Abstract
Using mRNA differential display analysis, we isolated a salt-induced transcript that showed a significant sequence homology with an EREBP/AP2 DNA binding motif from oilseed rape plants. With this cDNA fragment as a probe, cDNA clone Tsi1 (for Tobacco stress-induced gene1) was isolated from a tobacco cDNA library. RNA gel blot analysis indicated that transcripts homologous with Tsi1 were induced not only in NaCl-treated leaves but also in leaves treated with ethephon or salicylic acid. Transient expression analysis using a Tsi1::smGFP fusion gene in BY-2 cells indicated that the Tsi1 protein was targeted to the nucleus. Fusion protein of Tsi1 with GAL4 DNA binding domain strongly activated transcription in yeast, and the transactivating activity was localized to the 13 C-terminal amino acids of Tsi1. Electrophoretic mobility shift assays revealed that Tsi1 could bind specifically to the GCC and the DRE/CRT sequences, although the binding activity to the former was stronger than that to the latter. Furthermore, Agrobacterium-mediated transient expression and transgenic plants expressing Tsi1 demonstrated that overexpression of the Tsi1 gene induced expression of several pathogenesis-related genes under normal conditions, resulting in improved tolerance to salt and pathogens. These results suggest that Tsi1 might be involved as a positive trans-acting factor in two separate signal transduction pathways under abiotic and biotic stress.
INTRODUCTION
Plants respond to adverse environmental stress and pathogen attack by expressing specific genes and synthesizing a large number of stress proteins that have putative roles in stress adaptation and plant defense (Skriver and Mundy, 1990; Reymond and Farmer, 1998). The signals that mediate systemic responses must be transmitted rapidly throughout the plant and may involve cell-to-cell signaling. Putative systemic signals include ethylene (Ecker and Davis, 1987), salicylic acid (Durner et al., 1997), jasmonic acid (Farmer and Ryan, 1990), and abscisic acid (Zeevaart and Creelman, 1988). Communication between these plant hormones might modulate the expression of abiotic and biotic stress–responsive genes in plants. However, the interactions between these hormone-mediated signal pathways and molecular mechanisms governing their cross-regulation have remained generally unresolved.
Ethylene is a phytohormone that affects virtually all stages of plant development, including seed germination, cell elongation, cell fate, fruit ripening, senescence, and abscission. Ethylene also is a key regulator that mediates a plant's response to biotic and abiotic stresses (O'Donnell et al., 1996; Lund et al., 1998). Both ethylene and salicylic acid, a plant defense signal, accumulate in plants during pathogen infection (Penninckx et al., 1996). Among the different classes of ethylene-responsive genes, the most extensively studied are those activated by ethylene in response to pathogen attack. This class includes basic chitinase, β-1,3-glucanases, defensins, and other pathogenesis-related (PR) protein genes (Ohme-Takagi and Shinshi, 1995; Penninckx et al., 1996). Analysis of PR gene promoters has led to the identification of the 11-bp ethylene-responsive element TAAGAGCCGCC, which has been referred to as the GCC box (Ohme-Takagi and Shinshi, 1995). This element was shown to be necessary and sufficient for ethylene regulation in plants. Four ethylene-responsive element binding proteins (EREBPs) have been isolated in tobacco that bind specifically to the GCC box (Ohme-Takagi and Shinshi, 1995), and EREBP RNA levels are upregulated by ethylene, suggesting that EREBP expression also may be induced during the defense response. EREBPs contain a highly conserved novel DNA binding domain consisting of 58 or 59 amino acids (Ohme-Takagi and Shinshi, 1995), and that was found to be related to the AP2 domain (Weigel, 1995).
Levels of endogenous abscisic acid increase when plants are exposed to a saline environment (Downton and Loveys, 1981), water deficit (Zeevaart and Creelman, 1988), or low temperature stress. Also, exogenously applied abscisic acid can elicit the drought response without water deficit (Shinozaki and Yamaguchi-Shinozaki, 1996). From these results, abscisic acid is thought to be involved in osmotic stress signal transduction (Chandler and Robertson, 1994; Giraudat et al., 1994). However, analysis of water stress–inducible gene expression in abscisic acid–deficient (aba) and abscisic acid–insensitive (abi) Arabidopsis mutants showed that the expression of some water stress–inducible genes can act independently of abscisic acid (Thomashow, 1994; Shinozaki and Yamaguchi-Shinozaki, 1996). Promoter studies of some of these genes led to the identification of DRE/CRT, a novel cis-acting element that responds to cold or osmotic stress but not to abscisic acid (Yamaguchi-Shinozaki and Shinozaki, 1994). Recently, several cDNAs for DRE/CRT binding proteins were cloned independently from Arabidopsis by using the yeast one-hybrid screening system (Stockinger et al., 1997; Liu et al., 1998). All of the DRE/CRT binding proteins also contain a conserved EREBP/AP2 DNA binding motif. Ectopic expression of these genes, such as CBF1 or DREB1, in Arabidopsis improved tolerance to water stress (Kirsten et al., 1998; Kasuga et al., 1999).
Many genes that respond to osmotic stress at the transcriptional level have been described (Thomashow, 1994; Shinozaki and Yamaguchi-Shinozaki, 1996). Genes induced under water stress are thought to function not only to protect cells from water deficit by the production of important metabolic proteins but also to regulate the expression of genes required to mediate signal transduction in response to the stress. Several gene products, such as peroxidase, PR1, PR10, and osmotin (PR5), that are typically part of plant defense responses to wound and pathogen attack also are induced by water stress (Zhu et al., 1995; Ingram and Bartels, 1996). However, the roles of these gene products under water stress have not been determined.
In this study, a cDNA was cloned from tobacco that encodes an EREBP/AP2 DNA binding protein. The corresponding gene, designated Tsi1 (for Tobacco stress-induced gene1), was induced by salt, salicylic acid (SA), ethylene, methyl jasmonate, and wound treatments. The function of the Tsi1 protein as a trans-acting factor also was analyzed by using a GAL4 fusion protein in yeast and overexpressing it in transgenic tobacco plants.
RESULTS
Isolation and Sequence Analysis of the Tsi1 cDNA
We isolated several salt-induced cDNA fragments from oilseed rape by mRNA differential display. One of the fragments encoded a putative polypeptide exhibiting significant homology with a DNA binding domain of an EREBP/AP2–type transcription factor. Using this cDNA fragment as a probe, we screened a tobacco cDNA library and isolated a homologous Tsi1 gene. To examine the structure of the Tsi1 cDNA, we sequenced its 1.1-kb insert DNA. The Tsi1 cDNA clone contained a single open reading frame encoding a putative 251–amino acid residue with a predicted molecular mass of 28.6 kD (Figure 1A). Database searches revealed that the deduced amino acid sequence of Tsi1 contained a conserved DNA binding domain of 58 amino acids that is present in a large family of plant DNA binding proteins, including EREBPs of tobacco and AP2 of Arabidopsis. The rest of the amino acid sequences encoded by Tsi1 displayed only limited similarities to the tomato Pti6 (Zhou et al., 1997) and Arabidopsis putative proteins (Figure 1B). Overall, the predicted amino acid sequence of Tsi1 was highly homologous with that of tomato Pti6, with 73.3% identical and 81.3% conserved amino acid residues. The predicted Tsi1 protein contains a basic region in its N-terminal region that might function as a nuclear localization signal. Also, Tsi1 has an acidic C-terminal region that might act as an activation domain for transcription. These data suggest that the Tsi1 cDNA encoding a DNA binding protein might function as a transcriptional activator in plants.
Figure 1.
Analysis of Tobacco cDNA Sequence Encoding a Tsi1 Protein.
(A) Nucleotide and deduced amino acid sequences of the Tsi1 gene. The EREBP/AP2 DNA binding domain is underlined. The basic amino acids that potentially act as a nuclear localization signal are shown in boxes, and an acidic C-terminal region that might act as a transcriptional activation domain is shown in italics. The Tsi1 sequence was submitted to GenBank under the accession number AF058827.
(B) Comparison of the deduced amino acid sequences of Tsi1, Pti6, and a putative Arabidopsis protein (Ara). The amino acid sequence of Tsi1 was aligned with the amino acid sequences of tomato Pti6 (GenBank accession number O04682) and a putative Arabidopsis protein (GenBank accession number T02896). Boxes represent perfectly conserved amino acids, and dashes indicate gaps introduced to maximize alignment.
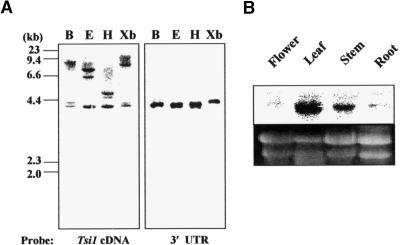
The copy number of the Tsi1 gene in the tobacco genome was estimated by DNA gel blot analysis. Tobacco genomic DNA was digested with various restriction endonucleases, fractionated, transferred, and hybridized with either the full-length cDNA or the 3′ untranslated region (UTR) of Tsi1 as a probe. As shown in Figure 2A, a small number of bands were observed for each DNA digest when the full-length Tsi1 cDNA was used as a probe, indicating that there may be a few Tsi1-related genes in the tobacco genome. When the 3′ UTR sequence of Tsi1 was used as a specific probe, only one band was detected. To monitor the expression pattern of the Tsi1 gene, we isolated total RNAs from different tissues of tobacco plants, such as flowers, stems, leaves, and roots. The RNAs were gel separated and blotted, and the blot was hybridized with the 3′ UTR sequence of Tsi1 as a specific probe. As shown in Figure 2B, transcripts corresponding to the Tsi1 gene were detected abundantly in leaves and weakly in stem tissue. In contrast, the transcripts were almost undetectable in flower and root. This result indicated the tissue specificity of Tsi1 gene expression.
Figure 2.
Genomic DNA Gel Blot Analysis and Organ-Specific Expression of Tsi1 in Tobacco.
(A) DNA gel blot analysis of the Tsi1 gene. Genomic DNA (10 μg) was digested with BamHI (B), EcoRI (E), HindIII (H), and XbaI (Xb) and gel separated. The DNA gel blots were hybridized with the full-length Tsi1 cDNA (left) or a 3′ UTR fragment probe (right).
(B) Organ-specific expression of the Tsi1 gene. In each lane, 20 μg of total RNAs prepared from flowers, roots, leaves, and stems was loaded. The blotted membrane was hybridized with the 3′ UTR fragment probe. Equal amounts of RNA were loaded per lane, as determined by ethidium bromide staining.
Expression Pattern of the Tsi1 Gene under Various Stress Conditions
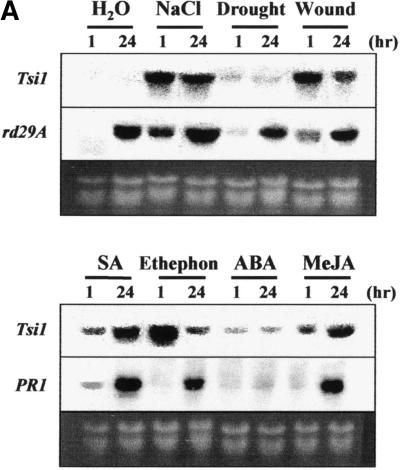
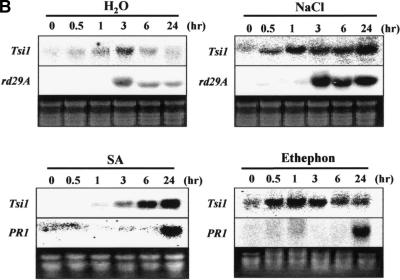
The expression pattern of the Tsi1 gene was analyzed under various abiotic stress conditions by using RNA gel blot analysis. As shown in Figure 3A, Tsi1 gene expression was rapidly induced after salt, wounding, SA, ethephon, or methyl jasmonate treatment. However, there was no significant Tsi1 mRNA accumulation under drought stress or after abscisic acid treatment. Expression of the rd29A and PR1 genes was followed as controls for abiotic and biotic stress, respectively. Expression of Tsi1 in response to salt, SA, and ethephon was further analyzed to investigate time-dependent induction patterns. As shown in Figure 3B, Tsi1 mRNA was accumulated within 30 min under high-salt treatment, and the gene was strongly expressed after 1 hr. When the detached leaves were transferred to water as a control, rather rapid but low-level accumulation of Tsi1 mRNA was detected. Similar results were obtained for the exogenous ethephon and SA treatments. In other words, the Tsi1 gene was induced within 30 min and peaked 1 hr after exposure to 1 mM ethephon. Tsi1 gene expression also was induced by SA treatment within 3 hr, and the gene was strongly expressed after 6 hr. On the other hand, strong expression of the rd29A gene was observed within 3 hr after salt treatment. Expression of PR1 was detected within 24 hr after initiation of ethephon treatment as well as SA treatment.
Figure 3.
Expression of the Tsi1 Gene in Response to Abiotic Stresses.
(A) Induction patterns of the Tsi1 gene after treatment with distilled water (H2O), NaCl, drought, wounding, SA, ethephon, abscisic acid (ABA), or methyl jasmonate (MeJA). Total RNA was prepared from tobacco leaves treated with distilled water, 200 mM NaCl, drought, wounding, 2 mM SA, 1 mM ethephon, 100 μM abscisic acid, or 100 μM methyl jasmonate for 1 or 24 hr as indicated.
(B) Time-course accumulation of Tsi1 transcripts detected upon water, NaCl, SA, or ethephon treatment. Each lane was loaded with 20 μg of total RNA extracted from detached tobacco leaves that had been treated with 200 mM NaCl, 1 mM ethephon, 2 mM SA, or distilled water for the designated times. PR1 and rd29A genes were used as positive controls for the SA, ethephon, or high-salt treatment. Ethidium bromide staining of the RNA gel was used to show equal loading. The RNA gel blots were hybridized with the 3′ flanking sequence of Tsi1 as a specific probe.
Targeting of Tsi1 to the Nucleus
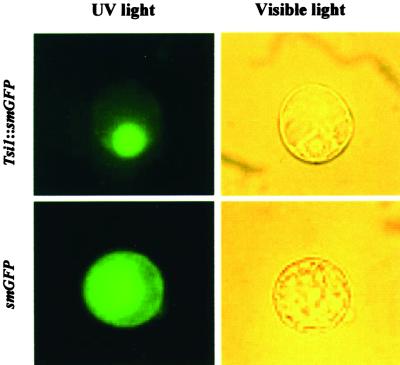
The N-terminal region of the putative Tsi1 protein contained a stretch of basic amino acids, KKRR, that resembled a simian virus 40–type nuclear localization signal (Kalderon et al., 1984). To investigate the cellular distribution of Tsi1, we performed an in vivo targeting experiment by using a Tsi1-fused green fluorescent protein (smGFP) as a fluorescent marker (David and Viestra, 1996). The Tsi1 coding region was fused to smGFP, and the resulting construct was introduced into tobacco BY-2 suspension cultured cells by the polyethylene glycol–mediated transformation method (Abel and Theologis, 1994). Localization of the fusion protein was determined by visualization with a fluorescence microscope. The introduced fused genes that were under the control of the cauliflower mosaic virus 35S promoter were found to be expressed strongly in the tobacco protoplasts. As shown in Figure 4, the fusion protein was localized to the nucleus of the BY-2 cell, whereas the control smGFP was uniformly distributed throughout the cell. These results are consistent with our observation that the Tsi1 sequence encodes a putative transcriptional regulator. They also suggest that the putative nuclear targeting sequence of Tsi1 was sufficient and that no additional post-translational modification was necessary for the Tsi1 protein to be targeted to the nucleus.
Figure 4.
Subcellular Localization of the Tsi1 Gene Product.
The Tsi1 coding region was fused in frame to the smGFP coding region in the plant transformation vector pMBP2. Constructs were introduced into tobacco BY-2 cell protoplasts by polyethylene glycol–mediated transformation. Expression of the introduced genes was examined after 12 hr by fluorescence and light microscopy.
The Tsi1 Protein Has Transcriptional Activation Activity in Yeast
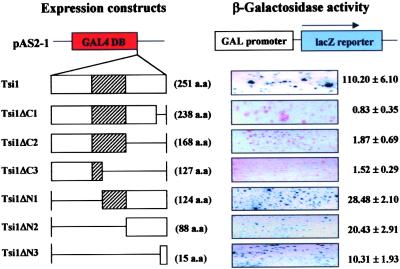
Tsi1 has an acidic C-terminal half (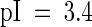 ) that might act as a transcriptional activator domain (Hahn, 1993). To investigate the functional role of this region of Tsi1, we fused the coding regions for Tsi1 and its mutants to the GAL4 DNA binding domain expression vector and examined the behavior of each construct as a potential transcriptional activator in yeast (Figure 5). In the absence of the GAL4 activation domain, the wild-type Tsi1 protein fused to the GAL4 DNA binding domain activated transcription of the lacZ reporter gene. This result indicates that the Tsi1 protein is capable of functioning as a transcriptional activator in yeast. To identify a minimal transcriptional activation domain of Tsi1, various deletion mutants of Tsi1 also were tested in the same manner. The Tsi1ΔC1, Tsi1ΔC2, and Tsi1ΔC3 mutants, which lacked 13, 83, and 124 C-terminal amino acids, respectively, demonstrated complete absence of β-galactosidase activity (Figure 5), indicating that the 13 C-terminal amino acids of Tsi1 play an important role in supporting the ability of Tsi1 as a potential transcriptional activator.
) that might act as a transcriptional activator domain (Hahn, 1993). To investigate the functional role of this region of Tsi1, we fused the coding regions for Tsi1 and its mutants to the GAL4 DNA binding domain expression vector and examined the behavior of each construct as a potential transcriptional activator in yeast (Figure 5). In the absence of the GAL4 activation domain, the wild-type Tsi1 protein fused to the GAL4 DNA binding domain activated transcription of the lacZ reporter gene. This result indicates that the Tsi1 protein is capable of functioning as a transcriptional activator in yeast. To identify a minimal transcriptional activation domain of Tsi1, various deletion mutants of Tsi1 also were tested in the same manner. The Tsi1ΔC1, Tsi1ΔC2, and Tsi1ΔC3 mutants, which lacked 13, 83, and 124 C-terminal amino acids, respectively, demonstrated complete absence of β-galactosidase activity (Figure 5), indicating that the 13 C-terminal amino acids of Tsi1 play an important role in supporting the ability of Tsi1 as a potential transcriptional activator.
Figure 5.
Transcriptional Activation Activity of Tsi1 in Yeast Cells.
The Tsi1 and Tsi1 deletion constructs were fused in frame to the GAL4 DNA binding domain (DB) expression vector and then transformed into yeast strain Y190. The transformants were selected by growth on Trp− synthetic dropout medium at 30°C for 3 days. The colony lift filter assay and liquid culture assay using o-nitrophenyl β-d-galactopyranoside as a substrate were subsequently performed to determine the ability of each translation product to activate transcription. Hatched boxes represent the EREBP/AP2 DNA binding domain. a.a., amino acids.
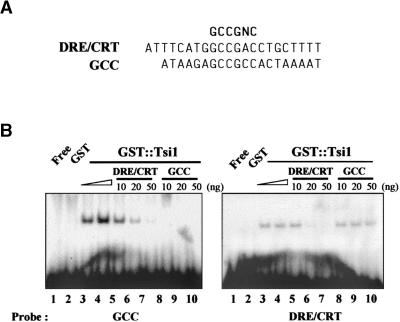
Tsi1 Binds Specifically to the GCC and DRE/CRT Sequences
It has been shown that the EREBP and DREB proteins, both of which contain the EREBP/AP2 DNA binding motif, bind to the GCC and DRE/CRT boxes, respectively (Büttner and Singh, 1997; Stockinger et al., 1997). To test the binding activity of Tsi1 to these cis-acting elements, we produced a recombinant Tsi1 protein as a fusion to glutathione S-transferase (GST) in Escherichia coli. The ability of the recombinant Tsi1 fusion protein to bind the cis-acting element was tested using a gel mobility shift assay. As shown in Figure 6, Tsi1 interacted with both the GCC and DRE/CRT boxes. The DNA binding specificity of Tsi1 also was confirmed by competition experiments (Figure 6). The specific interaction between Tsi1 and GCC box was effectively competed by 20 ng of unlabeled DRE/CRT sequences. Also, the native GCC box fragment inhibited the formation of the DRE/CRT–Tsi1 binding complex. In contrast, mutated GCC box (ATAAGATCC-TCCACTAAAAT; Büttner and Singh, 1997) or mutated DRE/CRT box (GATATACTATTTACATGAGTTCC; Liu et al., 1998) did not affect DNA binding activity of Tsi1 for GCC or DRE/CRT sequences, respectively (data not shown). When the binding intensities of Tsi1 to the GCC and DRE/CRT sequences were compared, the binding to the GCC box was stronger than that to the DRE/CRT box. These results suggest that Tsi1 may require post-translational modification or a putative cofactor for effective binding to the DRE/CRT sequence. In tomato, the interaction of the Pto kinase with Pti4/5/6 suggested that the phosphorylation of Pti4/5/6 and EREBPs may be required for their in vivo activity (Zhou et al., 1997). However, the binding of DREBs and CBF1 to the DRE/CRT box does not appear to require any post-translational modification, as shown by the DNA binding activity of the E. coli–expressed proteins (Stockinger et al. 1997; Liu et al., 1998).
Figure 6.
Characterization of the DNA Binding Affinity of the Recombinant Tsi1 Protein.
(A) Sequence of the oligonucleotides used in the DNA binding studies.
(B) Gel retardation assay showing sequence-specific binding of the recombinant Tsi1 protein. The radiolabeled probes were incubated in the presence or absence of the recombinant Tsi1 protein. Lane 1 contained only the free probes, and lane 2 contained GST only. Lane 3 contained 1 μg of bacterial extracts, and lane 2 and lanes 4 to 10 contained 2 μg of bacterial extracts. For competition assays, 2 μg of E. coli extract containing the Tsi1 protein was incubated with either the DRE/CRT or GCC oligonucleotide before the addition of the GCC or DRE/CRT probe. The amount of competitor DNA is indicated at the top of each lane.
Induction of PR Genes in Tobacco by Tsi1 Expression
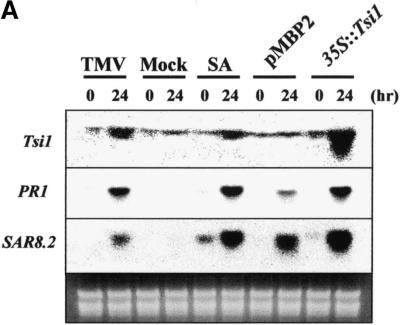
The GCC box, a DNA sequence element present in the upstream regulatory sequences of many ethylene-responsive genes, also is found in the corresponding sequences of PR genes, such as PR1, β-1,3-glucanase (PR2), chitinase (PR3), and osmotin (PR5) (Ohme-Takagi and Shinshi, 1995; Yang et al., 1997). As a means to analyze the role(s) of Tsi1 in vivo, the cDNA for Tsi1 was introduced into tobacco plants using the Agrobacterium-mediated transient expression method (Abel and Theologis, 1994). For this purpose, the entire coding sequence of Tsi1 cDNA was subcloned into a plant expression vector under the control of the cauliflower mosaic virus 35S double promoter. The resulting construct was transformed into Agrobacterium and subsequently infiltrated into tobacco leaves. Total RNA was isolated from the leaves after 24 hr of infiltration, and the effects of Tsi1 expression were monitored by RNA gel blot analysis. As shown in Figure 7A, transient expression of Tsi1 resulted in the accumulation not only of its own mRNA but also of the transcripts corresponding to PR1 and SAR8.2. The expression levels of these genes in response to Tsi1 expression were similar to those induced by SA or tobacco mosaic virus (TMV) treatment. Although Agrobacterium infiltration of unfused control plasmid also induced these genes, the levels of expression were substantially lower than those from the expression of Tsi1. These results demonstrate that transient expression of Tsi1 encoding a putative GCC box binding transcription factor effectively upregulated the expression of the PR1 and SAR8.2 genes. Apparently, the transient expression of Tsi1 in tobacco plants elicited cellular responses that might be functionally equivalent to those of a hormone (SA) or a pathogen (TMV) treatment.
Figure 7.
Transient and Stable Overexpression of the Tsi1 Gene.
(A) Induction of the PR1 and SAR8.2 genes in tobacco by Tsi1 expression. Seven-week-old soil-grown wild-type tobacco (cv Samsun NN) was infiltrated with Agrobacterium (strain LBA4404) bearing the expression plasmids pMBP2 and 35S::Tsi1 or treated with TMV and 1 mM SA. Plants were harvested at 0 or 24 hr after treatment, and RNA gel blots were hybridized with the probes indicated at left. Mock-inoculated plants were rubbed with phosphate buffer and carborundum only.
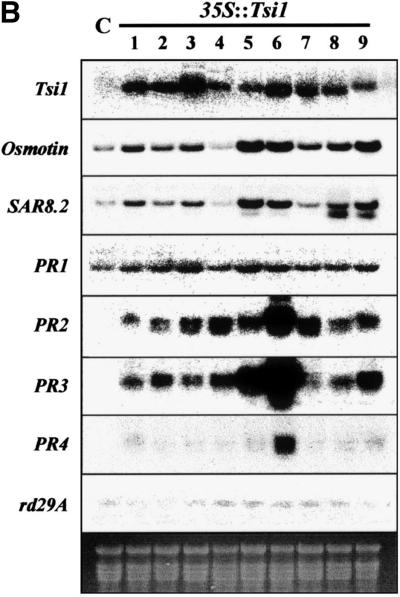
(B) Expression of Tsi1 target genes in 35S::Tsi1 transgenic plants and in a control plant under normal conditions. Total RNA was isolated from the plants and analyzed by RNA gel blot hybridization using the 3′ UTR Tsi1 cDNA as a specific probe; numbers indicate independent lines of transgenic T0 plants. RNA gel blotting also was conducted to measure the amount of osmotin, SAR8.2, PR1, PR2, PR3, PR4, or rd29A mRNA in control and transgenic tobacco plants carrying 35S::Tsi1. C indicates a pMBP2-transformed tobacco T0 plant; 1 to 9 indicate Tsi1-transformed independent tobacco T0 lines.
To understand the biological function of Tsi1 as a transcription factor in planta, we generated transgenic plants expressing the Tsi1 gene. Tobacco plants were transformed with binary vector carrying a fusion of the double 35S promoter and Tsi1 cDNA in the sense orientation by the Agrobacterium-mediated transformation method (Hoekema et al., 1983). Primary transformants were selected based on resistance against kanamycin. Polymerase chain reaction (PCR) analysis was performed subsequently using a pair of oligonucleotide primers specific for the neomycin phosphotransferase (nptII) gene. This analysis confirmed that ∼75% of the transformants were clearly positive and contained the transgene sequence. Transgenic plants were regenerated from these verified transformants and used for further analyses. The expression of Tsi1 and its possible target genes was analyzed in 35S::Tsi1 plants and compared with that of control plants transformed with pMBP2 (Han et al., 1999) (Figure 7B). The levels of Tsi1 mRNA were higher in 35S::Tsi1 plants than in the control plants. The expression of PR1, PR2, PR3, osmotin, and SAR8.2 genes was high in approximately eight of nine unstressed 35S::Tsi1 plants but low in the unstressed control plants. However, we found no difference in the expression of rd29A and PR4 between the 35S::Tsi1 and control plants except for induction of PR4 in one transgenic line (line 6). The levels of target gene expression under unstressed control conditions were approximately correlated with the levels of the Tsi1 transcripts in 35S::Tsi1 plants.
Tsi1 Overexpression Enhances Salt Tolerance and Resistance to Pathogens
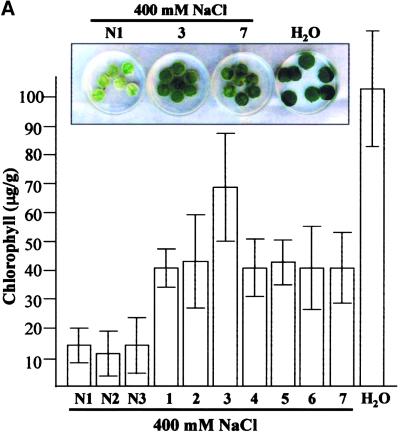
In this study, several PR genes that are characteristically transcribed in response to the invasion of pathogens were induced in tobacco leaves transiently expressing the Tsi1 gene. Stimulation of PR gene expression also has been demonstrated in transgenic tobacco plants overexpressing the Tsi1 gene. These observations strongly suggest that Tsi1 regulated the expression of a number of PR genes that are normally upregulated in response to not only pathogen attack but also osmotic stress. At first, to investigate whether the accumulation of these PR genes in plants enhances salt tolerance, we floated leaf discs of transgenic tobacco plants in NaCl solution for 72 hr and examined these by comparing phenotype and chlorophyll contents in transgenic and wild-type tobacco plants. Figure 8A shows the phenotypic differences among the wild-type and the sense transgenic plants after 72 hr of salt treatment. Measurement of the chlorophyll content in these plants after 48 hr of salt treatment further confirmed the observed phenotypic differences in the treated leaf discs (Figure 8A). These results provide evidence for a positive relationship between the expression of Tsi1 and tolerance to salt stress.
Figure 8.
Analysis of Salt-Induced Senescence and Disease Resistance of Tsi1 Transgenic Plants.
(A) Retardation of salt-induced senescence in transgenic tobacco plants. Leaf discs from the transgenic plants carrying the Tsi1 gene in the sense orientation (bars 1 to 7) and the wild-type tobacco plants (bars N1 to N3) were floated in 400 mM NaCl solution for 48 or 72 hr under continuous white light at 25°C. As a control, the wild-type leaf discs were floated in water (H2O). Phenotypic differences were observed (72 hr), and chlorophyll contents (mg/g fresh weight) were measured (48 hr) from NaCl-treated leaf discs of 35S::Tsi1 transgenic plants and wild-type tobacco. The experiments were repeated three times, each with seven leaf discs.
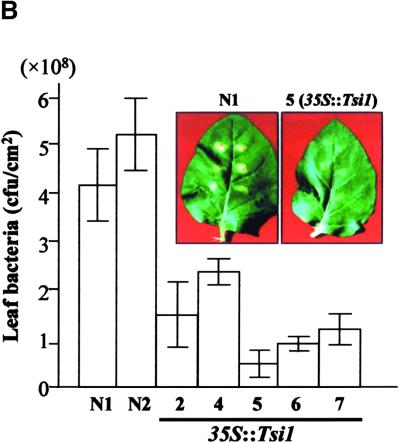
(B) Analysis of disease resistance against the bacterial pathogen P. s. tabaci in wild-type and 35S::Tsi1 transgenic plants. Disease symptoms caused by P. s. tabaci in wild-type and Tsi1 transgenic plants are shown in the inset. The photograph was taken 7 days after inoculation. The growth of P. s. tabaci strains in wild-type and 35S::Tsi1 transgenic plants was compared as follows. Fully expanded leaves of 7-week-old tobacco plants were inoculated with 107 colony-forming units (cfu)/mL of a P. s. tabaci strain. At 7 days after inoculation, the infected leaves were collected and the bacterial populations were determined. Bars N1 and N2 indicate wild-type tobacco, and bars 2 and 4 to 7 indicate Tsi1-transformed independent tobacco T0 lines. Values are means of three different experiments.
Error bars indicate ±se.
To determine whether overexpression of the Tsi1 gene enhanced resistance to pathogens, we inoculated wild-type and transgenic plants with Pseudomonas syringae pv tabaci, a virulent bacterial pathogen. As shown in Figure 8B, inhibition of pathogen growth was observed in 35S::Tsi1 plants. Seven days after inoculation, growth of the bacteria in the 35S::Tsi1 plants was inhibited threefold to fivefold compared with that in the wild-type plants. Although several lesions developed on leaves of 35S::Tsi1 plants, the size and density of the lesions were much smaller than those on wild-type plants. The enhanced resistance to P. s. tabaci in transgenic plants demonstrates that disease resistance conferred by Tsi1 overexpression is effective against bacterial pathogens.
DISCUSSION
Using a salt-induced oilseed rape cDNA fragment as a probe, we isolated a cDNA clone designated Tsi1 from a tobacco cDNA library. The Tsi1 encoded a DNA binding protein that interacted specifically with the GCC and DRE/CRT sequences, which are involved in defense- and drought-responsive gene expression, respectively. Overexpressed Tsi1 protein was demonstrated to function as a potential DNA binding protein in gel mobility shift assays (Figure 6). In vivo, the Tsi1 protein also displayed an ability to activate transcription, as shown by yeast one-hybrid analysis (Figure 5). Furthermore, both transient and stable expression of the Tsi1 gene in tobacco leaves and transgenic plants directly induced the expression of several PR genes (Figure 7). These results strongly suggest that the Tsi1 protein is involved in GCC- and/or DRE/CRT–dependent gene expression.
The Tsi1 protein contains a highly conserved EREBP/AP2–type DNA binding domain, but this domain is the only part of the protein that exhibits significant sequence homology with many known proteins. The EREBP/AP2 domain was first identified in the APETALA2 protein. The APETALA2 gene of Arabidopsis controls important processes during flower development (Jofuku et al., 1994). Recently, the EREBP/AP2 DNA binding motif has been found in various plant regulatory genes, such as Arabidopsis TINY (Wilson et al., 1996), CBF1 (Stockinger et al., 1997), AtEBP (Büttner and Singh, 1997), DREBPs (Liu et al., 1998), AINTEGUMENTA (Elliott et al., 1996; Klucher et al., 1996), maize Glossy15 (Moose and Sisco, 1996), and tomato Ptis (Zhou et al., 1997). These genes are divided into two classes based on the number of EREBP/AP2 motifs. One class includes AP2, AINTEGUMENTA, and Glossy15, each of which encodes a protein containing two EREBP/AP2 motifs. The other class includes EREBPs, TINY, CBF1, Ptis, AtEBP, DREBPs, and Tsi1, each of which encodes a protein with only one EREBP/AP2 motif. The EREBPs, Ptis, and AtEBP in the second class bind specifically to the GCC box sequence containing the core GCCGCC sequence, which is present in the promoter region of a large number of ethylene-inducible genes encoding PR proteins (Ohme-Takagi and Shinshi, 1995). The DREB1A, DREB2A, and CBF1 proteins bind specifically to the DRE/CRT sequence containing the core sequence C/GCCGAC. These sequences resemble the GCC box and contain CCGNC as a common core sequence. In this study, gel mobility shift assays showed that Tsi1 could bind to either the GCC box or the DRE/CRT box. However, the binding activity of Tsi1 to the GCC box was stronger than that to the DRE/CRT box. This is a novel observation, because other GCC box binding proteins, such as EREBPs, AtEBP, and Ptis, have not been tested for their ability to interact with the DRE/CRT box. These results suggested that Tsi1 could induce the expressions of either GCC or DRE/CRT box containing genes on their own promoter. However, transgenic plants overexpressing Tsi1 did not show expression of rd29A, which contained DRE/CRT box on its promoter region, under normal growth condition. There are at least two possibilities to explain this result. One is that the weak DNA binding activity of Tsi1 to DRE/CRT sequence is not enough to turn on rd29A gene expression. The other is that Tsi1 does not have a function for rd29A gene expres-sion. However, rd29A mRNA also did not accumulate in 35S::DREB2A transgenic plants under normal conditions although GST-DREB2A strongly bound to DRE/CRT sequences in vitro (Liu et al., 1998). It has been suggested that interaction between different transcription factors enhances binding to a specific cis-element; for example, OBP1 specifically increased the binding of the OBF proteins to ocs element sequences (Chen et al., 1996). Also, many experiments have shown that certain types of transcription factors required cofactor(s) or post-translational modification for their activation potential and DNA binding properties in plants (Despres et al., 2000; Gu et al., 2000). It is thus possible that Tsi1 may require post-translational modification or a putative cofactor for its function in response to osmotic stress.
The EREBP/AP2 motif contains an 18–amino acid core region that has been proposed to form an amphipathic α-helix; such a secondary structure has been implicated in protein–protein interactions to facilitate DNA binding (Okamuro et al., 1997). However, the present binding studies cannot determine whether Tsi1 binds to the GCC box or the DRE/CRT box as a monomer or some form of multimer.
Ethylene and SA are important inducers of defense-related genes in plants. Several PR proteins, such as PR1, PR2, PR4, and PR5, are expressed in an SA- and/or ethylene-dependent manner. In this study, the expression of Tsi1 was induced not only in NaCl-treated leaves but also in leaves treated with ethephon or SA. These results indicate that the Tsi1 gene is likely to be regulated by signaling pathways that are activated by high-salt stress, ethylene, and SA. It has been proposed that biotic and abiotic stresses induce overlapping sets of genes in plants (Zhu et al., 1995; Ingram and Bartels, 1996). This has led to the hypothesis that the biotic and abiotic signal pathways may interact to activate or repress biotic and abiotic response genes in plants. Cross-coupling of transcription factors is thought to play an important role in mediating responses to various signaling events; in animals, for example, cross-coupling is important in modulating the transcriptional activity of members of the steroid receptor family and Jun/Fos proteins (Schule and Evans, 1991). It will be important to determine whether Tsi1 and/or related EREBP proteins can interact with other transcription factors that are involved in distinct signal transduction pathways.
Results from the transient and stable expression of Tsi1 in tobacco leaves and transgenic plants strongly suggest that the Tsi1 protein is involved in the expression of several PR genes. It has been shown that PR gene expression is activated by a number of biotic or abiotic stresses, including infection by viral, bacterial, or fungal pathogens and treatment with plant hormones and elicitors such as ethylene, SA, jasmonic acid, H2O2, and UV light (Bol et al., 1990). Although the role of the PR proteins in plant disease resistance has not been established, their enzymatic functions indicate that they are well suited for defense against pathogens. However, the role of these PR genes under abiotic stress needs to be investigated (Zhu et al., 1995; Ingram and Bartels, 1996).
Transgenic tobacco plants carrying the 35S::Tsi1 fusion gene showed tolerance to high-salt stress and bacterial inoculation. This may be due to the increased expression of stress-inducible genes induced by the overexpression of Tsi1. The abundance of Tsi1 transcriptional activator most likely upregulated the transcription of several stress-inducible genes, so that their protein products could contribute to the increased levels of endurance under the stress conditions. It has been reported that DREB1A overexpression also enhances tolerance to drought, salt loading, and freezing (Kasuga et al., 1999). DREB1A interacts specifically with DRE/CRT and induces the expression of stress-tolerance genes, including rd29A, kin1, cor6.6, cor15a, and rd22. Overexpression of Tsi1 in transgenic tobacco activated the expression of the several PR genes, such as PR1, PR2, PR3, osmotin, and SAR8.2, under normal unstressed conditions.
It is a common observation that certain transcription factors serve as targets for different signaling pathways. For instance, the extensively studied animal transcription factor CREB (for cAMP response element binding protein) is phosphorylated and activated by protein kinases from a number of different signaling pathways (Hill and Treisman, 1995). Although Tsi1 binds to both DRE/CRT and GCC sequences, transgenic plants expressing Tsi1 induced only PR genes that contained the GCC box on their promoter region. These results suggest that the Tsi1 protein might have a role, as a positive transcription factor, in the induction of GCC gene transcripts in salt and pathogen signal pathways. However, they do not exclude the possibility that Tsi1 also regulates the expression of DRE/CRT genes in transmitting distinct sets of intracellular signals in plants. Further analyses of transgenic tobacco plants expressing Tsi1 will help to elucidate the role of Tsi1 or its gene product in PR gene regulation and in response to salinity.
METHODS
Plant Materials and Chemical Treatments
Tobacco plants (Nicotiana tabacum cv Samsun NN and cv Xanthi) were grown in soil in a growth chamber under a 16-hr-light/8-hr-dark photoperiod. Plants were treated and examined at 7 weeks after seed germination. For treatment with abscisic acid, salicylic acid (SA), ethephon, methyl jasmonate, and salts, detached tobacco leaves were placed in Falcon tubes filled with 100 μM abscisic acid, 2 mM SA, 1 mM ethephon, 100 μM methyl jasmonate, or 200 mM NaCl for various times and then frozen in liquid nitrogen for further analyses.
Viral Inoculation
Tobacco mosaic virus (TMV) strains were maintained in infected leaves of tobacco (cv Samsun) on a CaCl2-containing Petri dish. Plant saps containing TMV were prepared by grinding infected leaves in phosphate buffer (0.25 M Na2HPO4 and 5 mM EDTA). To inoculate plants, saps were applied to the surface of two or three fully expanded young leaves of tobacco seedlings (8- to 10-leaf stage) and rubbed with carborundum (Hayashi Chemical, Osaka, Japan). Mock-inoculated plants were rubbed with phosphate buffer and carborundum only.
cDNA Library Screening and DNA Sequence Analysis
A cDNA library was constructed from RNA extracted from tobacco leaves. Approximately 2.5 × 105 clones were screened from the cDNA library by using a 32P-labeled partial cDNA fragment obtained previously from oilseed rape as a probe. Hybridization was performed according to a published protocol (Church and Gilbert, 1984). After hybridization, the membranes were washed with 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) at room temperature for 10 min twice, with 0.1 × SSC containing 0.1% SDS at room temperature for 10 min, and with 0.1 × SSC containing 0.1% SDS at 65°C for 5 min. Filters were exposed to x-ray film with an intensifying screen at −70°C for 48 hr. Positive clones were excised with the helper phage and recircularized to generate a subclone in the pBluescript SK− phagemid vector (Stratagene, La Jolla, CA). The entire nucleotide sequence of the cDNA was determined by the dideoxy chain termination method (Sanger et al., 1977), using a sequencing kit (Amersham, Little Chalfont, UK). DNA sequence data were assembled and analyzed using the DNASTAR and Vector NTI analysis programs (InforMax, North Bethesda, MD). Database searches were performed with the National Center for Biotechnology Information (Bethesda, MD) BLAST search program.
DNA and RNA Analyses
Genomic DNA was isolated from 4-week-old tobacco leaves by the method of Ausubel et al. (1995). To prepare a DNA gel blot, 10 μg of genomic DNA was digested with 50 units of restriction enzymes (BamHI, EcoRI, HindIII, and XbaI) overnight. DNA fragments were separated by electrophoresis overnight on a 0.8% agarose gel in 1 × TBE buffer (45 mM Tris-borate and 1 mM EDTA) (Sambrook et al., 1989) and transferred to a Nytran Plus membrane (Schleicher and Schuell, Dassel, Germany) in 10 × SSC.
Total RNA was prepared from 4-week-old leaves as described by Ausubel et al. (1995). Twenty micrograms of total RNA was electrophoresed on a 1.5% agarose gel containing 6% formaldehyde in 3-(N-morpholino)-propanesulfonic acid buffer, pH 7.0, and transferred to a Nytran Plus membrane. Hybridization was performed with a 32P-labeled DNA probe according to a published protocol (Church and Gilbert, 1984), and the membranes were washed with 2 × SSC at room temperature for 10 min twice, with 0.1 × SSC containing 0.1% SDS at room temperature for 10 min, and with 0.1 × SSC containing 0.1% SDS at 65°C for 5 min. The membranes were dried and exposed to x-ray film or visualized directly with the BAS-2500 Phosphorimager (Fuji Photo Film, Tokyo, Japan).
For the control rd29A probe, a 503-bp fragment of Arabidopsis thaliana rd29A cDNA (GenBank accession number D13044) was amplified by polymerase chain reaction (PCR). Two specific primers, 5′-GACTCCGGTCAATGAGAAGGATCAA-3′ and 5′-ACAACAGTGGAG-CCAAGTGATTGT-3′, were designed based on the sequence in the database. Other control probes (PR1, PR2, PR3, PR4, osmotin, and SAR8.2 cDNA clones isolated previously from tobacco [Kang et al., 1998; Park et al., 1999]), were radiolabeled by PCR.
Construction of Various Tsi1 Deletion Mutants and Transactivation Activity Assays
The deletion mutant genes were generated using various restriction sites within the Tsi1 cDNA or by PCR. The plasmids pTsi1ΔC1 and pTsi1ΔC3 encode Tsi1 proteins with deletion of 13 and 124 C-terminal amino acids, respectively. They were constructed using internal restriction sites (EcoRI for pTsi1ΔC1 and SmaI for pTsi1ΔC3) in pTsi1. The same two restriction sites were used to construct the plasmids pTsi1ΔN1 and pTsi1ΔN3, which encode Tsi1 proteins with deletion of 127 and 236 N-terminal amino acids, respectively. The two other Tsi1 deletion derivatives, pTsi1ΔC2 and pTsi1ΔN2, were constructed by PCR using the synthetic oligonucleotide primers ΔC2 (5′-CAAGCTTCCGTTAATACTTTCGG-3′) and ΔN2 (5′-CGGATCCTATTAACGGCGGATATAAC-3′) in combination with the T7 (5′-AATACGACTCACTATAG-3′) and T3 (5′-ATTAACCCTCACTA AAG-3′) universal primers, respectively.
Coding regions for the deleted polypeptides were fused in frame to the yeast GAL4 DNA binding domain vector pAS2-1. The fusion plasmids were transformed into Saccharomyces cerevisiae strain Y190 (MATα, HIS3, lacZ, trp1, leu2, cyhr2) as described by the manufacturer (Clontech, Palo Alto, CA). The transformants were selected by growth on Trp− synthetic dropout medium at 30°C for 3 days. The colony lift filter assay and liquid culture assay using o-nitrophenyl β-d-galactopyranoside as a substrate were performed subsequently to determine the ability of each translation product to activate transcription.
Subcellular Localization of Tsi1
The termination codon of the Tsi1 cDNA was removed after PCR using a specific oligonucleotide primer, Tsi1-3′ (5′-ATGGATCCTCGAGCTTCGAGAGCAAA-3′) and the T3 universal primer of pBluescript SK− (5′-ATTAACCCTCACTAAAG-3′). The resulting fragment was fused in frame to the coding region of smGFP (David and Viestra, 1996). Transient expression of green fluorescent protein (GFP) fusion constructs was performed by introducing the DNAs into the BY-2 tobacco protoplasts using the polyethylene glycol–mediated transformation method (Abel and Theologis, 1994). The fluorescence photographs of protoplasts were taken using a Zeiss (Jena, Germany) Axiophot fluorescence microscope fitted with fluorescein isothiocyanate filters (excitation filter, 450 to 490 nm; emission filter, 520 nm; dichroic mirror, 510 nm) and Fuji 400 color film. The optimal exposure time was 1 sec.
Bacterial Expression of Tsi1 and Gel Mobility Shift Assays
A Tsi1 cDNA fragment was prepared by PCR and cloned into the NdeI site of the pGEX-3X vector (Amersham). The primer set used for the amplification was Tsi1-5′ (5′-GCGGATCCATGATAAGAAATCCAGTC-3′) and the T7 universal primer of pBluescript SK− (5′-AATACGACTCACTATAG-3′). The resulting in-frame fusion plasmid was transformed into Escherichia coli strain BL21(DE3). Overexpression of GST::Tsi1 was induced by 0.4 mM isopropyl-β-d-thiogalactoside at 30°C for 3 hr. Bacteria were pelleted after the induction, suspended in lysis buffer (50 mM Na2HPO4, pH 7.4, 300 mM NaCl, 10% glycerol, and 1% Triton X-100), and subjected to sonication on ice for 1 min with a Vibracell sonifier (Sonics and Materials, Danbury, CT). Bacterial lysates were then centrifuged, and the supernatant was used for gel mobility shift assays. The binding site DNAs used in gel mobility shift assays were derived from cloned oligonucleotides. The wild-type GCC box (GGATCCATAAGAGCCGCCACTAAA-ATAAGCTT) (Ohme-Takagi and Shinshi, 1995) and the DRE/CRT box (GGATCCATTTCATGGCCGACCTGCTTTTAAGCTTTTAAGCTT) (Stockinger et al., 1997) were synthesized and then subcloned into BamHI-HindIII sites of pBluescript SK− vector. BamHI-HindIII fragments were excised from plasmids and end labeled with α-32P-dATP as described (Hong et al., 1995). The assay mixtures contained E. coli extract (2 μg of protein), 0.5 ng of binding probe (2 × 104 cpm), 2 μg of poly(dI-dC)·(dI-dC), 20 mM Hepes-KOH, pH 7.9, 50 mM KCl, 0.1 mM EDTA, 15% glycerol, and 0.5 mM DTT in a 10-μL reaction volume. The mixtures were incubated at room temperature for 15 min and electrophoresed on a 5% polyacrylamide gel in 0.5 × TBE buffer. Subsequently, the gel was dried and exposed to x-ray film.
Agrobacterium tumefaciens–Mediated Transient Expression of Tsi1
A. tumefaciens–mediated transient expression assays were performed as described by Hoekema et al. (1983). The DNA fragment containing the Tsi1 coding region was cloned into polylinker sites of the plant expression vector pMBP2 under the control of the cauliflower mosaic virus 35S double promoter. Agrobacterium strain LBA4404 bearing the expression plasmids was grown to saturation in Luria-Bertani medium. After centrifugation, the cells were resuspended in infiltration buffer (60 mM sucrose, 55.5 mM glucose, 0.2 mM acetosyringone, and 20 mM Mes, pH 5.4) at a concentration of 1.0 OD600. This suspension was pressure infiltrated into leaves using a syringe.
Analysis of Transgenic Plants Overexpressing Tsi1
Plasmids used in the transformation of tobacco were constructed with Tsi1 full-length cDNA that was cloned into a polylinker site of a binary vector, pMBP2, in the sense orientation. After pMBP2 was digested with BamHI and ligated with the BamHI fragment of the Tsi1 cDNA, the orientation of the recombinant plasmids was determined by EcoRI digestion. The constructs were introduced into tobacco cv Xanthi using Agrobacterium-mediated transformation as described (Hoekema et al., 1983).
Healthy and fully expanded leaves from wild-type and transgenic plants were detached and washed briefly in distilled water. Leaf discs of 1 cm diameter were cut and floated in Murashige and Skoog (1962) medium containing 400 mM NaCl for 48 or 72 hr (Veena et al., 1999). The treatment was performed in continuous white light at 25°C. The chlorophyll content was measured as described by Aono et al. (1993).
Cells of Pseudomonas syringae pv tabaci strains were grown in King's medium B (Martin et al., 1993) and infiltrated by the method of Thilmony et al. (1995). The bacterial culture was washed and resuspended in 10 mM MgCl2. Bacterial density was determined by absorbance at OD600. Bacterial suspensions were infiltrated into fully expanded tobacco leaves using a 10-mL plastic syringe without a needle. Approximately 50 μL of inoculum was infiltrated per panel, forming an infiltrated area of ∼20 × 20 mm. The bacterial population of the leaves was measured by grinding four leaf discs in 10 mM MgCl2, plating serial dilutions on King's medium B plates, and counting colony-forming units.
Acknowledgments
We acknowledge Dr. Doil Choi (Korea Research Institute of Bioscience and Biotechnology, Taejon, Korea) for the gift of tobacco PR probes. We also thank Dr. Sangduk Kim for helpful discussion and comments on the manuscript. This work was funded by a G7 grant from the Korea Ministry of Science and Technology and grants from the Korea Science and Engineering Foundation to the Center for Plant Molecular Genetics and Breeding Research and the Research Center for the Development of Advanced Horticultural Technology.
References
- Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 5 421–427. [DOI] [PubMed] [Google Scholar]
- Aono, M., Kubo, A., Saji, H., Tanaka, K., and Kondo, N. (1993). Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant Cell Physiol. 34 129–136. [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Short Protocols in Molecular Biology. (New York: Wiley).
- Bol, J.F., Linthorst, H.J.M., and Cornelissen, B.J.C. (1990). Plant pathogenesis-related proteins induced by virus infection. Annu. Rev. Phytopathol. 28 113–138. [Google Scholar]
- Büttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP) and ethylene-inducible, GCC box DNA-binding protein interact with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., and Robertson, M. (1994). Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 113–141. [Google Scholar]
- Chen, W., Chao, G., and Singh, K.B. (1996). The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 10 955–966. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, S.J., and Viestra, R.D. (1996). Soluble derivatives of green fluorescent protein (GFP) for use in Arabidopsis thaliana. Weeds World 3 43–48. [Google Scholar]
- Despres, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. [PMC free article] [PubMed] [Google Scholar]
- Downton, W.J.S., and Loveys, B.R. (1981). Abscisic acid content and osmotic relations of salt-stressed grapevine leaves. Aust. J. Plant Physiol. 8 443–453. [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2 266–274. [Google Scholar]
- Ecker, J., and Davis, R.W. (1987). Plant defense genes are regulated by ethylene. Proc. Natl. Acad. Sci. USA 84 5202–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q.J., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1990). Interplant communication: Airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl. Acad. Sci. USA 87 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Parcy, F., Bertandre, N., Gosti, F., Leung, J., Morris, P.C., Bouvier-Durand, M., and Vartanian, N. (1994). Current advances in abscisic acid action and signaling. Plant Mol. Biol. 26 1557–1577. [DOI] [PubMed] [Google Scholar]
- Gu, Y.-Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, S. (1993). Structure(?) and function of acidic transcription activators. Cell 72 481–483. [DOI] [PubMed] [Google Scholar]
- Han, S.J., Cho, H.S., You, J.S., Nam, Y.W., Park, E.K., Shin, J.S., Park, Y.I., Park, W.M., and Paek, K.H. (1999). Gene silencing-mediated resistance in transgenic tobacco plants carrying potato virus Y coat protein gene. Mol. Cells 9 376–383. [PubMed] [Google Scholar]
- Hill, C.S., and Treisman, R. (1995). Transcriptional regulation by extracellular signals: Mechanisms and specificity. Cell 80 199–211. [DOI] [PubMed] [Google Scholar]
- Hoekema, A., Hirsch, P., Hooykaas, P.J.J., and Schilperoort, R. (1983). A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303 179–180. [Google Scholar]
- Hong, J.C., Cheong, Y.H., Nagao, R.T., Bahk, J.D., Key, J.L., and Cho, M.J. (1995). Isolation of two soybean G-box binding factors which interact with a G-box sequence of an auxin-responsive gene. Plant J. 8 199–211. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., den Boer, B.G.W., Van Montagu, M., and Okamura, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., Richardson, W.D., Markham, A.F., and Smith, A.E. (1984). Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311 33–38. [DOI] [PubMed] [Google Scholar]
- Kang, M.K., Park, K.S., and Choi, D. (1998). Coordinated expression of defense-related genes by TMV infection or salicylic acid treatment in tobacco. Mol. Cells 8 388–392. [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. [DOI] [PubMed] [Google Scholar]
- Kirsten, R., Jaglo-Ottosen, S.J., Gilmour, D.G., Zarka, O.S., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. [DOI] [PubMed] [Google Scholar]
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, S.T., Stall, R.E., and Klee, H.J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262 1432–1436. [DOI] [PubMed] [Google Scholar]
- Moose, S.P., and Sisco, P.H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10 3018–3027. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- O'Donnell, P.J., Calvert, C., Atzorn, R., Wasternack, C., Leyser, H.M.O., and Bowles, D.J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274 1914–1917. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, K.S., Suh, M.C., Cheong, J.J., and Choi, D. (1999). Isolation of defense-related genes from Nicotiana glutinosa infected by tobacco mosaic virus using a modified differential screening. Plant Pathol. J. 15 295–301. [Google Scholar]
- Penninckx, I.A., Eggermont, K., Terras, F.R., Thomma, B.P., De Samblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1 404–411. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule, R., and Evans, R.M. (1991). Cross-coupling of signal transduction pathways: Zinc finger meets leucine zipper. Trends Genet. 7 377–381. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1996). Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 7 161–167. [DOI] [PubMed] [Google Scholar]
- Skriver, K., and Mundy, J. (1990). Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain–containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony, R.L., Chen, Z., Bressan, R.A., and Martin, G.B. (1995). Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv tabaci expressing avrPto. Plant Cell 7 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1994). Arabidopsis thaliana as a model for studying mechanisms of plant cold tolerance. In Arabidopsis, E. Meyerowitz and C. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 807–834.
- Veena, Reddy, V.S., and Sopory, S.K. (1999). Glyoxylase I from Brassica juncea: Molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 17 385–395. [DOI] [PubMed] [Google Scholar]
- Weigel, D. (1995). The APETALATA2 domain is related to a novel type of DNA binding domain. Plant Cell 7 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11 1621–1639. [DOI] [PubMed] [Google Scholar]
- Zeevaart, J.A.D., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 439–473. [Google Scholar]
- Zhou, J., Tang, X., and Martin, G. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16 3207–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B., Chen, T.H., and Li, P.H. (1995). Activation of two osmotin-like protein genes by abiotic stimuli and fungal pathogen in transgenic potato plants. Plant Physiol. 108 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]