Abstract
Callose is synthesized on the forming cell plate and several other locations in the plant. We cloned an Arabidopsis cDNA encoding a callose synthase (CalS1) catalytic subunit. The CalS1 gene comprises 42 exons with 41 introns and is transcribed into a 6.0-kb mRNA. The deduced peptide, with an approximate molecular mass of 226 kD, showed sequence homology with the yeast 1,3-β-glucan synthases and is distinct from plant cellulose synthases. CalS1 contains 16 predicted transmembrane helices with the N-terminal region and a large central loop facing the cytoplasm. CalS1 interacts with two cell plate–associated proteins, phragmoplastin and a novel UDP-glucose transferase that copurifies with the CalS complex. That CalS1 is a cell plate–specific enzyme is demonstrated by the observations that the green fluorescent protein–CalS1 fusion protein was localized at the growing cell plate, that expression of CalS1 in transgenic tobacco cells enhanced callose synthesis on the forming cell plate, and that these cell lines exhibited higher levels of CalS activity. These data also suggest that plant CalS may form a complex with UDP-glucose transferase to facilitate the transfer of substrate for callose synthesis.
INTRODUCTION
Callose was first detected almost 100 years ago on sieve plates of phloem elements, around pollen mother cells, in pollen grains, and in pollen tubes on the basis of its specific staining with aniline blue. The chemical structure of callose was characterized by Aspinall and Kessler (1957). It is a linear 1,3-β-glucan with some 1,6- branches and differs from cellulose, the major component of the plant cell wall. Callose has been localized at other locations as well, including the cell plate, plasmodesmata, root hair, cotton seed hair, and spiral thickenings in tracheids (Stone and Clarke, 1992). The synthesis of callose also can be induced by wounding, pathogen infection, and physiological stress (Stone and Clarke, 1992; Kauss, 1996). The deposition of callose at the cell plate precedes the synthesis of cellulose (Kakimoto and Shibaoka, 1992; Samuels et al., 1995). It is believed that callose deposition into the tubulovesicular network during cell plate formation may provide the spreading force that widens the tubules and converts the network into a fenestrated sheet (Samuels et al., 1995; Staehelin and Hepler, 1996).
Attempts to purify callose synthase (CalS) from higher plants during the last two decades have generated variable data about the molecular mass and subunit composition of this enzyme. Partially purified CalS preparations have been shown to contain six to nine major polypeptides ranging in size from 25 to 92 kD (Kamat et al., 1992; Wasserman et al., 1992; Dhugga and Ray, 1994; McCormack et al., 1997). Using various affinity labeling techniques, it has been reported that the presumptive “catalytic subunit” of CalS from higher plants has a molecular mass of between 32 and 57 kD (Read and Delmer, 1987; Frost et al., 1990; Delmer et al., 1991; Li and Brown, 1993; Gibeaut and Carpita, 1994). Labeling techniques using UDP-glucose or its analogs as probes have identified one of the subunits that bind UDP-glucose. Our data suggest that this is not the catalytic subunit but an associated protein acting as a UDP-glucose transferase (Hong et al., 2001). Recently, density fractionation of membranes followed by product entrapment has revealed that the CalS activity in Nicotiana alata pollen tubes is associated with a 190-kD peptide (Turner et al., 1998). Several factors may have contributed to these discrepancies: CalS is likely to be a multisubunit and membrane-associated enzyme; extraction of the enzyme from membranes requires the use of detergents that may dissociate the complex or cause the loss of its activity; and the protein may be sensitive to protease degradation during purification. Moreover, the activity of this enzyme is highly regulated during plant development, and various biotic and abiotic factors may affect the activity of this enzyme.
CalS activity often is found to be associated with cellulose synthase (CelS) fraction in the plasma membrane of higher plants. It has been suggested that CelS and CalS might be the same enzyme, which changes the linkage specificity of its products after being modified around the active site by unidentified mechanisms that could include changes in phosphorylation state, binding to Ca2+, and interaction with other associated proteins (Jacob and Northcote, 1985; Delmer, 1999). Whereas CelS cDNAs have been cloned from plants (Pear et al., 1996; Arioli et al., 1998; Taylor et al., 1999), little is known about genes encoding plant CalS enzymes, and the complexity of this enzyme has not been elucidated in any detail.
Genes encoding the catalytic subunit of 1,3-β-glucan synthases have been identified and cloned in Saccharomyces, Schizosaccharomyces, Aspergillus, Candida, Cryptococcus, and Paracoccidioides (Douglas et al., 1994; Kelly et al., 1996; Ishiguro et al., 1997; Mio et al., 1997; Thompson et al., 1999; Pereira et al., 2000). The molecular data have demonstrated clearly that the catalytic subunit of the microbial 1,3-β-glucan synthases is ∼200 kD. This is consistent with the CalS (190 kD) purified recently from tobacco (Turner et al., 1998), although earlier data on higher plant CalS have been ambiguous. The work presented here describes the cloning of a CalS cDNA from Arabidopsis. The deduced 226-kD peptide shares homology with the catalytic subunits of 1,3-β-glucan synthases from other organisms. A green fluorescent protein (GFP)–tagged CalS1 copurified with the product-entrapped CalS complex and showed enhanced CalS activity, suggesting that CalS1 is a subunit of the CalS complex. Our data also demonstrate that higher plants encode multiple forms of CalS enzymes and that the Arabidopsis CalS1 is a cell plate–specific isoform. In addition, we demonstrate that CalS1 interacts with two other cell plate–specific proteins, phragmoplastin and a novel UDP-glucose transferase, and forms a large complex.
RESULTS
Cloning of an Arabidopsis cDNA Encoding a CalS Enzyme
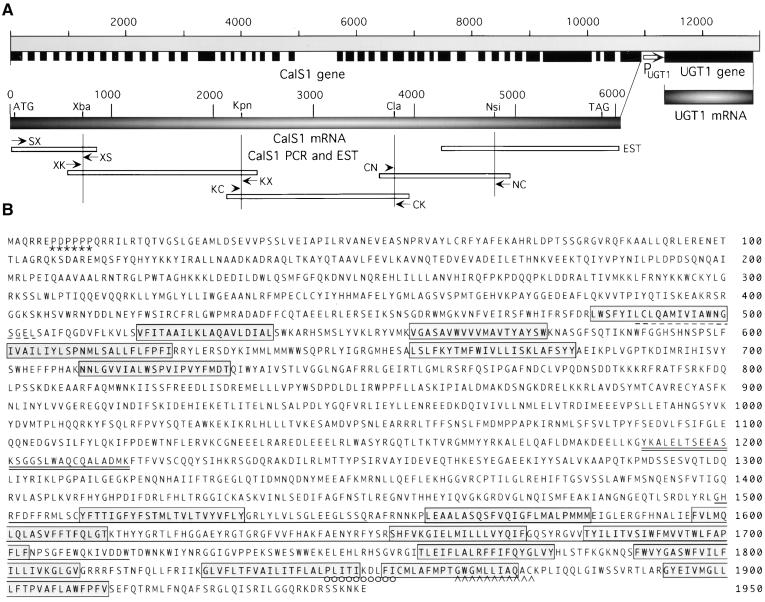
We have identified and cloned a novel, cell plate–specific UDP-glucose transferase (UGT1) that interacts with phragmoplastin and is targeted to the forming cell plate during cytokinesis (Hong et al., 2001). The UGT1 gene was found on two bacterial artificial chromosome clones, F3F20 and T25N20, which are mapped on chromosome 1 and share an overlapping region of 34 kb. Upstream of the UGT1 gene is a locus whose exons encode a putative CalS, termed CalS1, which shows significant overall sequence homology with the yeast 1,3-β-glucan synthase catalytic subunit FKS1 (Douglas et al., 1994). The distance between the coding regions of CalS1 and UGT1 is only 650 bp and consists of the 3′ untranslated region (UTR; 120 bp) of CalS1, the 5′ UTR of UGT1 (80 bp), and the putative promoter (450 bp) of the UGT1 gene.
We hypothesized that these two genes may be expressed coordinately during cytokinesis and that CalS1 may be a cell plate–specific enzyme. We synthesized oligonucleotide primers corresponding to the exon sequences of the CalS and performed reverse transcription–mediated polymerase chain reaction (RT-PCR) using RNA from shoot apical meristems of Arabidopsis seedlings. The 3′ end of CalS1 cDNA was taken from an expressed sequence tag (EST) clone (ATTS5466; GenBank accession number F15172). The PCR fragments and the EST clone were assembled into a 6-kb cDNA that contains a long open reading frame encoding a peptide of 1950 amino acid residues with a calculated molecular mass of 226 kD (Figure 1). This molecular mass is close to that of the CalS catalytic subunit isolated recently from the tobacco pollen tube plasma membrane by using a product-entrapment technique, which identified the presence of a 190-kD peptide as a catalytic subunit (Turner et al., 1998). That CalS1 is in fact a subunit of the CalS complex was demonstrated by tagging it with GFP and copurification of the chimeric protein with the product-entrapped highly purified CalS complex (see below) as well as by direct demonstration of an increase in CalS activity in BY-2 cells expressing CalS1 cDNA.
Figure 1.
Gene Structure and Peptide Sequence of the Cell Plate–Specific Callose Synthase (CalS1).
(A) Four fragments of the CalS1 cDNA were amplified by RT-PCR using specific primers (small arrows) and RNA from apical meristems of Arabidopsis seedlings. The cDNA was assembled from the PCR fragments and an EST clone (ATTS5466) through unique restriction enzyme sites. Exons are indicated by solid boxes. The larger arrow indicates the 450-bp promoter region (PUGT1) of UGT1.
(B) The deduced amino acid sequence of CalS1 contains 16 transmembrane helices (boxed). The region between positions 1499 and 1717 that shares homology with the yeast GNS1 protein is underlined. A G-protein–coupled receptor signature is marked by a dashed line, and an inner membrane protein signature of the “binding protein–dependent transport system” (Saurin et al., 1994) is double underlined. A proline-rich domain near the N terminus is marked by asterisks. An energy transfer protein signature is indicated by open circles, and a possible lipid attachment motif of membrane lipoproteins is indicated by carets. The GenBank accession number for UGT1 is AF237733.
CalS1 Is a Transmembrane Protein and Contains Several Motifs
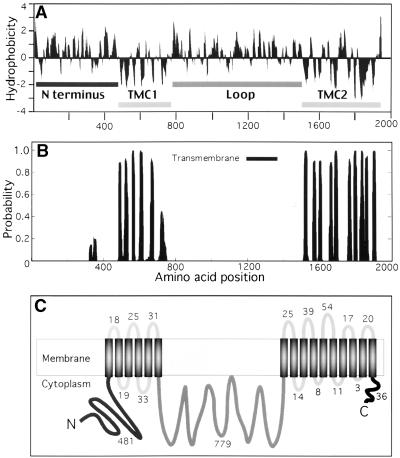
Hydropathy analysis of CalS1 indicated that it is an integral membrane protein with 16 transmembrane helices (Figure 2). The N terminus (481 amino acids) is hydrophilic and lacks an apparent cleavable signal sequence. According to the “positive inside” rule (i.e., the observation that more positively charged amino acids are found in the cytoplasmic than in the extracytoplasmic segments in transmembrane proteins [Sipos and von Heijne, 1993]), the data suggest that the hydrophilic N-terminal sequence of CalS1 is cytoplasmic (Figure 2). The transmembrane domains are clustered in two regions separated by a large hydrophilic domain (779 amino acids) that faces the cytoplasm. The rest of the loops connecting transmembrane helices are relatively short (3 to 54 residues; Figure 2). The overall topology of the CalS1 protein is very similar to that of the sequenced fungal 1,3-β-glucan synthases (Douglas et al., 1994; Kelly et al., 1996; Ishiguro et al., 1997; Mio et al., 1997; Thompson et al., 1999; Pereira et al., 2000). It is interesting that CelSs from plants also share similar topological arrangement (Pear et al., 1996; Arioli et al., 1998; Taylor et al., 1999), although generally they are smaller in size (∼1000 to 1200 amino acid residues compared with 1950 residues for CalS1) and do not share homology with CalS1 in the primary amino acid sequences.
Figure 2.
Predicted Membrane Topology Model of CalS1.
(A) Hydrophobicity plot by the Kyte-Doolittle method. TMC1 and TMC2, transmembrane clusters 1 and 2.
(B) Transmembrane helices predicted by the transmembrane hidden Markov model (TMHMM) program.
(C) Topology of CalS1 in membrane. The long rectangle indicates the membrane, and the vertical black bars represent the transmembrane helices of CalS1. The length of the peptide chain in each non-membrane-spanning segment is indicated by the number of amino acid residues.
The conserved D, D, D and QXXRW motifs implicated in the binding of UDP-glucose and the transfer of the glucosyl group in the bacterial and plant CelSs (Brown et al., 1996) are absent in the sequenced fungal FKS1 homologs (Ishiguro et al., 1997) and are not found in Arabidopsis CalS1 as well (Figure 1). A sequence (KSGG) matching the (R/K)XGG consensus that has been implicated in UDP-glucose binding in glycogen synthases (Farkas et al., 1990) is found in CalS1. However, this motif is not found in all fungal FKS1 homologs (Ishiguro et al., 1997) or in other members of the Arabidopsis CalSs. Moreover, the context of this sequence in CalS1 does not have any similarity to the surrounding regions of the (R/K)XGG consensus in the glycogen synthases.
Furthermore, a segment of 218 residues (underlined in Figure 1B) shows homology with the yeast GNS1 protein that acts as a regulatory subunit of 1,3-β-glucan synthase. Mutation in GNS1 resulted in a 90% reduction of 1,3-β-glucan synthase activity (el-Sherbeini and Clemas, 1995). GNS1 contains a site for binding to a neumocandin B0 analog (L-733,560), an inhibitor of 1,3-β-glucan synthase. It is not known whether plant CalS is sensitive to L-733,560, but the presence of a GNS1 homology sequence in CalS1 suggests that the two yeast genes (FKS1 and GNS1) may have been fused into one in plants during evolution. This situation is similar to that of Δ1-pyrroline-5-carboxylate synthetase, which we have shown to encode a bifunctional peptide in plants, although two separate peptides exist in bacteria (Hu et al., 1992). The N-terminal arm (481 residues) of CalS1 contains a proline-rich domain that may interact with SH3-containing proteins. A G-protein–coupled receptor signature that has been implicated in interaction with G-proteins (Strosberg, 1991) was found to partly overlap the first transmembrane domain (Figure 1B, dashed line). This domain may interact with phragmoplastin (see below) or Rho-like proteins, both of which are GTP binding proteins. In the middle of the CalS1 central loop is a signature (Figure 1B, double underlined) for the inner membrane component of “binding protein–dependent transport systems,” which include the active transport of arabinose, xylose, and maltose (Saurin et al., 1994). In CalS1, this domain may be involved in the transport of the glucosyl substrate or the polymer chain. An energy transfer protein signature that is involved in the ADP/ATP translocation across the mitochondrial membrane (Klingenberg, 1990) is also present in the short loop between transmembrane domains 12 and 13 (Figure 1B, open circles) and could be involved in the transfer of UDP back to the cytosol after transferring glucose residue from UDP-glucose. Near the C terminus is a potential lipid attachment site (Figure 1B, carets) that also has been reported to be present in plant proteins, including nodulin-24, a peribacteroid membrane protein (Cheon et al., 1994). Finally, CalS1 contains multiple consensus sites for serine/threonine/tyrosine phosphorylation, which may play a role in the regulation of CalS activity. The functionality of each of these putative domains needs to be further established.
Presence of Multiple CalSs in Plants
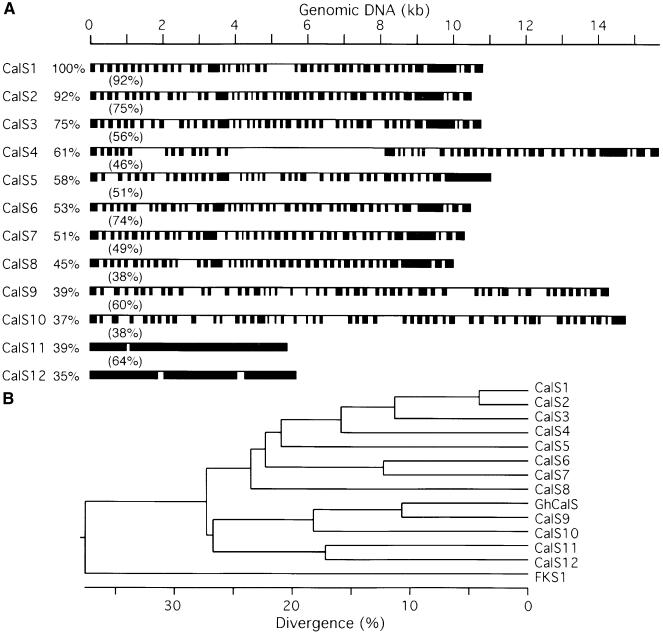
Using the deduced amino acid sequence of CalS1 to search various databases, we were able to identify 12 CalS1 homologous sequences in the Arabidopsis genome (Table 1). We assigned a number to each gene on the basis of homology with CalS1, with CalS2 being most closely related to CalS1. The 12 putative CalS genes are distributed over the five Arabidopsis chromosomes (Table 1) and can be divided into two groups based on gene structure. Genes in the first group contain 40 to 50 exons and encode peptides of 1923 to 1956 amino acid residues. Genes in the second group (CalS11 and CalS12) have only two to three exons (Figure 3A), and the encoded peptides are ∼170 amino acid residues shorter at their N-terminal ends than those in the first group. CalS11 and CalS12 are more closely related to each other than to other CalSs. CalS1 shares very high homology (92% identity) with CalS2, and it is possible that these two genes are functionally redundant. Recently, a putative CalS cDNA, CFL1 (GhCalS), was cloned from cotton, and its expression is detected in seed epidermal cells (Cui et al., 1999). Phylogenetic analysis suggests that GhCalS is most closely related to Arabidopsis CalS9 and shares relatively low homology (40%) with the cell plate–specific CalS1 (Figure 3B). Thus, higher plant CalSs are homologous overall with the yeast 1,3-β-glucan synthases. The region of highest homology between the plant and yeast enzymes (25% identity and 54% similarity over a segment of ∼700 amino acids) was found in the C-terminal half of the molecules in which the putative catalytic site is located. The lack of amino acid sequence homology with CelSs (Pear et al., 1996; Arioli et al., 1998; Taylor et al., 1999) indicates that CalS and CelS are distinct enzymes that retain similar overall structure and membrane topology (see below).
Table 1.
Multiple Forms of Putative CalSs in Arabidopsisa
| CalS | GenBank Accession Number |
Chromosome Location |
Amino Acids | No. of Exons | EST Clones |
|---|---|---|---|---|---|
| CalS1 |
AF237733AC007153 (AC005106) |
1 | 1950 | 42 |
F15172; AI996136; F15173; AV546456; AV646514 |
| CalS2 | AC006223 | 2 | 1950 | 42 | AV535768 |
| CalS3 | AL353013 | 5 | 1956 | 42 | AV534683 |
| CalS4 | AB025605 | 5 | 1911 | 42 | No EST |
| CalS5 | AC006436 | 2 | 1923 | 39 | No EST |
| CalS6 | AL163527 | 3 | 1933 | 42 | No EST |
| CalS7 | AC007592 | 1 | 1933 | 41 | No EST |
| CalS8 | AB023038 | 3 | 1935 | 41 | No EST |
| CalS9 | AC012395 | 3 | 1931 | 50 |
AI995793; AV522143; AV523506; AV529195; AV548414; W43833 |
| CalS10 | AC006922 | 2 | 1932 | 50 |
Z33984; F14234; F14233; AI997647; AI996091 |
| CalS11 |
AF162444 AL161502 (AC012392) |
4 | 1768 | 2 | N96260 |
| CalS12 |
AC005142 AF071527 AL161497 |
4 | 1780 | 3 | AV543953; AV546393; AV552411 |
CalS1 sequence was determined from cDNA, and CalS9, 11, and 12 were from GenBank deduced sequences. On the basis of CalS1, the sequences for CalS2 to CalS8 and CalS10 were deduced (see supplemental data for more details; http://www.aspp.org). Similar analysis has been done on this group of proteins, referred to as GSL (for glucan synthase-like) by C. Somerville's group (http://cellwall.stanford.edu/gsl/arabidopsis/structure.shtml). GenBank accession numbers within parentheses indicate genomic DNAs containing a partial CalS coding region. The underlined GenBank accession number (AF237733) represents the CalS1 cDNA from this report.
Figure 3.
Homologs of CalS1 in the Arabidopsis Genome.
(A) Genomic organization of Arabidopsis CalSs. Exons (solid boxes) of CalS1 were generated from the comparison of the cloned CalS1 cDNA and its genomic DNA (solid line). Exons of the rest of the CalSs were predicted by a combination of different software programs. The percentage of amino acid identity with CalS1 is shown before each gene, whereas the percentages in parentheses indicate amino acid identity between adjacent genes. A similar analysis has been performed independently by C. Somerville's group (see http://cellwall.stanford.edu/gsl/arabidopsis/structure.shtml).
(B) Phylogenetic tree of putative CalSs from Arabidopsis. Protein sequences of Arabidopsis CalSs were compiled using the Clustal method of the DNAStar MegAlign program. The putative cotton fiber–related CalS (GhCalS; GenBank accession number AF085717) and yeast 1,3-β-glucan synthase (FKS1; GenBank accession number SCU12893) are included. Numbers on the horizontal scale indicate percentage of divergence.
Expression of CalS Isoforms in Plants
We examined the expression of CalS isoforms in Arabidopsis by performing RT-PCR and searching for the presence of expressed sequences in the EST databases. The expression of CalS1, CalS2, and CalS5 was detected by RT-PCR followed by sequencing of the PCR fragments (data not shown), suggesting that these genes are expressed in shoot apical meristems of 3-week-old seedlings. Search of the databases resulted in the detection of expressed sequences corresponding to CalS1, CalS2, CalS3, CalS9, CalS10, CalS11, and CalS12 (Table 1), suggesting that these genes are expressed in the respective tissues used in the EST sequencing projects. We noticed that the CalS1, CalS9, CalS10, and CalS12 genes are highly represented in the EST database. The absence of CalS5, CalS6, CalS7, and CalS8 in the EST database suggests that these genes may be expressed at very low levels or that their expression may be induced under special conditions, such as pathogen infection. All CalS genes identified seem to be functional, because no CalS pseudogene or truncated gene was detected in the complete Arabidopsis genome.
Localization of CalS1 at the Forming Cell Plate
To determine the subcellular location of CalS1, we expressed a GFP–CalS1 fusion protein under the control of the cauliflower mosaic virus (CaMV) 35S promoter in transgenic tobacco BY-2 cells. We examined 20 transgenic lines for each construct and found that all lines expressed the GFP fusion protein. Green fluorescence was present in a punctate pattern throughout the cytoplasm in nondividing cells (Figure 4D). The fluorescence was found as a bright green line coinciding with the cell plate in cells undergoing cytokinesis (Figures 4C and 4D). Deletion of the last 10 transmembrane helices did not affect the association of CalS1 with the cell plate (data not shown), suggesting that the first half of the molecule is sufficient to target the fusion protein to the cell plate. This observation is similar to what has been seen with nodulin-26, an integral peribacteroid membrane protein of root nodules, which is targeted to the membrane by its first three transmembrane domains (Miao et al., 1992). As a control, GFP alone, expressed under the control of the CaMV 35S promoter, was found to be distributed throughout the nuclei and cytoplasm, and the fluorescence was not confined to the cell plate (Figure 4B) (Gu and Verma, 1997).
Figure 4.
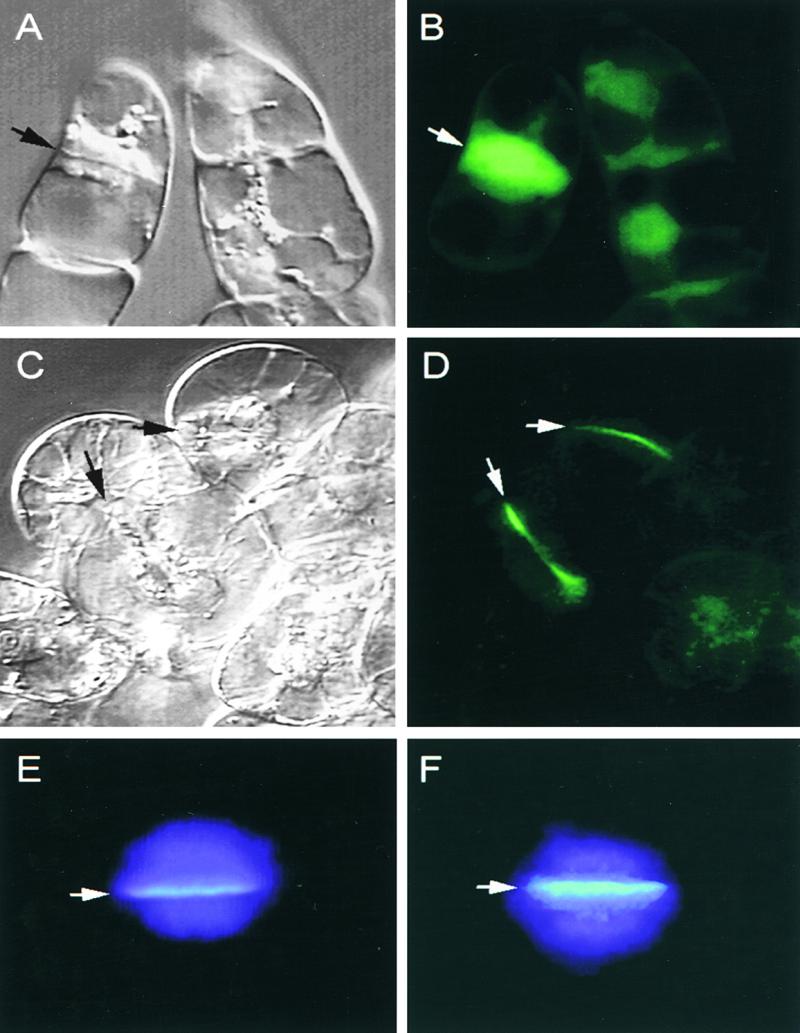
Subcellular Localization of CalS1 and Deposition of Callose on the Cell Plate.
(A) and (B) Control BY-2 cells expressing GFP alone shown as a bright-field image (A) and a fluorescent image (B).
(C) and (D) BY-2 cells expressing the GFP–CalS1 fusion protein shown as a bright-field image (C) and a fluorescent image (D).
(E) and (F) Callose deposition in the cell plate of control (E) and transgenic cells overexpressing GFP–CalS1 (F). The cells stained with aniline blue and 4′,6-diamidino-2-phenylindole were photographed with a fluorescence microscope with a UV filter set.
Arrows in (A) to (F) indicate the cell plate.
Accumulation of Callose in Transgenic Tobacco Cells Overexpressing CalS1
To determine if callose synthesis on the growing cell plate is affected by the overexpression of CalS1, we expressed GFP–CalS1 under the control of the CaMV 35S promoter and treated transgenic tobacco BY-2 cells with aniline blue, a callose indicator. The cells also were stained with 4′,6-diamidino-2-phenylindole for nuclear DNA. The staining of the cell plates at anaphase with aniline blue was much brighter in transgenic cells overexpressing GFP–CalS1 (Figure 4F) than in control cells (Figure 4E). As the growing cell plate fused to the parental cell wall and developed into mature cell wall, callose staining gradually became weaker and eventually faded completely in the control cells. In contrast, callose staining in transgenic cells persisted much longer and remained much brighter in young cell walls (data not shown). These results, along with the copurification of GFP–CalS1 with the product-entrapped CalS enzyme complex (see below), suggest that CalS1 is the catalytic subunit of the CalS complex responsible for the synthesis of callose on the forming cell plate. In a parallel experiment (data not shown), we found that overexpression of phragmoplastin also increased callose deposition, but expression of GFP alone, as a control, did not affect callose synthesis. This evidence supports the results of the interaction between phragmoplastin and CalS1 as demonstrated in the yeast two-hybrid system (see below).
Copurification of Chimeric GFP–CalS1 Protein with the CalS Complex
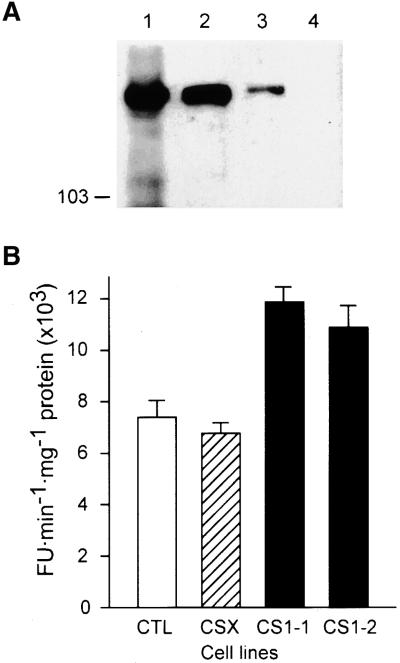
In an attempt to determine if CalS1 is part of the active CalS complex, we expressed CalS1 in an fks1 deletion mutant of yeast that displayed ∼90% depletion of the 1,3-β-glucan synthase activity (Douglas et al., 1994). The expression of recombinant CalS1 in yeast was confirmed using antibodies to the hemagglutinin (HA) tag (see below), but we were unable to detect an increase in CalS activity (data not shown). This result was not surprising considering the complexity of this enzyme both structurally and in terms of regulation. Using a product-entrapment procedure (Inoue et al., 1995; Turner et al., 1998; Hong et al., 2001), we purified CalS complex from the tobacco BY-2 cells expressing 35S::GFP–CalS1. We reasoned that if CalS1 is the active subunit of the CalS complex in plants, the GFP–CalS1 chimeric protein should be copurified with the CalS complex. Because there are no plant CalS antibodies available, we used a monoclonal antibody against GFP to follow the presence of GFP–CalS1 chimeric protein in different fractions during purification. It is known that CalS activity is associated with the membranes and can be extracted effectively from the membrane fraction by a zwitterionic detergent, 3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonic acid (Chaps) (Lawson et al., 1989; Meikle et al., 1991; Li et al., 1997). The use of the product-entrapment technique resulted in a 70- to 90-fold increase in CalS specific activity (data not shown) over the crude membrane extract, and a highly enriched fraction of the CalS enzyme was obtained. Protein gel blot analysis (Figure 5A) showed that GFP antibodies reacted with a peptide of ∼250 kD in total membranes, Chaps-extracted fraction, and the product-entrapped fraction from the GFP–CalS1 cells but not from control BY-2 cells. These data demonstrate that the GFP–CalS1 chimeric protein is part of the active CalS complex.
Figure 5.
Copurification of GFP–CalS1 with the CalS Complex and Increase in CalS Activity in Transgenic Cell Lines Expressing CalS1.
(A) Protein gel blot analysis of the presence of GFP–CalS1 in membrane fractions of transgenic BY-2 cells. Lane 1, total membranes isolated from tobacco BY-2 cells expressing the GFP–CalS1 chimeric protein; lane 2, Chaps-soluble fraction of the membranes; lane 3, product-entrapped fraction; lane 4, total membranes from control BY-2 cells. Proteins resolved by SDS-PAGE were transferred to a nitrocellulose filter and probed with a monoclonal antibody against GFP. The largest prestained molecular mass marker (103 kD; Bio-Rad) is indicated.
(B) Total membranes were isolated from tobacco BY-2 cells synchronized at the cytokinesis stage and extracted with a buffer containing 0.5% digitonin. CalS activity was assayed in the soluble fraction and expressed in fluorescence units per milligram per minute of protein (FU mg−1 min−1). CTL, control BY-2 cells; CSX, transgenic line (CalSX) expressing a deletion variant of CalS1 (only the first half of the molecule) fused with GFP (transgenic GFP- expressing control); CS1-1, 35S::GFP–CalS1 transgenic line 1; CS1-2, 35S::GFP–CalS1 transgenic line 2. Error bars indicate ±sd.
CalS Activity in Transgenic BY-2 Cells Expressing CalS1
To ascertain that CalS1 encodes the catalytic subunit of CalS, we measured CalS activity in two transgenic lines expressing 35S::GFP–CalS1, the same lines used in the callose deposition experiment (see above). An XhoI deletion construct (CalSX) carrying only the first half of the molecule was used as a transgenic control. We synchronized the GFP–CalS1 transgenic cell lines along with the CalSX line and nontransgenic BY-2 cells as described earlier (Gu and Verma, 1997) and prepared total membranes from cells at cytokinesis (1 hr after release from the propyzamide treatment). Measurement of the CalS activity in these preparations showed that both transgenic lines exhibited much higher levels (41 to 60% more) of CalS activity compared with those of the wild-type control and the deletion line (transgenic control). The lack of activity in the CalSX line is consistent with the suggestion that the C-terminal half carries the catalytic activity, as in the yeast enzyme (Douglas et al., 1994). These data also showed that the presence of GFP has no effect on the CalS in these experiments. Together, three lines of evidence (i.e., copurification of GFP–CalS1 with the CalS activity, cell plate–specific localization, and enhanced CalS activity) suggest that CalS1 is the catalytic subunit of the plant CalS enzyme.
CalS1 Interacts with Phragmoplastin and a Novel UDP-Glucose Transferase
Phragmoplastin is a cell plate–associated protein (Gu and Verma, 1996, 1997) that plays a pivotal role in cell plate formation (Verma, 2001). We have localized UGT1 at the cell plate and shown that it interacts with phragmoplastin (Hong et al., 2001). The cell plate–specific localization of UGT1 implies that it may function as a subunit of, or in association with, CalS on the forming cell plate and that it forms a complex with CalS1. We tested directly whether CalS1 interacts with UGT1 and phragmoplastin. The coding region of CalS1 was cloned in pJG4-5 in frame with the B42 activation domain. Phragmoplastin and UGT1 coding regions were inserted into pEG202 in frame with the LexA DNA binding domain. The expression of phragmoplastin and UGT1 in yeast EGY48 cells was detected using antibodies against phragmoplastin (Gu and Verma, 1996) and UGT1 (Hong et al., 2001), respectively (Figure 6B). Because CalS1 was expressed from pJG4-5 that encodes an epitope tag of HA, polyclonal antibodies to the HA tag were used to confirm the expression of recombinant CalS1 in yeast (Figure 6B).
Figure 6.
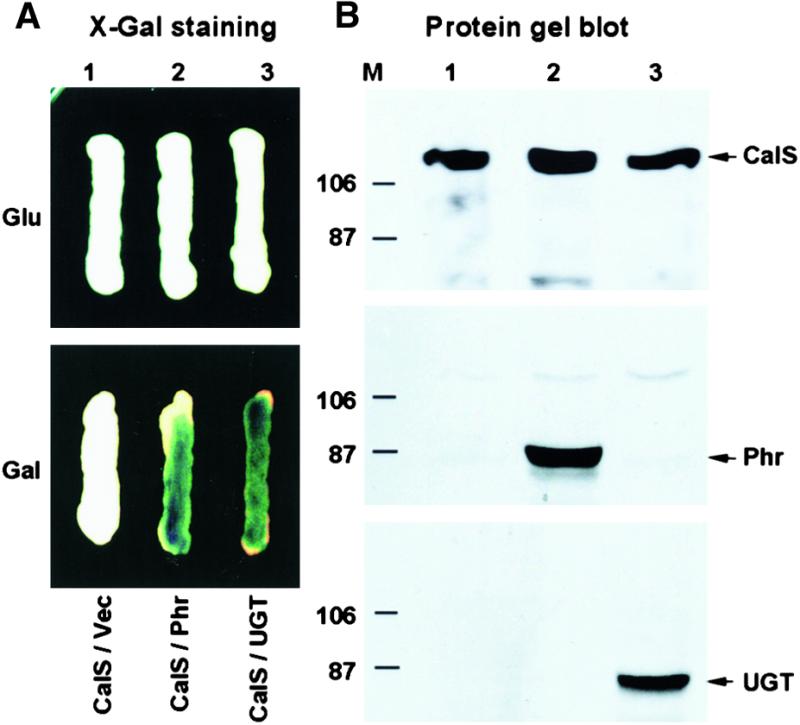
Protein–Protein Interaction of CalS1 with Phragmoplastin and UGT1 in a Yeast Two-Hybrid System.
(A) X-Gal assay of yeast two-hybrid cells. EGY48 cells containing pSH18-34 and pJG-CalS1 were transformed with vector pEG202 (lane 1, CalS/Vec), pEG-Phr (lane 2, CalS/Phr), or pEG-UGT1 (lane 3, CalS/UGT). The cells were grown on YNB medium containing X-Gal in the presence of glucose (Glu, top) or galactose (Gal, bottom). Note that the interaction occurred only when the expression of phragmoplastin (Phr) or UGT1 (UGT) was induced by galactose.
(B) Protein gel blots of proteins from yeast two-hybrid cells. Protein extracts of cells grown in the presence of galactose ([A], bottom) were resolved by SDS-PAGE, and the membrane was incubated with HA antibody that detects the chimeric protein of HA-tagged CalS1 (top). The same membrane was stripped and incubated with phragmoplastin (middle) or UGT1 antibody (bottom). Arrows indicate CalS1 (CalS), phragmoplastin (Phr), and UGT1 (UGT). Sizes (in kilodaltons) of prestained protein markers (M) from Bio-Rad are indicated.
When transferred to 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates, cells coexpressing CalS1/phragmoplastin or CalS1/UGT1 turned blue after an overnight incubation. Colonies that expressed only one of the proteins (controls) did not become blue even after an extended period of incubation, suggesting that these interactions are specific. This was confirmed in vitro (data not shown) using labeled peptides, as we have done with phragmoplastin (Zhang et al., 2000). These data show that CalS1 interacts with UGT1 and that both of these proteins interact with phragmoplastin, possibly forming a complex at the cell plate.
DISCUSSION
CalS1 Encodes a Cell Plate–Specific CalS
We have identified and cloned a CalS cDNA from Arabidopsis. Besides its homology with 1,3-β-glucan synthases from yeast and other fungi, three lines of evidence suggest that CalS1 is a cell plate–specific CalS. First, the GFP–CalS1 fusion protein was localized at the cell plate. Second, cells expressing this construct synthesized more callose at the cell plate. Third, they exhibited higher levels of CalS activity. Because of the fact that this enzyme forms a large complex with several proteins (Hong et al., 2001), it is not surprising that plant CalS1 could not complement the yeast fks mutant, but it did enhance the activity of CalS in BY-2 cells, confirming it as a catalytic subunit of the plant CalS.
Plant CalS and CelS Are Distinct Proteins but Share Similar Membrane Topology
The cloning of CalS and CelS cDNAs has demonstrated clearly that the encoded enzymes do not share homology in their primary amino acid sequences, disproving an earlier hypothesis that the two activities (β-1,3- and β-1,4-glucan synthesis) might be catalyzed by the same enzyme with a switch in the linkage specificity of the products (Delmer, 1999). Both CalS and CelS are large transmembrane proteins. They have very similar topology on the membrane, including the presence of a large hydrophilic loop, a long N-terminal segment, and a short C-terminal segment, all located on the cytoplasmic side. The transmembrane helices are arranged in two clusters separated by the hydrophilic central loop. The similarity in topology is consistent with their biological functions, that is, both use the same substrate (UDP-glucose) to synthesize similar products for deposition in the cell wall. Because the large central loop is relatively conserved among the CalS isoforms, it could act as a receptacle to hold other proteins that are essential for CalS catalytic activity (see below). On the other hand, the N-terminal segment differs between CalS isoforms (with an average of only 32% similarity). This region may contain subdomains for interaction with regulatory proteins such as Rop1, an Arabidopsis homolog of yeast Rho1 (Li et al., 1998, 1999), which is known to regulate 1,3-β-glucan synthase in yeast (Qadota et al., 1996).
Analysis of bacterial and plant CelSs has revealed the presence of a D, D, D35QXXRW motif distributed over two domains, A and B, in the repetitive transferases and only in domain A in the nonrepetitive transferases (Saxena et al., 1995). These motifs are not found in the fungal FKS1 homologs (Ishiguro et al., 1997) and are absent in CalS1 and other members of the Arabidopsis CalS family. Although CalS1 contains a sequence (KSGG) matching the consensus, (R/K)XGG, implicated in UDP-glucose binding in glycogen synthases (Farkas et al., 1990), it is absent in all other Arabidopsis CalSs and fungal FKS1 homologs (Ishiguro et al., 1997). Alternatively, CalS proteins may interact with another peptide that binds to UDP-glucose. This hypothesis is supported by data from our study of a novel UGT1 that interacts with CalS1 and copurifies with the CalS complex (Hong et al., 2001).
A recent study on the synthesis of curdlan, a 1,3-β-glucan, has provided the surprising result that the curdlan synthase (CrdS) from Agrobacterium (Stasinopoulos et al., 1999) is very similar to bacterial and plant 1,4-β-glucan synthases (Saxena et al., 1994; Pear et al., 1996; Arioli et al., 1998; Taylor et al., 1999) but is distinct from 1,3-β-glucan synthases from Saccharomyces, Schizosaccharomyces, Aspergillus, Candida, Cryptococcus, and Paracoccidioides (Douglas et al., 1994; Kelly et al., 1996; Ishiguro et al., 1997; Mio et al., 1997; Thompson et al., 1999; Pereira et al., 2000). Like most repetitive 1,4-β-glucan synthases, CrdS contains the D, D, D35QXXRW motif (Saxena et al., 1995), which is absent in the known 1,3-β-glucan synthases of yeasts and other fungi. It is interesting that the (1→3,1→4)-β-glucan hydrolases from plants (Hoj and Fincher, 1995) are distinct from those from bacteria (Heinemann et al., 1996), but their specificities and catalytic mechanisms are identical (Hoj and Fincher, 1995). These data suggest that CrdS and 1,3-β-glucan synthases may be products of convergent evolution (Stasinopoulos et al., 1999).
CalS1 Is a Cell Plate–Specific Isoform and Forms a Complex on the Cell Plate
We demonstrated that CalS1 interacts with both phragmoplastin and UGT1 in a yeast two-hybrid system as well as in vitro. The location of phragmoplastin and UGT1 on the cell plate and the juxtaposition of the CalS1 and UGT1 genes on chromosome 1 suggested that CalS1 and UGT1 may be induced coordinately and become associated on the forming cell plate. This was also suggested by the observation that the GFP–CalS1 fusion protein was localized on the cell plate and copurified with the product-entrapped CalS complex. UGT1 also was localized on the cell plate (Hong et al., 2001).
We have observed that UGT1 interacts with Rop1 (Hong et al., 2001), suggesting that plant CalS may be regulated similarly to the 1,3-β-glucan synthase in yeast (Qadota et al., 1996) and that UGT1 may act as a subunit of the CalS1 enzyme. Together, our data suggest that CalS1 may form a functional complex with phragmoplastin, UGT1, and Rop1. This hypothesis is consistent with an earlier observation that a peptide of 57 kD that binds radioactive UDP-glucose is associated with CalS activity (Frost et al., 1990). This peptide is likely to be the homolog of the cell plate–specific UGT1 (Hong et al., 2001), which has a molecular mass of ∼60 kD. Another possible component of the CalS1 complex is sucrose synthase (SuSy), which generates UDP-glucose from sucrose. SuSy has been shown to be associated with the developing cell plate (Amor et al., 1995), and the product of SuSy could be transferred to CalS1 through UGT1, forming a substrate channel. Such a structure could facilitate the rapid deposition of callose on the forming cell plate.
Multiple CalS Genes May Be Required for the Diverse Functions of Callose in Plants
In addition to the cell plate, callose is synthesized in a variety of specialized tissues and in response to mechanical and physiological stresses (Stone and Clarke, 1992). Multiple CalS isozymes may have evolved in higher plants to catalyze callose synthesis in different locations and in response to different physiological and developmental signals. Earlier kinetic studies on partially purified CalS have suggested the existence of CalS isoforms in higher plants. For example, the pollen tube CalS is Ca2+ independent and activated by protease or detergent treatment in vitro (Schlupmann et al., 1993; Li et al., 1997), whereas plasma membrane CalS is strictly Ca2+ dependent (Kudlicka and Brown, 1997). Ca2+ plays a key role in cell plate formation and may activate the cell plate–specific CalS1 (Verma, 2001), and Ca2+ also is known to affect CalS activity in vitro (Kudlicka and Brown, 1997).
In yeast, the catalytic subunit of 1,3-β-glucan synthase is encoded by two genes, FKS1 and FKS2 (Douglas et al., 1994; Inoue et al., 1995; Mazur et al., 1995). They share a high degree of homology (90%) and may act as alternative subunits with essentially overlapping functions. Mutations in the FKS1 gene significantly reduce 1,3-β-glucan synthase activity. Expression of the FKS1 and FKS2 genes is differentially regulated, the former being predominant during growth on glucose, whereas the latter is induced by pheromone and during sporulation (Mazur et al., 1995). Both peptides interact with GNS1 and Rho1, and the latter acts as a regulatory subunit of 1,3-β-glucan synthase (Qadota et al., 1996). The plant CalS1 has overall sequence homology with FKS1, including the topological arrangement, suggesting that the catalytic activity resides in this peptide. This is consistent with the size of the peptide (190 kD) purified by product entrapment from tobacco pollen tubes and BY-2 cells (Turner et al., 1998; Hong et al., 2001). However, to facilitate a rapid flux of substrate, CalS1 interacts with a novel UGT1 (Hong et al., 2001; Verma, 2001) that may transfer substrate (UDP-glucose) produced by SuSy (see below) to the catalytic site of CalS for rapid synthesis of copious amounts of callose needed for expanding cell plate.
Because CalS is involved in synthesizing a polymer, its mobility in the membrane could be limited. Moreover, callose is synthesized in various locations and in response to different environmental and developmental signals. It is likely that different forms of this enzyme are required for the proper growth and development of the plant. A xylem-specific CelS gene was isolated recently (Wu et al., 2000), suggesting that each of the CelS and CalS isoforms may have specific functions in a particular tissue. The formation of a functional CalS1 complex is vital to building a cell plate, which must be completed in a short time, and a delay in the synthesis of callose or its overproduction may alter the composition of the cell plate and produce daughter cells with altered cell walls (Verma, 2001). A detailed analysis of the CalS1 complex and the role of various components may reveal how the activity of this enzyme is regulated during cell plate formation and may lead to an understanding of the de novo callose synthesis machinery in plants. In addition, functions of CalS-associated proteins need to be determined in order to understand tissue-specific deposition of callose.
METHODS
Bacterial and Yeast Strains and Tobacco Cell Culture
Escherichia coli strain Top10F′ (Invitrogen, Carlsbad, CA) was used for plasmid manipulation. Saccharomyces cerevisiae strain EGY48 (MATα trp1 his3 ura3 leu2::6 LexAop-LEU2) containing pSH18-34 (β-galactosidase reporter plasmid; Gyuris et al., 1993) was grown in yeast nitrogen base (YNB) medium (0.67 g/L YNB [Sigma], 0.6 g/L amino acid dropout mix [Bio101, La Jolla, CA], and 20 g/L glucose or galactose). Tobacco BY-2 cells overexpressing soybean phragmoplastin (Gu and Verma, 1997) were maintained in Murashige and Skoog (1962) medium.
Protein Sequence Analysis
The Tblastp program (Stephen et al., 1997) was used to search the Arabidopsis thaliana database for homology with the yeast β-1,3-glucan synthase FKS1p (Douglas et al., 1994). The coding region of callose synthase1 (CalS1) was deduced from the cloned cDNA used in this study. Exons from genomic sequences for the rest of the CalSs were identified by software programs, including Procrustes (Gelfand et al., 1996; http://hto-13.usc.edu/software/procrustes), Grail (Informatics Group, Oak Ridge National Laboratory, Oak Ridge, TN; http://compbio.ornl.gov/section/index.html), GENSCAN (Burge and Karlin, 1997; http://gnomic.stanford.edu/~chris/GENSCANW.html), Fexa (V. Solovyev and A. Salamov, Sanger Centre, Cambridge, UK; http://genomic.sanger.ac.uk/), and NetPlantGene (Hebsgaard et al., 1996; http://www.cbs.dtu.dk/NetPlantGene.html). CalS1 protein sequence was compared, using TblastN (http://www.ncbi.nlm.nih.gov/blast), with genomic DNA sequences translated in all reading frames to identify more exons. Annotation of genomic sequences was finalized by combining exons that best fit in an alignment with CalS1 and other known CalS sequences. Alignment of peptide sequences and construction of the phylogenetic tree were performed using DNAStar software (DNAStar, Madison, WI). Motif analysis of CalS1 was performed using software on the World Wide Web, including ProfileScan (http://www.isrec.isb-sib.ch), PrositeScan (http://pbil.ibcp.fr), TMHMM (http://www.cbs.dtu.dk), and HMMTOP (http://www.enzim.hu).
Cloning of CalS1 cDNA
Four pairs of oligonucleotides corresponding to the sequences surrounding unique restriction enzyme sites in the deduced exons of CalS1 were synthesized (SX1-54, 5′-CGAGCTCGAATTCGAAAATAT- GGCTCAAAGAAGGGAACC-3′; XS1, 5′-CTGAAGCCAGTCTAGAAT- GTCTTCATCG-3′; XK1, 5′-CTCGATGAAGACATTCTAGACTGGC-3′; KX1, 5′-GTGGTACCAAGCAATCATTAAAGGCCCC-3′; KC1, 5′-GAT-TGCTTGGTACCACAAGATAACAGCG-3′; CK1, 5′-CTCATCGATATA-GGCGACACGAATAGACGG-3′; CN1, 5′-CGTGTCGCCTATATCGATGAGGTAGAGC-3′; and NC1, 5′-CTCAATCAATGCATTGTGGAA-CCCCC-3′). Total RNA was obtained from shoot apical meristems of 3-week-old Arabidopsis (ecotype Columbia) seedlings. Reverse transcription (RT) of the RNA was performed using oligo(dT) as a primer and Moloney murine leukemia virus reverse transcriptase (Gibco BRL). cDNA fragments were amplified by polymerase chain reaction (PCR) using Platinum Taq DNA polymerase (Gibco BRL) and cloned in the pT-Adv vector (Clontech, Palo Alto, CA). The DNA sequence was determined using an Applied Biosystems (Foster City, CA) model 373A DNA sequencer. The full-length cDNA (pCalS1) was assembled from the PCR fragments and an expressed sequence tag (EST) clone (ATTS5466) by digestion with unique restriction enzymes followed by ligation (Figure 1). To eliminate possible PCR errors, any inconsistency between cDNA and genomic sequences was verified by sequencing independent clones.
Yeast Two-Hybrid Assay
The CalS1 cDNA was cloned in the EcoRI-XhoI sites of pJG4-5 (Estojak et al., 1995; Golemis et al., 1996) with a human influenza (hemagglutinin [HA]) tag, generating pJG-CalS1. Plasmid pEG-Phr was constructed by cloning soybean phragmoplastin cDNA (Gu and Verma, 1996) in the BamHI-XhoI sites of pEG202. The coding region of UDP-glucose transferase1 (UGT1) (Hong et al., 2001) was cloned in the EcoRI-XhoI sites of pEG202, generating pEG-UGT1. Yeast EGY48 cells containing pSH18-34 were transformed with pJG-CalS1 and pEG-Phr or pEG-UGT1 using electroporation. Cloning vectors pJG4-5 and pEG202 were used as controls. Interaction assays were performed on YNB medium (YNB-His-Trp-Ura) containing 75 mg/L 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) in the presence of galactose (for induction) or glucose (as a control).
Protein Gel Blot Analysis
Yeast cells grown in YNB medium (YNB-His-Trp-Ura) in the presence of glucose were pelleted and washed with YNB containing galactose. The cells were resuspended in YNB-galactose medium to induce the expression of CalS1 under the control of the Gal1 promoter. Total proteins from the cells were resolved by SDS-PAGE, and protein gel blot analysis was performed using HA antibody (Santa Cruz Biotechnology, Santa Cruz, CA), phragmoplastin antibody (Gu and Verma, 1996), or Arabidopsis UGT1 antibody (Hong et al., 2001).
Expression of the GFP–CalS1 Fusion Protein and Fluorescence Detection
A 2.2-kb XhoI fragment was released from pCalS1, generating a partial CalS1 cDNA (CalSX) that lacks the coding region for the last 10 transmembrane helices. Both partial (CalSX) and full-length CalS1 cDNAs were fused with green fluorescent protein (GFP) under the control of the cauliflower mosaic virus (CaMV) 35S promoter and expressed in tobacco BY-2 cells (Gu and Verma, 1997). Localization of the GFP fusion protein was performed using an epifluorescence microscope.
Copurification of the GFP–CalS1 Chimeric Protein with the CalS Complex
Total membranes from transgenic tobacco BY-2 cells expressing GFP–CalS1 were isolated. Membranes were solubilized in 3-([3-cholamidopropyl]dimethylammonio)-1-propanesulfonic acid (Chaps) buffer, and the soluble fraction was subjected to a product-entrapment procedure, as described elsewhere (Hong et al., 2001). An aliquot of proteins from each step of purification as well as control membrane samples were analyzed by SDS-PAGE and processed for protein gel blot analysis using GFP monoclonal antibody (Clontech).
Synchronization of BY-2 Cells and Measurement of CalS Activity
Control and transgenic tobacco BY-2 cell lines were synchronized using aphidicolin and propyzamide, as described previously (Gu and Verma, 1997). Cells were harvested in liquid nitrogen 1 hr after the release from propyzamide and ground in a mortar with a pestle. The homogenized powders were transferred to extraction buffer (100 mM Hepes/KOH, pH 7.5, 5 mM EDTA, 5 mM mercaptoethanol, and 20 μM phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 9000g for 10 min, and the supernatant was subjected to ultracentrifugation at 100,000g for 40 min to obtain the total membrane fraction. The membrane pellet was extracted with 0.5% digitonin in 50 mM Hepes, pH 7.5, 1 mM DTT, 1 mM CaCl2, and 20 μM phenylmethylsulfonyl fluoride. The solubilized membrane fraction was used to measure CalS activity, as described elsewhere (Shedletzky et al., 1997; Hong et al., 2001).
Histological Detection of Callose
Tobacco BY-2 cells were fixed in FAA solution (formalin:acetic acid:50% ethanol, 5:5:90) for 2 hr followed by a rinse in wash solution (95% ethanol:37.5% HCl, 1:1) for 2 min. After an extensive wash with PBS, the cells were stained with aniline blue (1 mg/L) and ethidium bromide (1 mg/L). Micrographs were taken using a fluorescent Zeiss (Jena, Germany) microscope with appropriate filters.
Supplementary Material
Acknowledgments
We thank Dr. R. Dalbey for his comments on this work and Dr. S. Pang for the provision of GFP plasmids. This work was supported by the National Science Foundation (Grant No. IBN-9724014). EST clone was provided by the Ohio State University Arabidopsis Biological Resource Center.
References
- Amor, Y., Haigler, C.H., Johnson, S., Wainscott, M., and Delmer, D.P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92, 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Aspinall, G.O., and Kessler, G. (1957). The structure of callose from the grape vine. Chem. Ind. (London), p. 1296.
- Brown, R.M., Jr., Saxena, I.M., and Kudlicka, K. (1996). Cellulose biosynthesis in higher plants. Trends Plant Sci. 1, 149–156. [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Cheon, C.I., Hong, Z., and Verma, D.P. (1994). Nodulin-24 follows a novel pathway for integration into the peribacteroid membrane in soybean root nodules. J. Biol. Chem. 269, 6598–6602. [PubMed] [Google Scholar]
- Cui, X., Shin, H., and Brown, M. (1999). A novel gene from cotton shows homology to the yeast β-1,3-glucan synthase subunit FKS1. Am. Soc. Plant Physiol. Meeting, Baltimore, MD, Abstract, p. 62.
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 245–276. [DOI] [PubMed] [Google Scholar]
- Delmer, D.P., Solomon, M., and Read, S.M. (1991). Direct photolabeling with [32P]UDP-glucose for identification of a subunit of cotton fiber callose synthase. Plant Physiol. 95, 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., and Ray, P.M. (1994). Purification of 1,3-β-d-glucan synthase activity from pea tissue: Two polypeptides of 55 kDa and 70 kDa copurify with enzyme activity. Eur. J. Biochem. 220, 943–953. [DOI] [PubMed] [Google Scholar]
- Douglas, C.M., et al. (1994). The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-d-glucan synthase. Proc. Natl. Acad. Sci. USA 91, 12907–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Sherbeini, M., and Clemas, J.A. (1995). Cloning and characterization of GNS1: A Saccharomyces cerevisiae gene involved in synthesis of 1,3-b-glucan in vitro. J. Bacteriol. 177, 3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estojak, J., Brent, R., and Golemis, E.A. (1995). Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15, 5820–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas, I., Hardy, T.A., DePaoli-Roach, A.A., and Roach, P.J. (1990). Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J. Biol. Chem. 265, 20879–20886. [PubMed] [Google Scholar]
- Frost, D.J., Read, S.M., Drake, R.R., Haley, B.E., and Wasserman, B.P. (1990). Identification of the UDP-glucose-binding polypeptide of callose synthase from Beta vulgaris L. by photoaffinity labeling with 5-azido-UDP-glucose. J. Biol. Chem. 265, 2162–2167. [PubMed] [Google Scholar]
- Gelfand, M.S., Mironov, A.A., and Pevzner, P.A. (1996). Gene recognition via spliced sequence alignment. Proc. Natl. Acad. Sci. USA 93, 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut, D.M., and Carpita, N.C. (1994). Biosynthesis of plant cell wall polysaccharides. FASEB J. 8, 904–915. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., Gyuris, H., and Brent, R. (1996). Interaction trap/two hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley), pp. 20.1.1–20.1.28.
- Gu, X., and Verma, D.P.S. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1997). Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, U., Ay, J., Gaiser, O., Muller, J.J., and Ponnuswamy, M.N. (1996). Enzymology and folding of natural and engineered bacterial β-glucanases studied by X-ray crystallography. Biol. Chem. 377, 447–454. [PubMed] [Google Scholar]
- Hoj, P.B., and Fincher, G.B. (1995). Molecular evolution of plant β-glucan endohydrolases. Plant J. 7, 367–379. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Zhang, Z., Olson, J.M., and Verma, D.P.S. (2001). A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.A., Delauney, A.J., and Verma, D.P. (1992). A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc. Natl. Acad. Sci. USA 89, 9354–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S.B., Takewaki, N., Takasuka, T., Mio, T., Adachi, M., Fujii, Y., Miyamoto, C., Arisawa, M., Furuichi, Y., and Watanabe, T. (1995). Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231, 845–854. [DOI] [PubMed] [Google Scholar]
- Ishiguro, J., Saitou, A., Duran, A., and Ribas, J.C. (1997). cps1+, a Schizosaccharomyces pombe gene homolog of Saccharomyces cerevisiae FKS genes whose mutation confers hypersensitivity to cyclosporin A and papulacandin B. J. Bacteriol. 179, 7653–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, S.R., and Northcote, D.H. (1985). In vitro glucan synthesis by membranes of celery petioles: The role of the membrane in determining the type of linkage formed. J. Cell Sci. 2 (suppl.), 1.–11. [DOI] [PubMed] [Google Scholar]
- Kakimoto, T., and Shibaoka, H. (1992). Synthesis of polysaccharides in phragmoplasts isolated from tobacco BY-2 cells. Plant Cell Physiol. 33, 353–361. [Google Scholar]
- Kamat, U., Garg, R., and Sharma, C.B. (1992). Purification to homogeneity and characterization of a 1,3-β-glucan (callose) synthase from germinating Arachis hypogaea cotyledons. Arch. Biochem. Biophys. 298, 731–739. [DOI] [PubMed] [Google Scholar]
- Kauss, H. (1996). Callose synthesis. In Membranes: Specialized Functions in Plants, M. Smallwood, J.P. Knox, and D.J. Bowles, eds (Guildford, UK: Bios Scientific Publishers), pp. 77–92.
- Kelly, R., Register, E., Hsu, M.J., Kurtz, M., and Nielsen, J. (1996). Isolation of a gene involved in 1,3-β-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J. Bacteriol. 178, 4381–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg, M. (1990). Mechanism and evolution of the uncoupling protein of brown adipose tissue. Trends Biochem. Sci. 15, 108–112. [DOI] [PubMed] [Google Scholar]
- Kudlicka, K., and Brown, R.M. (1997). Cellulose and callose biosynthesis in higher plants. I. Solubilization and separation of (1→3)- and (1→4)-β-glucan synthase activities from mung bean. Plant Physiol. 115, 643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, S., Mason, T., Sabin, R., Sloan, M., Drake, R., Haley, B., and Wasserman, B. (1989). UDP-glucose:(1,3)-β-glucan synthase from Daucus carota L.: Characterization, photoaffinity labeling, and solubilization. Plant Physiol. 90, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Bacic, A., and Read, S.M. (1997). Activation of pollen tube callose synthase by detergents: Evidence for different mechanisms of action. Plant Physiol. 114, 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Lin, Y., Heath, R.M., Zhu, M.X., and Yang, Z. (1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Brown, R.M., Jr. (1993). β-Glucan synthesis in the cotton fiber. II. Regulation and kinetic properties of β-glucan synthases. Plant Physiol. 101, 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur, P., Morin, N., Baginsky, W., el-Sherbeini, M., Clemas, J.A., Nielsen, J.B., and Foor, F. (1995). Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 15, 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack, B.A., Gregory, A.C., Kerry, M.E., Smith, C., and Bolwell, G.P. (1997). Purification of an elicitor induced glucan synthase (callose synthase) from suspension cultures of French bean (Phaseolus vulgaris): Purification and immunolocation of a probable Mr-65000 subunit of the enzyme. Planta 203, 196–203. [DOI] [PubMed] [Google Scholar]
- Meikle, P.J., Ng, K.F., Johnson, E., Hoogenraad, N.J., and Stone, B.A. (1991). The β-glucan synthase from Lolium multiflorum: Detergent solubilization, purification using monoclonal antibodies, and photoaffinity labeling with a novel photoreactive pyrimidine analogue of uridine 5′-diphosphoglucose. J. Biol. Chem. 266, 22569–22581. [PubMed] [Google Scholar]
- Miao, G.-H., Hong, Z., and Verma, D.P.S. (1992). Topology and phosphorylation of nodulin-26 in the peribacteroid membrane of root nodules. J. Cell Biol. 118, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio, T., Adachi-Shimizu, M., Tachibana, Y., Tabuchi, H., Inoue, S.B., Yabe, T., Yamada-Okabe, T., Arisawa, M., Watanabe, T., and Yamada-Okabe, H. (1997). Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in β-1,3-glucan synthesis. J. Bacteriol. 179, 4096–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P., and Stalker, D.M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93, 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, M., Felipe, M.S., Brigido, M.M., Soares, C.M., and Azevedo, M.O. (2000). Molecular cloning and characterization of a glucan synthase gene from the human pathogenic fungus Paracoccidioides brasiliensis. Yeast 16, 451–462. [DOI] [PubMed] [Google Scholar]
- Qadota, H., Python, C.P., Inoue, S.B., Arisawa, M., Anraku, Y., Zheng, Y., Watanabe, T., Levin, D.E., and Ohya, Y. (1996). Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272, 279–281. [DOI] [PubMed] [Google Scholar]
- Read, S.M., and Delmer, D.P. (1987). Inhibition of mung bean UDP-glucose:(1→3)-β-glucan synthase by UDP-pyridoxal: Evidence for an active-site amino group. Plant Physiol. 85, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, A.L., Giddings, T.H., and Staehelin, L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, W., Koster, W., and Dassa, E. (1994). Bacterial binding protein–dependent permeases: Characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol. Microbiol. 12, 993–1004. [DOI] [PubMed] [Google Scholar]
- Saxena, I.M., Kudlicka, K., Okuda, K., and Brown, R.M., Jr. (1994). Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: Implications for cellulose crystallization. J. Bacteriol. 176, 5735–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, I.M., Brown, R.M., Jr., Fevre, M., Geremia, R.A., and Henrissat, B. (1995). Multidomain architecture of β-glycosyl transferases: Implications for mechanism of action. J. Bacteriol. 177, 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlupmann, H., Bacic, A., and Read, S.R. (1993). A novel callose synthase from pollen tubes of Nicotiana. Planta 191, 470–481. [Google Scholar]
- Shedletzky, E., Unger, C., and Delmer, D.P. (1997). A microtiter-based fluorescence assay for (1,3)-β-glucan synthases. Anal. Biochem. 249, 88–93. [DOI] [PubMed] [Google Scholar]
- Sipos, L., and von Heijne, G. (1993). Predicting the topology of eukaryotic membrane proteins. Eur. J. Biochem. 213, 1333–1340. [DOI] [PubMed] [Google Scholar]
- Staehelin, L.A., and Hepler, P.K. (1996). Cytokinesis in higher plants. Cell 84, 821–824. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos, S.J., Fisher, P.R., Stone, B.A., and Stanisich, V.A. (1999). Detection of two loci involved in (1→3)-β-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology 9, 31–41. [DOI] [PubMed] [Google Scholar]
- Stephen, A.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B.A., and Clarke, A.E. (1992). Chemistry and physiology of higher plant 1,3-β-glucans (callose). In Chemistry and Biology of (1,3)-β-Glucans, B.A. Stone and A.E. Clarke, eds (Bundoora, Australia: La Trobe University Press), pp. 365–429.
- Strosberg, A.D. (1991). Structure/function relationship of proteins belonging to the family of receptors coupled to GTP-binding proteins. Eur. J. Biochem. 196, 1–10. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.R., Douglas, C.M., Li, W., Jue, C.K., Pramanik, B., Yuan, X., Rude, T.H., Toffaletti, D.L., Perfect, J.R., and Kurtz, M. (1999). A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J. Bacteriol. 181, 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., Bacic, A., Harris, P.J., and Read, S.M. (1998). Membrane fractionation and enrichment of callose synthase from pollen tubes of Nicotiana alata Link et Otto. Planta 205, 380–388. [DOI] [PubMed] [Google Scholar]
- Verma, D.P.S. (2001). Cytokinesis and building of the cell plate in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 751–784. [DOI] [PubMed] [Google Scholar]
- Wasserman, B.P., Wu, A., and Harriman, R.W. (1992). Probing the molecular architecture of (1,3)-β-glucan (callose) synthase: Poly-peptide depletion studies. Biochem. Soc. Trans. 20, 18–22. [DOI] [PubMed] [Google Scholar]
- Wu, L., Joshi, C.P., and Chiang, V.L. (2000). A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J. 22, 495–502. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Hong, Z., and Verma, D.P.S. (2000). Phragmoplastin polymerizes into spiral coiled structures via intermolecular interaction of two self-assembly domains. J. Biol. Chem. 275, 8779–8784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.