Abstract
Transcript regulation in response to high salinity was investigated for salt-tolerant rice (var Pokkali) with microarrays including 1728 cDNAs from libraries of salt-stressed roots. NaCl at 150 mM reduced photosynthesis to one tenth of the prestress value within minutes. Hybridizations of RNA to microarray slides probed for changes in transcripts from 15 min to 1 week after salt shock. Beginning 15 min after the shock, Pokkali showed upregulation of transcripts. Approximately 10% of the transcripts in Pokkali were significantly upregulated or downregulated within 1 hr of salt stress. The initial differences between control and stressed plants continued for hours but became less pronounced as the plants adapted over time. The interpretation of an adaptive process was supported by the similar analysis of salinity-sensitive rice (var IR29), in which the immediate response exhibited by Pokkali was delayed and later resulted in downregulation of transcription and death. The upregulated functions observed with Pokkali at different time points during stress adaptation changed over time. Increased protein synthesis and protein turnover were observed at early time points, followed by the induction of known stress-responsive transcripts within hours, and the induction of transcripts for defense-related functions later. After 1 week, the nature of upregulated transcripts (e.g., aquaporins) indicated recovery.
INTRODUCTION
In agricultural systems, the abiotic stresses—salinity, low temperature, and drought in particular—are responsible for most of the reduction that differentiates yield potential from harvestable yield (Boyer, 1982). Salt stress, the focus of our work, can lead to changes in development, growth, and productivity, and severe stress may threaten survival. Many studies have examined the effects of these stress factors, with the aim of discovering mechanisms used by stress-tolerant species and the elements that might confer tolerance to sensitive plants. At the physiological level, the multitude of effects of salt stress indicates the importance of protecting the organism from damage by reactive oxygen species that inevitably increase as water deficit and increased ion uptake impair photosynthesis (Noctor and Foyer, 1998; Dat et al., 2000). Also implicated are the protective roles played by the accumulation of metabolites that seem to act in more than one function—preventing radical formation, acting as low-molecular-weight chaperones, contributing to redox control, and functioning as compatible solutes by decreasing the osmotic potential (Barkla et al., 1999; Hasegawa et al., 2000; Sakamoto and Murata, 2000). Work on the components involved in relative tolerance has identified several proteins that determine end points of physiological responses (Hasegawa et al., 2000). Studies with transgenic plants support the notion that altered gene expression can lead to improvements in tolerance (Van Camp et al., 1996; Shikanai et al., 1998; Apse et al., 1999).
In addition, components of abiotic stress signal recognition and transduction pathways and, in part, the transcription activators now have become known (Hasegawa et al., 2000; Mizoguchi et al., 2000; Zhu, 2001). These components initiate and control the expression of downstream biochemical reactions by, for example, the action of a calcium sensor on a protein kinase that affects the activity of an Na+/H+ antiporter (Halfter et al., 2000; Ishitani et al., 2000; Shi et al., 2000). It is not clear whether the mechanistic end points that until now have been targeted by the generation of transgenic additions will have significant effect on plants in the field. An approach that may hold more promise is the transgenic enhancement of stress signaling pathways (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). New tools are now available to further our understanding of the genetics of abiotic stress tolerance, allowing us to address the complexity of stress responses on a larger scale through genome-wide expression profiling (Reymond et al., 2000; Richmond and Somerville, 2000).
We have used DNA microarrays to monitor transcript abundance and expression patterns in rice (lines Pokkali and IR29) exposed to high salinity. On the basis of root cDNA libraries and expressed sequence tags (ESTs) from the moderately salt-tolerant rice line Pokkali, microarray elements were assembled to monitor changes in transcripts compared with unstressed plants, from 15 min to 1 week after salt shock. We compared the behavior of Pokkali with that of the salt-sensitive variety IR29 as part of a program to detect, describe, and understand plant responses to high salinity (www.stress-genomics.org). We have focused on the response to salt shock in rice in hybridizations to 1728 transcripts derived from the roots of stressed plants (salt shock). The results indicate a progression of regulated functions such that different categories of transcripts show regulation at different time points. A difference between the two lines existed with respect to the onset of the initial response, with IR29 responding more slowly than Pokkali. Also, after 3 to 6 hr of stress, changes in transcript abundance began to converge on the prestress level in Pokkali, whereas for IR29 a general decrease of transcript amounts started ∼3 hr into the period of stress. The latter led to the death of IR29 plants, whereas the plants of line Pokkali recovered.
RESULTS AND DISCUSSION
Physiological Analysis of Pokkali under Salt Stress Conditions
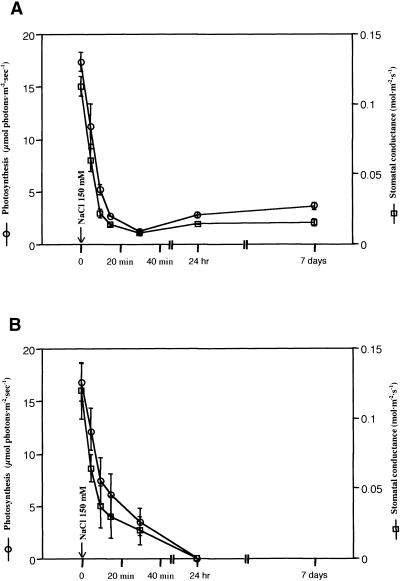
Photosynthesis, stomatal conductance, and transpiration were measured for Pokkali and IR29 seedlings (Figure 1) after the addition of 150 mM NaCl. Photosynthesis decreased within minutes and stabilized within 30 min at one tenth of the prestress level in line Pokkali (Figure 1A). The decrease was paralleled by decreased stomatal conductance and transpiration rates, indicating limited CO2 assimilation due to stomatal closure. Under long-term salt stress, Pokkali continued to grow at a low photosynthetic rate; after 7 days of salt stress, plant biomass had approximately doubled. It is known that Pokkali maintains water content in the shoot during a 6-week stress under the conditions used here (Moons et al., 1995). In contrast, salt-sensitive IR29 showed a slightly slower response to the shock treatment (Figure 1B), and the plants had wilted irreversibly after 24 hr. These data indicated that Pokkali achieved tolerance by rapidly expressing mechanisms that resist salt stress more efficiently than those available to the salt-sensitive IR29.
Figure 1.
Physiological Responses to High Salinity by Rice (var Pokkali).
Net CO2 assimilation and stomatal conductance in Pokkali (A) and IR29 (B) seedlings under salt-stress conditions (150 mM NaCl). Seedlings were grown in Hoagland's nutrient solution and exposed to salt stress (9 am) as indicated by the arrow.
Profiles of Transcript Abundance
A comparison of transcripts in Pokkali roots under control and salt stress conditions should identify differences pertinent to the tolerance “capacity” of the line. The most direct way to gather this information is to collect representative transcript profiles from the plants under control and stressed conditions for a comparison with sequences in public databases. From the root cDNA libraries (see Methods), ESTs were categorized and abundance profiles generated (library OC, no stress [1106 ESTs]; libraries OD, OE, and OF, stress for various times [1418 ESTs]; Table 1). The annotation process assembled 25% of the ESTs within contigs (based on stretches of overlapping sequences). Approximately 41% of the sequences from library OC were categorized as unclassified proteins, including those in the “no hits” category. This number increased to ∼58% in libraries from stressed plants. In the category no hits, 324 ESTs were found in the nonredundant database. Homologies were found for many of these ESTs, but 154 ESTs, mostly from stressed plants, did not show significant homology with ESTs in the databases. Other major distinctions in transcript composition between the control and stress library ESTs included, as a function of stress, a precipitous decrease in the protein synthesis category. In contrast, more transcripts were found for the transport facilitation and cell defense categories (Table 1).
Table 1.
Functional Categories of Transcripts in Rice (var Pokkali) Represented in Root cDNA Libraries
| No Stressa
|
NaCl Stressb
|
|||
|---|---|---|---|---|
| Major Functional Categories | No. | % | No. | % |
| Unclassified proteins | 354 | 32.0 | 590 | 41.6 |
| Protein synthesis | 253 | 22.9 | 152 | 10.7 |
| Metabolism | 109 | 9.9 | 142 | 10.0 |
| No hit | 94 | 8.5 | 230 | 16.2 |
| Cellular organization | 59 | 5.3 | 36 | 2.5 |
| Protein destination | 59 | 5.3 | 51 | 3.6 |
| Signal transduction | 45 | 4.1 | 41 | 2.9 |
| Energy | 29 | 2.6 | 39 | 2.8 |
| Transcription | 29 | 2.6 | 17 | 1.2 |
| Transport facilitation | 24 | 2.2 | 44 | 3.1 |
| Cell rescue, defense, and aging | 22 | 2.0 | 40 | 2.8 |
| Cell growth and division | 13 | 1.2 | 18 | 1.3 |
| Cellular biogenesis | 10 | 0.9 | 12 | 0.8 |
| Intracellular transport | 6 | 0.5 | 6 | 0.4 |
| Total | 1,106 | 100 | 1,418 | 100 |
OC library (no stress).
Combined OD, OE, and OF libraries (stress).
Microarray Assembly
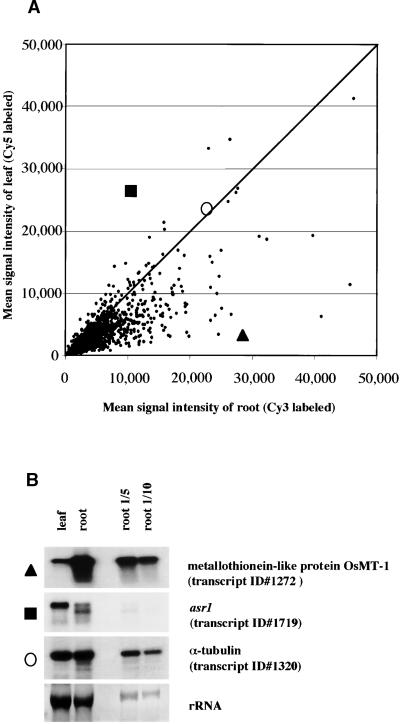
DNAs of selected clones containing inserts longer than 500 bp from the OC, OD, and OE libraries were amplified using T3/T7 primers. Amplicons longer than 400 bp were printed. The resulting microarrays comprised 1728 transcripts spotted in triplicate (5184 elements per slide). A table identifying all ESTs included in the array is provided at www.stress-genomics.org. After hybridization and washing, we analyzed the microarray data using commercial software. Signals from triplicate spots were averaged. The Cy3/Cy5 signal intensities were adjusted with the help of exogenously added control genes that had been placed in different sections of the microarray slides to compensate for variable background levels (see Methods). Also, ratios were calculated for the total Cy3 signal from all spots on a slide in relation to the total Cy5 signal. As an initial control, the microarrays were hybridized using fluorescent targets produced from mRNA taken from nonstressed root and leaf tissues (Figure 2A), indicating that most transcripts on the array, which derived from the root libraries, were more highly expressed in roots than in leaves. RNA gel blot analyses confirmed the tissue specificity of the array hybridization data (Figure 2B).
Figure 2.
Tissue Specificity by Microarray Hybridizations.
(A) Scatter plot comparing the spot intensities in hybridizations with probes from nonstressed roots (Cy3 labeled, x axis) and leaves (Cy5 labeled, y axis). Data from images of both Cy3 and Cy5 were plotted as the mean signal intensity after normalization of ESTs spotted in triplicate. The signal intensity of OsMT-1 (transcript 1272, triangle), asr1 (transcript 1719, square), and α-tubulin (transcript 1320, circle) are individually plotted and were confirmed by RNA gel blot analysis.
(B) RNA gel blot analysis for selected clones with total RNA (10 μg/lane) from control roots and leaves. RNA gel blot hybridizations were performed under the same condition used for the microarrays. The same membrane with total RNA from leaves and roots was used repeatedly for RNA gel blot hybridizations after washing with boiling water before reprobing. An RNA dilution series is included. Images exposed on x-ray film were analyzed by GelExpert software (version 3.5; Nucleotech, San Carlos, CA). The calculated log-10 ratios for root and leaf signals in RNA gel blot and microarrays are as follows: OsMT-1, 1.10/0.98; asr1, −0.4/−0.4; and α-tubulin, 0.02/−0.01.
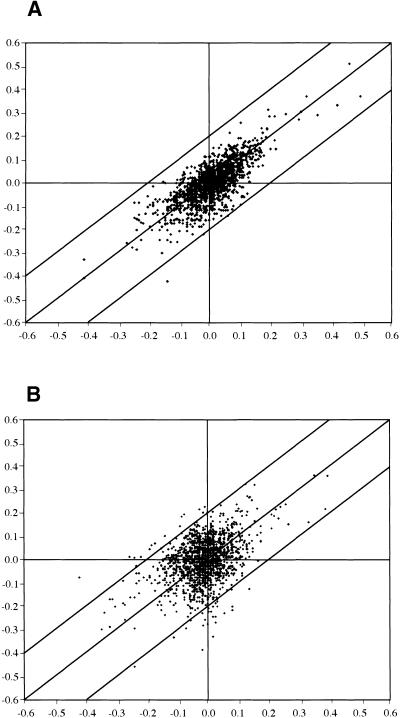
We tested variability within and between experiments (Figure 3). Changes in signal intensity were converted into log-10 expression ratios (LR) (LR [Cy3/Cy5]), for the Cy5/Cy3 signals of all clones on the arrays. Plotted are changes proportional to the scatter of data points around the mean as standard deviation from the population mean. Figure 3A compares the deviation from the mean of separate hybridizations with RNA from one experiment indicating 91% of the signals at LR 0.1 are equivalent to a deviation of ±1.25-fold. Within LR, 0.15 (±1.4-fold) were 98% of the signals and 0.18% of the signals at LR higher than 0.2 (±1.6-fold; Figure 3A). From technical repetitions for all time points, we concluded that the microarrays detected differential expression at greater than ±0.2 LR, similar to thresholds reported previously (Maleck et al., 2000). Higher variability was observed in repeat experiments using RNA from root tissues exposed to the same experimental conditions and harvested over a period of 1 year. Results are exemplified by a repeat experiment using RNA from 7-day stressed root tissue (Figure 3B). Within LR 0.1 were 76% of the signals, and 92% were within LR 0.15. Only 2.7% of the signals deviated greater than 0.2 LR, that is, 47 of 1728 ESTs, and this variation appeared to be random. Variation is, however, not the only parameter to consider, because a juxtaposition of the observed variations indicated that notwithstanding different absolute ratios of regulation, transcripts at greater than ±0.2 LR in one experiment typically were also among the most highly upregulated in independent experiments, although not always as high as the chosen threshold level. For significantly upregulated transcripts in the experiment listed in Tables 4 (Pokkali) and 6 (IR29), which we discuss below, we have listed regulation observed in a second, independent experiment.
Figure 3.
Comparison of Expression Ratios in Independent Microarray Hybridizations.
(A) Technical repetition. Scatter plot comparing the log-10 expression ratios (LR) from independent hybridizations using the same RNA. RNA of Pokkali root tissue from control and 150 mM NaCl- stressed Pokkali were labeled with Cy3 and Cy5, respectively, and the log ratios are compared. The repetition indicated 99% of the ESTs within ±0.2 LR.
(B) Scatter plot comparing the LR from independent hybridizations using RNA from different experiments with plants grown at different times under identical conditions in a greenhouse. Root RNA from control and 150 mM NaCl-stressed Pokkali extracted from the different sets of plants were labeled with Cy3 and Cy5 for microarray hybridization, and LRs were compared. This and other repetitions indicated 94 to 97% of the ESTs within ±0.2 LR. In the example, 2.7% of the ratios exceeded 0.2 LR from the mean; in all experiments, persistently variable elements were flagged and removed from the analysis.
Table 4.
Significantly Upregulated Transcripts in Rice Pokkali at Different Time Pointsa
| Pokkali
|
IR29b
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone No. | Accession No. | Annotation | 15 Min | 1 Hr | 3 Hr | 6 Hr | 24 Hr | 7 Days | 1 Hr | 3 Hr | 6 Hr |
| 15 min | |||||||||||
| 1723 | BE530977 | Glycine/serine-rich protein (1) | 0.46/.51c | 0.30 | Fd | −0.23 | F | F | −0.17 | −0.24 | −0.33 |
| 185 | BE039930 | Glycine/serine-rich protein (1) | 0.42/.33 | 0.24 | F | −0.17 | F | −0.30 | −0.15 | −0.19 | −0.13 |
| 446 | BE607361 | Glycine/serine-rich protein (2) | 0.36/.29 | 0.17 | F | −0.10 | F | −0.21 | −0.14 | −0.13 | −0.19 |
| 1539 | BE039583 | Glycine/serine-rich protein (2) | 0.32/.37 | 0.18 | 0.09 | −0.15 | −0.22 | 0.00 | −0.08 | −0.19 | −0.26 |
| 392 | BE607384 | Calcium-dependent protein kinase | 0.30/.30 | 0.48 | 0.26 | 0.18 | 0.04 | −0.13 | 0.12 | 0.31 | 0.18 |
| 1112 | BE039625 | Glycine/serine-rich protein (2) | 0.30/.27 | 0.09 | 0.01 | F | −0.19 | −0.09 | −0.19 | −0.34 | −0.38 |
| 1342 | BE607353 | β-Glucosidase, plastid type | 0.25/.30 | 0.13 | 0.29 | 0.08 | −0.05 | −0.20 | −0.05 | 0.08 | −0.03 |
| 1599 | BE607403 | Methionine synthase | 0.22/.14 | F | 0.09 | −0.11 | −0.07 | −0.09 | −0.12 | 0.08 | −0.02 |
| 662 | BE607330 | Unknown | 0.22/.18 | 0.08 | 0.07 | 0.07 | −0.09 | −0.10 | −0.06 | 0.07 | 0.08 |
| 1 hr | |||||||||||
| 392 | BE607384 | Calcium-dependent protein kinase | 0.30 | 0.48/.34 | 0.26 | 0.18 | 0.04 | −0.13 | 0.12 | 0.31 | 0.18 |
| 1133 | BE607429 | Unknown | 0.04 | 0.32/.17 | 0.13 | 0.05 | −0.09 | 0.00 | 0.07 | 0.25 | −0.03 |
| 1237 | BE607335 | Unknown | 0.06 | 0.32/.19 | −0.10 | −0.02 | −0.07 | F | −0.07 | −0.08 | −0.29 |
| 1089 | BE607327 | Peroxidase-1 | 0.03 | 0.32/.39 | 0.16 | 0.18 | 0.08 | −0.01 | 0.09 | 0.21 | −0.12 |
| 1723 | BE530977 | Glycine/serine-rich protein (1) | 0.46 | 0.30/.16 | F | −0.23 | F | F | −0.17 | −0.24 | −0.33 |
| 1463 | BE039983 | 40S ribosomal protein S9 | 0.11 | 0.28/.13 | 0.00 | 0.08 | −0.19 | −0.14 | −0.04 | 0.09 | 0.12 |
| 1014 | BE039953 | elongation factor-1α; EF-1α | 0.03 | 0.27/.22 | −0.03 | 0.11 | 0.00 | −0.05 | −0.04 | 0.03 | −0.12 |
| 1337 | BE607350 | 40S ribosomal protein S4 | 0.11 | 0.26/.29 | 0.06 | 0.11 | −0.02 | −0.10 | 0.05 | 0.12 | 0.06 |
| 1473 | BE039923 | 40S ribosomal protein S7 | 0.16 | 0.26/.24 | 0.00 | 0.06 | 0.05 | −0.09 | −0.06 | 0.12 | 0.06 |
| 598 | BE607369 | Nucleoside diphosphate kinase (NDPK-1) | 0.17 | 0.26/.26 | 0.12 | 0.05 | −0.22 | −0.24 | 0.00 | 0.11 | 0.08 |
| 3 hr | |||||||||||
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | 0.08 | F | 0.47/.36 | 0.47 | 0.35 | 0.05 | 0.05 | F | 0.56 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | 0.06 | 0.05 | 0.45/.31 | 0.31 | 0.22 | −0.02 | 0.07 | 0.41 | 0.38 |
| 1719 | BE530958 | asr1 (ABA and stress-induced protein) | 0.06 | −0.05 | 0.43/.33 | 0.54 | −0.21 | 0.17 | 0.16 | 0.48 | 0.32 |
| 167 | BE607370 | S-adenosylmethionine decarboxylase 2 | 0.12 | 0.13 | 0.35/.21 | 0.14 | 0.12 | 0.11 | 0.23 | 0.45 | 0.16 |
| 1220 | BE530895 | No hit | −0.01 | 0.05 | 0.33/.34 | 0.37 | 0.20 | F | 0.11 | 0.28 | 0.12 |
| 1043 | BE039925 | Trypsin inhibitor (1) | 0.06 | 0.19 | 0.32/.21 | 0.26 | 0.25 | 0.04 | 0.17 | 0.45 | 0.42 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.07 | 0.12 | 0.30/.29 | 0.29 | 0.16 | 0.25 | 0.13 | 0.39 | 0.26 |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.08 | 0.18 | 0.30/.16 | 0.44 | 0.11 | −0.10 | 0.22 | 0.35 | 0.41 |
| 1342 | BE607353 | β-Glucosidase, plastid type | 0.25 | 0.13 | 0.29/.23 | 0.08 | −0.05 | −0.20 | −0.05 | 0.08 | −0.03 |
| 1545 | BE039627 | LTI6B (low temperature, salt responsive) | 0.02 | −0.10 | 0.27/.21 | 0.00 | −0.06 | −0.04 | 0.12 | 0.09 | −0.07 |
| 6 hr | |||||||||||
| 1719 | BE530958 | asr1 (ABA- and stress-induced protein) | 0.06 | −0.05 | 0.43 | 0.54/.52 | −0.21 | 0.17 | 0.16 | 0.48 | 0.32 |
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | 0.08 | F | 0.47 | 0.47/.21 | 0.35 | 0.05 | 0.05 | F | 0.56 |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.08 | 0.18 | 0.30 | 0.44/.60 | 0.11 | −0.10 | 0.22 | 0.35 | 0.41 |
| 1220 | BE530895 | No hit | −0.01 | 0.05 | 0.33 | 0.37/.25 | 0.20 | F | 0.11 | 0.28 | 0.12 |
| 636 | BE040239 | Gigantea protein | 0.00 | 0.04 | 0.21 | 0.34/.56 | 0.03 | 0.06 | 0.06 | 0.44 | 0.35 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | 0.06 | 0.05 | 0.45 | 0.31/.31 | 0.22 | −0.02 | 0.07 | 0.41 | 0.38 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.07 | 0.12 | 0.30 | 0.29/.20 | 0.16 | 0.25 | 0.13 | 0.39 | 0.26 |
| 1043 | BE039925 | Trypsin inhibitor (1) | 0.06 | 0.19 | 0.32 | 0.26/.16 | 0.25 | 0.04 | 0.17 | 0.45 | 0.42 |
| 866 | BE607355 | Calmodulin (CaM1) | 0.03 | 0.13 | 0.26 | 0.26/.20 | 0.14 | 0.00 | 0.23 | 0.34 | 0.18 |
| 337 | BE039649 | Protein phosphatase 2C homolog | −0.01 | F | 0.00 | 0.24/.18 | 0.13 | 0.00 | −0.02 | 0.09 | −0.10 |
| 24 hr | |||||||||||
| 197 | BE607404 | Unknown | −0.24 | −0.30 | F | 0.02 | 0.41/.13 | 0.27 | 0.02 | 0.23 | 0.15 |
| 1110 | BE530893 | Glutathione S-transferase homolog | −0.07 | 0.16 | 0.04 | F | 0.39/.20 | 0.15 | 0.06 | 0.28 | 0.20 |
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | 0.08 | F | 0.47 | 0.47 | 0.35/.32 | 0.05 | 0.05 | F | 0.56 |
| 38 | BE607345 | Ascorbate peroxidase, cytosolic type | 0.01 | −0.02 | 0.10 | 0.14 | 0.31/.23 | 0.16 | 0.07 | 0.10 | 0.03 |
| 1043 | BE039925 | Trypsin inhibitor (1) | 0.06 | 0.19 | 0.32 | 0.26 | 0.25/.16 | 0.04 | 0.17 | 0.45 | 0.42 |
| 1011 | BE039950 | Unknown | 0.00 | 0.00 | 0.00 | F | 0.24/.25 | 0.23 | 0.04 | 0.08 | −0.02 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | 0.06 | 0.05 | 0.45 | 0.31 | 0.22/.25 | −0.02 | 0.07 | 0.41 | 0.38 |
| 840 | BE607389 | Osr40g2 (ABA and salt responsive) | 0.06 | −0.05 | 0.20 | 0.17 | 0.22/.16 | 0.05 | -0.03 | 0.21 | 0.15 |
| 7 days | |||||||||||
| 1625 | BE040365 | Metallothionein-like protein, OsMT-1 | −0.41 | −0.34 | −0.14 | 0.06 | 0.08 | 0.45/F | 0.08 | 0.01 | 0.01 |
| 577 | BE607365 | Water channel protein (WCP-I) | −0.25 | −0.44 | −0.11 | −0.21 | 0.09 | 0.40/.35 | 0.10 | 0.21 | −0.19 |
| 157 | BE607367 | Water channel protein (WCP-I, isoform) | −0.24 | −0.36 | 0.04 | −0.01 | 0.13 | 0.40/.21 | 0.10 | 0.22 | −0.04 |
| 1464 | BE607372 | Water channel protein (WCP-I) | −0.19 | −0.32 | 0.03 | −0.05 | 0.08 | 0.36/.36 | 0.10 | 0.10 | −0.10 |
| 1272 | BE607391 | Metallothionein-like protein, OsMT-1 | −0.41 | −0.46 | F | −0.01 | 0.00 | 0.34/.16 | −0.05 | −0.12 | −0.04 |
| 197 | BE607404 | Unknown | −0.24 | −0.30 | F | 0.02 | 0.41 | 0.27/.09 | 0.02 | 0.23 | 0.15 |
| 1534 | BE607333 | Ketol-acid reductoisomerase (KAR-1) | −0.14 | 0.06 | −0.20 | −0.07 | 0.20 | 0.26/.24 | 0.00 | 0.10 | −0.16 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.07 | 0.12 | 0.30 | 0.29 | 0.16 | 0.25/.23 | 0.13 | 0.39 | 0.26 |
| 1146 | BE607473 | Water channel protein (WCP-IV) | −0.14 | −0.22 | 0.07 | −0.21 | −0.14 | 0.24/.17 | 0.11 | 0.08 | −0.36 |
| 1011 | BE039950 | Unknown | 0.00 | 0.00 | 0.00 | F | 0.24 | 0.23/.19 | 0.04 | 0.08 | −0.02 |
In Tables 4 and 6, values of one repeat experiment are included for significantly upregulated ESTs for different time points. In some experiments, up- or downregulation of the ESTs in all repetitions varied by >0.2 LR, but these were not flagged.
For a comparison, IR29 LR for the regulated ESTs in Pokkali are provided.
Hybridization differences exceeding 0.2 LR (>1.6-fold induction of repression) are in boldface.
F, flagged ESTs represent ESTs for which the variability in repeated experiments deviated by greater than ±0.2 LR from the mean value.
Transcript Behavior in Pokkali under Salt Stress
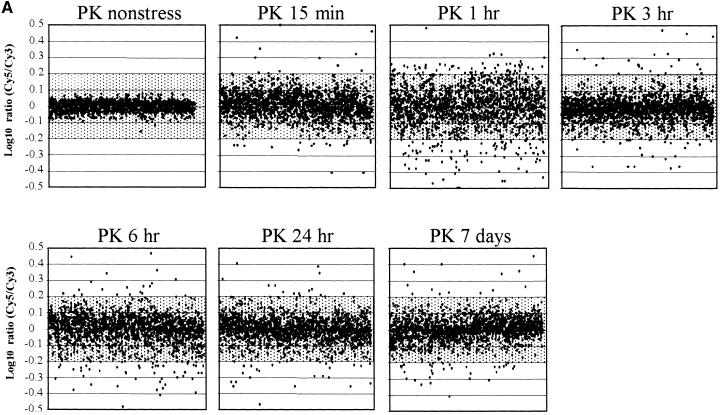
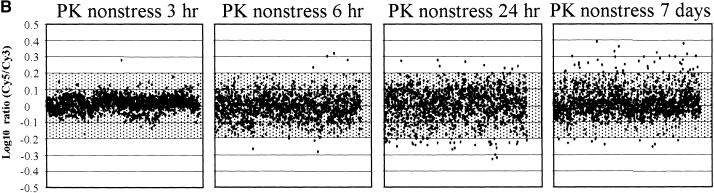
In all experiments, we compared the intensities of Cy3- and Cy5-labeled target (Figure 4A; labeled “PK nonstress”) to normalize variability. In this control, 95% of the spots were located within ±0.07 LR, providing a measure of experimental and systematic errors. Nonstressed 3-hr RNA (harvested 6 hr into the light period) hybridized against control RNA (nonstressed, harvested 3 hr into the light period) showed profiles very similar to those of control versus control RNA (98% of the spots located within ±0.1 LR), indicating that diurnal changes had little impact (Figure 4B, PK, nonstress 3 hr).
Figure 4.
Time Course of Transcript Expression in Rice (var Pokkali).
Transcripts expressed after 150 mM NaCl stress are shown. The expression ratios of transcripts (log-10 Cy5/Cy3) from a series of time course experiments using the microarray for Pokkali (PK) are plotted against the order of ESTs printed on the microarray (x axis).
(A) Stress responses during the 7-day period. RNA from control Pokkali roots (harvested at 9 am) labeled with Cy3- and Cy5-labeled RNA extracted from 150 mM NaCl-stressed roots for six time points (15 min, 1 hr, 3 hr, 6 hr, 24 hr, and 7 days) was used in hybridizations to ESTs deposited on microarray slides.
(B) Developmental changes during the 7-day period. RNA from control Pokkali roots (harvested at 9 am) labeled with Cy3- and Cy5-labeled RNA extracted from control (no stress) Pokkali roots from four time points (3, 6, and 24 hr and 7 days).
Altered regulation in gene expression after salt shock was detected by comparing probes from control times to probes at six RNA collection times after the imposition of stress (Figure 4A). Shaded in Figures 4 and 5 are LR at less than ±0.2 (less than ±1.6-fold change), which we considered not significant. The earliest time point after imposition of salt stress (15 min; Figure 4A, PK 15 min) indicated that Pokkali responded at the level of transcription within a time frame that corresponded to the changes in photosynthesis. After 15 min of salt stress, only 2% of the transcripts were upregulated or downregulated by greater than ±0.2 LR. An additional 14% were upregulated or downregulated by greater than ±0.1 LR (8% greater than +0.1 LR and 6% less than −0.1 LR). After 1 hr (Figure 4A, PK 1 hr), 33% of all transcripts were altered in transcription by a factor greater than ±0.1 LR (16% greater than +0.1 LR and 17% less than −0.1 LR), and 10% were regulated by greater than ±0.2 LR (4% greater than +0.2 LR and 6% less than −0.2 LR) (see www.stress-genomics.org).
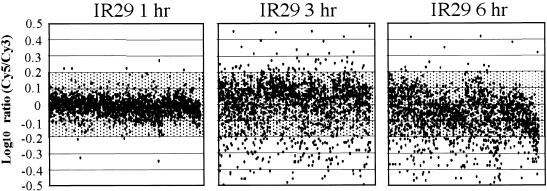
Figure 5.
Time Course of Transcript Expression in Rice (var IR29).
Transcripts expressed after 150 mM NaCl stress are shown. The expression ratios of transcripts (log-10 Cy5/Cy3) from a series of time course experiments are plotted against the order of ESTs printed on the microarray (x axis). RNA from control IR29 roots (harvested at 9 am) labeled with Cy3- and Cy5-labeled RNA extracted from 150 mM NaCl-stressed IR29 from three time points were hybridized to ESTs deposited on microarray slides.
In comparing stressed and control roots, the largest differences in transcript profiles were observed at the 1-hr time point (Figure 4A). Later, LRs increasingly converged to within ±0.1 LR of the control conditions (hatched areas). As one example, ribosomal proteins that were upregulated at the 1-hr time point (Table 2; see below) returned to the original expression levels after >3 hr of salt stress. Also, at 3 and 6 hr, transcripts homologous with abscisic acid–responsive genes or proteins were upregulated (see below). This finding coincides with a reported increase of abscisic acid levels in Pokkali roots (10-day-old seedlings) after salt stress (150 mM) with a peak after 8 hr (Moons et al., 1995, 1997). After 3 hr and up to 7 days, the number of transcripts that were either upregulated or downregulated by a factor of ±0.2 LR decreased to <5% of all transcripts. It is obvious that most transcripts in Pokkali returned within 24 hr and clearly after 7 days to their prestress expression levels.
Table 2.
ESTs for Ribosomal Proteins and Elongation Factor-1α Upregulated Early during Salt Stress in Rice Pokkali and IR29
| Time
|
||||||
|---|---|---|---|---|---|---|
| 15 Min | 1 Hr | 3 Hr | 6 Hr | 24 Hr | 7 Days | |
| Ribosomal proteinsa Pokkali | 0/2b | 36/47b | 1/0b | 2/0b | 0/0b | 0/0b |
| IR29 | —c | 0/1 | 2/0 | 2/0 | — | — |
| EF-1αd,e Pokkali | 0/0 | 9/7 | 0/0 | 0/0 | 0/0 | 0/0 |
| IR29 | — | 0/0 | 0/0 | 0/0 | — | — |
Upregulated transcripts (greater than +0.2 LR at 1 hr) in two experiments for proteins of 40S (S) and 60S (L) ribosome subunits: S4 (ID 12, 389, 875, 1337), S7 (ID 1473), S8 (ID 936, 1393), S9 (ID 1297), S10 (ID 1543), S19 (ID 1335), S26 (ID 995), L2 (ID 48), L5 (ID 502), L18 (ID 1454), L44 (ID 146), and PO (ID 384).
Number of significantly upregulated ESTs in two independent experiments.
Not included.
Upregulated transcripts (>0.2 LR at 1 hr) in two experiments for EF-1α: ID 27, 1014, 1015, 1033, and 1303.
EF-1α, elongation factor-1α.
An experiment was designed to monitor changes in transcripts in the absence of stress over the 1-week time period (Figure 4B) that would allow us to distinguish stress responses from developmental changes in gene expression. Of the 1728 ESTs printed, one (3 hr), five (6 hr), nine (24 hr), and 41 (1 week) showed higher than LR 0.2 compared with the expression at the start of the experiment. Only a few of these transcripts were also upregulated under stress conditions. One of these is a rice EST homologous (ID636; 67% identity) with Arabidopsis GIGANTEA (AJ133786). In Arabidopsis, this gene is regulated by the circadian clock with highest expression 8 to 10 hr after dawn (Fowler et al., 1999; Park et al., 1999). This compares with high signal intensity for the rice GIGANTEA-like transcript in unstressed and salt-stressed Pokkali (9 hr light). Salt stress, however, reduced the amount of the putative GIGANTEA transcripts from LR 0.55 to LR 0.35 (Table 3).
Table 3.
Upregulated Transcripts in Unstressed Plants at Different Time Points and Their Comparative Regulation during Salt Stress in Rice Pokkalia
| Clone No. | Accession No. | Annotation | Control | NaCl |
|---|---|---|---|---|
| 3-hr nonstress | ||||
| 834 | BG101703 | unknown | 0.28b | −0.05 |
| NaCl stress | ||||
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | 0.02 | 0.47 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | 0.04 | 0.45 |
| 1719 | BE530958 | asr1 (ABA- and stress-induced protein) | 0.13 | 0.43 |
| 167 | BE607370 | S-adenosylmethionine decarboxylase 2 | −0.17 | 0.35 |
| 1220 | BE530895 | No hit | 0.05 | 0.33 |
| 1043 | BE039925 | trypsin inhibitor (1) | 0.02 | 0.32 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.02 | 0.30 |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.00 | 0.30 |
| 1342 | BE607353 | β-Glucosidase, plastid type | 0.03 | 0.29 |
| 1545 | BE039627 | LTI6B (low temperature, salt responsive) | 0.04 | 0.27 |
| 6-hr nonstress | ||||
| 636 | BE040239 | Gigantea-like protein | 0.70 | 0.34 |
| 1432 | BG101702 | β-Glucosidase homolog | 0.29 | 0.20 |
| 1349 | BE607375 | No hit rice EST | 0.27 | −0.15 |
| 1570 | BE607450 | No hit rice EST | 0.25 | −0.05 |
| 669 | BG101699 | Similar to galactinol-raffinose galactosyltransferase | 0.21 | 0.00 |
| 1147 | BE607485 | Ascorbate peroxidase, cytosolic homolog | 0.21 | −0.12 |
| NaCl stress | ||||
| 1719 | BE530958 | asr1 (ABA- and stress-induced protein) | 0.11 | 0.54 |
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | −0.11 | 0.47 |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.20 | 0.44 |
| 1220 | BE530895 | No hit | 0.05 | 0.37 |
| 636 | BE040239 | Gigantea-like protein | 0.70 | 0.34 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | −0.08 | 0.31 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.13 | 0.29 |
| 1043 | BE039925 | Trypsin inhibitor (1) | 0.01 | 0.26 |
| 866 | BE607355 | Calmodulin (CaM1) | −0.11 | 0.26 |
| 337 | BE039649 | Protein phosphatase 2C homolog | 0.00 | 0.24 |
| 24-hr nonstress | ||||
| 168 | BE607371 | Unknown | 0.27 | −0.10 |
| 902 | BE607347 | Unknown | 0.27 | −0.11 |
| 1095 | BE607331 | Polyubiquitin 6 | 0.25 | 0.18 |
| 1534 | BE607333 | Ketol-acid reductoisomerase (KAR-1) | 0.24 | 0.20 |
| 1464 | BE607372 | Water channel protein (WCP-I) | 0.23 | 0.08 |
| NaCl stress | ||||
| 197 | BE607404 | Unknown | 0.12 | 0.41 |
| 1110 | BE530893 | Glutathione S-transferase homolog | 0.12 | 0.39 |
| 1129 | BE607422 | Osr40c1 (ABA and salt responsive) | −0.06 | 0.35 |
| 38 | BE607345 | Ascorbate peroxidase, cytosolic type | 0.02 | 0.31 |
| 1043 | BE039925 | Trypsin inhibitor (1) | −0.03 | 0.25 |
| 1011 | BE039950 | Unknown | 0.05 | 0.24 |
| 1427 | BE607408 | Osr40c1 (ABA and salt responsive) | −0.09 | 0.22 |
| 840 | BE607389 | Osr40g2 (ABA and salt responsive) | 0.02 | 0.22 |
| 7 days nonstress | ||||
| 1526 | BG101698 | No hit | 0.38 | −0.05 |
| 754 | BE607400 | Unknown | 0.36 | −0.07 |
| 691 | BE039604 | Soluble inorganic pyrophosphatase | 0.33 | −0.02 |
| 1214 | BE530894 | No hit | 0.33 | 0.02 |
| 1568 | BG101704 | No hit | 0.32 | 0.00 |
| 203 | BE040237 | Putative membrane protein | 0.30 | 0.03 |
| 1672 | BG101700 | Unknown | 0.30 | −0.02 |
| 699 | BE607421 | Unknown | 0.30 | −0.06 |
| 1053 | BG101701 | Unknown | 0.30 | 0.00 |
| 1251 | BE607383 | Unknown | 0.29 | 0.05 |
| 1001 | BE607410 | No hit | 0.27 | 0.00 |
| NaCl stress | ||||
| 1625 | BE040365 | Metallothionein-like protein, OsMT-1 | −0.01 | 0.45 |
| 577 | BE607365 | Water channel protein (WCP-I) | −0.24 | 0.40 |
| 157 | BE607367 | Water channel protein (WCP-I, isoform) | −0.23 | 0.40 |
| 1464 | BE607372 | Water channel protein (WCP-I) | −0.21 | 0.36 |
| 1272 | BE607391 | Metallothionein-like protein, OsMT-1 | 0.02 | 0.34 |
| 197 | BE607404 | Unknown | 0.00 | 0.27 |
| 1534 | BE607333 | Ketol-acid reductoisomerase (KAR-1) | −0.04 | 0.26 |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | −0.03 | 0.25 |
| 1146 | BE607473 | Water channel protein (WCP-IV) | −0.25 | 0.24 |
| 1011 | BE039950 | Unknown | −0.03 | 0.23 |
The table compares hybridization differences between control and 150-mM NaCl stress conditions and differences in the absence of stress that distinguish RNAs at the start of the experiments and after 7 days distinguishing developmental changes in gene expression from stress-related changes.
Differences in regulation by >0.2 LR (>1.6-fold difference) are indicated in boldface.
Expression Profiles in IR29 under Salt Stress Conditions
In contrast with the survival of the salt-tolerant Pokkali, the presence of 150 mM NaCl caused the salt-sensitive line IR29 to die within 24 hr. The expression profiles of transcripts during the initial phase reflect this process (Figure 4A). After 1 hr of salt stress, ∼7% (2% greater than +0.1 LR and 5% less than −0.1 LR) of the transcripts were regulated by a factor of ±0.1 LR compared with 33% for Pokkali. At this stage, no upregulation of ribosomal proteins or elongation factor-1α isoforms was observed (Table 2). Additional differences in transcript induction became apparent after 3 hr, at which time 38% of the transcripts were altered in transcription by a factor of greater than ±0.1 LR (20% greater than +0.1 LR and 18% less than −0.1 LR). At this stage, the transcripts that were upregulated in IR29 were upregulated similarly in Pokkali (Figure 5 and Tables 4 and 6). This increase in the number of upregulated transcripts was later replaced by a general downregulation. After 6 hr, 38% of the transcripts were downregulated to less than −0.1 LR; the comparable number for Pokkali was 13%. In contrast with Pokkali, the transcripts in IR29 that responded to salt stress by upregulation did not converge to the original expression level but became part of the general decrease. Thus, the differences in expression behavior between the two lines included a delayed initial response by IR29, and the fact that the recovery period observed for Pokkali after 3 hr of NaCl treatment was absent in IR29.
Table 6.
Significantly Upregulated Transcripts in Rice IR29 at Different Time Points
| Pokkalia
|
IR29
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone No. | Accession No. | Annotation | 15 Min | 1 Hr | 3 Hr | 6 Hr | 24 Hr | 7 Days | 1 Hr | 3 Hr | 6 Hr | |
| 1 hr | ||||||||||||
| 1259 | BE607385 | Translation initiation factor | −0.01 | −0.08 | 0.25 | b | 0.06 | −0.03 | 0.03 | 0.27/.35 | 0.31 | 0.04 |
| 167 | BE607370 | S-adenosylmethionine decarboxylase 2 | 0.12 | 0.13 | 0.35 | 0.14 | 0.12 | 0.11 | 0.23/.31 | 0.45 | 0.16 | |
| 866 | BE607355 | Calmodulin (CaM1) | 0.03 | 0.13 | 0.26 | 0.26 | 0.14 | 0.00 | 0.23/.37 | 0.34 | 0.18 | |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.08 | 0.18 | 0.30 | 0.44 | 0.11 | −0.10 | 0.22/.18 | 0.35 | 0.41 | |
| 3 hr | ||||||||||||
| 748 | BE607399 | Unknown | 0.09 | 0.08 | 0.29 | 0.21 | 0.10 | −0.01 | 0.05 | 0.51/.29 | 0.08 | |
| 1719 | BE530958 | asr1 (ABA- and stress-induced protein) | 0.06 | −0.05 | 0.43 | 0.54 | −0.21 | 0.17 | 0.16 | 0.48/F | 0.32 | |
| 167 | BE607370 | S-adenosylmethionine decarboxylase 2 | 0.12 | 0.13 | 0.35 | 0.14 | 0.12 | 0.11 | 0.23 | 0.45/.27 | 0.16 | |
| 1043 | BE039925 | Trypsin inhibitor (1) | 0.06 | 0.19 | 0.32 | 0.26 | 0.25 | 0.04 | 0.17 | 0.45/.14 | 0.42 | |
| 636 | BE040239 | Gigantea protein | 0.00 | 0.04 | 0.21 | 0.34 | 0.03 | 0.06 | 0.06 | 0.44/.22 | 0.35 | |
| 1427 | BE607408 | Osr40c1 (ABA- and salt-responsive) | 0.06 | 0.05 | 0.45 | 0.31 | 0.22 | −0.02 | 0.07 | 0.41/.24 | 0.38 | |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.07 | 0.12 | 0.30 | 0.29 | 0.16 | 0.25 | 0.13 | 0.39/.28 | 0.26 | |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.08 | 0.18 | 0.30 | 0.44 | 0.11 | −0.10 | 0.22 | 0.35/.23 | 0.41 | |
| 866 | BE607355 | Calmodulin (CaM1) | 0.03 | 0.13 | 0.26 | 0.26 | 0.14 | 0.00 | 0.23 | 0.34/.24 | 0.18 | |
| 392 | BE607384 | Calcium-dependent protein kinase | 0.30 | 0.48 | 0.26 | 0.18 | 0.04 | −0.13 | 0.12 | 0.31/.20 | 0.18 | |
| 6 hr | ||||||||||||
| 1129 | BE607422 | Osr40c1 (ABA- and salt-responsive) | 0.08 | Fc | 0.47 | 0.47 | 0.35 | 0.05 | 0.05 | F | 0.56/.46 | |
| 1043 | BE039925 | Trypsin inhibitor-(1) | 0.06 | 0.19 | 0.32 | 0.26 | 0.25 | 0.04 | 0.17 | 0.45 | 0.42/.23 | |
| 252 | BE039621 | Subtilisin-chymotrypsin inhibitor 2 | 0.08 | 0.18 | 0.30 | 0.44 | 0.11 | −0.10 | 0.22 | 0.35 | 0.41/.27 | |
| 1427 | BE607408 | Osr40c1 (ABA- and salt-responsive) | 0.06 | 0.05 | 0.45 | 0.31 | 0.22 | −0.02 | 0.07 | 0.41 | 0.38/.26 | |
| 636 | BE040239 | Gigantea protein | 0.00 | 0.04 | 0.21 | 0.34 | 0.03 | 0.06 | 0.06 | 0.44 | 0.35/.19 | |
| 1719 | BE530958 | asr1 (ABA- and stress-induced protein) | 0.06 | −0.05 | 0.43 | 0.54 | −0.21 | 0.17 | 0.16 | 0.48 | 0.32/.39 | |
| 470 | BE607346 | gda-1 (gibberellic acid–induced gene) | 0.07 | 0.12 | 0.30 | 0.29 | 0.16 | 0.25 | 0.13 | 0.39 | 0.26/.28 | |
| 898 | BE607343 | RCc3 protein (gibberellic acid–induced gene) | 0.12 | 0.09 | 0.24 | 0.16 | 0.04 | 0.00 | 0.06 | 0.22 | 0.25/.18 | |
| 1084 | BE607324 | Glycine hydroxymethyltransferase (SHM-2) | 0.05 | 0.03 | 0.20 | 0.11 | 0.07 | 0.06 | 0.04 | 0.28 | 0.24/.10 | |
| 1136 | BE607446 | Cyclophilin 2 | 0.01 | 0.06 | 0.11 | 0.28 | 0.12 | 0.09 | 0.03 | 0.07 | 0.23/.15 | |
For a comparison, Pokkali LRs for the regulated ESTs in IR29 are provided.
Hybridization differences exceeding 0.2 LR (>1.6-fold induction of repression) are in boldface.
F, flagged ESTs represent ESTs for which the variability in repeated experiments deviated by greater than ±0.2 LR from the mean value.
The Time-Specific Response of Transcripts in Pokkali
The expression profiles in Pokkali were further categorized into several patterns by using cluster analysis (Eisen et al., 1998). At least 10 different patterns of transcript regulation could be distinguished. The clusters are represented by transcripts that are upregulated at different times: 15 min (Figure 6A), 1 hr (Figure 6B), 3 and 6 hr (Figure 6C), 24 hr (Figure 6D), and 7 days (Figure 6E). These categories are in contrast with four clusters that showed different kinetics of downregulation (Figures 6F to 6K). RNA gel blot analyses confirmed the microarray results for selected clones representing different patterns (Figure 7). For different time points the most highly upregulated or downregulated transcripts are listed (Tables 4 to 7).
Figure 6.
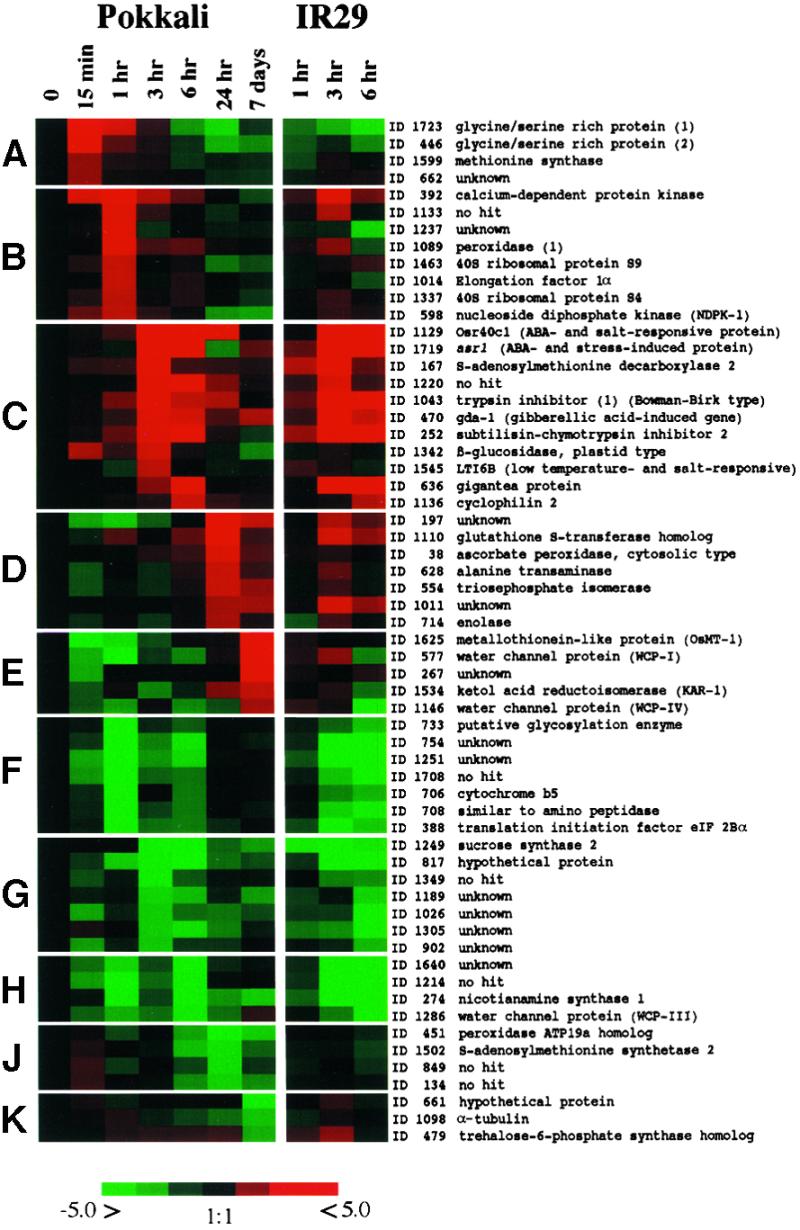
Cluster Analysis of 60 Transcripts in Different Response Categories in Pokkali and IR29 after Salt Stress.
Clustering was performed according to Eisen et al. (1998). The color saturation reflects the magnitude of the log-10 expression ratio (Cy5/Cy3) for each transcript with clone number (transcript number) and annotation. Transcripts are grouped into patterns (A) to (K) according to their expression profiles of upregulation or downregulation at different time points. ABA, abscisic acid.
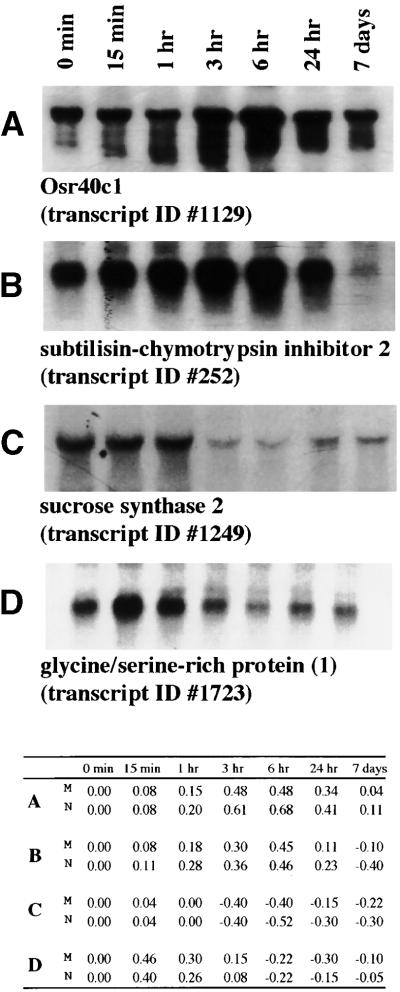
Figure 7.
RNA Gel Blot Analysis of Selected Transcripts.
(Top) Total RNA for control and stressed roots (15 min to 7 days) was used for RNA gel blot analysis (10 μg/lane) for four transcripts: Osr40c1 (transcript 1129 [A]), subtilisin-chymotrypsin inhibitor 2 (transcript 252 [B]), sucrose synthase 2 (transcript 1249 [C]), and glycine-serine–rich protein-1 (transcript 1723 [D]). Signal intensities were analyzed by GelExpert software version 3.5 (Nucleotech).
(Bottom) Expression ratios [M, microarray (stressed/nonstressed)/N, RNA gel blot analysis (Cy5, stressed/Cy3, nonstressed] for each clone [A] to [D] for 0 min, 15 min, 1 hr, 3 hr, 6 hr, 24 hr, and 7 days) are listed.
Table 7.
Downregulated Transcripts in Rice IR29 at Different Time Pointsa
| Clone No. | Accession No. | Annotation | 15 Min | 1 Hr | 3 Hr | 6 Hr | 24 Hr | 7 Days | 1 Hr | 3 Hr | 6 Hr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 hr | |||||||||||
| 1249 | BE607382 | Sucrose synthase 2 (sus2) | 0.03 | 0.00 | −0.38 | −0.41 | −0.17 | −0.23 | −0.35 | −0.41 | −0.34 |
| 347 | BE039674 | Sucrose synthase 2 (sus2) | 0.13 | 0.02 | −0.37 | −0.51 | −0.30 | −0.34 | −0.33 | −0.41 | −0.28 |
| 311 | BE040417 | α-Galactosidase | 0.11 | −0.06 | −0.09 | −0.07 | 0.04 | −0.05 | −0.21 | −0.29 | −0.46 |
| 838 | BE607388 | Enolase-phosphatase E-1 | 0.15 | −0.09 | −0.02 | −0.20 | −0.14 | −0.08 | −0.21 | −0.02 | −0.09 |
| 3 hr | |||||||||||
| 1001 | BE607410 | No hit | −0.20 | −0.22 | −0.18 | −0.24 | −0.04 | 0.00 | −0.11 | −0.80 | −0.77 |
| 1251 | BE607383 | Unknown | −0.24 | −0.46 | −0.22 | Fb | −0.03 | 0.05 | −0.14 | −0.72 | −0.70 |
| 1186 | BE040349 | Branched-chain amino acid aminotransferase | −0.19 | −0.24 | −0.21 | −0.17 | −0.01 | 0.02 | −0.13 | −0.57 | −0.77 |
| 274 | BE607438 | Nicotianamine synthase 1 | −0.05 | −0.38 | −0.13 | −0.38 | −0.23 | −0.31 | −0.16 | −0.54 | −0.47 |
| 1214 | BE530894 | No hit | −0.23 | −0.40 | −0.17 | −0.38 | −0.04 | 0.02 | −0.10 | −0.54 | −0.48 |
| 386 | BE607379 | Phosphoglycerate kinase | −0.12 | −0.33 | −0.17 | −0.26 | 0.05 | 0.00 | −0.09 | −0.51 | −0.43 |
| 1640 | BE039680 | Unknown | −0.17 | −0.32 | −0.14 | −0.40 | −0.10 | −0.04 | −0.09 | −0.49 | −0.55 |
| 754 | BE607400 | Unknown | −0.11 | −0.49 | −0.20 | F | −0.02 | −0.07 | −0.08 | −0.48 | −0.46 |
| 817 | BE607378 | Hypothetical protein | −0.11 | −0.30 | −0.37 | −0.48 | −0.19 | −0.14 | −0.10 | −0.46 | −0.46 |
| 277 | BE607454 | d-amino acid transaminase homolog | −0.12 | −0.27 | −0.16 | −0.25 | −0.04 | −0.04 | −0.10 | −0.46 | −0.43 |
| 6 hr | |||||||||||
| 1286 | BE530955 | Water channel protein (WCP-III) | −0.16 | −0.25 | −0.18 | −0.37 | −0.18 | 0.12 | −0.03 | −0.19 | −0.82 |
| 1001 | BE607410 | No hit | −0.20 | −0.22 | −0.18 | −0.24 | −0.04 | 0.00 | −0.11 | −0.80 | −0.77 |
| 1186 | BE040349 | Branched-chain amino acid aminotransferase | −0.19 | −0.24 | −0.21 | −0.17 | −0.01 | 0.02 | −0.13 | −0.57 | −0.77 |
| 1251 | BE607383 | Unknown | −0.24 | −0.46 | −0.22 | F | −0.03 | 0.05 | −0.14 | −0.72 | −0.70 |
| 954 | BE040178 | Putative seed imbibition protein | −0.04 | −0.08 | −0.17 | −0.31 | −0.10 | 0.06 | −0.12 | −0.24 | −0.68 |
| 1640 | BE039680 | Unknown | −0.17 | −0.32 | −0.14 | −0.40 | −0.10 | −0.04 | −0.09 | −0.49 | −0.55 |
| 1214 | BE530894 | No hit | −0.23 | −0.40 | −0.17 | −0.38 | −0.04 | 0.02 | −0.10 | −0.54 | −0.48 |
| 274 | BE607438 | Nicotianamine synthase 1 | −0.05 | −0.38 | −0.13 | −0.38 | −0.23 | −0.31 | −0.16 | −0.54 | −0.47 |
| 1290 | BE530963 | Hypothetical protein | −0.14 | −0.22 | −0.10 | −0.10 | −0.04 | 0.03 | −0.13 | −0.34 | −0.47 |
| 754 | BE607400 | Unknown | −0.11 | −0.49 | −0.20 | F | −0.02 | −0.07 | −0.08 | −0.48 | −0.46 |
Hybridization differences exceeding 0.2 LR (>1.6-fold induction of repression) are in boldface.
F, flagged ESTs represent ESTs for which the variability in repeated experiments deviated by greater than ±0.2 LR from the mean value.
An “instantaneous response” cluster (15 min) contained only a few transcripts (Figure 6A). Among these genes encoding several GRPs were the most highly upregulated transcripts. Two GRP isoforms, GRP-1 (transcripts 185 and 1723) and GRP-2 (transcripts 430, 446, 1112, and 1539), showed ∼60% amino acid sequence identity to the maize ZmGRP4 in overlapping regions. ZmGRP4 has been localized to the root cap, but its function is unknown (Matsuyama et al., 1999). One hypothesis is that this class of transcripts might function as an immediate defense to stress, for example, by strengthening cell walls.
The “early response” cluster (1 hr; Figure 6B) is exemplified by a sequence homologous with a calcium-dependent protein kinase (CDPK; transcript 392). This transcript shows 100% identity to rice CDPK7 (accession number AB042550) and to Arabidopsis ATCDPK1 (71% identity; Q06850). These CDPKs are reported as salt-stress– and low temperature–induced transcripts (Urao et al., 1994; Berberich and Kusano, 1997). Overexpression of the rice CDPK7 gene in rice recently has been shown to increase tolerance to low temperature, drought, and high salt (Saijo et al., 2000). The CDPK-like sequence was threefold upregulated in Pokkali but was not regulated significantly in IR29 1 hr after salt shock. At the 3- and 6-hr time points, CDPK was upregulated in both lines, suggesting a difference between the two lines in signal transduction at the early stages of stress.
One hour into the salt treatment, many transcripts homologous with ribosomal proteins were upregulated (Table 2). Their upregulation already was evident at the 15-min time point but peaked at 1 hr. Within the category of protein synthesis, nine isoforms of the elongation factor-1 were found, all identical in sequence to rice EF-1α (D63580). All EF-1α ESTs showed an induction profile matching that of the ribosomal protein. It is tempting to assign a crucial role in defense signaling to the 1-hr time point, especially because the salt-sensitive line IR29 does not show similar responses. The second upregulated category in this cluster, the synthesis of ribosomal proteins, and EF-1α (Table 2) could have a function in restructuring the protein synthesis apparatus.
Upregulated sequences at 3 and 6 hr of salt stress (Figure 6C) constitute an “early recovery” cluster. Several of these transcripts have been identified by two-dimensional electrophoresis as salt-stress– and growth regulator–induced proteins (abscisic acid and jasmonic acid; Moons et al., 1995, 1997). One of the transcripts is the rice 40-kD protein Osr40c1, the induction of which is related to the increase of endogenous and exogenously applied abscisic acid. Osr40c1 is suggested to perform a structural role in preventing water loss and preserving the rigidity of cell walls (Moons et al., 1997). Transcripts 1129 and 1427 are identical to Osr40c1 (X95402), with peak expression at the 3-hr time points. Also included in the cluster (Figure 6C) is transcript 840, which encodes a protein with 85 to 92% identity in overlapping regions with Osr40c1, Osr40g2 and Osr40g3. The latter two proteins are induced by salt stress and abscisic acid (Moons et al., 1995). Also included is transcript 1719, which is identical to an abscisic acid–responsive mRNA (asr1) that encodes a functionally unknown protein (AF039573 [Vaidyanathan et al., 1999]). The upregulated transcript 470 showed 77% identity with the functionally unknown gda-1, which is induced by gibberellic acid (T06822 [Li et al., 1998]). Transcripts 167 and 1190, both of which are upregulated, are identical to rice S-adenosylmethionine decarboxylase 2 (SAMDC2; AJ251899). Transcript 1342 is identical to rice β-glucosidase (U28047), transcript 1043 showed 91% identity to a rice trypsin inhibitor in the region of overlap (U57640), and transcript 252 showed 79% identity with barley subtilisin-chymotrypsin inhibitor 2 (Y08625).
A function for an upregulated SAMDC could be in polyamine synthesis, in which this enzyme catalyzes a rate-limiting step (Kumar et al., 1997). Polyamines, which play important roles in plant development, are known to increase under abiotic stress (Evans and Malmberg, 1989). In rice seedlings, polyamine increases have been reported as early as 6 hr after salt stress (Basu and Maitra, 1988). Three other ESTs (transcripts 217, 229, and 409) with less identity to rice SAMDC2, likely encoding isoforms, showed no significant change in hybridization intensity, indicating the specificity of the microarrays and the stress-specific induction of isoforms.
Again within the 3- to 6-hr time frame, upregulation of protease inhibitors (trypsin inhibitor and subtilisin-chymotrypsin inhibitor) was observed. These have been reported as inducible by stresses such as high aluminium, fungal infection, and wounding (Cordero et al., 1994; Richards et al., 1998). With the same kinetics, β-glucosidases were found that catalyse the hydrolysis of 1,3-β-d-glucosidic linkages in 1,3-β-d-glucans. These have been implicated in three processes. First, they are involved in the alteration of specific β-linked polysaccharides during cell expansion in development (Leah et al., 1995). Second, they are involved in pathogen defense reactions by cyanogenesis, wherein the enzymes catalyse the hydrolysis of glucosides after pathogen attack (Hughes et al., 1992). Third, β-glucosidases could release active cytokinins, gibberellins, and auxins from biologically inactive hormone-glucoside conjugates (Brzobohaty et al., 1993). Transcript 1342 displayed 40 to 60% identity to biologically characterized β-glucosidases, but its function(s) in salt stress is not known. The transcripts from Pokkali in this category, which include yet other growth factor– and stress-regulated mRNAs (Tables 4 to 7), showed similar responses in IR29 (Figure 6C), suggesting that hormonal effects on the regulation of transcripts could be similar in Pokkali and IR29.
Downregulation was observed for transcripts 347 and 1249, which are 100% identical to rice sucrose synthase-2 (sus2; X59046), and both remained downregulated after 3 hr of salt stress (Figures 6G and 6H). In IR29, the sus2 transcripts had decreased already after 1 hr of stress. Other ESTs (transcripts 221, 657, and 729) are homologous with rice sus1 (X64770). All are slightly downregulated after the 3-hr time point. Sucrose synthase (UDP glucose: d-fructose 2-α-glucosyltransferase) catalyzes the reversible conversion of sucrose uridine-diphosphate to fructose and UDP glucose, a key reaction in carbohydrate metabolism (Koch, 1996). In rice, the presence of at least three sucrose synthase isoforms, which are differentially regulated during development, is known (Wang et al., 1999). Glucose and sucrose (Wang et al., 1999) modulate these transcripts. For example, maize Sh1, an ortholog of rice sus2, is downregulated by increased glucose (Koch, 1996), suggesting that root carbohydrate metabolism may be altered by salt stress in Pokkali. In the period of 3 to 6 hr after stress, the divergence between Pokkali and IR29 became apparent, whereas before that time IR29 simply showed a “no response” reaction.
Transcripts induced 24 hr after the imposition of stress (Figure 6D) fall into a “stress compensation” cluster, although a more appropriate description might be “back to normal but with a difference.” Upregulated transcripts for the defense against reactive oxygen species dominate this cluster. Included are transcripts homologous with glutathione S-transferase (transcript 1110, 45% identical to wheat glutathione-S-transferase [AAD10129]) and cytoplasmic ascorbate peroxidase (transcript 38, 100% identical to rice ascorbate peroxidase [D45423]). Glutathione S-transferases contribute antioxidation functions (Edwards et al., 2000), and some isoforms have been reported to be stress inducible (Van der Kop et al., 1996; Riechers et al., 1997). Ascorbate peroxidases, which exist in compartment-specific isoforms and are induced by reactive oxygen species, are important scavengers of cytosolic hydrogen peroxide produced under conditions of stress, and their functions have been documented both biochemically and in transgenic studies (Smirnoff, 2000). The relationship of the encoded proteins with the defense against reactive oxygen species (Gueta-Dahan et al., 1997; Chen and Singh, 1999) suggests a function for these upregulated transcripts in salt-stressed roots.
Transcripts 156, 1067, and 1502 were downregulated after 24 hr of salt stress (Figure 6J). They are identical in the sequenced regions to rice S-adenosylmethionine synthetase (SAMS) mRNA (ACC05590). S-adenosyl-l-methionine serves as the major methyl-group donor, for example, in the synthesis of secondary products (Yang and Hoffman, 1984; Heby and Persson, 1990). Transcript 1465, which is identical to a second rice SAMS (Z29867), was not downregulated, indicating that SAMS isoforms are differentially regulated by salt stress, as has been seen for other species (Espartero et al., 1994; Schröder et al., 1997).
After 7 days the plants resumed faster growth and could be considered as adapted to the stress conditions. The transcript profiles (Figure 6E), when compared with the prestress control and early stress time points, confirm this adaptation. The behavior of many transcripts reflects this recovery and adaptation process. For example, the transcript encoding a putative water channel protein (WCP-I) was upregulated threefold over the prestress level after 7 days, whereas the other WCP transcripts were not regulated or were insignificantly affected during the entire stress period. The nine ESTs included in the microarray represent five different WCP transcripts. The upregulated WCP-I (transcripts 157, 577, and 1464) was 80% identical to an ice plant water channel transcript (AAD31846). WCP-2 (transcripts 428, 1134, 1466) was identical to an unnamed rice aquaporin mRNA (AJ224327); WCP-3 (transcript 1286) was similar to wheat PIP2 (AF139815); WCP-4 (transcript 1146) was similar to wheat PIP1 (AF139814); and WCP-5 (transcript 357) was identical to rice aquaporin PIP2a (AF06393).
Only a few other transcripts showed increased expression compared with the control in 7-day stressed plants (Figure 6E). Transcripts homologous with rice metallothioneins are one example: MTP-1 (transcripts 1272 and 1625) was identical to OSMT-1 (U43529), and MTP-2 (transcript 689) was identical to rice metallothionein Mte transcript (AF048750). MTP-3 (transcript 1605) showed homology with metallothioneins in general but represents a novel rice metallothionein. Only MTP-1 was upregulated at this time.
Distinguishing Response Categories in Salt-Sensitive and Salt-Tolerant Rice
Most transcripts in Pokkali displayed constant expression levels (within 0.1 LR) during all phases of salt stress. Particularly, the expression of transcripts essential for cellular homeostasis such as those in the categories of energy supply (respiration, tricarboxylic-acid pathway), transcription (including mRNA processing), transport facilitation (transport ATPases), cellular biogenesis (biogenesis of cell wall), and DNA synthesis were not affected by the chosen stress conditions. Only during the 15-min to 6-hr time periods, with a peak at 1 hr, did global expression changes affect up to 33% (at 1 hr) of the transcripts. During this transition phase, the nature of the most highly upregulated (3.6%) and most drastically downregulated (5.5%) transcripts are indicative of a succession of responses that seem to prepare Pokkali to raise its defensive mechanisms. In IR29, many transcripts, including those unaffected in Pokkali, showed little change early, began to decrease at 3 hr, and were mostly downregulated within 6 hr of salt treatment. In addition, many transcripts that responded to the initial salt shock in Pokkali by downregulation returned to the original expression level, or exceeded it, as early as 3 hr into the salt treatment but IR29 transcripts did not behave similarly.
Another distinguishing characteristic revealed by microarray analysis was the timing of otherwise similar responses. Thus, the expression profile in Pokkali after 15 min of treatment was similar to that seen for IR29 after 1 hr, suggesting that a delay in the processing of signals could underlie the ineffective response of IR29 to salt stress. Essentially, the initial response of individual upregulated transcripts in Pokkali (e.g., ribosomal protein, CDPK, and several functionally unknown ESTs) was absent in IR29 (Figure 6B). These results indicate that the subtle regulation of a subpopulation of transcripts in functional categories intuitively associated with stress defense contributes essential elements for maintaining cellular homeostasis under salinity stress conditions. The importance of the early phase of the response is underscored by the different behavior of the two lines. It seems that Pokkali can overcome the stress due to its ability to induce transcripts that, among other functions, stimulate protein synthesis and components of signaling circuits. In contrast, IR29 shows a delay in responding by upregulation and fewer responses in total. This delay may bring about a general decrease of transcription and death with 24 hr.
The succession of transcript induction in different categories that is revealed by this microarray analysis can be juxtaposed to physiological reactions that have been reported during the adaptation of rice and several other grass crops to salinity stress. A succession of responses has been distinguished (Munns, 1993) in a time frame of hours, days, and weeks after stress. These phases could be correlated with several mechanisms. Several biochemical pathways, ion and metabolite transport processes, hormonal control, cell structure and organ growth, and signaling have been identified (Yeo et al., 1990; Flowers and Yeo, 1995; Garcia et al., 1995; Price et al., 1997; Hasegawa et al., 2000). It is not possible to correlate the physiological terms and only partially understood mechanisms in the context of this microarray analysis of plant salinity stress. The nature of functionally known transcripts that change in response to stress corroborate earlier data (Hardwick et al., 1999; Hasegawa et al., 2000; Mizoguchi et al., 2000). The microarrays from this salt shock experiment, however, provide a significantly finer resolution than previous experiments. The new aspects—upregulation of signal transduction elements, protein synthesis, and changes in the transcripts associated with hormonal changes only later—accentuate the importance of responses that take place at the very early times of stress. The description of this succession of changes in gene expression over time begins to establish connections between and among pathways. This large-scale analysis reveals patterns that will allow us eventually to pinpoint the underlying gene expression changes that are at the basis of a plethora of physiological responses.
METHODS
Plants and Growth Conditions
The International Rice Research Institute (Los Baños, Philippines) supplied rice (Oryza sativa) seeds (lines Pokkali and IR29). After imbibition, seeds were transplanted into Hoagland solution, with the amount of iron doubled, in hydroponic tanks. Plants were grown at 28°C/25°C (50% humidity, 12-hr-light/dark cycle; 700 μmol photons m−2 sec−1). Plants were used when the roots and shoot measured ∼7 and 10 cm, respectively. Salt stress was initiated 3 hr after the start of the light period by adding 150 mM NaCl in dilute (one-fourth) Hoagland solution, which provided external calcium at 1 mM. Roots and leaves were harvested, frozen in liquid nitrogen, and kept at −80°C.
Physiological Analyses
Net CO2 assimilation, stomatal conductance, and transpiration rates were measured with attached leaves at saturating light intensity (1000 μmol photons m−2 sec−1) at 28°C using an infrared gas analyzing system (Li-6400; Li-Cor, Lincoln, NE). Data were collected twice for each time course experiment. Physiological parameters were measured before and after salt additions.
RNA Isolation and cDNA Library Construction
Four cDNA libraries were constructed from Pokkali roots. The OC library used RNA from 10-day-old unstressed roots. The libraries OD, OE, and OF were made from root RNA harvested every 30 min for a period of 12 hr (OD), and similarly for the time periods 24 hr to 72 hr (OE), and 1 week (OF) after salt stress. Total root RNA and poly(A)+ RNA were isolated and cDNA libraries were generated (Stratagene) with Escherichia coli XL1-Blue MRF as the host. Inserts cloned into pBluescript SK+ were sequenced from the 5′ ends. Sequences were annotated accepting rice expressed sequence tags (ESTs) included in public databases for transcripts that showed >95% identity to the Pokkali ESTs. All EST sequences included have been deposited in the databases.
Preparation of DNA Microarrays
Polymerase chain reaction (PCR) amplification (40 cycles, annealing at 55°C) was performed in 96-well format with individual colonies or 1 μL of plasmid DNA as templates, using T3 and T7 primers with amino-modified ends. PCR products were combined with 100 μL of binding solution (150 mM potassium acetate, pH 4.8, and 7 M guanidine hydrochloride), and filtered. PCR products were eluted with 10 mM Tris-EDTA, and 1.2 to 2.4 μg was dried, and the pellets were dissolved in 6 μL of 1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) for printing. Microarrays were produced by using the Omnigrid spotter (GeneMachines, San Carlos, CA). Slides, coated with either polylysine or aminoalkylsilane, contained 1728 cDNAs spotted in triplicate. Only amplicons longer than 400 bp were printed.
Preparation of Labeled Probes
Fluorescence-labeled probes were prepared from RNAs by incorporation of fluorescent nucleotide analogs during first-strand reverse transcription. Each reaction (50 μL) consisted of 1 μg of mRNA, 200 ng of in vitro transcripts as human control mixture, 2 μg of oligo(T) primers, 0.5 mM each of dATP, dCTP, and dGTP, 0.2 mM dTTP, and 0.5 units reverse transcriptase (Superscript II; GIBCO) in 1 × reaction buffer and 2 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham, Pharmacia). RNA and primers were heated to 65°C (10 min) and quenched on ice before the remaining reaction components were added. The reverse transcription reaction proceeded for 10 min at 42°C preincubation followed by 90 min at 42°C. Buffer exchange, purification, and concentration of cDNA products were achieved by microfiltration (Qiagen, Valencia, CA). Labeled targets were collected by centrifugation after the addition of 0.1 volumes of 3 M potassium acetate and 1 volume of isopropanol. The dried pellets were reconstituted in 20 μL of 5 × SSC, 0.1% SDS, and 50% formamide and denatured (95°C) before use in the hybridizations.
Microarray Hybridization and Data Analysis
Hybridizations were performed overnight at 42°C in humidified chambers. The slides then were washed in 1 × SSC and 0.2% SDS (10 min) and in 0.1 × SSC (10 min). Slides were rinsed for 1 min in 0.01 × SSC and dried by centrifugation. The fluorescent signatures were captured using a ScanArray 3000 (GSI; Luminomics, Billercia, MA) and analyzed using ImaGene III software (BioDiscovery, Los Angeles, CA). Local background was subtracted from the value of each spot on the array. Spots covered by dust particles, missing spots, spots with low signal intensity, and spots in high background areas were flagged as candidates for exclusion after further analysis. Normalization between the Cy3 and Cy5 fluorescent dye emission channels initially was achieved by adjusting the level of both image intensities of the signal intensity of exogenously added nonplant control genes and internal control genes (Ruan et al., 1998). Transcript regulation is expressed as the ratio of intensities between stress and control (log-10 ratio, termed LR). Five human cDNA clones, accession numbers AA418251 (zinc finger protein PLAG1), AA464627 (intestinal membrane protein A4), H28469 (IGa-2 chain C), AA456109 (scaffold protein Pbp1), and AA485668 (integrin b-4 subunit) were transcribed in vitro. The five cRNAs were diluted each to a different degree (order as listed above) to provide 2, 1, 0.2, 0.1, and 0.02 ng of cRNA, respectively. The mixture of cRNAs was added to the plant target RNA before the incorporation of the Cy3 and Cy5 dyes. Microarray slides were printed to generate four major segments into which individual elements were printed multiple times (triplicates in these experiments). In some experiments, it was necessary to use different normalization factors in segments due to differences in background or variable target intensity. To reduce area-specific effects, we achieved normalization between the Cy3 and Cy5 fluorescent dye channels by calculating the ratio between the total Cy3 signal from all spots in relation to the total Cy5 signal from all spots in each segment (Hardwick et al., 1999).
RNA Gel Blot Analysis
Total RNA (10 μg) for each sample was loaded on 1.0% agarose/formaldehyde gels and blotted onto nylon membranes (Stratagene). Filters were hybridized overnight at 42°C with randomly primed 32P-labeled cDNA probes in 2 × SSC buffer containing 50% formamide (as for microarrays), washed twice in 2 × SSC and 0.1% SDS and twice in 0.1 × SSC and 0.1% SDS, both at 42°C, and exposed to x-ray films at −80°C. Intensities were determined by phosphorimaging.
Table 5.
Downregulated Transcripts in Rice Pokkali at Different Time Points
| Pokkali
|
IR29
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone No. | Accession No. | Annotation | 15 Min | 1 Hr | 3 Hr | 6 Hr | 24 Hr | 7 Days | 1 Hr | 3 Hr | 6 Hr | |
| 15 min | ||||||||||||
| 1625 | BE040365 | Metallothionein-like protein, OsMT-1 | −0.41 | a | −0.34 | −0.14 | 0.06 | 0.08 | 0.45 | 0.08 | 0.01 | 0.01 |
| 1272 | BE607391 | Metallothionein-like protein, OsMT-1 | −0.41 | −0.46 | Fb | −0.01 | 0.00 | 0.34 | −0.05 | −0.12 | −0.04 | |
| 880 | BE607362 | Heat shock protein 70 | −0.27 | 0.02 | F | −0.07 | −0.10 | 0.15 | 0.06 | 0.04 | −0.14 | |
| 577 | BE607365 | Water channel protein (WCP-I) | −0.25 | −0.44 | −0.11 | −0.21 | 0.09 | 0.40 | 0.10 | 0.21 | −0.19 | |
| 267 | BE607420 | Unknown | −0.24 | 0.02 | −0.01 | 0.05 | 0.01 | 0.35 | 0.07 | 0.00 | 0.10 | |
| 1026 | BE039974 | Unknown | −0.24 | −0.08 | −0.30 | −0.17 | −0.19 | −0.14 | −0.10 | −0.11 | −0.48 | |
| 197 | BE607404 | Unknown | −0.24 | −0.30 | F | 0.02 | 0.41 | 0.27 | 0.02 | 0.23 | 0.15 | |
| 157 | BE607367 | Water channel protein (WCP-I, isoform) | −0.24 | −0.36 | 0.04 | −0.01 | 0.13 | 0.40 | 0.10 | 0.22 | −0.04 | |
| 1407 | BE607374 | Nramp1 protein | −0.24 | F | F | −0.09 | −0.01 | 0.08 | −0.03 | −0.05 | −0.15 | |
| 1251 | BE607383 | Unknown | −0.24 | −0.46 | −0.22 | F | −0.03 | 0.05 | −0.14 | −0.72 | −0.70 | |
| 1 hr | ||||||||||||
| 733 | BE607402 | Putative glycosylation enzyme | −0.05 | −0.54 | −0.08 | −0.15 | −0.01 | −0.02 | −0.05 | −0.15 | −0.26 | |
| 754 | BE607400 | Unknown | −0.11 | −0.49 | −0.20 | F | −0.02 | −0.07 | −0.08 | −0.48 | −0.46 | |
| 1272 | BE607391 | Metallothionein-like protein, OsMT-1 | −0.41 | −0.46 | F | −0.01 | 0.00 | 0.34 | −0.05 | −0.12 | −0.04 | |
| 577 | BE607365 | Water channel protein (WCP-I) | −0.25 | −0.44 | −0.11 | −0.21 | 0.09 | 0.40 | 0.10 | 0.21 | −0.19 | |
| 1708 | BE530916 | No hit | −0.22 | −0.43 | −0.16 | −0.18 | 0.00 | −0.02 | −0.06 | −0.27 | −0.43 | |
| 706 | BE607439 | Cytochrome b5 | −0.09 | −0.41 | 0.03 | −0.19 | −0.01 | 0.05 | −0.01 | −0.19 | −0.24 | |
| 708 | BE607443 | Amino peptidase homolog | −0.08 | −0.41 | −0.12 | −0.19 | −0.01 | −0.05 | −0.03 | −0.29 | −0.35 | |
| 388 | BE607380 | Putative translation initiation factor eIF-2Ba | −0.07 | −0.41 | −0.17 | F | −0.09 | 0.02 | −0.12 | −0.42 | −0.28 | |
| 1214 | BE530894 | No hit | −0.23 | −0.40 | −0.17 | −0.38 | 0.04 | 0.02 | −0.10 | −0.54 | −0.48 | |
| 746 | BE040330 | TMK (gibberellic acid induced) | 0.08 | −0.39 | −0.10 | 0.01 | −0.06 | −0.07 | 0.02 | 0.05 | −0.11 | |
| 3 hr | ||||||||||||
| 1249 | BE607382 | Sucrose synthase-2 (sus2) | 0.03 | 0.00 | −0.38 | −0.41 | −0.17 | −0.23 | −0.35 | −0.41 | −0.34 | |
| 347 | BE039674 | Sucrose synthase-2 (sus2) | 0.13 | 0.02 | −0.37 | −0.51 | −0.30 | −0.34 | −0.33 | −0.41 | −0.28 | |
| 1189 | BE040360 | Unknown | −0.06 | −0.04 | −0.32 | −0.28 | −0.08 | −0.23 | −0.11 | −0.04 | −0.30 | |
| 1026 | BE039974 | Unknown | −0.24 | −0.08 | −0.30 | −0.17 | −0.19 | −0.14 | −0.10 | −0.11 | −0.48 | |
| 1305 | BE607359 | Receptor-like protein | 0.09 | 0.02 | −0.30 | −0.35 | −0.21 | −0.07 | −0.17 | −0.20 | −0.37 | |
| 902 | BE607347 | Unknown | −0.22 | −0.07 | −0.29 | −0.11 | −0.11 | 0.03 | −0.03 | −0.07 | −0.25 | |
| 168 | BE607371 | Unknown | −0.13 | −0.05 | −0.26 | −0.06 | −0.10 | 0.03 | 0.01 | 0.01 | −0.15 | |
| 1251 | BE607383 | Unknown | −0.24 | −0.46 | −0.22 | F | −0.03 | 0.05 | −0.14 | −0.72 | −0.70 | |
| 657 | BE607326 | Sucrose synthase-1 (sus1) | −0.04 | −0.05 | −0.22 | −0.09 | 0.01 | −0.11 | −0.05 | 0.00 | −0.19 | |
| 1186 | BE040349 | Branched-chain amino acid aminotransferase | −0.19 | −0.24 | −0.21 | −0.17 | −0.01 | 0.02 | −0.13 | −0.57 | −0.77 | |
| 6 hr | ||||||||||||
| 347 | BE039674 | Sucrose synthase-2 (sus2) | 0.13 | 0.02 | −0.37 | −0.51 | −0.30 | −0.34 | −0.33 | −0.41 | −0.28 | |
| 1249 | BE607382 | Sucrose synthase-2 (sus2) | 0.03 | 0.00 | −0.38 | −0.41 | −0.17 | −0.23 | −0.35 | −0.41 | −0.34 | |
| 1640 | BE039680 | Unknown | −0.17 | −0.32 | −0.14 | −0.40 | −0.10 | −0.04 | −0.09 | −0.49 | −0.55 | |
| 1214 | BE530894 | No hit | −0.23 | −0.40 | −0.17 | −0.38 | −0.04 | 0.02 | −0.10 | −0.54 | −0.48 | |
| 274 | BE607438 | Nicotianamine synthase 1 | −0.05 | −0.38 | −0.13 | −0.38 | −0.23 | −0.31 | −0.16 | −0.54 | −0.47 | |
| 1305 | BE607359 | Receptor-like protein | 0.09 | 0.02 | −0.30 | −0.35 | −0.21 | −0.07 | −0.17 | −0.20 | −0.37 | |
| 954 | BE040178 | Putative seed imbibition protein | −0.04 | −0.08 | −0.17 | −0.31 | −0.10 | 0.06 | −0.12 | −0.24 | −0.68 | |
| 1502 | BE040447 | S-adenosyl-l-methionine synthetase | 0.10 | 0.05 | −0.05 | −0.29 | −0.36 | −0.17 | −0.05 | −0.04 | −0.12 | |
| 1189 | BE040360 | Unknown | −0.06 | −0.04 | −0.32 | −0.28 | −0.08 | −0.23 | −0.11 | −0.04 | −0.30 | |
| 847 | BE530921 | No hit | −0.03 | −0.33 | −0.14 | −0.27 | −0.06 | 0.00 | −0.04 | −0.36 | −0.38 | |
| 24 hr | ||||||||||||
| 1502 | BE040447 | S-adenosyl-l-methionine synthetase | 0.10 | 0.05 | −0.05 | −0.29 | −0.36 | −0.17 | −0.05 | −0.04 | −0.12 | |
| 156 | BE039957 | S-adenosyl-l-methionine synthetase | 0.10 | 0.05 | −0.01 | −0.19 | −0.36 | −0.21 | −0.06 | 0.04 | −0.04 | |
| 134 | BE607409 | No hit | 0.14 | 0.03 | −0.03 | −0.22 | −0.33 | −0.22 | −0.04 | −0.02 | −0.04 | |
| 347 | BE039674 | Sucrose synthase-2 (sus2) | 0.13 | 0.02 | −0.37 | −0.51 | −0.30 | −0.34 | −0.33 | −0.41 | −0.28 | |
| 1067 | BE040436 | S-adenosyl-l-methionine synthetase | 0.09 | 0.06 | −0.03 | −0.19 | −0.29 | −0.14 | −0.06 | 0.01 | −0.12 | |
| 1197 | BE607401 | Unknown | 0.10 | 0.06 | −0.01 | −0.16 | −0.29 | −0.16 | −0.01 | 0.00 | −0.02 | |
| 266 | BE607418 | No hit | 0.04 | 0.01 | −0.01 | −0.10 | −0.28 | −0.13 | −0.06 | 0.01 | −0.04 | |
| 274 | BE607438 | Nicotianamine synthase 1 | −0.05 | −0.38 | −0.13 | −0.38 | −0.23 | −0.31 | −0.16 | −0.54 | −0.47 | |
| 359 | BE039715 | Adenine phosphoribosyltransferase form 3 | 0.00 | F | −0.19 | −0.26 | −0.23 | −0.13 | −0.17 | −0.29 | −0.38 | |
| 7 days | ||||||||||||
| 347 | BE039674 | Sucrose synthase-2 (sus2) | 0.13 | 0.02 | −0.37 | −0.51 | −0.30 | −0.34 | −0.33 | −0.41 | −0.28 | |
| 451 | BE607363 | Peroxidase ATP19a | 0.07 | −0.14 | 0.00 | F | −0.47 | −0.33 | 0.04 | −0.01 | −0.08 | |
| 274 | BE607438 | Nicotianamine synthase 1 | −0.05 | −0.38 | −0.13 | −0.38 | −0.23 | −0.31 | −0.16 | −0.54 | −0.47 | |
| 1098 | BE607332 | α-tubulin | 0.06 | 0.10 | −0.08 | −0.14 | −0.19 | −0.31 | −0.09 | 0.09 | 0.01 | |
| 889 | BE607340 | α2-tubulin | 0.08 | 0.02 | −0.05 | −0.18 | −0.14 | −0.30 | −0.09 | 0.04 | 0.01 | |
| 479 | BE607351 | Trehalose-6p-phosphatase, fission yeast | 0.01 | 0.10 | 0.12 | 0.12 | F | −0.27 | 0.13 | 0.20 | −0.04 | |
| 598 | BE607369 | Nucleoside diphosphate kinase (NDPK-1) | 0.17 | 0.26 | 0.12 | 0.05 | −0.22 | −0.24 | 0.00 | 0.11 | 0.08 | |
| 1249 | BE607382 | Sucrose synthase-2 (sus2) | 0.03 | 0.00 | −0.38 | −0.41 | −0.17 | −0.23 | −0.35 | −0.41 | −0.34 | |
| 851 | BE530928 | High mobility group protein HMGd1 | 0.09 | 0.05 | −0.17 | F | −0.15 | −0.23 | −0.11 | −0.24 | −0.39 | |
| 1189 | BE040360 | Unknown | −0.06 | −0.04 | −0.32 | −0.28 | −0.08 | −0.23 | −0.11 | −0.04 | −0.30 | |
Hybridization differences exceeding 0.2 LR (>1.6-fold induction of repression) are in boldface.
F, flagged ESTs represent ESTs for which the variability in repeated experiments deviated by greater than ±0.2 LR from the mean value.
Acknowledgments
We thank Chris Michalowski for advice, and we are grateful to Ariana Call, Heather Ferrea, Chris Kirman, Andrew McCullough, Chris Palacio, and Mark Wheeler for help with the DNA work. Supplemental materials have been deposited at www.stress-genomics.org. Financial support from the National Science Foundation (Grant No. DBI 98-13360) is gratefully acknowledged.
References
- Apse, M.P., Aharon, G.S., Snedden, W.A., and Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Barkla, B.J., Vera-Estrella, R., and Pantoja, O. (1999). Towards the production of salt-tolerant crops. Adv. Exp. Med. Biol. 464, 77–89. [DOI] [PubMed] [Google Scholar]
- Basu, S., and Maitra, U. (1988). Salinity results in polyamine accumulation in early rice (Oryza sativa L.) seedlings. Aust. J. Plant Physiol. 15, 777–786. [Google Scholar]
- Berberich, T., and Kusano, T. (1997). Cycloheximide induces a subset of low temperature-inducible genes in maize. Mol. Gen. Genet. 254, 275–283. [DOI] [PubMed] [Google Scholar]
- Boyer, J.S. (1982). Plant productivity and environment. Science 218, 443–448. [DOI] [PubMed] [Google Scholar]
- Brzobohaty, B., Moore, I., Kristoffersen, P., Bako, L., Campos, N., Schell, J., and Palme, K. (1993). Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262, 1051–1054. [DOI] [PubMed] [Google Scholar]
- Chen, W., and Singh, K.B. (1999). The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 19, 667–677. [DOI] [PubMed] [Google Scholar]
- Cordero, M.J., Raventos, D., and San-Segundo, B. (1994). Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: Systemic wound-response of a monocot gene. Plant J. 6, 141–150. [DOI] [PubMed] [Google Scholar]
- Dat, J., Vandenabeele, S., Vranova, E., Van Montagu, M., Inze, D., and Van Breusegem, F. (2000). Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 57, 779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R., Dixon, D.P., and Walbot, V. (2000). Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5, 193–198. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espartero, J., Pintor-Toro, J.A., and Pardo, J.M. (1994). Differential accumulation of S-adenosylmethionine synthetase transcripts in response to salt stress. Plant Mol. Biol. 25, 217–227. [DOI] [PubMed] [Google Scholar]
- Evans, P.T., and Malmberg, R.L. (1989). Do polyamines have roles in plant development? Annu. Rev. Plant Physiol. 40, 235–269. [Google Scholar]
- Flowers, T.J., and Yeo, A.R. (1995). Breeding for salinity resistance in crop plants: Where next? Aust. J. Plant Physiol. 22, 875–884. [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 17, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, A., Senadhira, D., Flowers, T.J., and Yeo, A.R. (1995). The effects of selection for sodium transport and of selection for agronomic characteristics upon salt resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 90, 1106–1111. [DOI] [PubMed] [Google Scholar]
- Gueta-Dahan, Y., Yaniv, Z., Zilinskas, B.A., and Ben-Hayyim, G. (1997). Salt and oxidative stress: Similar and specific responses and their relation to salt tolerance in citrus. Planta 203, 460–469. [DOI] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, J.S., Kuruvilla, F.G., Tong, J.K., Shamji, A.F., and Schreiber, S.L. (1999). Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96, 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, P.M., Bressan, R.A., Zhu, J.-K., and Bohnert, H.J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. [DOI] [PubMed] [Google Scholar]
- Heby, O., and Persson, L. (1990). Molecular genetics of polyamine synthesis in eukaryotic cells. Trends Biochem. Sci. 15, 153–158. [DOI] [PubMed] [Google Scholar]
- Hughes, M.A., Brown, K., Pancoro, A., Murray, B.S., Oxtoby, E., and Hughes, J. (1992). A molecular and biochemical analysis of the structure of the cyanogenic beta-glucosidase (linamarase) from vassava (Manihot esculenta Cranz). Arch. Biochem. Biophys. 295, 273–279. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W., and Zhu, J.-K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12, 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Altabella, T., Taylor, M.A., and Tiburcio, A.F. (1997). Recent advances in polyamine research. Trends Plant Sci. 4, 124–130. [Google Scholar]
- Leah, R., Kigel, J., Svendsen, I., and Mundy, J. (1995). Biochemical and molecular characterization of a barley seed β-glucosidase. J. Biol. Chem. 270, 15789–15797. [DOI] [PubMed] [Google Scholar]
- Li, H.Y., Guo, Z.F., and Zhu, Y.X. (1998). Molecular cloning and analysis of a pea cDNA that is expressed in darkness and very rapidly induced by gibberellic acid. Mol. Gen. Genet. 259, 393–397. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Matsuyama, T., Satoh, H., Yamada, Y., and Hashimoto, T. (1999). A maize glycine-rich protein is synthesized in the lateral root cap and accumulates in the mucilage. Plant Physiol. 120, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi, T., Ichimura, K., Yoshida, R., and Shinozaki, K. (2000). MAP kinase cascades in Arabidopsis: Their roles in stress and hormone responses. Results Probl. Cell Differ. 27, 29–38. [DOI] [PubMed] [Google Scholar]
- Moons, A., Bauw, G., Prinsen, E., Van Montagu, M., and Van der Straeten, D. (1995). Molecular and physiological responses to abscisic acid and salts in roots of salt-sensitive and salt-tolerant Indica rice varieties. Plant Physiol. 107, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons, A., Gielen, J., Vandekerckhove, J., Van der Straeten, D., Gheysen, G., and Van Montagu, M. (1997). An abscisic-acid– and salt-stress–responsive rice cDNA from a novel plant gene family. Planta 202, 443–454. [DOI] [PubMed] [Google Scholar]
- Munns, R.E. (1993). Physiological processes limiting plant growth in saline soils: Some dogmas and hypotheses. Plant Cell Environ. 16, 15–24. [Google Scholar]
- Noctor, G., and Foyer, C.H. (1998). Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. [DOI] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Price, A.H., Young, E.M., and Tomos, A.D. (1997). Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (Oryza sativa L.). New Phytol. 137, 83–91. [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, K.D., Schott, E.J., Sharma, Y.K., Davis, K.R., and Gardner, R.C. (1998). Aluminum induces oxidative stress genes in Arabidopsis thaliana L. Plant Physiol. 116, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, T., and Somerville, S. (2000). Chasing the dream: Plant EST microarrays. Curr. Opin. Plant Biol. 3, 108–116. [DOI] [PubMed] [Google Scholar]
- Riechers, D.E., Irzyk, G.P., Jones, S.S., and Fuerst, E.P. (1997). Partial characterization of glutathione S-transferases from wheat (Triticum spp.) and purification of a safener-induced glutathione S-transferase from Triticum tauschii. Plant Physiol. 114, 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., and Murata, N. (2000). Genetic engineering of glycinebetaine synthesis in plants: Current status and implications for enhancement of stress tolerance. J. Exp. Bot. 51, 81–88. [PubMed] [Google Scholar]
- Schröder, G., Eichel, J., Breinig, S., and Schröder, J. (1997). Three differentially expressed S-adenosylmethionine synthetases from Catharanthus roseus: Molecular and functional characterization. Plant Mol. Biol. 33, 211–222. [DOI] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Takeda, T., Yamauchi, H., Sano, S., Tomizawa, K.I., Yokota, A., and Shigeoka, S. (1998). Inhibition of ascorbate peroxidase under oxidase stress in tobacco having bacterial catalase in chloroplasts. FEBS Lett. 428, 47–51. [DOI] [PubMed] [Google Scholar]
- Smirnoff, N. (2000). Ascorbic acid: Metabolism and functions of a multi-facetted molecule. Curr. Opin. Plant Biol. 3, 229–235. [PubMed] [Google Scholar]
- Urao, T., Katagiri, T., Mizoguchi, T., Yamaguchi-Shinozaki, K., Hayashida, N., and Shinozaki, K. (1994). Two genes that encode Ca2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol. Gen. Genet. 244, 331–340. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan, R., Kuruvilla, S., and Thomas, G. (1999). Characterization and expression pattern of an abscisic acid and osmotic stress responsive gene from rice. Plant Sci. 140, 25–36. [Google Scholar]
- Van Camp, W., Capiau, K., Van Montagu, M., Inze, D., and Slooten, L. (1996). Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiol. 112, 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kop, D.A., Schuyer, M., Scheres, B., Zaal, V.D., and Hooykaas, P.J. (1996). Isolation and characterization of an auxin-inducible glutathione S-transferase gene of Arabidopsis thaliana. Plant Mol. Biol. 30, 839–844. [DOI] [PubMed] [Google Scholar]
- Wang, A.Y., Kao, M., Yang, W., Sayion, Y., Liu, L., Lee, P., and Su, J.C. (1999). Differentially and developmentally regulated expression of three rice sucrose synthase genes. Plant Cell Physiol. 40, 800–807. [DOI] [PubMed] [Google Scholar]
- Yang, S.F., and Hoffman, N.E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. [Google Scholar]
- Yeo, A.R., Yeo, M.E., Flowers, S.A., and Flowers, T.J. (1990). Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 79, 377–384. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. [DOI] [PubMed] [Google Scholar]