGTGTTCAGATCGGAGCTG | |
| Forward nested | GCGACGAGGAATCGTACGTTGCTG | ||
| Reverse | GACATCGACCTCTGACTATCTAT | ||
| Reverse nested | GAGATGACAAGCACGGATGCGCAC | ||
| syp41 | dSpm primers are same as listed above for syp21-2 | ||
| SYP42 | Forward | CGTCTTTGCGCCACCGTGTTAAAACGTGTG | |
| Forward nested | CACACACTTATATTGACCCTATCGCGCAAG | ||
| Reverse | TCATGTGTTATTGAATTTGGAGGACTAGTG | ||
| Reverse nested | ATACTTCTACAGATTTGGTAAAACGATTG | ||
| syp42 | T-DNA primers are same as listed for syp22 |
Primers specific to the DNA inserted in these alleles (i.e., for the particular T-DNA or transposon).
syp21-1 T-DNA locus is inserted with two left borders (see text), so no right border primers are listed.
syp22 was identified using only left border primers, and the listed right border primer was used only to verify the opposite end of the insertion.
Seeds of the heterozygous syntaxin mutants can be acquired from the authors after completion of a Materials Transfer Agreement with Mendel Biotechnology Inc. Contact the authors for further information.
Genomic DNA Extraction and Purification
Genomic DNA was extracted from single leaves taken from young soil-grown Arabidopsis plants by using a CTAB extraction method modified from Rogers and Bendich (1994). Briefly, single leaves were placed in microcentrifuge tubes and ground to a powder with liquid nitrogen. Four hundred microliters of CTAB extraction buffer (1% [w/v] hexadecyltrimethylammonium bromide [CTAB; Sigma], 50 mM Tris-HCl, pH 8.0, 0.7 M NaCl, 10 mM EDTA, 0.5% [w/v] polyvinylprylidone, and 0.1% [v/v] 2-mercaptoethanol) was added to each tube, and the tube was incubated at 60°C for 60 min. An equal volume of chloroform was added, and the tube was vortexed and then centrifuged in a microcentrifuge for 5 min. Genomic DNA was precipitated from the aqueous phase by addition of 1 volume of isopropanol, incubating at −20°C for 10 min, and centrifuging for 5 min. Genomic DNA was resuspended in RNase buffer (25 mM Tris-HCl, pH 7.5, 10 mM EDTA, and 100 μg/mL RNase A) and incubated at 37°C for 30 min. Two volumes of ethanol were added, and the genomic DNA was recovered by centrifugation. Finally, the DNA was resuspended in 50 μL of TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), quantified by A260, and stored at 4°C.
Growth and Selection of Plants
The T-DNA of syp21-1 and the dSpm of syp21-2 and syp41 each carries a gene that results in resistance to glufosinate (Cresent Chemical Co., Hauppauge, NY). The T-DNA of syp22 and syp42 each carries a gene leading to resistance to kanamycin (Sigma). Seedlings were selected on Murashige Minimal Organics Medium (Life Technologies, Grand Island, NY) containing 1% (w/v) sucrose solidified with 0.8% (w/v) Phytagar (Life Technologies) by using either 10 mg/L glufosinate ammonium or 50 mg/L kanamycin sulfate. Selection also was performed by sowing seeds on soil containing 100 mg/L glufosinate (Basta; AgrEvo, Frankfurt/Main, Germany). Plants carrying the T7-SYP21 transgene (previously called T7-AtPEP12; Sanderfoot et al., 1999) also encode a gene for resistance to hygromycin B (Sigma). Selection of plants carrying both syp21-1 and T7-SYP21 was performed in the presence of 5 mg/L glufosinate ammonium and 30 mg/L hygromycin B hydrochloride.
Pollen was germinated in vitro as described in Li et al. (1999). Pollen collected from three independent plants of each genotype was examined by light microscopy for the presence of a pollen tube 12 hr after plating. At least 300 pollen grains were scored for each plant in each experiment.
SYP21 Chicken Antiserum
SYP21[1-129]-H6 (produced and purified as described by Sanderfoot et al. [1999]) was injected into a chicken at Cocalico Biologicals (Reamstown, PA). Cross-reactivity with SYP22 was removed (Sanderfoot et al., 1999), and the antibodies were affinity purified (as described by Bassham et al. [2000]) by using the cytosolic domain of SYP21 fused to glutathione S-transferase (Sanderfoot et al., 1999). These antibodies show staining patterns identical to those of the previously characterized rabbit SYP21 antisera (Conceição et al., 1997; Sanderfoot et al., 1999; data not shown).
Acknowledgments
We gratefully acknowledge the help of many researchers involved with the production of the T-DNA and dSpm insertion lines, including the Arabidopsis Biological Resource Center, the Arabidopsis Functional Genomics Center, the Sainsbury Laboratory, the Institut National de la Recherche Agronomique, and the members of the Ken Feldmann, Thomas Jack, and Detlef Weigel laboratories. We also thank Diane Bassham, John Froehlich, Linda Fitzpatrick, and Farhah Assaad for comments on the manuscript. A.A.S. was supported by a National Institutes of Health postdoctoral fellowship (GM 18861). N.V.R. was supported by grants from the Department of Energy (No. DE-FG02-91ER-20021) and the National Science Foundation (No. MCB-9507030).
References
- Aalto, M.K., Ronne, H., and Keranen, S. (1993). Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12, 4095–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich, H., Grote, E., Novick, P., and Ferro-Novick, S. (1998). Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273, 11719–11727. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Sequence and analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bassham, D.C., Gal, S., Conceição, A.S., and Raikhel, N.V. (1995). An Arabidopsis syntaxin homologue isolated by functional complementation of a yeast pep12 mutant. Proc. Natl. Acad. Sci. USA 92, 7262–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D.C., Sanderfoot, A.A., Kovaleva, V., Zheng, H., and Raikhel, N.V. (2000). AtVPS45 complex formation at the trans-Golgi network. Mol. Biol. Cell 11, 2251–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer, K.A., Reider, S.E., Emr, S.D., and Jones, E.W. (1996). Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole of yeast. Mol. Biol. Cell 7, 579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, R.W., Deitcher, D.L., and Schwarz, T.L. (1997). The synaptic protein syntaxin1 is required for cellularization of Drosophila embryos. J. Biol. Chem. 138, 861–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição, A.S., Marty-Mazars, D., Bassham, D.C., Sanderfoot, A.A., Marty, F., and Raikhel, N.V. (1997). The syntaxin homolog AtPEP12p resides on a late post-Golgi compartment in plants. Plant Cell 9, 571–582. [PMC free article] [PubMed] [Google Scholar]
- De, D.N. (2000). Plant Cell Vacuoles: An Introduction. (Collingwood VIC, Australia: CSIRO Publishing).
- De Buck, S., Jacobs, A., Van Montagu, M., and Depicker, A. (1997). The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J. 20, 295–304. [DOI] [PubMed] [Google Scholar]
- Holthuis, J.C.M., Nichols, B.J., Dhruvakumar, S., and Pelham, H.R. (1998). Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17, 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jurgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- McNew, J.A., Parlati, F., Fukuda, R., Johnston, J., Paz, K., Paumet, F., Söllner, T.H., and Rothman, J.H. (2000). Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature 407, 153–159. [DOI] [PubMed] [Google Scholar]
- Ogawa, H., Harada, S., Sassa, T., Yamamoto, H., and Hosono, R. (1998). Functional properties of the unc-64 gene encoding a Caenorhabditis elegans syntaxin. J. Biol. Chem. 273, 2192–2198. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R. (1999). SNAREs and the secretory pathway—Lessons from yeast. Exp. Cell Res. 247, 1–8. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Yang, B., Oorschot, V., Klumperman, J., and Scheller, R.H. (1999). Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell 10, 3891–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1994). Extraction of total cellular DNA from plants, algae and fungi. In Plant Molecular Biology Manual, 2nd ed, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–8.
- Sanderfoot, A.A., Kovaleva, V., Zheng, H., and Raikhel, N.V. (1999). The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 121, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot, A.A., Assaad, F.F., and Raikhel, N.V. (2000). The Arabidopsis thaliana genome: An abundance of l soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 124, 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M.H., Nakamura, N., Ohsumi, Y., Kouchi, H., Kondo, M., Hara-Nishimura, I., Nishimura, M., and Wada, Y. (1997). The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J. Biol. Chem. 272, 24530–24535. [DOI] [PubMed] [Google Scholar]
- Schulze, K.L., and Bellen, H.J. (1996). Drosophila syntaxin is required for cell viability and may function in membrane formation and stabilization. Genetics 144, 1713–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séron, K., Tieaho, V., Prescianotto-Baschong, C., Aust, T., Blondel, M.-O., Guillaud, P., Devilliers, G., Rossanese, O.W., Glick, B.S., Riezman, H., Keränen, S., and Hauguenauer-Tsapis, R. (1998). A yeast t-SNARE involved in endocytosis. Mol. Biol. Cell 9, 2873–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, A., Bremnes, B., Rønning, E., Aasland, R., and Stenmark, H. (1998). Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol. 75, 223–231. [DOI] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Bassham, D.C., Conceição, A.S., and Raikhel, N.V. (1999). The syntaxin family of proteins in Arabidopsis: A new syntaxin homologue shows polymorphism between two ecotypes. J. Exp. Bot. 50, 915–924. [Google Scholar]