Abstract
Fibers are one of the mechanical tissues that provide structural support to the plant body. To understand how the normal mechanical strength of fibers is regulated, we isolated an Arabidopsis fragile fiber (fra2) mutant defective in the mechanical strength of interfascicular fibers in the inflorescence stems. Anatomical and chemical analyses showed that the fra2 mutation caused a reduction in fiber cell length and wall thickness, a decrease in cellulose and hemicellulose contents, and an increase in lignin condensation, indicating that the fragile fiber phenotype of fra2 is a result of alterations in fiber cell elongation and cell wall biosynthesis. In addition to the effects on fibers, the fra2 mutation resulted in a remarkable reduction in cell length and an increase in cell width in all organs, which led to a global alteration in plant morphology. The FRA2 gene was shown to encode a protein with high similarity to katanin (hence FRA2 was renamed AtKTN1), a protein shown to be involved in regulating microtubule disassembly by severing microtubules. Consistent with the putative function of AtKTN1 as a microtubule-severing protein, immunolocalization demonstrated that the fra2 mutation caused delays in the disappearance of perinuclear microtubule array and in the establishment of transverse cortical microtubule array in interphase and elongating cells. Together, these results suggest that AtKTN1, a katanin-like protein, is essential not only for normal cell wall biosynthesis and cell elongation in fiber cells but also for cell expansion in all organs.
INTRODUCTION
The plant cell wall, as an exocytoskeleton, provides structural support to the cells and to the entire plant body. Plant cells can be grouped, according to their wall thickening, into three basic types: parenchyma, collenchyma, and sclerenchyma. Both parenchyma and collenchyma cells, consisting of the primary wall, provide the main structural support in growing regions of the plant body. Sclerenchyma cells, having both primary wall and thick secondary wall, provide the major mechanical support in non-elongating regions of the plant body (Carpita and McCann, 2000). The molecular mechanisms that control the deposition of cell wall materials and that determine cell wall mechanical strength are not yet known.
Because cell walls delimit the boundaries of individual cells, the shapes of individual cell walls determine cell morphology and whole plant morphology. Thus, understanding how cells make walls will help us to discover the molecular mechanisms that control cell morphology. The essential roles of cell walls in regulating cell morphology and cell elongation have been demonstrated in mutants defective in genes involved in cell wall biosynthesis or modification. It has been shown that disruption of crystalline cellulose biosynthesis in the rsw1 mutant results in a swollen cell phenotype, indicating the direct role of cellulose in maintaining cell morphology (Arioli et al., 1998). The importance of normal cell wall biosynthesis or modification in regulating cell elongation has been demonstrated by the kor mutants defective in a gene encoding endo-1,4-β-glucanase (Nicol et al., 1998; Zuo et al., 2000). The kor1 mutation causes abnormal cell wall structure and aberrant cell plate formation. Cells in the kor1 mutants cannot elongate normally, resulting in an extremely dwarf phenotype. This finding suggests that proper modification of cell wall materials during cytokinesis and cell expansion is essential for normal cell morphology.
Because cellulose microfibrils must be oriented properly for directional cell expansion, it is obvious that cellulose microfibril deposition must be regulated by cellular machinery. Several lines of evidence have shown that the cytoskeletal microtubules regulate exoskeletal cellulose microfibril orientation (Giddings and Staehelin, 1991; Baskin, 2000). First, cortical microtubule arrays are aligned inside the plasma membrane parallel with newly synthesized cellulose microfibrils, both of which are oriented transversely to the axis of cell elongation. Second, pharmacological studies have proven the direct role of microtubules in determining cellulose microfibril orientation and cell elongation. Disruption of microtubule orientation in parenchyma cells with colchicin, a microtubule-destabilizing drug, effectively disrupts the orientation of cellulose microfibrils and changes the direction of cell expansion. On the basis of these observations, it has long been accepted that microtubules direct the orientation of cellulose microfibrils, which controls cell elongation. However, it is not known whether microtubules influence cell wall biosynthesis (Seagull and Falconer, 1991).
Fibers have traditionally been used as a model for the study of cell differentiation, cell elongation, and cell wall biosynthesis (Aloni, 1987). Fiber initial cells typically undergo considerable elongation at both ends. For example, in Boehmeria nivea, fiber initials ∼20 μm long can elongate up to 550 mm. At maturity, a massive amount of secondary wall is laid down inside the primary wall, which enables fiber cells to function as an excellent mechanical tissue (Mauseth, 1988). Therefore, fiber cells are a remarkable example of the coordinated regulation of cell elongation and cell wall biosynthesis. It is conceivable that studying the mechanisms that control fiber formation will help us understand the mechanisms that regulate cell elongation and cell wall biosynthesis in general.
In this article, we describe the characterization of the Arabidopsis fra2 mutant with a defect in fiber strength and the molecular cloning of the FRA2 gene. We show that the reduction in the mechanical strength of fibers in fra2 inflorescence stems is associated with alterations of fiber cell elongation and cell wall biosynthesis. We present evidence that the fra2 mutation not only alters fiber cell elongation but also changes cell expansion in all organs, thereby leading to a global alteration in plant morphology. We demonstrate that the FRA2 gene encodes a protein with high similarity to katanin that is involved in severing microtubules. Our results provide direct evidence that a katanin-like protein is essential for cell wall biosynthesis and cell elongation.
RESULTS
Isolation of the fra2 Mutant with a Defect in the Mechanical Strength of Fibers
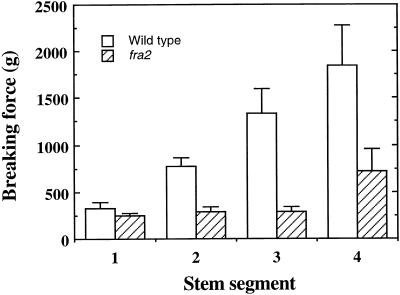
Our previous work has shown that the mechanical strength of the inflorescence stems of Arabidopsis is conferred mainly by interfascicular fibers. Elimination of interfascicular fibers, as in the ifl1 mutants, dramatically reduces the breaking strength of the inflorescence stems, which results in a pendent shoot phenotype (Zhong et al., 1997; Zhong and Ye, 1999). To further investigate the mechanisms controlling fiber differentiation and fiber mechanical strength, we screened ethyl methanesulfonate–mutagenized Arabidopsis populations for mutants with reduced normal mechanical strength in the inflorescence stems. We found several mutants with a dramatic reduction in the breaking strength of the inflorescence stems. One of them was chosen for this study because of its alteration in cell elongation. Quantitative analysis showed that the force required to break the mutant stems was two- to threefold less than that required for the wild-type stems (Figure 1). The mutant stems (Figure 2B) developed interfascicular fiber cells like those in the wild type (Figure 2A), indicating that the reduction in the breaking strength of mutant stems was due to an alteration in the mechanical strength of fibers rather than to an absence of interfascicular fibers. Thus, we designated the mutant locus as the fra2 (fragile fiber) locus.
Figure 1.
Breaking Force Measurement in Stems of the Wild Type and the fra2 Mutant.
The main inflorescence stems of 8-week-old plants were divided into four equal segments and measured for the force required to break the stems. Segments were numbered in order from the top to the bottom of the stems. Data are mean values ±se for 15 plants.
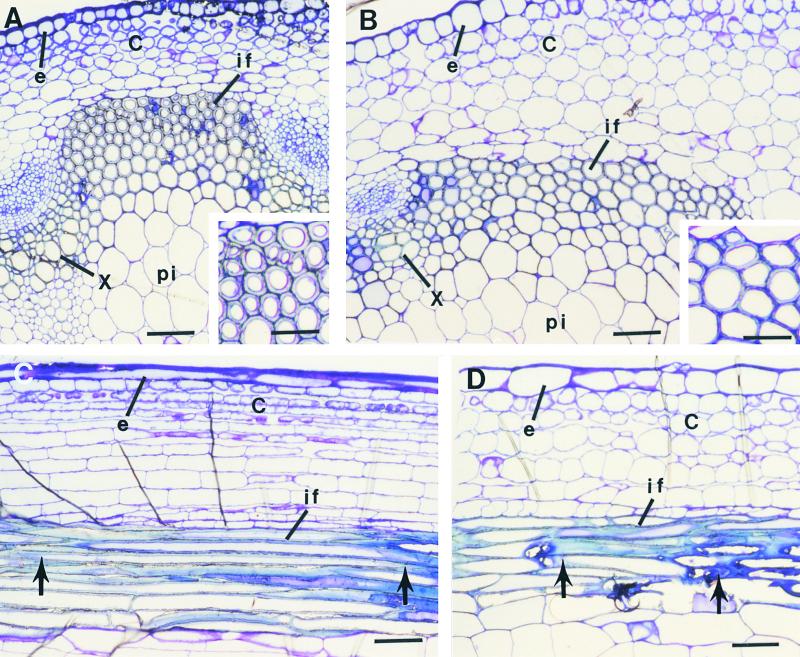
Figure 2.
Anatomy of Interfascicular Fibers in Stems of the Wild Type and the fra2 Mutant.
(A) and (B) Cross-sections of the stems of the wild type (A) and fra2 (B) showing the presence of interfascicular fibers. Insets show enlarged fiber cells. It is evident that the fiber cell wall in the wild type (A) is much thicker than that in fra2 (B).
(C) and (D) Longitudinal sections of the stems of the wild type (C) and fra2 (D). Fiber cells in fra2 (D) are much shorter and wider than those in the wild type (C). Arrows mark the ends of a fiber cell.
C, cortex; e, epidermis; if, interfascicular fiber; pi, pith; x, xylem. Bars in (A) to (D) = 55 μm; bars in insets in (A) and (B) = 28 μm.
Anatomical Examination of Fiber Morphology
Reduction in the mechanical strength of fibers could result from alterations in cell wall composition, cell wall structure, fiber cell length, or adhesion of fiber cells. To investigate which specific alterations occurred in the fra2 fibers, we first examined fiber cell length and fiber cell wall thickness. Longitudinal sections of mature inflorescence stems showed a dramatic alteration in fiber cell length in fra2 (Figure 2). In the wild type, fiber cells were narrow and long with two tapered ends (Figure 2C). However, fiber cells in fra2 (Figure 2D) were much shorter than those in the wild type. Furthermore, fiber cell walls in fra2 (Figure 2A, inset) appeared to be much thinner than those in the wild type (Figure 2B, inset). These results indicate that the fra2 mutant is defective in both fiber cell elongation and fiber cell wall thickening, which most likely contributes to the reduction in the mechanical strength of fra2 fibers.
Chemical Analysis of Cell Wall Composition
The finding that the fra2 mutation caused reduced thickness of the fiber cell wall prompted us to investigate whether it caused any alterations in cell wall biosynthesis. Analysis of crystalline cellulose in stems showed that the amount of cellulose in fra2 stems was reduced to 80% of that in the wild type (Table 1). The reduction of cellulose level in the fra2 mutant was further demonstrated by sugar composition analysis of stem cell walls. Glucose levels were reduced by 24% in fra2 compared with those of the wild type (Table 1). It is intriguing that the amounts of other sugars such as xylose, arabinose, and rhamnose were also decreased significantly. However, the fra2 mutation did not affect the levels of sugars such as mannose and galactose. Because glucose and xylose are the two sugars that constitute cellulose and hemicellulose, respectively, in the secondary cell wall, the fra2 mutation caused a reduction in cellulose and hemicellulose in fibers.
Table 1.
Sugar Composition of Stems of the Wild Type and the fra2 Mutant (mg/g)
| Sample | Rhamnose | Arabinose | Xylose | Mannose | Galactose | Glucose | Fucose | Cellulose |
|---|---|---|---|---|---|---|---|---|
| Wild typea | 5.0 ± 0.7 (100%) |
17.2 ± 1.6 (100%) |
149.7 ± 15.6 (100%) |
14.6 ± 1.2 (100%) |
12.8 ± 0.7 (100%) |
186.7 ± 9.7 (100%) |
1.7 ± 0.5 (100%) |
230 ± 7 (100%) |
| fra2b | 3.9 ± 0.5 (78%) |
11.5 ± 2.1 (68%) |
105.5 ± 15.5 (70%) |
13.9 ± 1.9 (90%) |
13.0 ± 1.2 (102%) |
141.7 ± 4.7 (76%) |
1.8 ± 0.2 (106%) |
184 ± 5 (80%) |
Data in parentheses are the sugar contents in the wild type taken as 100.
Data in parentheses are the sugar contents in fra2 expressed as a percentage of that in the wild type.
Lignin is another main component in the secondary cell wall in addition to cellulose and hemicellulose. Because the fra2 mutation resulted in reductions in both cellulose and hemicellulose, we investigated whether it caused any changes in lignin content or structure. Analysis of Klason lignin showed a slight decrease in lignin content (Table 2). Surprisingly, the levels of base-extractable guaiacyl and syringyl lignin units in fra2 were decreased 58 and 35%, respectively, compared with the levels in the wild type (Table 2). Because guaiacyl and syringyl lignin units in fra2 are much less extractable by base than are those in the wild type, this finding indicates that lignin in fra2 is much more condensed than that in the wild type.
Table 2.
Lignin Content and Composition in the Wild Type and the fra2 Mutant
| Klason Lignina
|
Lignin Composition (mg/g Cell Wall)b
|
||||
|---|---|---|---|---|---|
| Plant | % of Cell Wallc | % of Wild-Type Klason Lignin | Guaiacyl Ligninc | Syringyl Ligninc | S/Gd |
| Wild type | 15.8 ± 0.4 | 100 | 34.0 ± 0.3 (100%) | 15.6 ± 1.4 (100%) | 0.46 |
| fra2 | 14.4 ± 0.6 | 91 | 14.2 ± 4.2 (42%) | 10.2 ± 3.6 (65%) | 0.72 |
Klason lignin was assayed according to Kirk and Obst (1988).
Lignin monolignol composition was analyzed accroding to Akin et al. (1993).
Each data point is the mean ±se of two separate assays. Data in parentheses are guaiacyl or syringyl lignin units taken as either 100 in the wild type or a percentage of the wild type in fra2.
G (guaiacyl) is the sum of vanillin, acetovanillin, and vanillic acid; S (syringyl) is the sum of syringaldehyde, acetosyringaldehyde, and syringic acid.
In-Source Pyrolysis Mass Spectrometry of Cell Walls
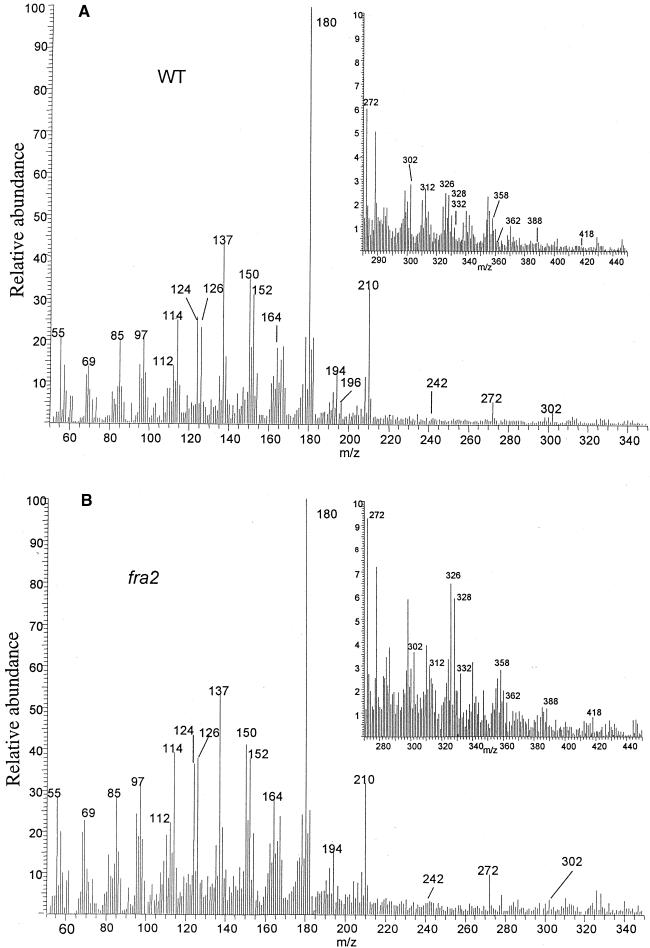
To confirm the results from the chemical analysis, we applied in-source pyrolysis mass spectrometry to examine the relative levels of different cell wall materials. The results showed that the relative intensities of the mass markers for cellulose, hemicellulose, and lignin were altered significantly, with the carbohydrate markers making a greater contribution to the spectrum of fra2 (Figure 3B) than to the spectrum of the wild type (Figure 3A). Examination of the mass markers for dimeric lignin (van der Hage et al., 1993) showed that the mass spectrum of fra2 cell walls exhibited a higher proportion of these markers than did that of the wild type (Figures 3A and 3B, insets), suggesting that the lignin in fra2 is more thermal stable than that in the wild type. This is consistent with the results from the chemical analysis indicating that lignin in fra2 is more condensed than that in the wild type.
Figure 3.
In-Source Pyrolysis Mass Spectrometry of Cell Walls.
(A) and (B) Mass spectra of cell walls of the wild type (WT) (A) and the fra2 mutant (B). Mass peaks of guaiacyl lignin had mass–charge ratio (m/z) values of 124, 137, 138, 150, 152, 164, 166, 178, and 180. Mass peaks of syringyl lignin had m/z values of 154, 167, 168, 180, 182, 194, 196, 208, and 210. Mass peaks of cellulose and amylose had m/z values of 57, 60, 73, 85, 86, 96, 98, 100, 102, 110, 112, 126, and 144. Mass peaks of hemicellulose had m/z values of 58, 85, 86, and 114. Insets show the expanded portions of the spectra of mass markers for dimeric lignin, with m/z values of 272, 302, 312, 320, 326, 328, 332, 358, 388, and 418. The relative intensities of mass peaks for cell wall polysaccharides and lignin were altered significantly between the wild type and the fra2 mutant.
Plant Morphology
In addition to the defects in fiber cell elongation and cell wall biosynthesis, the fra2 mutant exhibited a dramatic alteration in plant morphology (Figure 4A). The height of mature inflorescence stems in fra2 reached only ∼50% of the height of the wild type (Table 3). Measurement of internode number and length showed no change in internode number but a dramatic reduction in internode length (Figures 4B and 4C, and Table 3), indicating that the decrease in plant height in fra2 was caused by a reduction in internode length.
Figure 4.
Morphology of the Wild Type and the fra2 Mutant.
(A) Morphology of 8-week-old plants. The main inflorescence stem of a fra2 plant (right) is much shorter than that of a wild-type plant (left).
(B) and (C) The main inflorescence stems. The fra2 stem (C) has reduced internode length compared with the wild-type stem (B).
(D) and (E) Siliques of wild type (D) and fra2 (E).
(F) and (H) The rosette leaves of a 5-week-old fra2 plant (H) are more compact than those of a wild-type plant (F).
(G) and (I) Individual leaves of 5-week-old plants. The lengths of both blades and petioles in fra2 (I) are reduced compared with those of the wild type (G). From left to right, the leaves are arranged according to the order from cotyledons to the youngest leaves.
Table 3.
Morphology of Inflorescence Stems and Trichomes of the Wild Type and the fra2 Mutant
| Morphology | Wild Type | fra2 |
|---|---|---|
| Main inflorescence stema | ||
| Height (cm) | 27.2 ± 3.3 | 13.6 ± 1.5 |
| Diameter (mm)b | 1.7 ± 0.4 | 2.5 ± 0.4 |
| Cauline branch number | 7.3 ± 1.8 | 6.9 ± 1.8 |
| Internode length (mm) | ||
| First internode | 16.5 ± 10.2 | 5.0 ± 4.8 |
| Second internode | 20.2 ± 7.3 | 4.8 ± 4.1 |
| Third internode | 22.1 ± 9.7 | 6.4 ± 2.5 |
| Trichome branch points (%)c | ||
| No points | 0.11 | 7.5 |
| One point | 2.41 | 64.9 |
| Two points | 79.7 | 25.6 |
| Three points | 17.8 | 1.9 |
Data are mean values ±se from 10 (wild type) or 20 (fra2) plants.
The first internode was used for measurement of the stem diameter.
A total of 1000 trichomes on rosette leaves were counted. The nomenclature of trichome branch point is according to Luo and Oppenheimer (1999).
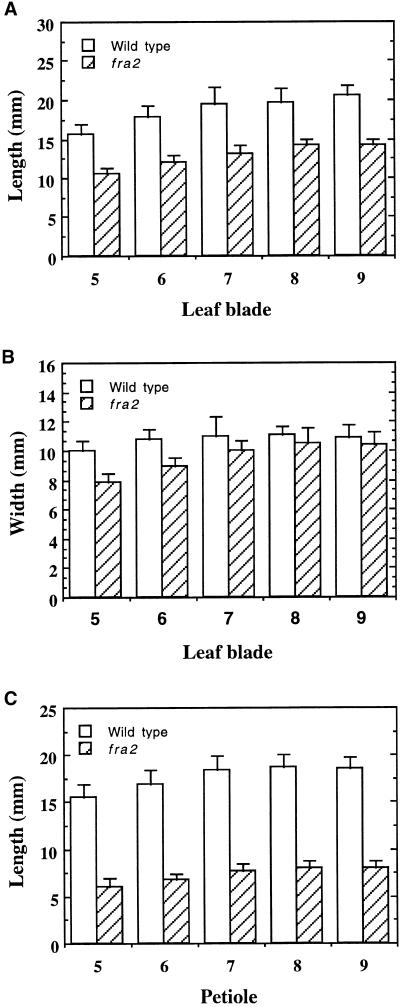
It was noted that the length of all other organs was reduced in fra2. The leaves of fra2 plants were much shorter than those of the wild type (Figures 4G and 4I), which resulted in more compact rosette leaves compared with those of the wild type (Figures 4F and 4H). The length of leaf blades and petioles of fra2 plants was reduced to 70 and 40%, respectively, of those in the wild type (Figures 5A and 5C). However, the width of fra2 leaf blades did not show significant changes (Figure 5B).
Figure 5.
Measurement of the Length and Width of Leaves.
The fifth to ninth leaves of 5-week-old plants were measured for their length and width. Data are means ±se of 10 leaves.
(A) Length of leaf blades, showing a reduction in fra2 compared with the wild type.
(B) Width of leaf blades, showing little change in fra2 compared with the wild type.
(C) Length of petioles, showing a dramatic reduction in fra2 compared with the wild type.
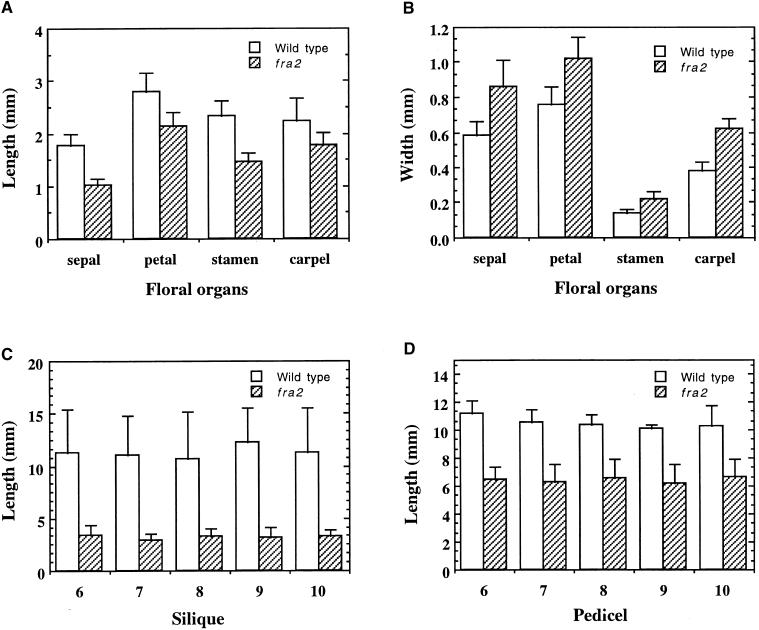
The length of floral organs was also reduced significantly in the fra2 mutant. Quantitative examination showed that the lengths of all four floral organs in fra2—including sepal, petal, stamen filament, and carpel—were reduced to 50 to 80% of those of the wild type (Figure 6A). However, the widths of all four floral organs in fra2 plants were increased 30 to 40% over wild-type values (Figure 6B). The siliques and pedicels were also much shorter than those in the wild type (Figures 4D and 4E). Quantitative measurements showed that the lengths of siliques and pedicels of fra2 mutants were reduced to 30 to 80%, respectively, of those in the wild type (Figures 6C and 6D). The reduction in organ length was also obvious in young seedlings. One-week-old fra2 seedlings had much shorter hypocotyls and roots than did wild-type seedlings (Figures 7A and 7D). Together, these results demonstrate that the fra2 mutation causes a common phenotype: reduction in organ length.
Figure 6.
Measurement of the Length and Width of Floral Organs.
The sixth to tenth siliques of 8-week-old plants were measured for their length and width. Data are means ±se of 30 samples.
(A) and (B) Length and width of floral organs, showing reduced length (A) but increased width (B) in fra2 compared with the wild type.
(C) and (D) Length of silique and pedicel, showing a reduction in fra2 compared with the wild type.
Figure 7.
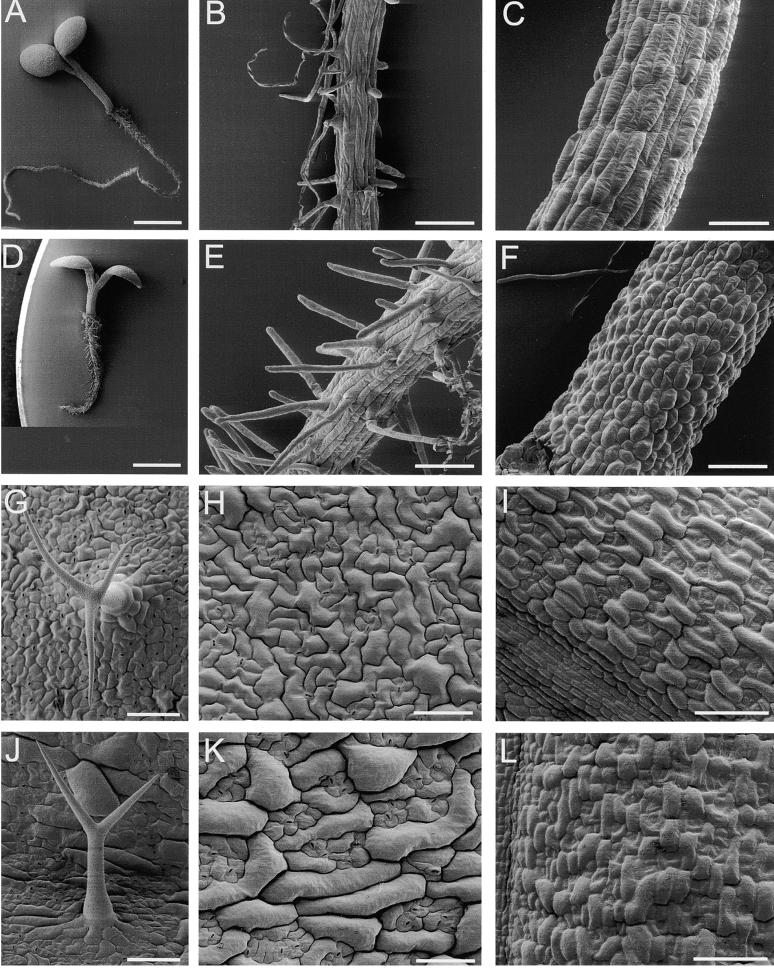
Scanning Electron Micrographs of Epidermal Cell Morphology.
(A) and (D) Five-day-old seedlings showing that fra2 (D) has shorter hypocotyl and root than does the wild type (A).
(B) and (E) Roots of 5-day-old seedlings showing that root hair cells in fra2 (E) are more compact than are those in the wild type (B).
(C) and (F) Hypocotyls of 5-day-old seedlings showing that epidermal cells in fra2 (F) have reduced length compared with those in the wild type (C).
(G) and (J) Trichomes showing that the wild type (G) has two branch points and fra2 (J) has one branch point.
(H) and (K) Upper epidermal cells of leaves showing that fra2 (K) has less convolution than does the wild type (H).
(I) and (L) Epidermal cells of carpels showing that fra2 (L) has reduced length compared with that of the wild type (I).
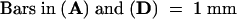 ;
; 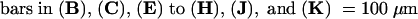 ;
; 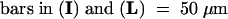 .
.
Scanning Electron Microscopy of the Morphology of Cells and Organs
To investigate whether the reduction in organ length resulted from a reduction in cell length, we examined epidermal cell lengths of different organs. Scanning electron microscopy revealed a significant reduction in the length of epidermal cells of hypocotyls (Figures 7C and 7F) and carpels (Figures 7I and 7L) in fra2 compared with those of the wild type. Root hair cells in fra2 appeared to be more compact than those in the wild type (Figures 7B and 7E), due to shortening of root epidermal cells. The shapes of epidermal cells in fra2 leaves were also altered. Ordinary epidermal cells in wild-type leaves exhibited fairly convoluted shapes (Figure 7H), whereas many of those in fra2 leaves became tubular in shape with little convolution (Figure 7K). Furthermore, most trichomes on fra2 leaves had one branch point instead of two branch points, as seen in the wild type (Figures 7J and 7G, and Table 3). These results indicate that the fra2 mutation disrupts the normal cell expansion process, which leads to alterations in cell and organ morphology.
Anatomical Examination of Cellular Morphology in Different Organs
To ascertain whether the fra2 mutation affected the morphology of other cells in addition to fibers and epidermis, we examined the cellular morphology in both transverse and longitudinal sections of fra2 organs. In inflorescence stems, both epidermal cells and cortical cells beyond the interfascicular regions were much shorter but wider in fra2 (Figure 2D) than those in the wild type (Figure 2C). Similar changes were seen in the epidermis and cortex beyond the fascicular regions (Figures 8A and 8B). It was interesting to note that the tracheary elements in fra2 also were distorted in shape (Figure 8B) compared with the long, straight columns seen in the wild type (Figure 8A). The most visible changes occurred in pith. In contrast to the pith cells in the wild type, which were long and tubular and arranged in regular files (Figure 8C), pith cells in fra2 were short and wide and exhibited irregular cell files (Figure 8D). It was evident that the appearance of some randomly arranged pith cells resulted from irregular placement of cell division planes (Figure 8D), a phenotype similar to that of the tangled1 mutant, in which leaf cell divisions are oriented nearly randomly (Smith et al., 1996; Cleary and Smith, 1998). The number of pith cells from one vascular bundle to another in a cross-section remained the same as in the wild type. Therefore, the increase in the width of pith cells apparently accounted for the increase of the stem diameter in fra2 (Table 3).
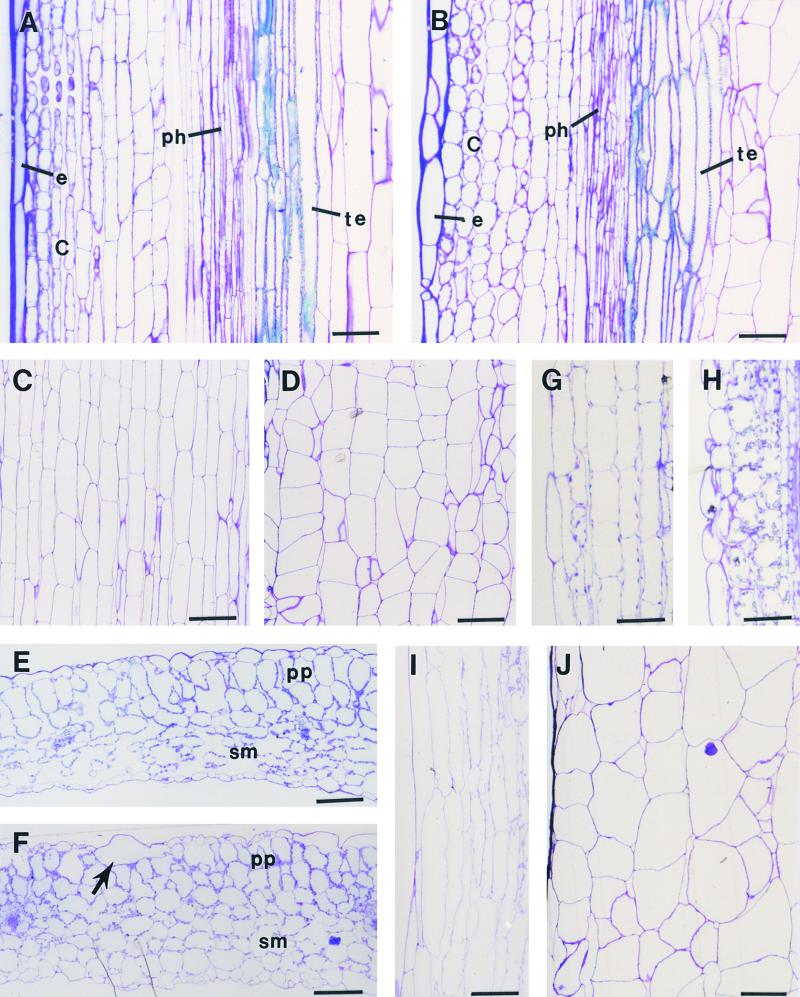
Figure 8.
Cellular Morphology in Different Organs of the Wild Type and the fra2 Mutant.
(A) and (B) Longitudinal sections of stems. fra2 (B) has shorter epidermal and cortical cells and altered shape of tracheary elements compared with those of the wild type (A). C, cortex; e, epidermis; ph, phloem; te, tracheary element.
(C) and (D) Longitudinal sections of pith showing reduced cell length and less organized cell files in fra2 (D) compared with those in the wild type (C).
(E) and (F) Cross-sections of leaves showing enlarged spongy mesophyll cells (sm) in fra2 (F) compared with those in the wild type (E). The arrow points to a giant epidermal cell in fra2 (F). pp, palisade parenchyma.
(G) and (H) Longitudinal sections of hypocotyls. Cortical cells in fra2 (H) have reduced length compared with those in the wild type (G).
(I) and (J) Longitudinal sections of petioles showing reduced cell length and less organized cell files in fra2 (J) compared with those in the wild type (I).
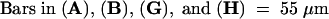 ;
; 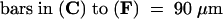 ;
; 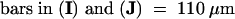 .
.
Examination of sections from leaves, hypocotyls, and roots also showed a reduction in cell length. In cross-sections of leaf blades, columnar palisade cells became shorter and less regular in shape in fra2 (Figure 8F) than in the wild type (Figure 8E). It was also obvious that some epidermal cells in fra2 (Figure 8F) were much wider than those in the wild type (Figure 8E), which most likely corresponded to the long, tubular cells revealed by electron scanning microscopy (Figure 7K). The most noticeable change was seen in the spongy mesophyll region. The spongy mesophyll cells in wild-type leaves were typically loosely distributed, leaving large air spaces between them (Figure 8E), whereas those in the fra2 mutant were closely packed with little space between. It was apparent that the spongy mesophyll cells in fra2 were much larger than those in the wild type, thus filling the arenchymatous spaces.
In leaf petioles, the parenchyma cells in fra2 (Figure 8J) were enlarged radially but shortened longitudinally compared with those in the wild type (Figure 8I). It was also noted that cells in the wild type were arranged in regular files, whereas some cells in fra2 were placed irregularly. The alteration in the cell file pattern in fra2 appeared to be caused by irregular orientation of cell division planes, which was similar to what occurred in pith cells (Figure 8D). Significant reduction in cell length was also evident in the cortical cells of fra2 hypocotyls (Figures 8G and 8H). Together, the anatomical examination of cell morphology in different organs clearly demonstrated that the fra2 mutation causes reduced cell length but increased cell width in all organs. It also indicated that the fra2 mutation might cause an irregular placement of cell division planes in some of the parenchyma cells of pith and petioles.
Positional Cloning of the FRA2 Gene
The phenotypic characterization of the fra2 mutant clearly showed its main defects in cell wall biosynthesis and cell elongation. Because the molecular mechanisms that control cell wall biosynthesis and cell elongation are still poorly understood, the isolation of genes such as FRA2 will be instrumental in dissecting the molecular control of these processes. We used a positional cloning approach to isolate the FRA2 gene. We first determined whether the fra2 mutation was monogenic and recessive. All F1 plants that resulted from the cross of fra2 and wild-type Columbia exhibited wild-type phenotypes, indicating that the fra2 mutation was recessive. In a population of 210 F2 plants from the cross, 159 plants exhibited wild-type phenotypes and 51 plants showed fra2 mutant phenotypes. This gave a segregation ratio of 3:1 (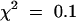 , P > 50%), indicating that the fra2 mutation was monogenic.
, P > 50%), indicating that the fra2 mutation was monogenic.
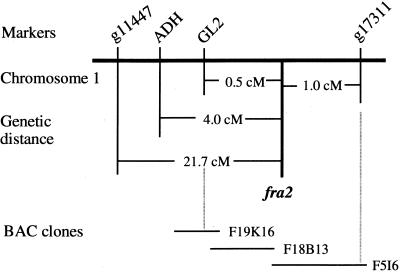
To map the fra2 locus, we crossed fra2 mutants in a Columbia background with wild-type Landsberg erecta. The F2 plants that resulted from the cross were screened for the fra2 mutant phenotype and used for mapping. Codominant amplified polymorphic sequence (CAPS) markers were used to map the fra2 locus to an individual chromosome. When CAPS markers from chromosomes 2 to 5 were used for mapping, no linkages between those markers and fra2 were found, indicating that the fra2 mutation did not occur on these chromosomes. When the CAPS marker ADH (Konieczny and Ausubel, 1993) from chromosome 1 was used, a close linkage was found between ADH and fra2. Of 538 F2 mutant plants used for mapping, 43 had crossovers between ADH and fra2, which placed the fra2 locus 4.0 centimorgans (cM) away from ADH (Figure 9). To determine on which side of ADH the fra2 locus was located, we used markers located on both sides of ADH for further mapping. Use of the CAPS marker g11447, which is located to the left of ADH, and marker g17311, which is located to the right of ADH, gave results that suggested that the fra2 locus was located between ADH and g17311 (Figure 9). Of 192 F2 mutant plants used for mapping, four plants had crossovers between g17311 and fra2, which placed fra2 at 1.0 cM away from g17311. Further mapping with GL2 showed four crossovers out of 444 plants used for mapping. Because these four plants showing crossovers with GL2 also had crossovers with ADH but not with g17311, fra2 appeared to be located 0.5 cM away on the right side of GL2 (Figure 9).
Figure 9.
Fine Mapping of the fra2 Locus.
F2 mutant plants segregating from the cross of fra2 and Landsberg erecta were used for mapping with CAPS markers. The fra2 locus was mapped to a 230-kb region located between GL2 and g17311, which was covered by three overlapping BAC clones. Markers are not positioned on the scale.
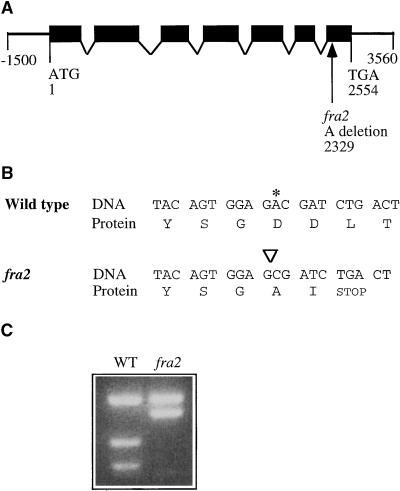
Having mapped the fra2 locus to a small region between GL2 and g17311, we searched the Arabidopsis database and located three overlapping bacterial artificial chromosome (BAC) clones (F19K16, F18B13, and F5I6) covering a 230-kb region between GL2 and g17311 (Figure 9). All three of these BAC clones had been sequenced by the Arabidopsis sequencing project, and putative genes had been annotated. This allowed us to tentatively select and test possible candidates responsible for the fra2 mutant phenotypes. Because FRA2 was a global regulator of cell wall biosynthesis and cell expansion, we reasoned that FRA2 candidates most likely could be cell wall–modifying enzymes, proteins involved in cytoskeletal regulation, or transcription factors. Of 57 putative genes within these three BAC clones, four genes fell into these categories. These putative genes showed high sequence similarities to microtubule-severing proteins, cyclins, auxin-responsive proteins, or ring finger proteins. Considering the known roles of microtubules in directing the deposition of cell wall materials, the most likely candidate lay in the F5I6.10 gene encoding a putative microtubule-severing protein located between nucleotide residues 33,019 and 35,570 in BAC clone F5I6. Sequencing of this gene in the fra2 mutant identified a deletion mutation that occurred at nucleotide residue 33,241 of F5I6 (Figure 10A), suggesting that the gene encoding the putative microtubule-severing protein most likely represents the FRA2 gene. This was confirmed by complementation analysis showing that the wild-type DNA fragment covering this gene (Figure 10A) was able to rescue the fra2 mutant phenotypes when it was transferred into the fra2 mutant (data not shown).
Figure 10.
Structure of the FRA2 Gene and the Nature of the fra2 Mutation.
(A) Exon and intron organization of the FRA2 gene. The FRA2 gene has 2554 nucleotides from the start codon (designated nucleotide 1) to the stop codon (designated nucleotide 2554). A single nucleotide deletion was found at nucleotide 2329 in fra2. Black boxes indicate exons, and lines between boxes indicate introns.
(B) Effect of the deletion mutation in fra2 on the translation of the predicted protein. Shown are nucleotide sequences and their amino acid sequences around the deletion site. Deletion of the nucleotide A (marked with an asterisk in the wild type and with an inverted triangle in fra2) leads to a frameshift of codons, thereby generating a premature stop codon at the second codon after the mutation site.
(C) Elimination of a BsmAI site in the mutant fra2 cDNA. The deletion mutation in fra2 happens to occur at a BsmAI site. This is readily revealed by digesting polymerase chain reaction (PCR)–amplified cDNA fragments with BsmAI, which shows that one BsmAI site is missing in fra2 cDNA compared with the wild-type (WT) FRA2 cDNA.
Sequence Analysis of the FRA2 Gene
To confirm the predicted exons and introns of the FRA2 gene as shown in the annotation of BAC clone F5I6, we isolated and sequenced the full-length FRA2 cDNA fragment. The longest open reading frame was 1572 nucleotides in length, encoding a polypeptide of 523 amino acids with a predicted molecular mass of 57 kD. Comparison of the FRA2 gene and its cDNA identified seven exons and six introns in the gene sequence. The fra2 mutation occurred in the seventh exon, in which the A at nucleotide residue 2329 was deleted (Figure 10A). This resulted in a frameshift of the coding sequence, which led to the creation of a premature stop codon in the fra2 cDNA (Figure 10B). The mutant fra2 protein lacked 78 amino acids at the C-terminal region. The deletion mutation in fra2 was further confirmed by sequencing of the fra2 cDNA and by detection of a polymorphism between the wild-type and mutant fra2 cDNAs (Figure 10C).
Sequence comparison of FRA2 with proteins in the GenBank database revealed a high sequence similarity to a group of microtubule-severing proteins called katanins. FRA2 exhibited the highest amino acid sequence identity (43% identity and 56% similarity in the entire open reading frame) to sea urchin katanin (Figure 11), which was the first katanin cDNA isolated and confirmed to have microtubule-severing activity (Hartman et al., 1998). The highest sequence similarity (62% identity and 75% similarity) was seen in the putative ATP binding module between amino acid residues 236 and 454 of fra2. The amino acid sequence of fra2 at the N-terminal region before the ATP binding module had 26% sequence identity (38% similarity) with sea urchin katanin. No significant amino acid sequence similarity was found between FRA2 and other proteins except katanins. These analyses strongly suggest that FRA2, a gene regulating cell wall biosynthesis and cell expansion, encodes a katanin-like protein. Hence, FRA2 was renamed AtKTN1.
Figure 11.
Alignment of the Deduced Amino Acid Sequences of AtKTN1 and Katanin (SuKTN) from Sea Urchin.
AtKTN1 exhibits 56% sequence similarity in the entire open reading frame with katanin from sea urchin. The ATP binding module, which shares 75% sequence similarity between AtKTN1 and katanin, is underlined. The GenBank accession number for the AtKTN1 cDNA sequence data is AF358779.
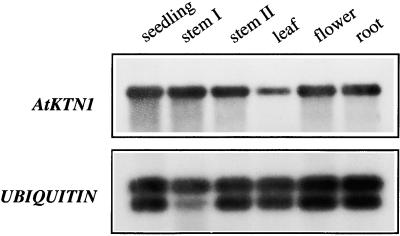
Examination of AtKTN1 expression patterns showed that the AtKTN1 gene was expressed in all organs examined, including stems, leaves, flowers, and roots (Figure 12). This is consistent with the fra2 phenotypes of alterations in cell elongation in all organs.
Figure 12.
Analysis of AtKTN1 Gene Expression in Arabidopsis Organs.
Total RNA was isolated from different organs of Arabidopsis plants and used for reverse transcription–PCR. The ubiquitin gene was used as an internal control for PCR. The seedlings were 3 weeks old. Leaves, roots, and flowers came from 8-week old plants. Stems I and II were from 4- and 8-week-old plants, respectively.
Microtubule Organization in fra2 Cells
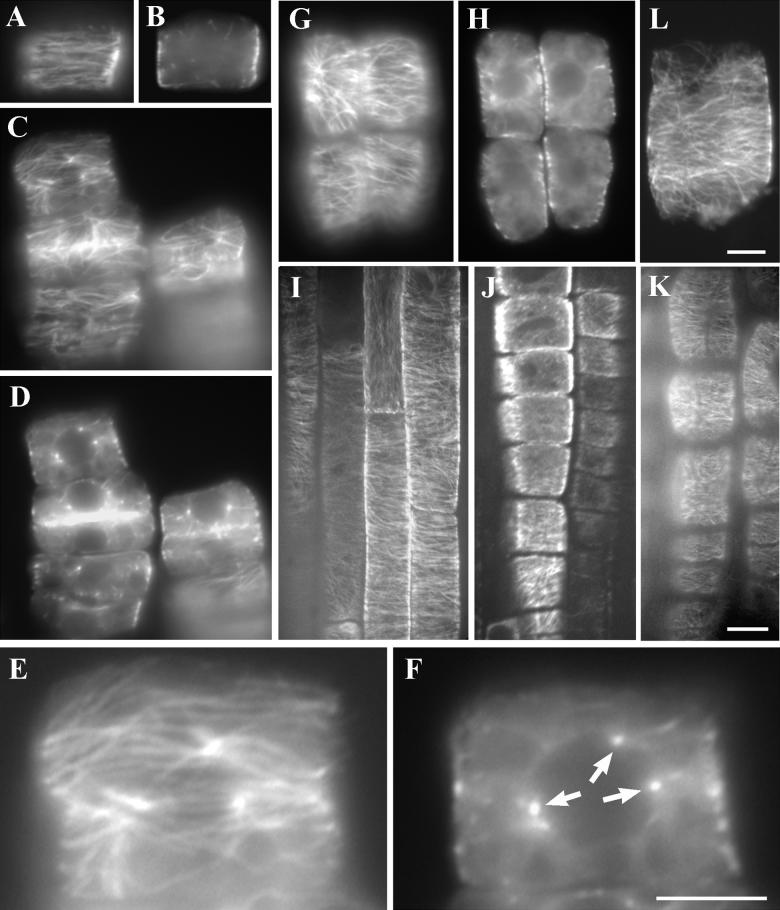
The finding that the AtKTN1 gene encodes a katanin-like protein prompted us to examine microtubule organization in the fra2 cells because microtubules are essential for cell morphogenesis. In wild-type root tip cells, a cortical array of parallel microtubules was always detected in interphase cells (Figure 13A). Microtubules were not detected when we focused away from the cell cortex (Figure 13B). In the fra2 mutant, abnormal microtubule distribution was detected in flat cells, which were considered to be cells exited from cell division recently (Figures 13C to 13F). In these cells, microtubules started to appear at the cell cortex in a converged pattern (Figures 13C and 13E). However, on the nuclear envelope, microtubule converging points could be clearly detected (Figures 13D and 13F). Such converging points clearly accounted for an aster-like microtubule organization pattern toward the cell cortex (Figure 13E). Such a microtubule configuration has never been detected in interphase cells among higher plants. In most of elongating cells, the microtubule converging pattern could no longer be detected on the nuclear envelope, and microtubules gradually organized into parallel pattern in the cell cortex (Figures 13G and 13H). However, some elongating cells (Figures 13G and 13H) with the microtubule converging pattern still remain, indicating that disappearance of the microtubule aggregation points during cell elongation was not synchronous. In wild-type cells undergoing elongation, cortical microtubules retained a parallel pattern mostly perpendicular to the growth axis (Figure 13I). Although the fra2 cells had very limited elongation activity, cortical microtubules gradually became parallel to the growth axis (Figures 13J and 13K). In the fra2 cells that had become highly vacuolated (Figure 13L), typical cortical microtubules could be still visualized.
Figure 13.
Immunostaining of Microtubules in Root Cells of the Wild Type and fra2.
Young roots from 5-day-old seedlings were treated with cell wall–digesting enzymes, probed with the antibodies against α-tubulin and fluorescein isothiocyanate–conjugated secondary antibodies, and visualized with an epifluorescence microscope or a confocal microscope.
(A) and (B) A surface view (A) of a wild-type interphase cell that has yet to undergo elongation, showing the parallel cortical microtubule network. A midplane view (B) of the same cell, showing no obvious microtubules.
(C) and (D) A surface view (C) of the fra2 interphase cells, displaying microtubules in a converged pattern near the cell cortex. A midplane view (D) of the same cells, showing microtubule aggregation points.
(E) Close-up of a surface view of the uppermost cell in (C), showing the converging microtubule organization pattern.
(F) Close-up of a midplane view of the uppermost cell in (D), showing the microtubule aggregation points (arrows), which are the centers of three microtubule asters facing toward the cell cortex (E).
(G) and (H) Surface (G) and midplane (H) views of the fra2 elongating cells, showing the disappearance of microtubule aggregation patterns, except in the top left cell, which retains the microtubule converging pattern.
(I) A whole-mount view of the wild-type root, showing cortical microtubules in a transverse pattern in the cell cortex of elongating epidermal cells.
(J) and (K) A whole-mount view of the fra2 root, showing cortical microtubules aligned in the cell cortex of elongating epidermal cells. Note that the fra2 epidermal cells are much shorter in length compared with those of the wild type (I).
(L) A surface view of a highly vacuolated fra2 cell with well-organized cortical microtubules.
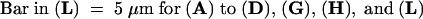 ;
; 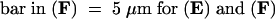 ;
; 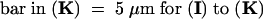 .
.
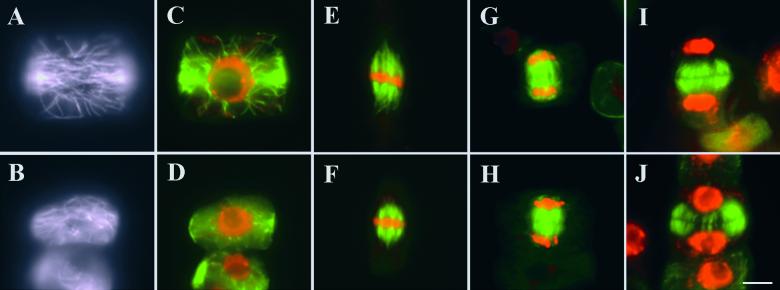
Because mammalian katanin is largely responsible for microtubule severing during mitosis or meiosis (McNally and Thomas, 1998; Srayko et al., 2000), we investigated whether abnormal microtubule organization could be detected when fra2 cells undergo cell division. In somatic cells, a cortical microtubule band—the preprophase band—is present during late G2 phase to prophase to predict the future division plane. Both wild-type and fra2 had clear preprophase bands starting in a broad ring in the cell cortex (Figures 14A to 14D). During later stages, narrower microtubule bands could be detected in both wild-type and mutant cells (data not shown). Before the nuclear envelope breaks down, the preprophase band microtubules are depolymerized while spindle microtubules get organized. We detected normal metaphase spindles in wild-type and fra2 mutant (Figures 14E and 14F). Spindle microtubules undergo rapid reorganization (both depolymerization and polymerization) during anaphase. Concomitant with the reorganization of spindle microtubules, sister chromatids are separated. In both wild-type and fra2, cells at late anaphase presented a similar microtubule configuration, with microtubules starting to accumulate in the central spindle (Figures 14G and 14H). After the nuclear division, the phragmoplast, which contains two sets of antiparallel microtubules oriented perpendicular to the cell plate, is formed to guide vesicles bound for the division site to give rise to the new plasma membrane and the new cell wall. During the centrifugal expansion of cell plate, the phragmoplast microtubules depolymerize centrifugally as well. Identical phragmoplast microtubule arrays were found in a wild-type cell (Figure 14I) and a fra2 cell (Figure 14J). Therefore, our data indicate that in the fra2 cells, microtubule reorganization during mitosis and cytokinesis is not affected.
Figure 14.
Immunostaining of Microtubules in Dividing Root Cells of the Wild Type and fra2.
Microtubules were pseudocolored in green, and DNA was pseudocolored in red in (C) to (J).
(A) to (D) The microtubules in early broad preprophase bands in a wild-type cell ([A] and [C]) and a fra2 mutant cell ([B] and [D]). (A) and (B) were in the focus at the cell cortex, and (C) and (D) were in the middle.
(E) and (F) Metaphase spindles in wild-type (E) and fra2 mutant (F) cells.
(G) and (H) Microtubule spindles between two sets of segregated chromatids of a wild-type cell (G) and a fra2 mutant cell (H) during late anaphase.
(I) and (J) Typical phragmoplast microtubule arrays in a wild-type cell (I) and a fra2 mutant cell (J). Microtubules have already started to depolymerize in the central region in the phragmoplast shown in (J).
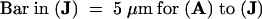 .
.
DISCUSSION
It has long been accepted that cortical microtubule arrays regulate cellulose microfibril orientation. Cortical microtubules undergo dynamic rearrangement in direction during different stages of cell expansion and differentiation. Cellulose microfibril orientation accompanies the dynamic alterations of cortical microtubules, thereby regulating the direction of cell expansion. A variety of mechanisms have been proposed as regulators of the dynamic changes of cortical microtubules in plants, such as assembly and disassembly and microtubule translocation (Cyr and Palevitz, 1995). Our finding that a katanin-like protein is essential for cell wall biosynthesis and cell elongation suggests that microtubule-severing proteins also might contribute to the dynamic changes of cortical microtubule arrays.
The AtKTN1 Gene Encodes a Katanin-like Protein
Katanin, a microtubule-severing protein, couples ATP hydrolysis to disassemble microtubules into tubulin subunits. The microtubule-severing activity was first identified in the mitotic extracts of Xenopus laevis eggs (Vale, 1991). Biochemical characterization of katanin from sea urchin oocytes has shown that katanin is a heterodimer of 60- and 80-kD subunits and requires ATP hydrolysis for its microtubule-severing activity (McNally and Vale, 1993). Subsequently, the genes encoding the 60- and 80-kD subunits were cloned from sea urchin (Hartman et al., 1998). The katanin 60-kD subunit was found to be a novel member of the AAA (ATPase associated with diverse cellular activity) family of ATPases. The 60-kD subunit alone possesses both microtubule-stimulating ATPase activity and microtubule-severing activity. The katanin 80-kD subunit contains WD40 repeats and is suggested to target katanin to the centrosome. Katanins have been localized to the centrosome in interphase cells and to the spindle poles in sea urchin, X. laevis, and human cells, indicating their roles in the cell cycle regulation of microtubule arrays (McNally et al., 1996; McNally and Thomas, 1998; Ahmad et al., 1999). Katanin-like microtubule-severing activity is also implicated in flagellar excision in Chlamydomonas reinhardtii (Lohret et al., 1998) and meiotic spindle organization in Caenorhabditis elegans (Srayko et al., 2000). However, no katanin-like microtubule-severing protein is known to be involved in any cellular activities in higher plants.
Sequence analysis of AtKTN1 revealed that it shares significant amino acid sequence similarity with katanins from sea urchin and C. elegans (Hartman et al., 1998; Srayko et al., 2000). The highest sequence identity between AtKTN1 and katanin from sea urchin resides in the ATP binding module located in the C-terminal region. This similarity pattern was also found between sea urchin katanin and C. elegans MEI-1, which was shown to possess microtubule-severing activity (Srayko et al., 2000). However, unlike MEI-1, in which the N-terminal region has no similarity to sea urchin katanin, AtKTN1 exhibited significant sequence similarity to sea urchin katanin in the N-terminal region outside the ATP binding module. These lines of evidence strongly indicate that AtKTN1 encodes a katanin-like protein, a possible ortholog in plants. Definite proof of whether AtKTN1 possesses microtubule-severing activity awaits further investigation.
The fra2 Mutation Delays the Establishment of Cortical Microtubule Array
Microtubule orientations undergo dynamic changes during cell division, expansion, and differentiation (Hush et al., 1994; Yuan et al., 1994; Marc et al., 1998; Mathur and Chua, 2000). For example, during the transition from isotropic to anisotropic cell expansion, cortical microtubules reorient from a random pattern to a transversely positioned pattern along the elongation axis. After transverse positioning, cortical microtubules must reorient constantly at different angles to direct cellulose microfibril deposition. Although a number of possible mechanisms to control the dynamic changes in microtubules have been proposed, none has been demonstrated to be critical during cell elongation (Cyr and Palevitz, 1995). The finding that AtKTN1, a gene that regulates cell elongation, encodes a katanin-like protein suggests that microtubule-severing activity may play an important role in regulating the dynamic changes in microtubules during the initiation and subsequent maintenance of cell elongation.
Our results indicate that the fra2 mutation renders a clear phenotype in microtubule organization in early stages during establishment of the cortical microtubule array. However, the mutation appears not to affect the organization of microtubule arrays when cells are undergoing mitosis and cytokinesis. During late stages of cytokinesis in somatic plant cells, a perinuclear microtubule array can be detected transiently (Hasezawa et al., 1991). When cells exit from cytokinesis, perinuclear microtubule array rapidly depolymerizes while cortical microtubule array arises, and two arrays do not overlap (Hasezawa et al., 1991). Therefore, a rapid microtubule depolymerization event takes place during the transition between two arrays. We propose that the plant katanin-like protein AtKTN1 is required for such a rapid microtubule depolymerization activity. Tubulins from the depolymerization of the perinuclear array could supply subunits for the cortical array. Because the perinuclear microtubules cannot be rapidly depolymerized on the nuclear envelope, these microtubules may be aggregated together possibly by microtubule motors and/or proteins of microtubule-organizing centers, for example, γ-tubulin. Because of the phenotypes of microtubule organization and cell elongation defects in the fra2 mutant cells, we further hypothesize that the defects in cell wall biosynthesis and cell elongation caused by the fra2 mutation are due to the inefficiency in converting the perinuclear array to the cortical array. Our finding that the plant katanin-like protein AtKTN1 plays a role in severing microtubules after cells exit from cell division is in contrast to animal katanins, which play a role in severing microtubules during mitosis. Interestingly, the interphase-specific microtubule-severing activity has also been detected in the green alga C. reinhardtii during deflagellation (Lohret et al., 1999; Quarmby, 2000).
Although the establishment of the cortical microtubule array is delayed in the fra2 mutant cells, the cortical array eventually emerges. We do not rule out the possibility that AtKTN1 also affects the dynamics of cortical microtubules during cell elongation. It is likely that the fra2 mutation might also impair the microtubule turnover rate, thereby contributing to a decrease in cell elongation but an increase in radial expansion, although a direct link between microtubule turnover and cell elongation has not been established in the cells of higher plants. Further study is required to directly establish the effects of the fra2 mutation on microtubule turnover rate.
The defects in the microtubule organization observed in the fra2 mutant are very different from those of the fass mutants, which also are defective in cell elongation (Torres-Ruiz and Jurgens, 1994; Traas et al., 1995; McClinton and Sung, 1997). The fass mutation causes disorganization of cortical microtubule arrays and the absence of a preprophase band, suggesting that the mutation affects the organization of microtubules. Therefore, the defect in cell elongation in fass mutants is caused by an inability to form cortical microtubule arrays in transverse orientation (McClinton and Sung, 1997). The effect of the fra2 mutation on plant morphology also is distinguishable from fass mutant phenotypes. fra2 plants have an almost normal shoot architecture except for shortening of all organs (Figure 4), whereas fass plants exhibit a ball-like shape (McClinton and Sung, 1997).
The fra2 Mutation Alters Cell Wall Biosynthesis
It is generally accepted that the orientation of cellulose microfibrils synthesized from the cellulose synthase complex is regulated by underlying microtubules beneath the plasma membranes. The trafficking of vesicles containing noncellulosic materials synthesized in the Golgi body also may be guided by microtubules. Thus, it is conceivable that alteration of the normal dynamic changes in cortical microtubules might result in an inability of microtubules to guide efficiently the deposition of cell wall materials, thereby leading to a delay of cell wall biosynthesis. Our finding that the fra2 mutation reduces cellulose and hemicellulose contents provides strong evidence to support this possibility. It suggests that microtubules not only guide the orientation of cellulose microfibril deposition, which in turn regulates cell elongation, but also might influence cell wall biosynthesis in general.
It is intriguing that the fra2 mutation caused the formation of more condensed lignin. In particular, the guaiacyl lignin unit is much more highly condensed than the syringyl lignin unit. This is consistent with the structure of the guaiacyl lignin unit, which has an additional 5-C on the aromatic ring exposed for cross-linking. Reduction of cellulose in secondary walls and possible alteration of cellulose microfibril orientation may cause a change in the distribution of monolignols across the secondary walls. This might result in an accumulation of monolignols in certain wall areas, thereby leading to the formation of a highly condensed lignin structure. It will be interesting to determine whether there is any alteration in the distribution of lignin within the walls of fiber cells.
The fra2 Mutation Dramatically Reduces the Mechanical Strength of Fibers
The fra2 mutant was isolated based on its marked reduction in the breaking strength of the stems. Because the mechanical strength of the mature stems is conferred largely by the presence of interfascicular fibers, the fra2 mutant must have a dramatic decrease in the mechanical strength of interfascicular fibers. Anatomical and chemical analyses indicate that the reduction in the mechanical strength of fibers is caused by alterations in fiber cell length and cell wall composition. Because AtKTN1 shows high similarity to microtubule-severing proteins, microtubule-severing activity might be essential for normal fiber cell elongation and normal synthesis of fiber walls with strong mechanical strength.
In conclusion, we have shown that the fra2 mutation results in a reduction in cellulose and hemicellulose contents, the formation of highly condensed lignin, a decrease in fiber cell length and fiber mechanical strength, a global alteration in cell elongation and plant morphology, and a delay in the establishment of the normal cortical microtubule array. On the basis of the finding that the AtKTN1 gene encodes a protein similar to katanin that severs microtubules, we propose that AtKTN1 regulates the dynamic changes of microtubules during early stages of establishment of the cortical microtubule array as well as during cell elongation, which in turn influences cell morphogenesis, the biosynthesis and deposition of cell wall materials, and cell wall strength.
METHODS
Mutant Screening
Ethyl methanesulfonate–mutagenized M2 Arabidopsis thaliana (ecotype Columbia) plants were grown in a greenhouse. The inflorescence stems of 8-week-old plants were measured for their mechanical strength. Plants showing a dramatic reduction in normal stem strength were selected for further analysis. Mutant lines were backcrossed with the wild type three times to reduce background mutations.
Breaking Strength Measurement
The main inflorescence stems of 8-week-old plants were used for breaking force measurement (Reiter et al., 1993) with a digital force/length tester (model DHT 4-50; Larson Systems, Minneapolis, MN). Each stem was divided into four equal segments. The ends of stem segments with a space of 1 cm between were clamped, and a force was applied manually until the stem segments were broken. Stems from 15 plants were tested for the breaking force.
Histology
Tissues were fixed in 2% glutaraldehyde in PBS (33 mM Na2HPO4, 1.8 mM NaH2PO4, and 140 mM NaCl, pH 7.3) at 4°C overnight. After fixation, segments were dehydrated through a gradient of ethanol, cleared in propylene oxide, and embedded in Araldite resin (Electron Microscopy Sciences, Fort Washington, PA). One-micrometer-thick sections were cut with a microtome and stained with toluidine blue for observation of anatomy.
Scanning Electron Microscopy
Tissue samples were fixed in 2% glutaraldehyde, dehydrated in ethanol, and then dried in a Samdri critical point dryer (Tousimis, Rockville, MD) before being mounted on stubs with carbon paste. Samples were coated with gold using an Edwards 306 vacuum evaporator (Edwards High Vacuum International, Wilmington, MA) and viewed with a LEO 982 FE scanning electron microscope (LEO, Thornwood, NY).
Cellulose Content Assay
Stem materials were ground into powder and extracted twice with 70% ethanol at 70°C for 1 hr. After vacuum drying, cell wall materials were used for cellulose content assays according to Updegraff (1969). The cellulose content was determined with the anthrone reagent. Whatman (Clifton, NJ) 3MM paper was used as standard cellulose for quantitation.
Cell Wall Sugar Composition Analysis
Sugars (as alditol acetates) were measured as described by Hoebler et al. (1989), with the initial digestion time increased from 30 to 90 min.
Lignin Content and Composition Analysis
Ethanol-extracted cell wall materials were used for measurement of Klason lignin according to Kirk and Obst (1988). Lignin content was expressed as a percentage of the original weight of cell wall residues.
Lignin composition was analyzed according to Morrison et al. (1996). Cell wall materials were hydrolyzed in 4 N NaOH at 170°C for 2 hr. The hydrolysate was acidified with 2 N HCl to pH 2.0. Lignin monomers released from base hydrolysis were extracted into diethylether and vacuum dried. The residue was dissolved in 10 μL of pyridine and 10 μL of N,O-bis(trimethylsilyl)trifluoroacetamide and analyzed by gas-liquid chromatography. Compounds were identified by comparing their mass spectra with published spectra or with those of the authentic compounds. All samples were run in duplicate.
In-Source Pyrolysis Mass Spectrometry
In-source pyrolysis mass spectrometry was performed on a Finngan GCQ mass spectrometer equipped with a direct exposure probe (rhenium loop) (Thermoquest, San Jose, CA), as described by Morrison and Archibald (1998). Analysis conditions were as follows: ionization energy of 20 electron volts; mass range of 50 to 500 m/z; scan time of 1 sec; temperature increase of ∼10°C per sec to 700°C; and ion source temperature of 175°C. All samples were run in triplicate.
Genetic Analysis
The mutant line was crossed with the wild-type Arabidopsis ecotype Landsberg erecta for mapping study. F2 plants showing mutant phenotypes were selected for genetic mapping. Genomic DNA was isolated from the F2 mapping plants and used for polymerase chain reaction (PCR) with codominant amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993). The information on CAPS markers used in this study came from the Arabidopsis database.
Gene Expression Analysis
The level of AtKTN1 mRNA in different organs was analyzed with reverse transcription–PCR. One-tenth microgram of total RNA was used for the synthesis of the first strand cDNA, which was further PCR-amplified for 20 cycles with gene-specific primers. The PCR products were run on an agarose gel and transferred to a nylon membrane. The membrane was then hybridized with a digoxigenin-labeled AtKTN1 gene probe, and the hybridized signals were detected with a chemiluminescence detection kit (Reche Molecular Biochemicals, Indianapolis, IN), according to the manufacturer's protocol.
Localization of Microtubules
Seed of the wild type and the fra2 mutant were germinated on wet filter paper at room temperature for 5 days. Two different protocols were used for microtubule localization. A whole-mount staining protocol was adopted from Sugimoto et al. (2000) to observe cells undergoing elongation. To observe cells undergoing cell division, we subjected root tip cells to tubulin staining according to Liu and Palevitz (1992). In brief, seedlings were fixed for 60 min with 4% paraformaldehyde in PME buffer (50 mM Pipes, 5 mM EGTA, and 2 mM MgSO4, pH 6.9). After being rinsed extensively with the PME buffer, the seedlings were treated with 1% cellulase RS (Yakult, Tokyo, Japan) and 0.1% Macerozyme R-10 (Karlan Research Products, Santa Rosa, CA) in PME for 15 min to partially digest the cell wall. Root tips were excised from the rest of the seedlings and squashed between a cover slip and a microscopic slide precoated with gelatin and chrom-alum. Root cells were treated with 0.5% Triton in PME for 10 min, followed by a 10-min treatment with −20°C methanol. The cells were then rehydrated in PBS. Immunolocalization was performed with an anti-α-tubulin antibody (DM1A; Sigma) diluted at 1:400 followed by a fluorescein isothiocyanate–conjugated goat anti-mouse IgG antibody (Sigma) diluted at 1:400. Cells were then mounted on a slide in a mounting medium containing 100 mM Tris, pH 9.2, 50% glycerol, 1 mg/mL phenylenediamine, and 1 μg/mL 4′,6-diamidino-2-phenylindole. Roots stained with the whole-mount method were observed either under a Wallac UltraVIEW LCI confocal microscope with an Olympus 60X Plapo objective or under a Nikon E600 epifluorescence microscope with a 60X Plan-Apo objective. Root tip cells were observed under a Nikon E600 epifluorescence microscope with a 100X Plan Fluor objective. Conventional epifluorescence images were acquired by a CCD camera (model ORCA100; Hamamatsu Photonics, Hamamatsu City, Japan) with the ImageProPlus software package (Media Cybernetics, Silver Spring, MD). Final assembly of images in Figures 13 and 14 was performed with Adobe Photoshop software (Adobe, San Jose, CA).
Acknowledgments
We thank M.H. Zhou and J.M. Scholey for their help with the confocal microscopy, Jan Nadeau for information on CAPS markers, G. Freshour and E. Richardson for their assistance in sectioning, J. Shields and M. Farmer for their help on scanning electron microscopy, B. Palevitz for stimulating discussions and helpful suggestions, and the editor and reviewers for their comments and suggestions. D.H.B. was supported by a Plant Evolution Training Grant from the National Science Foundation. This work was supported in part by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture.
References
- Ahmad, F.J., Yu, W., McNally, F.J., and Baas, P.W. (1999). An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akin, D.E., Morrison, W.H., and Himmelsbach, D.S. (1993). Characterization of digestion residues of alfalfa and orchardgrass leaves by microscopic, spectroscopic and chemical analysis. J. Sci. Food Agric. 63, 339–347. [Google Scholar]
- Aloni, R. (1987). Differentiation of vascular tissues. Annu. Rev. Plant Physiol. 38, 179–204. [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2000). The cytoskeleton. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 202–258.
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Cleary, A.L., and Smith, L.G. (1998). The tangled1 gene is required for spatial control of cytoskeletal arrays associated with cell division during maize leaf development. Plant Cell 10, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, R.J., and Palevitz, B.A. (1995). Organization of cortical microtubules in plant cells. Curr. Opin. Cell Biol. 7, 65–71. [DOI] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego, CA: Academic Press), pp. 85–99.
- Hartman, J.J., Mahr, J., McNally, K., Okawa, K., Iwamatsu, A., Thomas, S., Cheesman, S., Heuser, J., Vale, R.D., and McNally, F.J. (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93, 277–287. [DOI] [PubMed] [Google Scholar]
- Hasezawa, S., Marc, J., and Palevitz, B.A. (1991). Microtubule reorganization during the cell cycle in synchronized BY-2 tobacco suspensions. Cell Motil. Cytoskeleton 18, 94–106. [Google Scholar]
- Hoebler, C., Barry, L.D., and Delort-Laval, J. (1989). Rapid hydrolysis of plant cell wall polysaccharides by gas-liquid chromatography. J. Agric. Food Chem. 37, 360–367. [Google Scholar]
- Hush, J.M., Wadsworth, P., Callaham, D.A., and Hepler, P.K. (1994). Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107, 775–784. [DOI] [PubMed] [Google Scholar]
- Kirk, T.K., and Obst, J.R. (1988). Lignin determination. Methods Enzymol. 161, 87–101. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Liu, B., and Palevitz, B.A. (1992). Organization of cortical microfilaments in dividing root cells. Cell Motil. Cytoskeleton 23, 252–264. [Google Scholar]
- Lohret, T.A., McNally, F.J., and Quarmby, L.M. (1998). A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Mol. Biol. Cell 9, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohret, T.A., Zhao, L.F., and Quarmby, L.M. (1999). Cloning of Chlamydomonas p60 katanin and localization to the site of outer doublet severing during deflagellation. Cell Motil. Cytoskeleton 43, 221–231. [DOI] [PubMed] [Google Scholar]
- Luo, D., and Oppenheimer, D.G. (1999). Genetic control of trichome branch number in Arabidopsis: The roles of the FURCA loci. Development 126, 5547–5557. [DOI] [PubMed] [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T.-H., McCubbin, A.G., and Cyr, R.J. (1998). A GFP–MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.-H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauseth, J.D. (1988). Plant Anatomy. (Menlo Park, CA: Benjamin/Cummings Publishing).
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. [DOI] [PubMed] [Google Scholar]
- McNally, F.J., and Thomas, S. (1998). Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol. Biol. Cell 9, 1847–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, F.J., and Vale, R.D. (1993). Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell 75, 419–429. [DOI] [PubMed] [Google Scholar]
- McNally, F.J., Okawa, K., Iwamatsu, A., and Vale, R.D. (1996). Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci. 109, 561–567. [DOI] [PubMed] [Google Scholar]
- Morrison, W.H., and Archibald, D.D. (1998). Analysis of graded flax fiber and yarn by pyrolysis mass spectrometry and pyrolysis gas chromatography mass spectrometry. J. Agric. Food Chem. 46, 1870–1876. [Google Scholar]
- Morrison, W.H., Akin, D.E., Ramaswamy, G., and Baldwin, B. (1996). Evaluating chemically retted kenaf using chemical, histochemical, and microspectrophotometric analyses. Text. Res. J. 66, 651–656. [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane–bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby, L. (2000). Cellular samurai: Katanin and the severing of microtubules. J. Cell Sci. 113, 2821–2827. [DOI] [PubMed] [Google Scholar]
- Reiter, W.-D., Chapple, C.C.S., and Somerville, C.R. (1993). Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261, 1032–1035. [DOI] [PubMed] [Google Scholar]
- Seagull, R.W., and Falconer, M.M. (1991). In vitro xylogenesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego, CA: Academic Press), pp. 183–194.
- Smith, L.G., Hake, S., and Sylvester, A.W. (1996). The tangled1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122, 481–489. [DOI] [PubMed] [Google Scholar]
- Srayko, M., Buster, D.W., Bazirgan, O.A., McNally, F.J., and Mains, P.E. (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072–1084. [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz, R.A., and Jurgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677. [Google Scholar]
- Updegraff, D.M. (1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. (1991). Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell 64, 827–839. [DOI] [PubMed] [Google Scholar]
- van der Hage, E.R.E., Mulder, M.M., and Boon, J.J. (1993). Structural characterization of lignin polymers by temperature-resolved in-source pyrolysis–mass spectrometry and Curie-point pyrolysis–gas chromatography/mass spectrometry. J. Anal. Appl. Pyrolysis 25, 149–183. [Google Scholar]
- Yuan, M., Shaw, P.J., Warn, R.M., and Lloyd, C.W. (1994). Dynamic reorientation of cortical microtubules, from transverse to longitudinal, in living plant cells. Proc. Natl. Acad. Sci. USA 91, 6050–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.-H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.-W., Nishizawa, N., Wu, Y., Kost, B., and Chua, N.-H. (2000). KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]