Abstract
Jasmonates (JAs) inhibit plant growth and induce plant defense responses. To define genes in the Arabidopsis JA signal pathway, we screened for mutants with constitutive expression of a luciferase reporter for the JA-responsive promoter from the vegetative storage protein gene VSP1. One mutant, named constitutive expression of VSP1 (cev1), produced plants that were smaller than wild type, had stunted roots with long root hairs, accumulated anthocyanin, had constitutive expression of the defense-related genes VSP1, VSP2, Thi2.1, PDF1.2, and CHI-B, and had enhanced resistance to powdery mildew diseases. Genetic evidence indicated that the cev1 phenotype required both COI1, an essential component of the JA signal pathway, and ETR1, which encodes the ethylene receptor. We conclude that cev1 stimulates both the JA and the ethylene signal pathways and that CEV1 regulates an early step in an Arabidopsis defense pathway.
INTRODUCTION
Jasmonates (JAs) are a family of cyclopentanone derivatives synthesized from linolenic acid via the octadecanoic pathway that regulate plant growth, plant defense responses, and aspects of development. They inhibit plant growth generally, but in addition, they promote diverse processes including fruit ripening, senescence, tuber formation, tendril coiling, pollen formation, and defense responses against pests and pathogens (reviewed in Creelman and Mullet, 1997). In Arabidopsis, JAs inhibit root elongation (Staswick et al., 1992) and are required for pollen development, anther dehiscence (Feys et al., 1994; McConn and Browse, 1996; Sanders et al., 2000), and defense against insects (McConn et al., 1997) and necrotrophic pathogens (Thomma et al., 1999). JAs also induce many genes, including those for vegetative storage proteins (VSPs; Benedetti et al., 1995), a thionin (Thi2.1; Epple et al., 1995; Vignutelli et al., 1998), and a plant defensin (PDF1.2; Penninckx et al., 1996).
JAs interact with ethylene in the regulation of plant wound and defense responses. Both JA and ethylene are formed when plants are wounded or attacked by pests or pathogens (Creelman et al., 1992; O'Donnell et al., 1996; Kuc, 1997). Mutants defective in either the production or the perception of ethylene or JA have increased susceptibility to several pests and pathogens (McConn et al., 1997; Pieterse et al., 1998; Vijayan et al., 1998; Thomma et al., 1999), indicating that both signal pathways are required for resistance. A possible mechanism for JA- and ethylene-dependent defenses is through their synergistic interaction in the induction of many defense-related genes, including PR5, PDF1.2, and basic chitinase (CHI-B), and of a hevein-like protein (Xu et al., 1994; Penninckx et al., 1998; Norman-Setterblad et al., 2000). However, expression of the JA-responsive VSP gene is increased in ethylene-insensitive mutants (Rojo et al., 1999; Norman-Setterblad et al., 2000), suggesting that the ethylene signal pathway represses the induction of VSP. JA and ethylene therefore cooperate synergistically in the activation of wound-related defense responses, but some JA responses are antagonized by ethylene.
Mutants in hormone signal transduction pathways are of two main types: mutants that cause loss of response to the hormone identify genes that normally function as positive regulators, and mutants that cause constitutive activation of the pathway identify negative regulators. Examples of positive regulators include the ethylene receptor ETR1 (Chang et al., 1993), the ABI1 and ABI2 protein phosphatases, which are involved in abscisic acid response (Leung et al., 1994, 1997), and the F-box proteins TIR1 and COI1, which are involved in auxin and JA responses (Ruegger et al., 1998; Xie et al., 1998). Examples of negative regulators include the Raf-like protein kinase CTR1, which suppresses ethylene-activated genes (Kieber et al., 1993), and SPY1, which suppresses the gibberellin pathway (Jacobsen et al., 1996).
The only reported mutants in the JA pathway have lost JA responsiveness and therefore are likely to define positive regulators (Staswick et al., 1992; Feys et al., 1994; Berger et al., 1996). We have developed a screen for mutations in negative regulators of the JA signal pathway based on constitutive expression of a JA-responsive promoter. For this purpose, we constructed transgenic lines containing the promoter of the JA-responsive gene VSP1 (Utsugi et al., 1996) fused to the firefly luciferase gene as reporter. The transgenic seed were mutagenized and screened for plants that expressed the reporter construct in the absence of exogenously applied methyl jasmonate (MeJA). Here, we describe a mutant isolated in this screen that we have named constitutive expression of VSP1 (cev1).
RESULTS
Isolation of cev1
Arabidopsis ecotype Columbia-glabrous (Col-gl1) seed homozygous for the VSP1::luciferase reporter transgene were mutagenized with ethyl methanesulfonate. M2 seed were germinated individually in wells of 96-well microtiter plates. After 8 days, seedlings were sprayed with luciferin, and luminescence was measured. Under these conditions, the parental plants did not exhibit luciferase activity. Of the ∼20,000 seedlings screened, 359 displayed high luciferase activity, and of these, 240 survived to produce seed. The 240 M3 families were rescreened, and only seven had constitutively high luciferase activity. Test crosses made between these seven mutants gave F1 progeny with wild-type luciferase activity, and F2 populations that segregated for individuals with wild-type luciferase activity and with constitutive luciferase activity. This indicated that the mutants were recessive alleles at different loci that we named cev1 to cev7. cev1 was chosen for further characterization. It was backcrossed to the parental line, and 56 of 261 F2 progeny tested had constitutive luciferase activity (Figure 1A), indicating that cev1 segregated as a single recessive allele (P > 0.1). cev1 plants were smaller and were darker green (Figure 1B), and their roots were shorter with longer root hairs than parental plants (Figures 1C and 1D). The cev1 plants also accumulated purple anthocyanins in their leaves. These morphological traits are characteristic of plants treated with JA (Feys et al., 1994).
Figure 1.
Phenotypes of cev1 Plants.
(A) Low-light images of Col-gl1 and cev1 seedlings containing the VSP1::luciferase reporter transgene. Seedlings were grown for 12 days on MS (Murashige and Skoog, 1962) agar. The figure shows the positions of seedlings (left), and a low-light image of the same seedlings sprayed with luciferin solution reveals luciferase activity (right).
(B) Four-week-old Col-gl1 (left) and cev1 (right) plants grown in soil.
(C) Seven-day-old Col-gl1 and cev1 seedlings germinated on MS medium in the presence (+) and absence (−) of 20 μM MeJA.
(D) Root phenotypes of 7-day-old seedlings of (left to right) Col-gl1 and the mutants cev1, etr1, the double mutant cev1/cev1;etr1/etr1, and cev1 grown in 17 μM Ag+.
cev1 was crossed to Arabidopsis ecotype Landsberg erecta, and the genotypes of cev1 mutants in the F2 progeny were analyzed with cleaved-amplified polymorphic sequence markers and with simple sequence-linked polymorphic markers (Bell and Ecker, 1994). cev1 was mapped genetically to an 11-cM interval at the top of chromosome 5 between nga225 and nga249. This region contains no experimentally characterized gene for response to JA, ethylene, salicylic acid, or for spontaneous lesions. cev1 therefore defines a recessive mutation responsible for the complex mutant phenotype described in Figure 1.
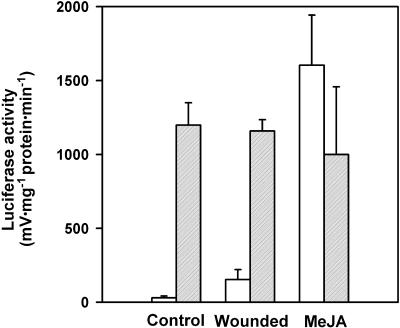
Luciferase Activity Reflects VSP1 Transcript Abundance in Wild-Type and cev1 Plants Containing the VSP::Luciferase Transgene
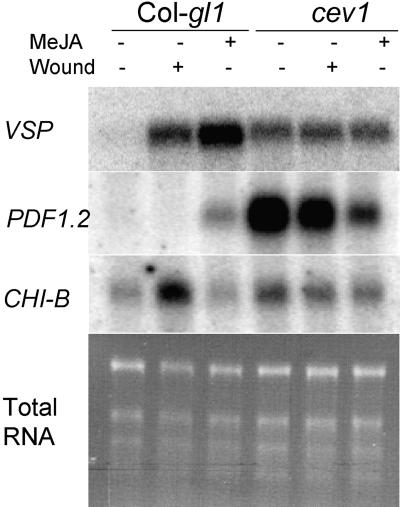
The VSP::luciferase transgene faithfully reported VSP transcription, as monitored by RNA gel blot analysis. In untreated wild-type seedlings, the transgene was not active (Figure 2), and VSP transcripts could not be detected (Figure 3). In seedlings that were wounded or treated with MeJA, however, the transgene was active and VSP transcripts were abundant (Figures 2 and 3), in agreement with previous reports (Staswick et al., 1992; Benedetti et al., 1995). Significantly, in untreated cev1 plants, luciferase activity was high, and VSP transcripts were abundant (Figures 2 and 3), which confirmed that the constitutive luciferase activity was not due to a mutation in the transgene. The VSP1 and VSP2 genes have 87% DNA sequence identity (Utsugi et al., 1996), and the probe used in this study (Benedetti et al., 1995) likely detects both genes.
Figure 2.
VSP1 Promoter Activity in Col-gl1 and cev1 Plants.
Col-gl1 and cev1 plants containing the VSP1::luciferase transgene were harvested and assayed for luciferase activity 12 days after germination. Treated seedlings were wounded with blunt forceps 16 hr before harvest or were transferred to MS plates containing 20 μM MeJA for 48 hr before harvest. Seedlings then were assayed individually for luciferase activity in vitro. Error bars show standard deviations of five replicates for Col-gl1 (white bars) and cev1 (gray bars).
Figure 3.
RNA Gel Blot Analysis of Col-gl1 and cev1 RNA Extracted from 12-Day-Old Seedlings.
Treated seedlings had been wounded with blunt forceps 16 hr before harvest or were transferred to MS plates containing 20 μM MeJA for 48 hr before harvest. Total RNA (0.5 μg) was electropho-resed, blotted, and probed with a 32P-labeled DNA fragment of the VSP gene, the PDF1.2 gene, or the CHI-B gene.
cev1 Seedlings Expressed PDF1.2 and CHI-B Constitutively
RNA gel blot analysis of Col-gl1 seedlings indicated that PDF1.2 was induced by MeJA but not by wounding and that CHI-B was induced by wounding but not by MeJA (Figure 3). In mature Arabidopsis plants, however, wounding does not induce CHI-B (Samac et al., 1990). Untreated cev1 seedlings contained substantially more PDF1.2 mRNA than Col-gl1 plants treated with MeJA. Surprisingly, wounding or treatment with MeJA reduced the amount of PDF1.2 mRNA in cev1 seedlings (Figure 3). Untreated cev1 seedlings also expressed CHI-B constitutively at a level slightly greater than in wild-type plants; however, unlike PDF1.2, this gene was not greatly decreased in plants that were wounded or treated with MeJA.
The VSP1, VSP2, and Thi2.1 Promoters Were Activated Constitutively in the Lamina Tissues of cev1 Plants
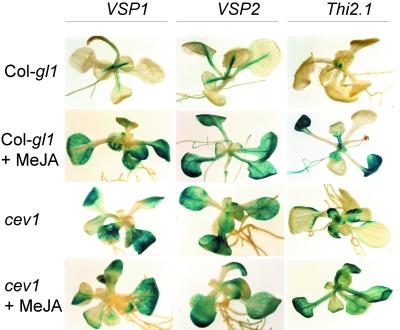
MeJA induces expression of VSP1, VSP2, and Thi2.1 in Arabidopsis (Benedetti et al., 1995; Epple et al., 1995; Vignutelli et al., 1998). To investigate the effect of cev1 on the pattern of expression of these genes, we fused their promoters to β-glucuronidase (GUS) as a reporter, introduced the constructs into Col-gl1 plants by Agrobacterium-mediated transformation, and crossed the transgenes into the cev1 mutant. In untreated Col-gl1 plants, the VSP1::GUS, VSP2::GUS, and Thi2.1::GUS transgenes were expressed only in the apical meristem and the midveins, and treatment with MeJA induced expression in the leaf lamina (Figure 4). In cev1 plants, the GUS transgenes were expressed constitutively in the leaf lamina but not obviously so in the midveins, and their expression was not visibly altered by treatment with MeJA (Figure 4).
Figure 4.
GUS Expression Driven by the VSP1, VSP2, and Thi2.1 Promoters in Col-gl1 and cev1 Seedlings.
Seedlings were grown for 12 days on MS medium or for 10 days on MS medium and transferred to medium containing 20 μM MeJA for 2 days and then stained for GUS activity.
cev1 Has Stunted Roots with Reduced Responsiveness to MeJA
The roots of 10-day-old Col-gl1 seedlings were 33.6 ± 8.2 mm long and were reduced by 74% to 8.9 ± 1.2 mm by growth on MeJA, confirming previous reports (Staswick et al., 1992; Feys et al., 1994). In contrast, the roots of cev1 seedlings were 8.5 ± 1.5 mm long and were reduced by 18% to 7.0 ± 1.0 mm by growth on MeJA (Figure 2C). Thus, cev1 roots were stunted and had reduced sensitivity to inhibition by MeJA.
Part of the cev1 Mutant Phenotype Is Dependent on COI1
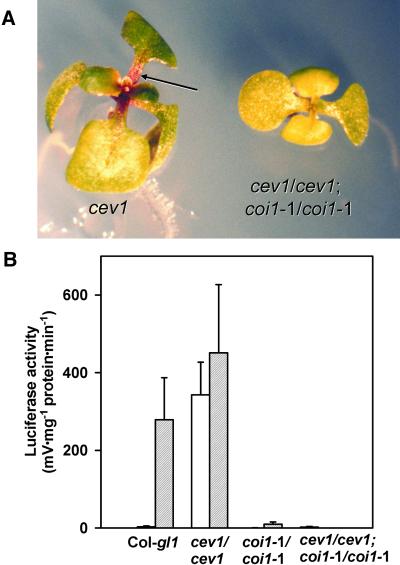
COI1 is required for Arabidopsis responses to JAs (Feys et al., 1994). Therefore, we examined whether COI1 was required for the cev1 phenotype by analysis of the double mutant cev1/cev1;coi1-1/coi1-1. Significantly, the cev1/cev1;coi1-1/coi1-1 double mutants did not accumulate anthocyanin as did cev1 plants (Figure 5A), nor did they express luciferase in the presence or absence of MeJA (Figure 5B). Like cev1 plants, however, the double mutants had short roots with prolific root hairs. Therefore, COI1 was required for cev1-dependent anthocyanin formation and for VSP activity but not for cev1-dependent short roots and prolific long root hairs.
Figure 5.
Effect of the coi1-1 and etr1 Mutations on the cev1 Phenotype.
(A) Ten-day-old seedlings of cev1/cev1 (left) showing anthocyanin in petioles (arrow) and the cev1/cev1; coi1-1/coi1-1 (right) double mutant.
(B) VSP1 promoter activity in wild-type seedlings, the cev1/cev1 and coi1-1/coi1-1 mutants, and the cev1/cev1;coi1-1/coi1-1 double mutant. Ten-day-old seedlings containing the VSP1::luciferase transgene in these mutant backgrounds were transferred to MS medium or MS supplemented with 20 μM MeJA for 2 days. Seedlings were extracted individually, and luciferase activity was measured. Error bars show standard deviations of 10 replicates without MeJA (white bars) and with MeJA (gray bars).
Part of the cev1 Mutant Phenotype Is Dependent on ETR1
The cev1 mutant had thickened roots with prolific root hairs and a thickened hypocotyl (Figure 1). Similar features appear in Arabidopsis seedlings exposed to ethylene during germination. To determine whether ethylene contributed to this phenotype in cev1, we used etr1/etr1 plants, which are insensitive to ethylene (Chang et al., 1993), to make the double mutant cev1/cev1;etr1/etr1. The double mutants were similar to cev1 plants in that they were slightly stunted with small dark-green leaves, but they differed from cev1 plants in that they lacked prolific root hairs, had thinner roots (Figure 1D) and thinner hypocotyls, and had two to five times greater constitutive luciferase activity from the VSP1::luciferase transgene. cev1 seedlings germinated on 17 μM Ag+, which antagonizes the perception of ethylene (Chang et al., 1993), had thin roots, and root hairs were absent (Figure 1D). To test the effect of perturbations to the ethylene signal pathway alone on expression of the VSP:: luciferase transgene, we crossed this construct into the etr1/etr1 and ctr1/ctr1 (which has constitutive ethylene responses; Kieber et al., 1993) mutant backgrounds. Ten-day-old transgenic seedlings were transferred to medium containing 20 μM MeJA for 2 days, and luciferase activity was measured. In the wild type, etr1/etr1, and ctr1/ctr1 backgrounds, luciferase activity was 90 ± 24, 1097 ± 210, and 5 ± 3 mV · mg−1 protein · min−1, respectively. This indicated that activation of the ethylene signal pathway suppressed expression of the VSP::luciferase transgene. Arabidopsis seedlings germinated in the dark in the presence of ethylene have stunted roots with prolific root hairs, a thickened hypocotyl, and an exaggerated apical hook. cev1 seedlings germinated in the dark had stunted roots with prolific root hairs and a thickened hypocotyl, but they lacked an apical hook. We observed that 20 μM JA in the growth medium (Murashige and Skoog [MS] agar) suppressed formation of the exaggerated apical hook in dark-grown ctr1 seedlings, and in dark-grown wild-type seedlings germinated in the presence of 5 parts per million (ppm) ethylene, but not in dark-grown coi1-1 seedlings germinated in the presence of 5 ppm ethylene. Apparently, JA suppressed the ethylene-induced apical hook in a COI1-dependent manner.
cev1 Plants Exhibit Increased Resistance to Three Powdery Mildew Pathogens
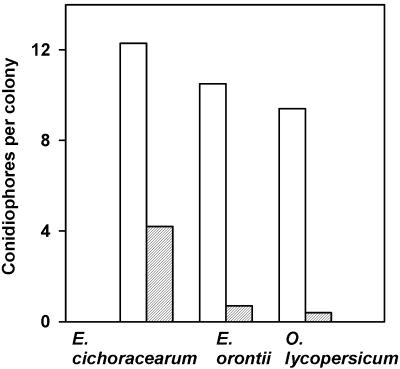
Col-gl1 plants are susceptible to several species of powdery mildew, including Erysiphe cichoracearum UCSC1 (Xiao et al., 1997), Erysiphe orontii MGH, and Oidium lycopersicum Oxford. We examined whether the cev1 mutation affected colonization by these pathogens. Seven days after inoculation, hyphal growth and conidiophore development on Col-gl1 plants were significantly greater than on cev1 plants (Figure 6). Fungal colonies on the cev1 plants were smaller and fewer in number than those on Col-gl1 plants.
Figure 6.
Resistance of cev1 to Powdery Mildews.
Four-week-old Col-gl1 and cev1 plants were inoculated with E. cichoracearum UCSC1, E. orontii MGH, or O. lycopersicum Oxford. After 7 days (for E. cichoracearum) or 5 days (for E. orontii and O. lycopersicum), leaves were fixed and cleared in a lactophenol solution and stained with trypan blue, and the number of conidiophores per colony was recorded. Numbers are the means from 50 colonies for Col-gl1 (white bars) and cev1 (gray bars). An unpaired t test analysis indicated that for each fungus, differences between the means for the Col-gl1 and cev1 plants were statistically significant (P < 0.001).
DISCUSSION
JAs regulate both plant development and defense. Signaling pathways for other plant hormones, including ethylene, auxin, and abscisic acid, have been defined through the characterization of mutants, some of which reveal negative regulators. However, only four JA response mutants have been isolated, on the basis of JA-insensitive root growth, and these are presumed to define positive regulators (Creelman and Mullet, 1997). We sought mutants with constitutive JA responses that, we reasoned, would define negative regulators of the JA pathway, and isolated cev1.
The cev1 mutant was identified by the constitutive expression of a reporter gene controlled by the JA-responsive VSP1 promoter. It also exhibited other phenotypes: the plants were stunted, had short roots with an excess of root hairs, accumulated anthocyanins, and constitutively expressed the JA-regulated genes VSP1, VSP2, and Thi2.1 (Feys et al., 1994; Bohlmann et al., 1998) and the ethylene- and JA-regulated genes PDF1.2 (Penninckx et al., 1998) and CHI-B (Norman-Setterblad et al., 2000). cev1 also altered the tissue-specific pattern of expression of VSP1, VSP2, and Thi2.1 in Arabidopsis. In soybean, VSPs are localized in the vacuoles of paraveinal mesophyll and bundle sheath cells of leaves (Franceschi et al., 1983). Similarly, Col-gl1 plants expressed GUS gene fusions to the VSP1, VSP2, and Thi2.1 promoters only in the leaf midveins and the apical meristem region. However, in Col-gl1 plants treated with MeJA, these promoters were expressed at high levels in the leaf lamina. In untreated cev1 plants, the same GUS transgenes also were expressed at high levels in the leaf lamina, and their activity and pattern of expression were not altered detectably by treatment with MeJA. Therefore, the activity of the VSP1, VSP2, and Thi2.1 promoters in the cev1 mutant resembles their activity in Col-gl1 plants treated with MeJA. This finding indicated that the cev1 mutant had a constitutively active JA signal pathway, and we investigated whether this could account for its other phenotypes.
COI1 is required for Arabidopsis responses to JA, including increased expression of VSP, accumulation of anthocyanin, and inhibition of root growth (Feys et al., 1994; Xie et al., 1998). We tested whether these characteristics in cev1 also required COI1. The cev1/cev1;coi1-1/coi1-1 double mutant had low activity of the VSP1 promoter and reduced anthocyanin formation (Figure 5), indicating that COI1 was required for the constitutive expression of these phenotypes. cev1 therefore regulates a step before COI1 in the JA pathway. However, the double mutant had short roots, indicating that COI1 was not required for this phenotype in cev1. We conclude that there is a constitutively active JA signal pathway in cev1 that causes the enhanced formation of anthocyanin and the constitutive expression of VSP. However, JA alone cannot account for the inhibited root growth in this mutant.
Genetic evidence indicated that the ethylene signal pathway was constitutively expressed in cev1. cev1-dependent prolific root hairs and thickened roots and hypocotyls were suppressed by etr1 in the cev1/cev1;etr1/etr1 double mutant, revealing that the ethylene signal pathway was activated by the cev1 mutation. Roots of the double mutant were shorter than those of wild-type plants, however, indicating that ethylene alone cannot account for this phenotype of cev1. It is possible that JA and ethylene act independently to cause short roots in cev1. The enhanced luciferase activity from the VSP1::luciferase transgene in the cev1/cev1;etr1/etr1 double mutant compared with that of the cev1 mutant indicated that an ethylene-dependent signal suppressed the induction of the VSP promoter by JA. This finding was confirmed by our observation that MeJA-induced activity of the VSP::luciferase transgene in wild-type plants was greater in the etr1 mutant background and suppressed in the ctr1 background, and it supports a previous report that ethylene antagonizes the JA induction of VSP (Rojo et al., 1999). Seedlings germinated in the dark in the presence of ethylene display the triple-response that includes an exaggerated apical hook (Kieber et al., 1993). We observed that cev1 displayed part of the triple response but lacked the apical hook, and that JA suppressed the ethylene-induced apical hook in a COI1-dependent manner. This indicated that the JA signal pathway also suppresses an ethylene-induced response. Previous studies have shown that JA and ethylene jointly regulate transcription of 36 of 2375 genes tested (Schenk et al., 2000). Our results indicate that these signal pathways also have some mutually antagonistic effects.
Expression of defense-related genes was consistent with the activation of the ethylene and JA pathways in cev1. JA and ethylene interact synergistically to induce PDF1.2 (Penninckx et al., 1998) and CHI-B (Norman-Setterblad et al., 2000), which could account for the observed constitutive expression of PDF1.2 and CHI-B in untreated cev1 plants (Figure 3). We conclude that constitutive activation of the JA and ethylene signal pathways in cev1 plants is sufficient to account for the phenotype of these plants, and for the observed pattern of gene induction, and we summarize our findings in a model in Figure 7.
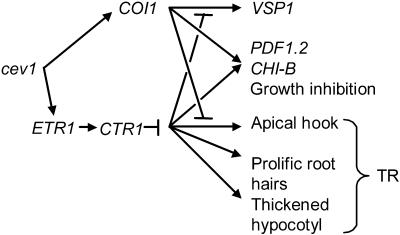
Figure 7.
Model for the cev1 Mutant Phenotype.
The cev1 mutation activates the JA and ethylene signal pathways, possibly by inducing the synthesis of JA and ethylene, or by regulating COI1 and ETR1 directly. The model accounts for the observed cev1 mutant phenotype through independent effects of the activated JA and ethylene signal pathways, and through cross-talk between these pathways causing activation and suppression of different responses. TR indicates triple response, arrows indicate activation, and bars indicate suppression.
JAs are required, alone or in combination with ethylene, for defense against insects and necrotrophic pathogens (McConn et al., 1997; Vijayan et al., 1998; Thomma et al., 2000). We demonstrate that cev1 plants have enhanced resistance to the biotrophic, obligately pathogenic powdery mildew pathogens E. cichoracearum UCSC1, E. orontii MGH, and O. lycopersicum Oxford, to which Col-gl1 plants are normally susceptible (Xiao et al., 1997). Many antifungal agents are synthesized in response to JA, alone or in combination with ethylene, including thionins (Epple et al., 1995) and defensins (Penninckx et al., 1998). Although genes for these proteins are not induced by powdery mildew infection (Reuber et al., 1998), they are constitutively expressed in cev1 plants and therefore could contribute to the observed resistance.
The cev1 mutation causes constitutive activation of the JA and ethylene signal pathways and CEV1, therefore, may function at an early step in these pathways, either as regulator of the levels of JA and ethylene or as regulator of the flux through the JA and ethylene signaling pathways. Its characterization should increase our understanding of early steps in plant wound and defense responses.
METHODS
Construction of Transgenic Lines
The VSP1 promoter was isolated from a λFIX library of Arabidopsis thaliana Landsberg genomic DNA. Polymerase chain reaction (PCR) was used to amplify a 1.5-kb DNA fragment from the 5′ untranslated region of VSP1 using the primers 5′-CTCTCTAGAGGGCGAACT-CGAGCTCC-3′ and 5′-AGGATTTTCATAAGCTTTTGTATGGT-3′. The luciferase gene from pGEMluc (Promega) was inserted into a pBI101 vector (Clontech, Palo Alto, CA) in place of the β-glucuronidase (GUS) gene using the BamHI and SacI restriction endonuclease sites. The VSP1 upstream region was digested with HindIII and inserted 5′ to the luciferase gene. This construct was introduced via electroporation into Agrobacterium tumefaciens GV3101 pMP90, which in turn was used to transform Columbia-glabrous (Col-gl1) plants (Bechtold et al., 1993). A 1.5-kb DNA fragment containing the 5′ untranslated region of VSP2 was isolated via PCR using the primers 5′-CTTCTTAATTAAGCTTATATCTTC-3′ and 5′-GAGGATTTT-CATGGATCCTAATGG-3′. The 5′ untranslated regions of VSP1 and VSP2 also were cloned into the HindIII and the HindIII and BamHI sites of pBI101, respectively. These constructs were introduced into Col-gl1 by Agrobacterium-mediated transformation to create the VSP1::GUS and VSP2::GUS lines. The Thi2.1::GUS line has been described previously (Xie et al., 1998).
Transgenic lines were analyzed by DNA gel blotting to determine the number of copies of the transgenes and by measurement of luciferase activity to determine reporter gene activity. Lines that contained a single copy of the transgene and exhibited a high level of reporter gene expression were chosen for use in this study.
Luciferase and GUS Assays
A. thaliana Col-gl1 seed homozygous for the VSP1::luciferase transgene were mutagenized by immersion in 0.3% ethyl methanesulfonate for 15 hr, M1 plants were raised, and M2 seed were germinated individually in the wells of 96-well microtiter plates containing 150 μL of Murashige and Skoog (1962) (MS) agar. Eight-day-old seedlings were sprayed with 1 mM luciferin in 0.01% Triton X-100, and luminescence was measured in an EG&G Wallac Victor Multilabel Counter (Perkin-Elmer, Boston, MA). In vitro luciferase assays were performed with an LKB Wallac 1251 Luminometer (LKB, Stockholm, Sweden) using the method of Mudge et al. (1996) and luciferase assay reagent (Promega). Low-light images of plants were taken with a liquid nitrogen–cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ) using Metamorph imaging software (Universal Imaging, West Chester, PA). Protein concentration was measured by the Bradford (1976) method. GUS staining was according to Xie et al. (1998).
Isolation of Double Mutants
To isolate double mutants, we crossed cev1 plants homozygous for the VSP1::luciferase transgene to coi1-1 plants also homozygous for the VSP1::luciferase transgene, and F2 progeny plants were scored for male sterility (a marker for coi1-1) and for short roots (a marker for cev1). Of 787 F2 plants examined, 455 had long roots (>15 mm) and were fertile, 153 had long roots and were male sterile, 132 had short roots (<12 mm) and were fertile, and 47 had short roots and were male sterile, conforming to an expected ratio of 9:3:3:1 (P > 0.5) for two independently segregating genes. The presumed double mutants (male sterile and short roots) were confirmed to be coi1-1/coi1-1 by a cleaved-amplified polymorphic sequence marker for the coi1-1 allele (Xie et al., 1998) and were confirmed to be cev1/cev1 by backcrossing to cev1 plants, which gave F1 progeny that were fertile and had short roots.
Approximately one-quarter of the F2 population from the cross cev1/cev1 to etr1/etr1 had small dark-green leaves (a marker for cev1), and the roots of approximately three-quarters of these lacked prolific root hairs (a marker for etr1). This indicated that etr1 abolished the root hair phenotype of cev1 plants and was consistent with etr1 segregating as a dominant mutation. Test backcrosses of these candidate double mutants to their cev1 and etr1 parents identified lines with the double mutation cev1/cev1;etr1/etr1.
RNA Gel Blot Analysis
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Crawley, UK) from 12-day-old seedlings that had been wounded or treated with methyl jasmonate (MeJA), and VSP mRNA was determined by RNA gel blot analysis. RNA gel blotting was performed as described previously (Benedetti et al., 1995). Blots were probed with random primed 32P-labeled DNA fragments (MegaPrime kit; Amersham). The VSP probe has been described previously (Benedetti et al., 1995). The PDF1.2 probe was amplified from genomic DNA by PCR using the primers 5′-GCATGTCGATAGTCCATTACGT-3′ and 5′-ACATGGGACGTAACAGATACAC-3′ (Penninckx et al., 1998). The CHI-B probe was amplified from genomic DNA by PCR using the primers 5′-GATGGGCTACAGCACCAGAC-3′ and 5′-GTAACAATC-AAGATTACCACCAGG-3′ (Samac et al., 1990).
Conidiophore Determinations
Plants were grown for 4 weeks under short-day conditions, and then ∼10 plants were inoculated with powdery mildew as described previously (Xiao et al., 1997). After 7 days (for Erysiphe cichoracearum) or 5 days (for Erysiphe orontii and Oidium lycopersicum), leaves were fixed and cleared in a lactophenol solution and stained with trypan blue (Reuber et al., 1998). The number of conidiophores per colony then was recorded.
Acknowledgments
We thank Shunyuan Xiao for assistance with powdery mildew infections and the Biotechnology and Biological Sciences Research Council for financial support.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316 1194–1199. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Benedetti, C.E., Xie, D., and Turner, J.G. (1995). COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlmann, H., Vignutelli, A., Hilpert, B., Miersch, O., Wasternack, C., and Apel, K. (1998). Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defence thionin, via the octadecanoid pathway. FEBS Lett. 437 281–286. [DOI] [PubMed] [Google Scholar]
- Bradford, M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 355–381. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., Tierney, M.L., and Mullet, J.E. (1992). Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc. Natl. Acad. Sci. USA 89 4938–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol. 109 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi, V.R., Wittenbach, V.A., and Giaquinta, R.T. (1983). Paraveinal mesophyll of soybean leaves in relation to assimilate transfer and compartmentation. III. Immunohistochemical localization of specific glycopeptides in the vacuole after depodding. Plant Physiol. 72 586–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Binkowski, K.A., and Olszewski, N.E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 93 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J., Rothenberg, M., Roman, G., Feldmann, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the ref family of protein kinases. Cell 72 427–441. [DOI] [PubMed] [Google Scholar]
- Kuc, J. (1997). Molecular aspects of plant responses to pathogens. Acta Physiol. Plant. 19 551–559. [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.-C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense. Proc. Natl. Acad. Sci. USA 94 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge, S.R., Lewis-Henderson, W.R., and Birch, R.G. (1996). Comparison of Vibrio and firefly luciferases as reporter gene systems for use in bacteria and plants. Aust. J. Plant Physiol. 23 75–83. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Norman-Setterblad, C., Vidal, S., and Palva, E.T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall–degrading enzymes from Erwinia carotovora. Mol. Plant-Microbe Interact. 4 430–438. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Layser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Metraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Wees, S.C.M., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defence gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16 473–485. [DOI] [PubMed] [Google Scholar]
- Rojo, E., Leon, J., and Sanchez-Serrano, J.J. (1999). Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J. 20 135–142. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac, D.A., Hironaka, C.M., Yallaly, P.E., and Shah, D.M. (1990). Isolation and characterization of genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 93 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Broekaert, W.F., and Cammue, B.P.A. (2000). Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol. Biochem. 38 421–427. [Google Scholar]
- Utsugi, S., Sakamoto, W., Ogura, Y., Murata, M., and Motoyoshi, F. (1996). Isolation and characterization of cDNA clones corresponding to the genes expressed preferentially in floral organs of Arabidopsis thaliana. Plant Mol. Biol. 32 759–765. [DOI] [PubMed] [Google Scholar]
- Vignutelli, A., Wasternack, C., Apel, K., and Bohlmann, H. (1998). Systemic and local induction of an Arabidopsis thionin gene by wounding and pathogens. Plant J. 14 285–295. [DOI] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., Ellwood, S., Findlay, K., Oliver, R.P., and Turner, J.G. (1997). Characterization of three loci controlling resistance of Arabidopsis thaliana accession Ms-0 to two powdery mildew diseases. Plant J. 12 757–768. [DOI] [PubMed] [Google Scholar]
- Xie, D., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Chang, L., Liu, D., Narasimhan, M.L., Raghothama, K.G., Hasewaga, P.M., and Bressan, R.A. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]