Abstract
DNA microarrays bearing nearly all of the genes of the unicellular cyanobacterium Synechocystis sp PCC 6803 were used to examine the temporal program of gene expression during acclimation from low to high light intensity. A complete pattern is provided of gene expression during acclimation of a photosynthetic organism to changing light intensity. More than 160 responsive genes were identified and classified into distinct sets. Genes involved in light absorption and photochemical reactions were downregulated within 15 min of exposure to high light intensity, whereas those associated with CO2 fixation and protection from photoinhibition were upregulated. Changes in the expression of genes involved in replication, transcription, and translation, which were induced to support cellular proliferation, occurred later. Several unidentified open reading frames were induced or repressed. The possible involvement of these genes in the acclimation to high light conditions is discussed.
INTRODUCTION
Photosynthetic organisms must acclimate to changing light intensity in their environment. The acclimation process includes changes in the photosynthetic apparatus, presumably to balance energy input and consumption (Boardman, 1977). If energy supply (light harvesting and electron transport) exceeds its dissipation (by CO2 fixation and other energy-demanding processes), particularly under high light (HL) conditions, the photosynthetic electron transport components could become relatively reduced. This may result in excess production of reactive oxygen species (ROS) leading to severe damage to many cellular processes (Asada, 1994). Absorption of excess light energy therefore must be avoided by reduction of both antenna size and photosystem content. Furthermore, under HL conditions the capacity for CO2 fixation increases (Björkman, 1981; Anderson, 1986) and protection from ROS is enhanced (Grace and Logan, 1996).
Acclimation from low light (LL) to HL conditions can be divided into short-term and long-term processes (Anderson, 1986; Anderson et al., 1995). Short-term acclimation includes state transitions, protective energy dissipation (Campbell et al., 1998; Niyogi, 1999), changes in the efficiency of energy transfer from the harvesting complex to photosystem II (PSII; Hassidim et al., 1997), and the formation of nonfunctional PSII reaction centers (Chow, 1994; Anderson et al., 1997). These responses occur rather rapidly and usually are completed within several minutes. Conversely, the long-term acclimation to HL is much slower because it involves changes in the composition, function, and structure of the photosynthetic apparatus as well as other photosynthesis-related components. Completion of the long-term processes may take hours or even days.
Physiological responses to changing light intensity have been examined extensively (Foy and Gibson, 1982a, 1982b; Anderson, 1986; Neale and Melis, 1986), and some of the relevant molecular mechanisms have been described (Anderson et al., 1995; Hihara, 1999; Niyogi, 1999). Nevertheless, the mechanisms that enable the cells to acclimate to the changing light field are still poorly understood. By means of DNA microarray analyses, the expression of thousands of genes can be monitored simultaneously (Schena et al., 1995; DeRisi et al., 1996, 1997). In this study, we surveyed time-dependent gene expression in the unicellular cyanobacterium Synechocystis sp PCC 6803 as affected by transfer from LL to HL. This organism is well suited for such study because the entire genomic sequence has been determined (Kaneko et al., 1996), and DNA microarrays representing all open reading frames (ORFs) are now available. Mutants impaired in the ability of this organism to acclimate to HL are available in our laboratory including a pmgA mutant that could not modulate the photosystem stoichiometry during acclimation to HL (Hihara and Ikeuchi, 1997; Hihara et al., 1998). We examined the mRNA levels change at 15 min, 1 hr (initial stressed phase), 6 hr, and 15 hr (acclimated phase) after the shift to HL conditions. We found that the expression of 84 ORFs was upregulated and the expression of 80 ORFs was downregulated after these HL exposures.
RESULTS
Light Intensity and Duration of Exposures
Synechocystis cells can grow under a wide range of light intensities, up to approximately 1000 μmol photons m−2 sec−1 (Mohamed and Jansson, 1989; Yokoyama et al., 1991). However, to avoid excessive damage under very HL intensities (Constant et al., 1997), we examined the response of gene expression to a shift of light intensity from 20 μmol photons m−2 sec−1 (LL) to 300 μmol photons m−2 sec−1 (HL). This intensity was sufficient to induce large changes in the gene expression profile. Our earlier studies on the effect of light intensity on the content of chlorophyll and phycocyanin per cell showed that the cells responded in three different phases: (1) drastic reduction to approximately two thirds within 3 hr; (2) moderate decrease at a slower rate for up to 12 hr; and (3) gradual recovery after 12 hr (Hihara et al., 1998). Hence, in the present study, we sampled the cells for RNA isolation after 15 min, 1 hr, 6 hr, and 15 hr of exposure to the HL treatment. The measurements at 15 min and 1 hr represent the initial stressed phase, whereas the 15-hr point represents the acclimated phase. The 6-hr point was selected because it was the doubling time of the culture under the HL regimen and may indicate the change from the stressed phase to the acclimated phase.
Reliability of the DNA Microarray Analysis
Total RNA from LL and HL cells was labeled with fluorescent dye, either Cy3 or Cy5. Generated cDNA probes were mixed and hybridized to CyanoCHIP version 0.8 (see Methods). Figure 1 shows a typical overlay image of Cy3 and Cy5 fluorescence on the microarray after hybridization. In the case presented here, the microarray was hybridized with Cy3-labeled cDNA from LL-grown cells and Cy5-labeled cDNA from cells grown in HL for 6 hr. In this image, some red spots (preferentially expressed in LL) and green spots (preferentially expressed in HL) are seen, in addition to many yellow spots (equally expressed). To evaluate such differences in gene expression, we quantified the fluorescence intensity of each spot in both Cy3 and Cy5 images (see Methods), and the expression of each gene under HL conditions was shown as a percentage of the LL levels.
Figure 1.
Two-Color Overlaid Fluorescent Image of a DNA Microarray of HL-Induced and HL-Repressed Genes in Synechocystis sp PCC 6803.
Two hybridization data sets from the same microarray were converted into pseudocolor images and superimposed to visualize differential gene expression between LL (Cy3, red) and HL of 6 hr (Cy5, green). ORFs unaffected by the HL shift appear as yellow spots. The upper part of the microarray is shown.
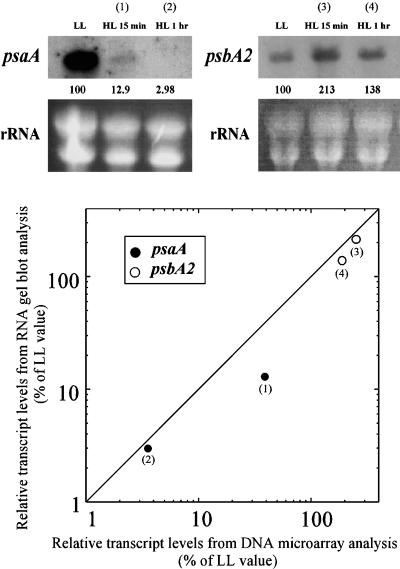
To determine the reliability of these data, we compared the expression levels of psaA and psbA2 between the DNA microarray and the RNA gel blot analyses shown in Figure 2. It is clear that relative transcript levels assessed by RNA gel blot analysis correlated well over a wide range with those obtained by the DNA microarray analyses. In some cases, RNA levels deduced by the DNA microarray analyses were higher than those assessed by the RNA gel blot analysis (Figure 2, spot 1), probably because rapidly degrading RNA was being detected by dot blot analysis such as the microarray technique. Furthermore, in the DNA microarray methodology, the ratio of HL to LL signals tended to be lower than in the RNA gel blot analysis. This was most pronounced in the case of “weak spots” due to the masking effects of background hybridization (data not shown). In fact, background fluorescence levels or signal intensities of negative control spots (human transferrin receptor gene) often scored at approximately 2500 of 3079 spots. In the case of genes with expression levels lower than 2000 of 3079, it would be difficult to reproducibly detect changes in the transcript levels even if they were markedly affected by HL. Therefore, in this study, we focused mainly on genes with expression levels that were >2000 of 3079 in both LL and HL conditions.
Figure 2.
Comparison of the Results Obtained by DNA Microarray Analysis and RNA Gel Blot Analysis.
The top panel shows the RNA gel blot analysis of psaA and psbA2 transcripts under LL and HL conditions. Values below the blots show the relative levels of transcripts expressed as a percentage of values under LL conditions. The profile of rRNAs stained with ethidium bromide is shown as a control for equal RNA loading. The bottom panel shows the correlation between the relative transcript levels obtained by DNA microarray analysis and RNA gel blot analysis. The same RNA sample was used for the DNA microarray analysis and the RNA gel blot analysis.
Changes in the Transcript Abundance of Genes Encoding Thylakoid-Located Complexes
PSII-Related Genes
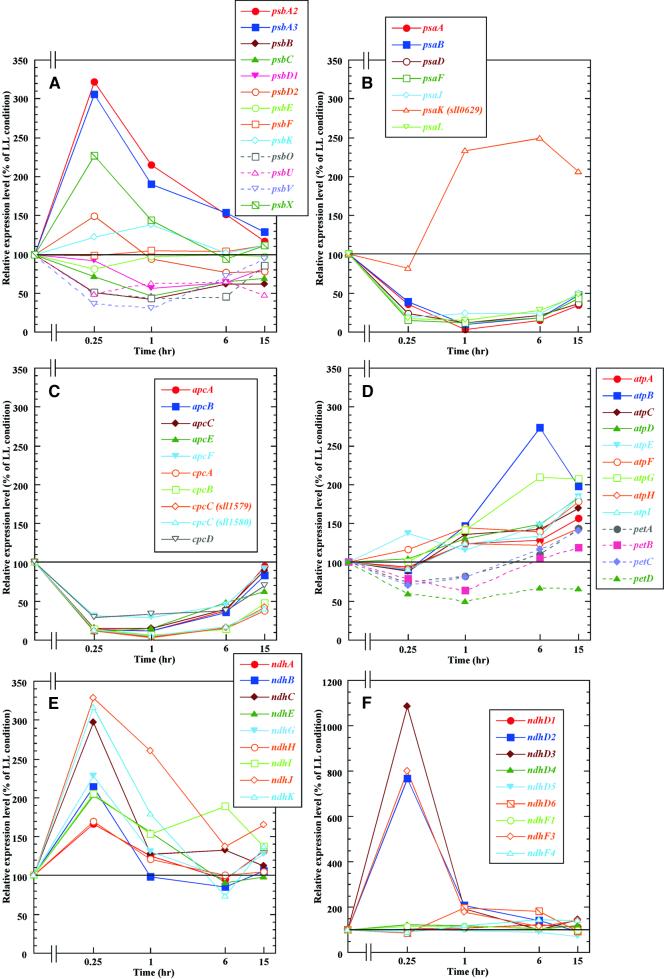
Among the psb genes, which encode subunits of PSII, the transcript abundance of psbA genes encoding the D1 protein increased approximately threefold within 15 min and subsequently decreased (Figure 3A). Because the nucleotide sequences of the coding regions of psbA2 and psbA3 are almost identical, we could not distinguish their transcripts. Similar induction pattern by HL treatment was observed in the case of psbX (Figure 3A), which encodes the small subunit localized to the vicinity of the reaction center (Shi et al., 1999). In contrast, the transcript levels of psbD1 that encodes reaction center D2, psbB-encoding CP47, and psbC that encodes CP43 decreased within the first period of HL and reached a minimum value 1 hr after the shift to HL. However, longer exposure to HL resulted in an increased level of transcripts originating from these genes. After 15 hr of HL treatment, their abundance was 50 to 100% of the level observed under LL. Transcripts of genes involved in the oxygen-evolving complex, psbO, psbU, and psbV, were all downregulated to half the level in LL within 15 min. The transcript levels of many other genes that encode small subunits of PSII, including psbE, psbF, and psbK, were not affected by the HL treatment. We did not observe significant induction of the psbD2 gene, which has been reported to be inducible by HL in Synechococcus sp PCC 7942 (Bustos and Golden, 1992; Anandan and Golden, 1997).
Figure 3.
Accumulation Profile of Transcripts for Protein Complexes in Thylakoid Membranes upon the Shift to HL.
The transcript abundance as affected by time after transfer of Synechocystis sp PCC 6803 from LL to HL. Transcript levels for subunits of PSII (A), subunits of PSI (B), phycobiliproteins (C), subunits of ATP synthase and the cytochrome b6/f complex (D), subunits of NADPH dehydrogenase in which genes exist as a single copy in the genome (E), and subunits of NADPH dehydrogenase that are encoded by multiple genes (F) are shown. Each time point is shown logarithmically on the horizontal axis. Accumulation levels of transcripts are expressed on the vertical axis as a percentage of values under LL conditions. Values at 15 min and 1 hr are averages of duplicate results for three independent experiments (six data points in total), and values at 6 and 15 hr are averages of duplicate results for two experiments (four data points in total). The weakly expressed genes encoding thylakoid proteins are omitted from the figures. Identification numbers of the depicted genes are as follows: psbA2, slr1311; psbA3, sll1867; psbB, slr0906; psbC, sll0851; psbD1, sll0849; psbD2, slr0927; psbE, ssr3451; psbF, smr0006; psbK, sml0005; psbO, sll0427; psbU, sll1194; psbV, sll0258; psbX, sml0002; psaA, slr1834; psaB, slr1835; psaD, slr0737; psaF, sll0819; psaJ, sml0008; psaK, sll0629; psaL, slr1655; apcA, slr2067; apcB, slr1986; apcC, ssr3383; apcE, slr0335; apcF, slr1459; cpcA, sll1578; cpcB, sll1577; cpcC, sll1579 and sll1580; cpcD, ssl3093; atpA, sll1326; atpB, slr1329; atpC, sll1327; atpD, sll1325; atpE, slr1330; atpF, sll1324; atpG, sll1323; atpH, ssl2615; atpI, sll1322; ndhA, sll0519; ndhB, sll0223; ndhC, slr1279; ndhE, sll0522; ndhG, sll0521; ndhH, slr0261; ndhI, sll0520; ndhJ, slr1281; ndhK, slr1280; ndhD1, slr0331; ndhD2, slr1291; ndhD3, sll1733; ndhD4, sll0027; ndhD5, slr2007; ndhD6, slr2009; ndhF1, slr0844; ndhF3, sll1732; and ndhF4, sll0026.
Photosystem I–Related Genes
In contrast with the case of genes encoding subunits of PSII, some of which were upregulated and others downregulated (Figure 3A), almost all of the photosystem I (PSI) genes were downregulated by five- to 10-fold within 1 hr of the transfer from LL to HL (Figure 3B). Notably, the level of psaAB transcripts for the reaction center of PSI declined by 30-fold of its original value after 1 hr of HL treatment. An exception was one of the two psaK genes (Nakamoto and Hasegawa, 1999; Naithani et al., 2000) that was markedly induced within 1 hr and stayed at a high level throughout the exposure to HL (Figure 3B).
Phycobilisome-Related Genes
Figure 3C shows that the transcription of apc genes related to allophycocyanin and of cpc genes related to phycocyanin is regulated differently. The apc genes were downregulated to one-tenth of their original level within 1 hr of HL but then recovered to the initial level within 15 hr. Conversely, the abundance of the cpc transcript decreased by 20-fold within 1 hr of HL and recovered after 15 hr of HL to a level 50% of the initial. The cpc genes may be downregulated more strictly than apc genes during exposure to HL because phycocyanin, which is located at the terminal regions of the phycobilisome rods, is the primary target for the reduction of antenna size. It is interesting that cpcD, which is part of the cpcBACCD operon, apparently was not downregulated as much as the other cpc genes. It is possible that this was due to the relatively weak signal of the cpcD spot. As indicated above, changes in transcript levels of weakly expressed genes tend to be underestimated because of background hybridization. Furthermore, cpcD may be transcribed not only polycistronically but also monocistronically. Belknap and Haselkorn (1987) reported that, in Anabaena sp PCC 7120, the cpcBACDE operon was downregulated by HL, whereas the level of the transcript that encodes the last two ORFs, cpcDE, was relatively insensitive to light intensity.
The ndh Genes
Synechocystis sp PCC 6803 possesses many ndh genes encoding subunits of NADPH dehydrogenase. Some of the ndh genes are present in single copy (Figure 3E), and some are present in multiple copies (Figure 3F). The transcript abundance of ndhC, ndhK, and ndhJ, which constitutes an operon structure (slr1279 to slr1281), increased by threefold within 15 min of exposure to HL (Figure 3E), whereas the extent of induction of ndhA, ndhI, ndhG, and ndhE (which constitutes another operon, sll0519 to sll0522) was smaller (Figure 3E). There are six copies of the ndhD gene and three copies of the ndhF gene in Synechocystis sp PCC 6803 (Price et al., 1998). Surprisingly, ndhD2, ndhD3, and ndhF3 showed transient and prominent induction by eightfold or even more within 15 min of HL, whereas transcription of the other genes did not respond to the HL treatment (Figure 3F). ndhF3, ndhD3, and an ORF encoded by sll1734 form an operon thought to be involved in CO2 uptake in Synechocystis sp PCC 6803 (Ohkawa et al., 1998, 2000). The putative protein encoded by sll1734 also was induced by HL (Table 1). Ohkawa et al. (1998) observed that ndhD2, which is transcribed monocistronically, and the ndhF3 operon also are induced by low CO2. These ndh genes may be induced by common signals shared by low CO2 and HL (Kaplan and Reinhold, 1999).
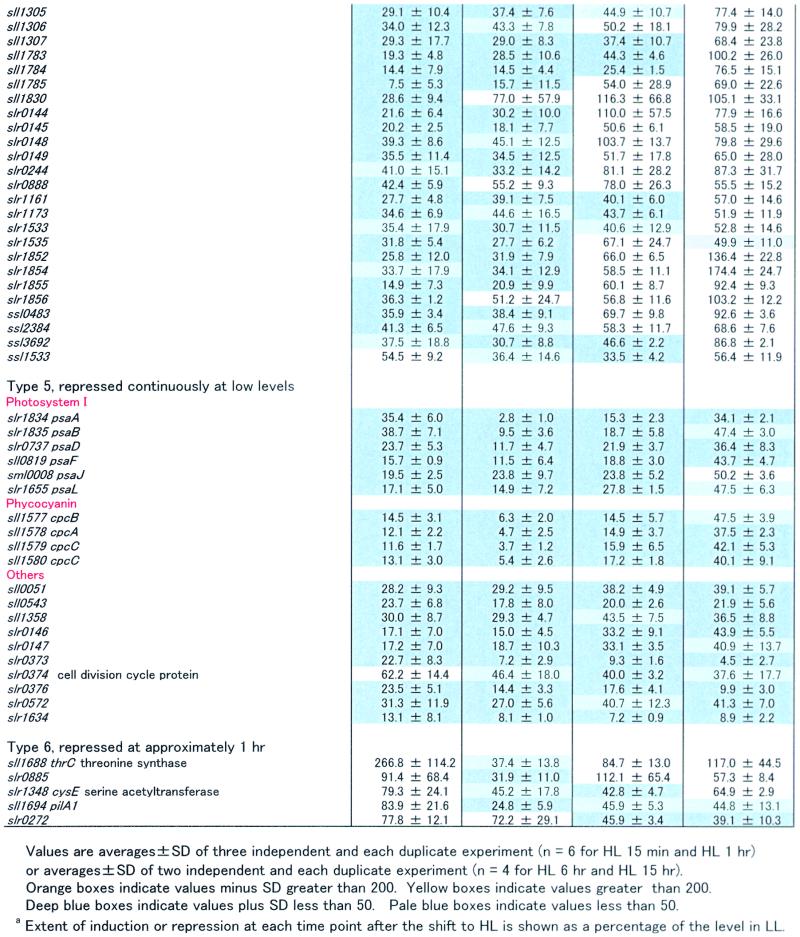
Table 1.
Complete List of the HL-Induced and HL-Repressed Genes
Other Thylakoid-Related Genes
The genes encoding subunits of ATP synthase, atp, and cytochrome b6/f complex, pet, were little affected by HL. The level of transcripts originating from atp increased gradually by 1.5- to twofold, whereas those from pet decreased slightly at the beginning of exposure to HL and then returned to the original level (Figure 3D). We could not reproducibly detect changes in transcript levels of cta genes, which encode subunits of cytochrome c oxidase (data not shown).
Changes in the Transcript Abundance of the Other Genes
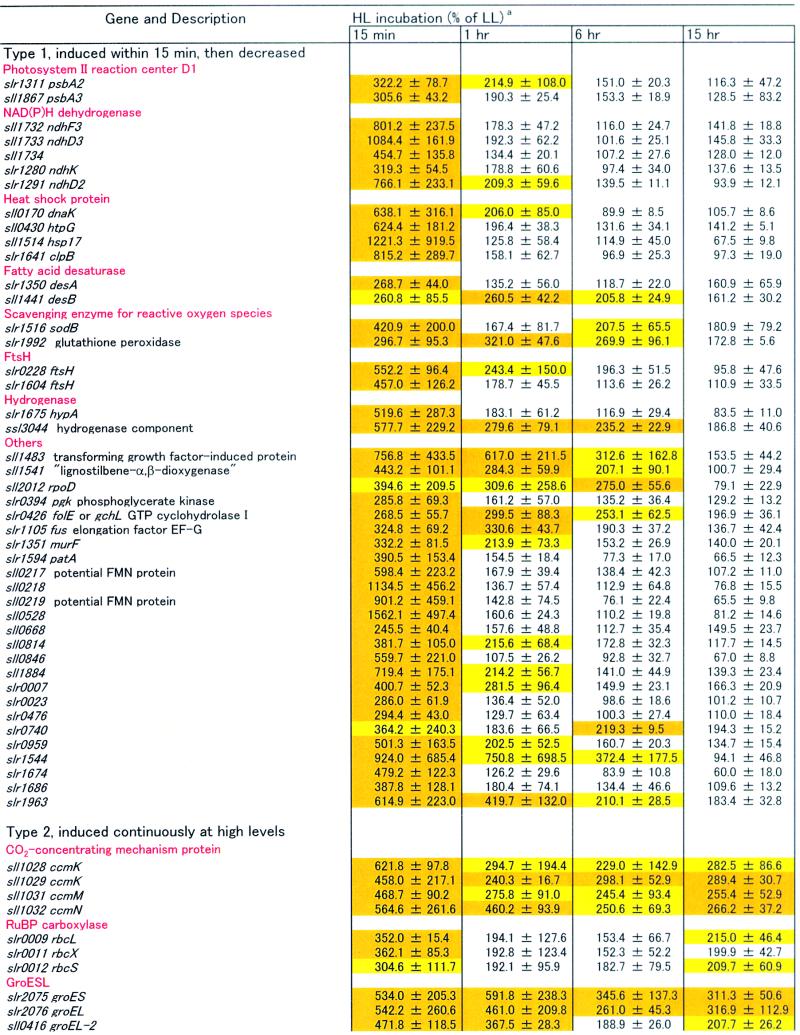
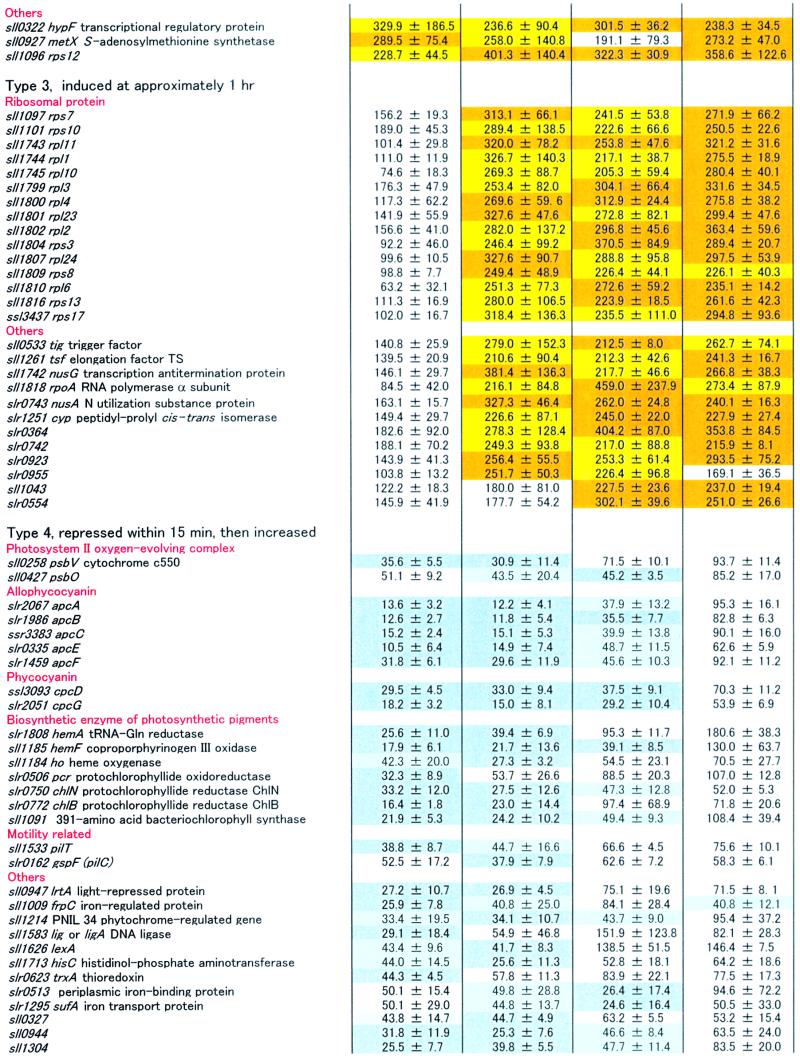
The expression of many genes not directly related to light harvesting or thylakoid structure also was affected by the light treatment. Although our microarray analysis included all 3079 ORFs of Synechocystis PCC 6803, we present in Table 1 only those for which the transcript levels were altered by at least twofold during the exposure to HL (15-min, 1-hr, 6-hr, and 15-hr time points). In view of these criteria, 84 ORFs were induced by HL, and 80 ORFs were repressed in the genome of Synechocystis sp PCC 6803. For simplicity, we subdivided these ORFs into six groups according to the changing patterns of their transcription after the shift from LL to HL: type 1, induction observed within 15 min, then decreased; type 2, induced at a high level throughout the HL period; type 3, induction observed after 1 hr; type 4, transcript level decreased within 15 min, then increased; type 5, repressed throughout the HL period; and type 6, repressed at approximately 1 hr. Below, we mention some of the ORFs that were strongly up- or downregulated by HL.
Many genes homologous with heat shock proteins from other organisms have been identified in Synechocystis sp PCC 6803, including clpB, htpG, three dnaKs, groES, groEL, groEL-2, four dnaJs, grpE, and hsp17 (Kaneko et al., 1996; Glatz et al., 1999). Notably, many of them were markedly induced by the shift to HL. For example, clpB, htpG, dnaK (sll0170), groES, groEL, groEL-2, and hsp17 were clearly upregulated by HL. The dnaJ (sll0897) also was induced, but in this case the standard deviation was high. Interestingly, most of the transcripts for heat shock proteins were only transiently induced within 15 min (type 1), with the exception of groES, groEL, and groEL-2, which stayed at high transcript levels even 15 hr after the shift to HL and thus must be included in the type 2 category.
Of the four genes for fatty acid desaturase, desA, desB, and desD were induced within 15 min of exposure to HL (type 1, but the standard deviation of desD was relatively high), whereas desC was constitutive. Note that the same three des genes were induced by low temperature in Synechocystis sp PCC 6803 (Los et al., 1997).
Genes that encode scavenging enzymes for ROS were expected to be upregulated under HL conditions in which production of ROS may be accelerated (Asada, 1994). However, only sodB and slr1992, which encodes glutathione peroxidase, were induced (type 1). Transcription of katG, slr1171, which encodes another glutathione peroxidase, and sll0755, which encodes thioredoxin peroxidase, was not affected significantly.
There are four ftsH homologs that encode the AAA-type protease in Synechocystis sp PCC 6803. Among them, slr0228 and slr1604 were induced upon the shift to HL. It has been observed that disruption of slr0228 caused a 60% reduction in the amount of functional PSI (Mann et al., 2000). This ftsH gene may play a chaperone-like role in the regulation of PSI content under HL conditions.
Some hydrogenase-related genes, hypA, ssl3044 (type 1), and hypF (type 2), were upregulated within 15 min, although hox genes, which encode the main structural components of hydrogenase (Appel and Schulz, 1996), were unaffected by the shift to HL.
Other ORFs belonging to the transiently induced group (type 1) might include ssr2595 and ssl2542, which encode HL-inducible proteins (Dolganov et al., 1995), and two nblA genes (ssl0452 and ssl0453) involved in the degradation of phycobilisome (Collier and Grossman, 1994). However, because in these cases the standard deviations were relatively high, conclusions regarding the effect of the HL treatment on their transcription must await confirmation.
Genes that encode components of the carboxysome (ccm; Price et al., 1998) and rbc genes, which encode ribulose bisphosphate (RuBP) carboxylase, were induced within 15 min, and their transcripts levels remained high even at 15 hr after the shift to HL (type 2).
Many genes involved in DNA replication, transcription, and translation were not induced within 15 min but were induced after approximately 1 hr (type 3). For example, three ribosomal operons (sll1096 to sll1101, sll1740 to sll1747, and sll1799 to sll1822) seemed to be actively transcribed at this time point. This observation is not unexpected given that cells divided faster under HL.
Among genes that were transiently downregulated within 15 min, some were related to the biosynthesis of photosynthetic pigments (type 4). These include hem genes involved in tetrapyrrole biosynthesis, a heme oxygenase gene for phycobilin biosynthesis, and various genes for chlorophyll biosynthesis. Because the cellular contents of chlorophyll and phycocyanin decreased rapidly in HL, it is not surprising that these genes were downregulated upon transfer to HL.
Of interest, some genes related to cell motility were repressed by HL. Not only the pilT (sll1533) and pilC (slr0162) listed in Table 1 (type 4) but also another pilT (slr0161) and pilC (slr0163) were downregulated within 15 min (but standard deviations were relatively high). Moreover, the pilA1 gene, which encodes a major subunit of filamentous appendages, type IV pili (Bhaya et al., 2000; Yoshihara et al., 2001), also was repressed by HL (type 6).
DISCUSSION
Relationship between the Regulation of Transcript Levels and Acclimation Responses to HL
Photosynthetic organisms show various acclimation responses to changing light intensity in their environment. Using DNA microarray technology, we revealed the dynamics and regulation of transcript abundance of known and unknown genes during acclimation from LL to HL. Although changes in the transcript levels are not necessarily reflected by changes in the amount of corresponding proteins, our results may help to elucidate the mechanisms whereby Synechocystis sp PCC 6803 acclimate to the rise in light intensity. One may distinguish the responses of genes involved in the photosynthetic machinery from those engaged in “housekeeping” processes (see Table 1), leading to, for example, shortening of the generation time from 20 under LL to 6 hr at HL.
Generally, short-term acclimation responses that took place upon the shift to HL mainly included changes in the efficiency of energy transfer to the reaction center (see Introduction). For example, Bissati et al. (2000) reported that nonphotochemical quenching developed within 5 min after the shift to HL in Synechocystis sp PCC 6803. It was proposed that rpaA (slr0115), rpaB (slr0947), and rpaC (sll1926) are involved in energy transfer from the phycobilisomes to PSII in Synechocystis sp PCC 6803 (Ashby and Mullineaux, 1999; Emlyn-Jones et al., 1999). However, our analysis indicated that rpaA and rpaB were not affected by HL, whereas rpaC was downregulated throughout the exposure to HL, although the standard deviation was high. Therefore, possible involvement of rpa in short-term acclimation to HL has not been established. Similarly, our analysis identified some genes (listed in Table 1), the transcription of which altered strongly after the transfer to HL, but their possible involvement in processes of short-term acclimation must await further experimental verification.
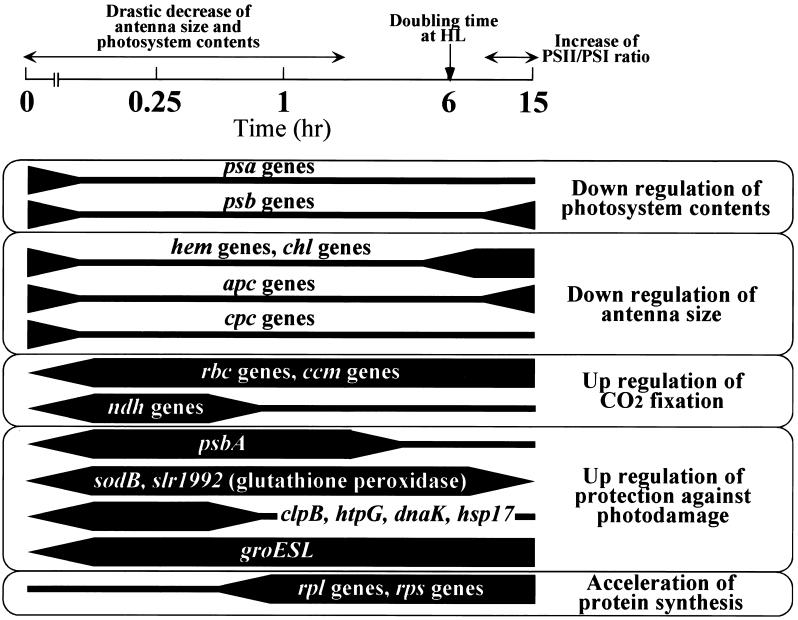
Generally, there is closer agreement between long-term acclimation responses of photosynthesis and upregulation and downregulation of genes detected in this study. Figure 4 shows the time-dependent changes in transcript levels that are involved in HL acclimation. In some cases, the induction or repression of genes involved in long-term acclimation could be completed within 15 min, whereas changes in protein levels were observed for hours or even days. Transcripts of housekeeping genes for DNA replication, transcription, and translation accumulated approximately 1 hr after the shift to HL. Acclimation of photosynthetic machinery to cope with the sudden shift to HL seems to come first, followed by changes in the housekeeping processes to support the faster cell division under HL.
Figure 4.
A Scheme Representing the Relationship between Changes of Transcript Levels and Acclimation to HL.
The physiological changes of cells during acclimation to HL (300 μmol photons m−2 sec−1) are shown above the time scale expressed logarithmically. Changes of transcript levels shown by DNA microarray analyses are indicated below the time scale. The thickness of lines roughly represents the amount of transcripts.
In cyanobacteria, the PSII/PSI ratio generally increases upon the shift to HL (Kawamura et al., 1979; Murakami and Fujita, 1991; Hihara et al., 1998). Declining PSI content would be expected to lower the susceptibility of the cells to HL damage particularly under prolonged exposure (Hihara and Ikeuchi, 1998; Hihara et al., 1998). Data presented here provide evidence that, in Synechocystis sp PCC 6803, alteration of the transcript levels of genes that encode various subunits of the photosystems may be involved in the determination of photosystem stoichiometry under changing light conditions. Repression of psa genes after the shift to HL was far more pronounced and for longer duration than that of psb genes. After 15 hr of HL, the level of psa transcripts was 50% lower than for LL, whereas that of psb was close to the LL level.
Chlorophyll and phycocyanin content per cell declined drastically within 3 hr of HL. Simultaneously, phycobilisome size and photosystem content probably were reduced to avoid absorption of excess light energy. These changes may originate from the downregulation of genes that encode enzymes for the biosynthesis of photosynthetic pigments (hem and chl genes), structural components of phycobilisome (apc and cpc genes), and subunits of photosystems (psa and psb genes). Although many psb genes were downregulated or did not change after the HL, psbA genes, which encode D1, were strongly upregulated. It is likely that the elevated level of psbA transcripts (Mohamed and Jansson, 1989) makes the increasing turnover rate of the D1 protein under HL conditions possible.
In contrast with photochemical reactions downregulated by HL, CO2 fixation is upregulated. Because the capacity for CO2 fixation by RuBP carboxylase is rate limiting under saturating light intensity, it is not surprising that rbc genes that encode large and small subunits of RuBP carboxylase and ccm genes encoding components of the CO2-concentrating mechanism were induced. Upregulation of ndh genes, especially the ndhF3 operon involved in high affinity CO2 uptake (Ohkawa et al., 1998, 2000) may help to increase the availability of CO2 under HL.
Despite the increasing energy consumption during HL, the cells could suffer damage from ROS at HL. Genes encoding ROS scavenging enzymes and genes that have chaperonin-like roles such as heat shock proteins were upregulated.
Existence of Common Signal Transduction Pathway(s) for Different Environmental Changes
The HL treatment led to the induction of several genes that, apparently, are not directly involved with the photosynthetic activity such as heat and cold shock–related genes (Table 1). This is in agreement with the report (Glatz et al., 1997) that induction of groEL-2 by heat shock was largely suppressed by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) or dark conditions. Similarly, the cold shock–inducible des genes were upregulated by a shift from dark to light but not in the presence of DCMU (Kis et al., 1998). These observations may indicate that different environmental changes may lead to the induction of similar sets of genes via common signaling pathway(s). In fact, it may enable one to distinguish between a specific response to a certain environmental stimuli and general responses to a number of stressors (Schwarz and Grossman, 1998), possibly mediated by the redox state of component(s) on the photosynthetic electron transport chain. Further application of DNA microarray methodology on RNA isolated after exposure to various environmental conditions may help to clarify stress-sensing mechanisms and networks of gene expression for acclimation responses.
Advantages and Limitations of the DNA Microarray Method
Substantial amount of information on the modulation of transcript levels during acclimation from LL to HL was obtained using the DNA microarray, providing an entire profile of gene expression in Synechocystis sp PCC 6803. However, we find it necessary to discuss some of the difficulties involved with the application of this technique and possible artifacts. First, we could not detect genes induced or repressed by HL in ∼1000 weak spots. Some of these genes such as those encoding for regulatory components (e.g., signal transduction and transcription factors) are of great interest but are present in low abundance. They may have responded to the changing light regimen, but we could not detect them due to inevitable false hybridization with probes derived from abundant rRNA. The extent of masking may vary depending on the fortuitous sequence similarity of each gene with rRNA. To increase the sensitivity of the DNA microarray, it might be necessary to remove rRNA before the reverse transcription reaction by, for example, subtraction with biotin-labeled antisense rRNA (Su and Sordillo, 1998). Second, we could not distinguish between highly homologous genes with the current version of CyanoCHIP. For example, although we obtained independent data for psbA2 and psbA3 spots, they seem to be identical (Figure 3) probably because only the coding regions of ORFs were fixed on the CyanoCHIP, and psbA2 and psbA3 are 99% identical. Highly homologous genes may result in cross-hybridization and wrong interpretation of the results. Third, although both synthesis and degradation determine the abundance of a specific transcript, we could not distinguish between them with the DNA microarray analysis. It might be necessary to perform microarray analyses in the presence or absence of inhibitors of RNA synthesis.
Naturally, we could not mention all the ORFs and many hypothetical genes are indeed transcribed. Some of them responded strongly to the HL treatment, suggesting that they may have a significant role, yet to be unraveled, in further studies.
METHODS
Strains and Culture Conditions
A glucose-tolerant wild-type strain of Synechocystis sp PCC 6803 was grown at 32°C in BG-11 medium (Stanier et al., 1971) with 20 mM Hepes-NaOH, pH 7.0, under continuous illumination provided by fluorescent lamps. Cells were grown in volumes of 50 mL in test tubes (3 cm in diameter) and bubbled by air containing 1.0% (v/v) CO2. Cell density was estimated as A730 with a spectrophotometer (model UV-160A; Shimadzu, Kyoto, Japan).
For the low light (LL) samples, cells were grown at 20 μmol photons m−2 sec−1 for at least 1 day and harvested at a cell density of 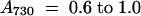 . For the exposure to high light (HL) for 15 min and 1 hr, cells grown at 20 μmol photons m−2 sec−1 to a cell density of
. For the exposure to high light (HL) for 15 min and 1 hr, cells grown at 20 μmol photons m−2 sec−1 to a cell density of 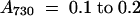 were transferred to HL (300 μmol photons m−2 sec−1) without dilution. For the exposure to HL for 6 and 15 hr, cells grown in LL were diluted to adjust the cell density to 0.1 (for 6-hr samples) or 0.05 (for 15-hr samples) and transferred to HL. After incubation in HL for 6 or 15 hr, the cell density was
were transferred to HL (300 μmol photons m−2 sec−1) without dilution. For the exposure to HL for 6 and 15 hr, cells grown in LL were diluted to adjust the cell density to 0.1 (for 6-hr samples) or 0.05 (for 15-hr samples) and transferred to HL. After incubation in HL for 6 or 15 hr, the cell density was 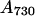 ≈
≈ 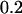 ; thus, self-shading of cells was minimized.
; thus, self-shading of cells was minimized.
Isolation of Total RNA
Total RNA was isolated using the RNeasy Midi kit (Qiagen, Hilden, Germany). The standard protocol for breakage of cells was modified as follows. In brief, approximately 100 mL of LL-grown cells (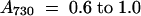 ) or 200 mL of HL-grown cells (
) or 200 mL of HL-grown cells (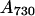 ≈
≈ 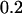 ) was broken with a Mini-Bead Beater (Biospec, Bartlesville, OK) with zircon beads (100 μm in diameter; Biospec) for three pulses of 50 sec at 4°C. After removal of the beads by centrifugation, the volume of cell lysate was adjusted to 3.8 mL with the buffer provided with the kit. After a brief centrifugation, 2.8 mL of 100% ethanol was added to the supernatant. Total RNA then was isolated using a spin column (provided as part of the kit) according to the manufacturer's instructions (Qiagen) and used for the labeling reaction.
) was broken with a Mini-Bead Beater (Biospec, Bartlesville, OK) with zircon beads (100 μm in diameter; Biospec) for three pulses of 50 sec at 4°C. After removal of the beads by centrifugation, the volume of cell lysate was adjusted to 3.8 mL with the buffer provided with the kit. After a brief centrifugation, 2.8 mL of 100% ethanol was added to the supernatant. Total RNA then was isolated using a spin column (provided as part of the kit) according to the manufacturer's instructions (Qiagen) and used for the labeling reaction.
Preparation of Labeled cDNA
Fluorescently labeled cDNA probes were prepared from the total RNA pool by direct incorporation of fluorescent nucleotide analogs during the first-strand reverse transcriptase reaction. Each 40 μL of labeling solution consisted of 20 μg of total RNA, 2 ng of λRNA as an internal control, 300 pmol of random hexamer, 0.5 mM each of dATP, dCTP, and dGTP, 0.2 mM dTTP, 100 units of RNase inhibitor (TaKaRa, Kyoto, Japan), 8 μL of 5 × reaction buffer provided with reverse transcriptase (TaKaRa), and 3 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia, Uppsala, Sweden). The solution was incubated at 65°C for 5 min and then at 25°C for 2 min, and then 38 units of reverse transcriptase (AMV Reverse Transcriptase XL; TaKaRa) was added, and the reverse transcription was performed at 42°C for 2 hr. Another 38 units of reverse transcriptase was added 1 hr after the beginning of reverse transcription. Unincorporated fluorescent nucleotides were removed using Centri-Sep spin columns (Princeton Separations, Adelphia, NJ). After ethanol precipitation, pellets were dissolved in 8 and 7 μL of water for probes labeled with Cy3 and Cy5, respectively.
Features of DNA Microarrays
DNA microarrays (CyanoCHIP version 0.8) were provided by TaKaRa. On this microarray, polymerase chain reaction (PCR) fragments of the C-terminal 1-kb coding regions of 3079 open reading frames (ORFs) were fixed, which covered all ORFs in the genome of Synechocystis sp PCC 6803 except some genes that encode transposase. In cases of genes smaller than 1 kb, the full length of the ORF was amplified and fixed. Approximately 90% of the PCR products fulfilled the requirement for concentration of 0.1 mg mL−1 and sufficient purity. In addition to the ORFs mentioned above, there were four spots each of 16S rRNA, 23S rRNA, λ phage DNA (as an internal control), and human transferrin receptor gene (as a negative control) and 44 spots of diluted solution of rRNAs used as positional markers. These spots (approximately 150 μm in diameter) were aligned in an area of 18.0 × 18.0 mm. Duplicate sets of DNA spots were fixed on the upper and lower parts of the DNA microarray to verify the reproducibility of experiments.
Hybridization and Washing Conditions of the DNA Microarray
Hybridization buffer (22 μL) containing 4 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.2% SDS, 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), and 100 ng μL−1 salmon sperm DNA was spread onto the microarray surface, covered with a cover slip, and sealed with glue. Prehybridizations were performed at 65°C for 1 hr followed by washing of the microarrays with 0.2 × SSC and drying by brief centrifugation. Hybridization was performed with 22 μL of hybridization solution containing 4 μL of Cy3 probe, 7 μL of Cy5 probe, and 11 μL of 2 × hybridization buffer at 65°C overnight. Washing was done at 65°C in 2 × SSC for 5 min, in 0.2 × SSC, and 0.1% SDS for 5 min, followed by three washes in 0.2 × SSC at room temperature, and then the microarrays immediately were spun dry.
Image Acquisition and Analysis
Microarrays were scanned with two wavelengths for Cy3 (570 nm) and Cy5 (660 nm) by using a laser fluorescent scanner (418 Array Scanner; Affymetrix, Santa Clara, CA) with three different photomultiplier gains. Data analysis was performed using Imagene version 3.0 software (BioDiscovery, Los Angeles, CA). The raw data obtained with the highest photomultiplier gain were routinely used for quantification. For spots in which signal intensity was saturated, the raw data of the lower photomultiplier gain were reanalyzed. The fluorescence intensity of each spot in both Cy3 and Cy5 images was quantified, and fluorescence levels of the local background were subtracted. Normalization of Cy3 and Cy5 images were performed by adjusting the total signal intensities of two images (“global normalization”). Because extraneously added controls such as λRNA could not correct errors in steps that proceeded probe labeling, inaccuracy in the quantification of RNA samples had a substantial effect on the results normalized by such methods. Thus, more reliable global normalization was used, although λDNA spots were provided on CyanoCHIP. After normalization, the expression of each gene under HL conditions was shown as a percentage of the LL levels. For example, the induction rate in HL was calculated from the ratio of Cy5 to Cy3 signals in the experiment represented in Figure 1. To obtain reliable data, we performed three independent experiments (cell culture, RNA isolation, labeling of probes, and hybridization all were performed independently) for the time points of 15 min and 1 hr and two experiments for the time points of 6 and 15 hr. Because the upper and lower parts on CyanoCHIP gave us duplicate results from single hybridizations, we had six, six, four, and four results for the time points of 15 min, 1 hr, 6 hr, and 15 hr, respectively. All induction rates described here are shown as averages and standard deviations of six or four results. The analyzed data are available at http://www. genome.ad.jp/kegg/expression.
RNA Gel Blot Analysis
Total RNA (15 μg) extracted as described above was fractionated on a 1.2% denaturing agarose gel, blotted, and probed with PCR-amplified DNAs. Labeling of probes and detection was performed using enhanced chemiluminescence direct nucleic acid labeling and detection systems (Amersham Pharmacia) according to the manufacturer's instructions.
Acknowledgments
We thank Dr. M. Takayama and M. Rokushima (TaKaRa) for providing the protocol for hybridization. We also thank Prof. K. Sonoike (University of Tokyo) for helpful advice. This work was supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (to Y.H. and A.K), by the Genome Frontier Project “Genetic and Molecular Networks” (to M.K.), by the Program for Promotion of Basic Research Activities for Innovative Biosciences of Japan (to M.I.), by Grants-in-Aid for Scientific Research (to M.I.), and by a Grant for Scientific Research from the Human Frontier Science Program (to M.I.).
References
- Anandan, S., and Golden, S.S. (1997). cis-acting sequences required for light-responsive expression of the psbDII gene in Synechococcus sp. strain PCC 7942. J. Bacteriol. 179, 6865–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.M. (1986). Photoregulation of the composition, function, and structure of thylakoid membranes. Annu. Rev. Plant Physiol. 37, 93–136. [Google Scholar]
- Anderson, J.M., Chow, W.S., and Park, Y.-I. (1995). The grand design of photosynthesis: Acclimation of the photosynthetic apparatus to environmental cues. Photosynth. Res. 46, 129–139. [DOI] [PubMed] [Google Scholar]
- Anderson, J.M., Park, Y.-I., and Chow, W.S. (1997). Photoinactivation and photoprotection of photosystem II in nature. Physiol. Plant. 100, 214–223. [Google Scholar]
- Appel, J., and Schulz, R. (1996). Sequence analysis of an operon of a NAD(P)-reducing nickel hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803 gives additional evidence for direct coupling of the enzyme to NAD(P)H-dehydrogenase (complex I). Biochim. Biophys. Acta 1298, 141–147. [DOI] [PubMed] [Google Scholar]
- Asada, K. (1994). Production and action of active oxygen species in photosynthetic tissues. In Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants, C.H. Foyer and P.M. Mullineaux, eds (Boca Raton, FL: CRC Press), pp. 77–104.
- Ashby, M.K., and Mullineaux, C.W. (1999). Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol. Lett. 181, 253–260. [DOI] [PubMed] [Google Scholar]
- Belknap, W.R., and Haselkorn, R. (1987). Cloning and light regulation of expression of the phycocyanin operon of the cyanobacterium Anabaena. EMBO J. 6, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya, D., Bianco, N.R., Bryant, D., and Grossman, A.R. (2000). Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37, 941–951. [DOI] [PubMed] [Google Scholar]
- Bissati, K.E., Delphin, E., Murata, N., Etienne, A.-L., and Kirilovsky, D. (2000). Photosystem II fluorescence quenching in the cyanobacterium Synechocystis sp. PCC6803: Involvement of two different mechanisms. Biochim. Biophys. Acta 1457, 229–242. [DOI] [PubMed] [Google Scholar]
- Björkman, O. (1981). Responses to different quantum flux densities. In Encyclopedia of Plant Physiology, Vol. 12A: Physiological Plant Ecology I, O.L.L. Lange, P.S. Nobel, C.B. Osmond, and H. Ziegler, eds (Berlin: Springer-Verlag), pp. 57–107.
- Boardman, N.K. (1977). Comparative photosynthesis of sun and shade plants. Annu. Rev. Plant Physiol. 28, 355–377. [Google Scholar]
- Bustos, S.A., and Golden, S.S. (1992). Light-regulated expression of the psbD gene family in Synechococcus sp. strain PCC 7942: Evidence for the role of duplicated psbD genes in cyanobacteria. Mol. Gen. Genet. 232, 221–230. [DOI] [PubMed] [Google Scholar]
- Campbell, D., Hurry, V., Clarke, A.K., Gustafsson, P., and Öquist, G. (1998). Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol. Mol. Biol. Rev. 62, 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, W.S. (1994). Photoprotection and photoinhibitory damage. Adv. Mol. Cell. Biol. 10, 151–196. [Google Scholar]
- Collier, J.L., and Grossman, A.R. (1994). A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 13, 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant, S., Perewoska, I., Alfonso, M., and Kirilovsky, D. (1997). Expression of psbA gene during photoinhibition and recovery in Synechocystis PCC 6714: Inhibition and damage of transcriptional and translational machinery prevent the restoration of photosystem II activity. Plant Mol. Biol. 34, 1–13. [DOI] [PubMed] [Google Scholar]
- DeRisi, J., Penland, L., Brown, P.O., Bittner, M.L., Meltzer, P.S., Ray, M., Chen, Y., Su, Y.A., and Trent, J.M. (1996). Use of a cDNA microarray to analyze gene expression patterns in human cancer. Nat. Genet. 14, 457–460. [DOI] [PubMed] [Google Scholar]
- DeRisi, J., Vishwanath, R.L., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Dolganov, N.A.M., Bhaya, D., and Grossman, A.R. (1995). Cyanobacterial protein with similarity to the chlorophyll a/b binding proteins of higher plants: Evolution and regulation. Proc. Natl. Acad. Sci. USA 92, 636–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlyn-Jones, D., Ashby, M.K., and Mullineaux, C.W. (1999). A gene required for the regulation of photosynthetic light harvesting in the cyanobacterium Synechocystis 6803. Mol. Microbiol. 33, 1050–1058. [DOI] [PubMed] [Google Scholar]
- Foy, R.H., and Gibson, C.E. (1982. a). Photosynthetic characteristics of planktonic blue-green algae: The response of twenty strains grown under high and low light. Br. Phycol. J. 17, 169–182. [Google Scholar]
- Foy, R.H., and Gibson, C.E. (1982. b). Photosynthetic characteristics of planktonic blue-green algae: Changes in photosynthetic capacity and pigmentation of Oscillatoria redekei van Goor under high and low light. Br. Phycol. J. 17, 183–193. [Google Scholar]
- Glatz, A., Horváth, I., Varvasovszki, V., Kovács, E., Török, Z., and Vígh, L. (1997). Chaperonin genes of the Synechocystis PCC 6803 are differentially regulated under light-dark transition during heat stress. Biochem. Biophys. Res. Commun. 239, 291–297. [DOI] [PubMed] [Google Scholar]
- Glatz, A., Vass, I., Los, D.A., and Vígh, L. (1999). The Synechocystis model of stress: From molecular chaperones to membranes. Plant Physiol. Biochem. 37, 1–12. [Google Scholar]
- Grace, S.C., and Logan, B.A. (1996). Acclimation of foliar antioxidant systems to growth irradiance in three broad-leaved evergreen species. Plant Physiol. 112, 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim, M., Keren, N., Ohad, I., Reinhold, L., and Kaplan, A. (1997). Acclimation of Synechococcus strain WH7803 to the ambient CO2 concentration and to elevated light intensity. J. Phycol. 33, 811–817. [Google Scholar]
- Hihara, Y. (1999). The molecular mechanism for acclimation to high light in cyanobacteria. Curr. Top. Plant Biol. 1, 37–50. [Google Scholar]
- Hihara, Y., and Ikeuchi, M. (1997). Mutation in a novel gene required for photomixotrophic growth leads to enhanced photoautotrophic growth of Synechocystis sp. PCC 6803. Photosynth. Res. 53, 243–252. [Google Scholar]
- Hihara, Y., and Ikeuchi, M. (1998). Toward the elucidation of physiological significance of pmgA-mediated high-light acclimation to adjust photosystem stoichiometry: Effect of the prolonged high-light treatment on pmgA mutants. In Photosynthesis: Mechanism and Effects, G. Garab, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 2929–2932.
- Hihara, Y., Sonoike, K., and Ikeuchi, M. (1998). A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis sp. PCC 6803 in response to high light. Plant Physiol. 117, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, T., et al. (1996). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3, 109–136. [DOI] [PubMed] [Google Scholar]
- Kaplan, A., and Reinhold, L. (1999). CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 539–570. [DOI] [PubMed] [Google Scholar]
- Kawamura, M., Mimuro, M., and Fujita, Y. (1979). Quantitative relationship between two reaction centers in the photosynthetic system of blue-green algae. Plant Cell Physiol. 20, 697–705. [Google Scholar]
- Kis, M., Zsiros, O., Farkas, T., Wada, H., Nagy, F., and Gombos, Z. (1998). Light-induced expression of fatty acid desaturase genes. Proc. Natl. Acad. Sci. USA 95, 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los, D.A., Ray, M.K., and Murata, N. (1997). Temperature-dependent expression of four desaturase genes in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 25, 1167–1175. [DOI] [PubMed] [Google Scholar]
- Mann, N.H., Novac, N., Mullineaux, C.W., Newman, J., Bailey, S., and Robinson, C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp PCC 6803. FEBS Lett. 479, 72–77. [DOI] [PubMed] [Google Scholar]
- Mohamed, A., and Jansson, C. (1989). Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13, 693–700. [DOI] [PubMed] [Google Scholar]
- Murakami, A., and Fujita, Y. (1991). Regulation of photosystem stoichiometry in the photosynthetic system of the cyanophyte Synechocystis PCC 6714 in response to light-intensity. Plant Cell Physiol. 32, 223–230. [Google Scholar]
- Naithani, S., Hou, J.-M., and Chitnis, P.R. (2000). Targeted inactivation of the psaK1, psaK2 and psaM genes encoding subunits of photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 63, 225–236. [DOI] [PubMed] [Google Scholar]
- Nakamoto, H., and Hasegawa, M. (1999). Targeted inactivation of the gene psaK encoding a subunit of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 40, 9–16. [DOI] [PubMed] [Google Scholar]
- Neale, P.J., and Melis, A. (1986). Algal photosynthetic membrane complexes and the photosynthesis-irradiance curve: A comparison of light-adaptation responses in Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 22, 531–538. [Google Scholar]
- Niyogi, K.K. (1999). Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 333–359. [DOI] [PubMed] [Google Scholar]
- Ohkawa, H., Sonoda, M., Katoh, H., and Ogawa, T. (1998). The use of mutants in the analysis of the CO2-concentrating mechanism in cyanobacteria. Can. J. Bot. 76, 1035–1042. [Google Scholar]
- Ohkawa, H., Price, G.D., Badger, M.R., and Ogawa, T. (2000). Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803. J. Bacteriol. 182, 2591–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, G.D., Sültemeyer, D., Klughammer, B., Ludwig, M., and Badger, M.R. (1998). The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: A review of general physiological characteristics, genes, proteins, and recent ad-vances. Can. J. Bot. 76, 973–1002. [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schwarz, R., and Grossman, A.R. (1998). A response regulator of cyanobacteria integrates diverse environmental signals and is critical for survival under extreme conditions. Proc. Natl. Acad. Sci. USA 95, 11008–11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L.-X., Kim, S.J., Marchant, A., Robinson, C., and Schröder, W.P. (1999). Characterisation of the PsbX protein from photosystem II and light regulation of its gene expression in higher plants. Plant Mol. Biol. 40, 737–744. [DOI] [PubMed] [Google Scholar]
- Stanier, R.Y., Kunisawa, R., Mandel, M., and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green alga (order Chroococcales). Bacteriol. Rev. 35, 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, C., and Sordillo, L.M. (1998). A simple method to enrich mRNA from total prokaryotic RNA. Mol. Biotechnol. 10, 83–85. [DOI] [PubMed] [Google Scholar]
- Yokoyama, E., Murakami, A., Sakurai, H., and Fujita, Y. (1991). Effect of supra-high irradiation on the photosynthetic system of the cyanophyte Synechocystis PCC 6714. Plant Cell Physiol. 32, 827–834. [Google Scholar]
- Yoshihara, S., Geng, X.X., Okamoto, S., Yura, K., Murata, T., Go, M., Ohmori, M., and Ikeuchi, M. (2001). Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 42, 63–73. [DOI] [PubMed] [Google Scholar]