Abstract
Some plant cytoplasms express novel mitochondrial genes that cause male sterility. Nuclear genes that disrupt the accumulation of the corresponding mitochondrial gene products can restore fertility to such plants. The Texas (T) cytoplasm mitochondrial genome of maize expresses a novel protein, URF13, which is necessary for T cytoplasm–induced male sterility. Working in concert, functional alleles of two nuclear genes, rf1 and rf2, can restore fertility to T cytoplasm plants. Rf1 alleles, but not Rf2 alleles, reduce the accumulation of URF13. Hence, Rf2 differs from typical nuclear restorers in that it does not alter the accumulation of the mitochondrial protein necessary for T cytoplasm–induced male sterility. This study established that the rf2 gene encodes a soluble protein that accumulates in the mitochondrial matrix. Three independent lines of evidence establish that the RF2 protein is an aldehyde dehydrogenase (ALDH). The finding that T cytoplasm plants that are homozygous for the rf2-R213 allele are male sterile but accumulate normal amounts of RF2 protein that lacks normal mitochondrial (mt) ALDH activity provides strong evidence that rf2-encoded mtALDH activity is required to restore male fertility to T cytoplasm maize. Detailed genetic analyses have established that the rf2 gene also is required for anther development in normal cytoplasm maize. Hence, it appears that the rf2 gene was recruited recently to function as a nuclear restorer. ALDHs typically have very broad substrate specificities. Indeed, the RF2 protein is capable of oxidizing at least three aldehydes. Hence, the specific metabolic pathway(s) within which the rf2-encoded mtALDH acts remains to be discovered.
INTRODUCTION
Maternally inherited cytoplasmic male sterility (CMS) occurs in many plant species and is widely used to facilitate the production of hybrid seed because it eliminates the need for emasculation by hand. Mitochondrial defects account for all instances in which the nature of the lesion responsible for CMS has been identified (reviewed in Mackenzie et al., 1994; Schnable and Wise, 1998). In many species, the deleterious effects of these mitochondrial defects can be avoided or overcome by the action of nuclear genes, termed nuclear restorers. However, the specific mechanisms by which restoration can occur are only poorly understood.
The male-sterile Texas (T) cytoplasm (cms-T) was used to produce ∼85% of U.S. hybrid maize seed until the 1970 epidemic of southern corn leaf blight (Ullstrup, 1972; Pring and Lonsdale, 1989). cms-T maize is highly sensitive to a host-selective toxin (T toxin) produced by race T of Cochliobolus heterostrophus, the causal organism of southern corn leaf blight (Hooker et al., 1970; Comstock and Scheffer, 1973; Yoder, 1973). The genomes of T cytoplasm mitochondria contain a unique mitochondrial gene, urf13, which encodes the URF13 protein. URF13 accumulates in the inner membrane of the mitochondria (Forde and Leaver, 1980; Dewey et al., 1986; Wise et al., 1987a; Hack et al., 1991; Korth et al., 1991; Levings and Siedow, 1992) and is believed to be responsible for both the sensitivity to T toxin and the male sterility of cms-T maize (reviewed in Levings, 1990, 1993; Wise et al., 1999). The URF13 protein accumulates in many tissues of cms-T maize plants. However, in the absence of T toxin, the only severely affected tissue is the tapetal cell layer of the anthers, which undergoes a premature degeneration at the early microspore stage, resulting in pollen abortion (Warmke and Lee, 1977).
The combined action of two dominant alleles of two nuclear genes, rf1 and rf2, restores the male fertility of cms-T maize (reviewed in Wise et al., 1999). Neither of these restorers alters the sensitivity of cms-T maize to T toxin. The function of the rf1 gene in restoration relates to its ability to modify the expression of urf13, thereby reducing the accumulation of URF13 (Dewey et al., 1987; Kennell and Pring, 1989).
As a first step toward determining its function in fertility restoration, the rf2 gene was cloned via transposon tagging (Cui et al., 1996). The rf2-encoded protein, RF2, contains a predicted mitochondrial targeting sequence and exhibits ∼60% identity and 75% similarity to class II mammalian mitochondrial aldehyde dehydrogenases (mtALDHs). ALDHs are a family of NAD(P)+-dependent enzymes that catalyze the oxidation of numerous aldehydes (reviewed in Lindahl, 1992; Yoshida et al., 1998). A large number of ALDHs have been characterized in mammals, yeast, insects, and bacteria (reviewed in Perozich et al., 1999). In mammals and yeast, the class I (a cytosolic form) and class II (a mitochondrial form) isozymes have been particularly well characterized (Lindahl, 1992; Wang et al., 1998). According to recently revised nomenclature (Vasiliou et al., 1999), the rf2 gene is equivalent to ALDH2B1. Only a few studies of plant class II mtALDHs have been reported (Asker and Davies, 1985; Osakovskii et al., 1992; op den Camp and Kuhlemeier, 1997). Although plant betaine ALDHs have been subjected to fairly intensive study (Vojtechova et al., 1997), these enzymes are only distantly related to class II mtALDHs.
The primary functions of ALDHs are believed to be the detoxification of ethanol-derived acetaldehyde and the oxidization of aldehydes derived from biogenic polyamines (Lindahl and Petersen, 1991). On the basis of the presence of indoleacetaldehyde dehydrogenase activity in cell-free extracts from mung bean seedlings (Wightman and Cohen, 1968), it has been suggested that ALDHs may be involved in the production of the plant hormone indole-3-acetyl acetate (Marumo, 1986). However, this biochemical reaction also can be catalyzed by an aldehyde oxidase (Rajagopal, 1971).
Most maize inbred lines carry functional Rf2 alleles, even though they have never been exposed to T cytoplasm. This suggests that the RF2 protein has an important physiological role other than restoring male fertility to plants that carry T cytoplasm (Schnable and Wise, 1994). In this report, we demonstrate that the RF2 protein is, as predicted (Cui et al., 1996), an mtALDH that accumulates in most organs. In addition, we demonstrate that this mtALDH activity is required for normal anther development not only in T cytoplasm maize but also in normal (N) cytoplasm maize.
RESULTS
RF2 Accumulation in Maize
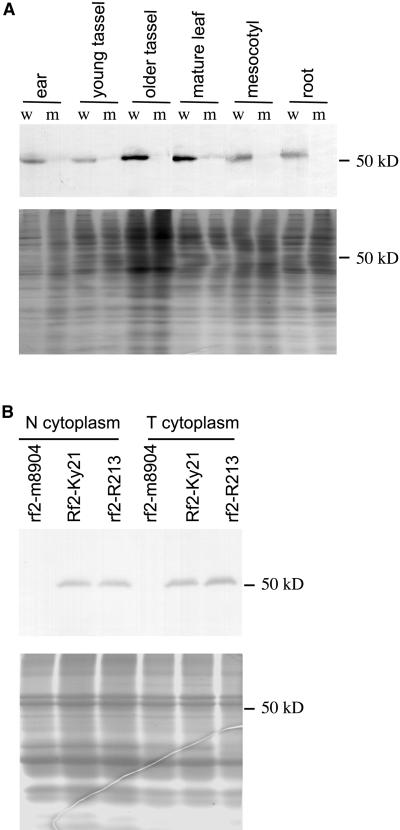
The rf2 cDNA was cloned into the expression vector pET-30b, and the resulting plasmid, pLB333, was expressed in Escherichia coli. The His-tagged recombinant RF2 protein was purified to homogeneity and used as an antigen to inject rabbits. Affinity-purified anti-RF2 polyclonal antibodies were used for immunoblot analyses. These analyses revealed that the RF2 protein accumulates in most organs examined, including seedling and mature leaves, mesocotyls, roots, tassels, ears, and stems (Figure 1A). However, the levels of RF2 accumulation varied among these organs, and it was not detectable in mature dried kernels (data not shown). A weak hybridization signal can be detected in protein preparations from some, but not all, tissues from plants homozygous for the rf2-m8904 allele, suggesting either that this allele is not completely null or that the purified antibody can recognize a closely related protein. There is no significant difference in the accumulation of RF2 protein between seedlings that carry N or T cytoplasm (Figure 1B), nor is there a significant difference in RF2 protein accumulation between seedlings homozygous for rf1 or Rf1 (data not shown). The inbred line R213 is homozygous for the rf2 reference allele (rf2-R213). Even though this allele does not restore male fertility to T cytoplasm plants, the inbred line R213 accumulates RF2 protein at levels indistinguishable from those seen in the inbred line Ky21 (Figure 1B).
Figure 1.
Immunoblot Analysis of RF2 Protein Accumulation.
(A) Different organs from mutant and wild-type maize plants. w, Rf2-Ky21/Rf2-Ky21; m, rf2-m8904/rf2-m8904. Both maize strains carried T cytoplasm and had been backcrossed to the inbred line Ky21 for three generations. The lower panel is a duplicated gel stained with Coomassie Blue and serves as a protein loading control. Ear, a young unpollinated ear ∼10 cm long; young tassel, premeiotic stage of microsporogenesis; older tassel, early microspore stage of microsporogenesis; mesocotyl and root, both from 7-day-old seedlings.
(B) Seedlings of different cytoplasmic and nuclear backgrounds. Total protein extracts were prepared from 7-day-old seedlings homozygous for the indicated alleles in the indicated cytoplasmic background. Rf2-Ky21 and rf2-m8904 are in a Ky21 genetic background, and the rf2-R213 allele is carried by the inbred line R213 with the indicated cytoplasm. The lower panel is a duplicated gel stained with Coomassie blue and serves as a protein loading control.
RF2 Is a Mitochondrial Protein
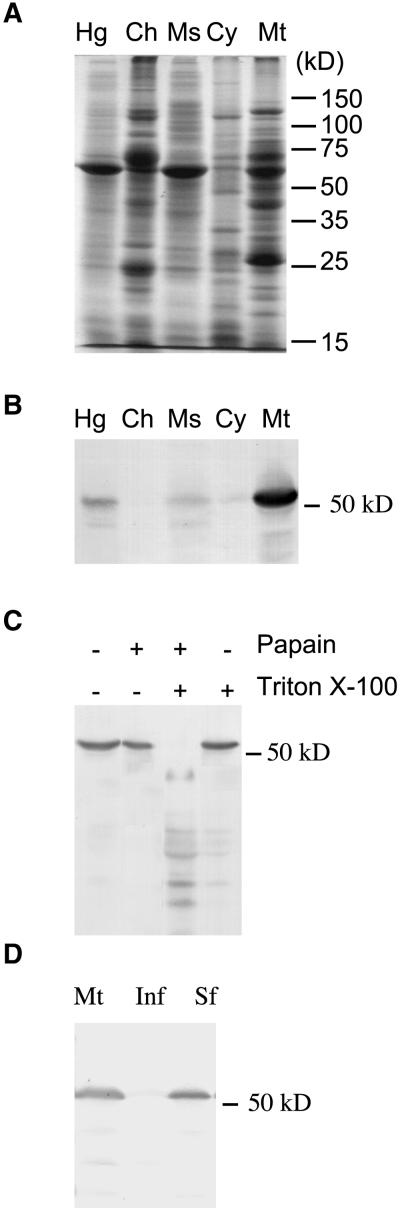
Various computational analyses (Nakai, 1991) of the predicted RF2 protein suggest that it is targeted to mitochondria. To determine the subcellular localization of the RF2 protein, we prepared chloroplastic, microsomal, cytosolic, and mitochondrial fractions by differential centrifugation or by Percoll density centrifugation. Chlorophyll content and catalase, alcohol dehydrogenase, and cytochrome c oxidase activities were used as markers for these four fractions. These assays demonstrated that the mitochondrial fraction was not significantly contaminated by proteins from other organelles or the cytosol (Table 1). Immunoblot analyses revealed that the RF2 protein is present primarily in the mitochondrial fraction (Figures 2A and 2B). Furthermore, when isolated mitochondria were incubated with papain, in the presence of Triton X-100, the RF2 protein was digested; in the absence of Triton X-100, the RF2 protein was resistant to papain digestion (Figure 2C). This result indicates that the RF2 protein is located within the mitochondrion and not simply associated with its outer surface. Purified mitochondria were further fractionated into soluble and insoluble fractions. Cytochrome c oxidase (Errede et al., 1967) and malate dehydrogenase (Rocha and Ting, 1970) activities were used as markers for mitochondria membrane and matrix proteins, respectively. More than 98% of the cytochrome c oxidase activity was found in the insoluble fraction, whereas malate dehydrogenase activity was found exclusively in the soluble fraction. These results indicate that the insoluble fraction contains mitochondrial membrane proteins and that it has no detectable level of contamination by matrix proteins. Similarly, the soluble fraction contains mitochondrial matrix proteins and is not significantly contaminated by mitochondrial membrane proteins. Immunoblot analyses of these soluble and insoluble mitochondrial fractions revealed that RF2 is a matrix protein (Figure 2D).
Table 1.
Markers for Cell Fractionationa
| Chloroplastic Fraction
|
Microsomal, Cytosolic, and Mitochondrial Fractions
|
|||||
|---|---|---|---|---|---|---|
| Marker | Totalb | Chloroplastic | Totalc | Microsomal | Cytosolic | Mitochondrial |
| COXd | 0.392 ± 0.032e | 0 | 0.480 ± 0.018 | 0.348 ± 0.048 | 0.072 ± 0.003 | 2.712 ± 0.084 |
| ADHd | NDf | ND | 0.290 ± 0.013 | 0.011 ± 0.007 | 0.526 ± 0.017 | 0 |
| Catalased | 0.742 ± 0.013 | 0 | 1.231 ± 0.132 | 8.743 ± 0.520 | 0.650 ± 0.100 | 0.339 ± 0.015 |
| Chlorophyllg | 2.069 ± 0.126 | 10.396 ± 0.039 | ND | ND | ND | ND |
Data represent the average of three measurements from one typical experiment.
The homogenate of green seedlings that was used for the isolation of the chloroplastic fraction.
The homogenate of etiolated seedlings that was used for the isolation of microsomal, cytosolic and mitochondrial fractions.
The enzyme activities are expressed as arbitrary units per minute per milligram of protein. COX, cytochrome c oxidase; ADH, alcohol dehydrogenase.
Standard deviation.
ND, not determined.
Chlorophyll concentration is expressed as milligrams of chlorophyll per milligram of protein.
Figure 2.
RF2 Protein Accumulates in Mitochondria.
(A) Homogenate (Hg), purified chloroplastic (Ch), microsomal (Ms), cytosolic (Cy), and mitochondrial (Mt) fractions of Ky21 seedlings were subjected to SDS-PAGE and stained with Coomassie blue.
(B) A duplicate gel was transferred to a nitrocellulose membrane and reacted with affinity-purified anti-RF2 antibodies.
(C) Protein extracts from isolated mitochondria of the inbred line Ky21 were treated with Triton X-100 and papain as indicated, subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and reacted with affinity-purified anti-RF2 antibodies.
(D) Purified mitochondria from Ky21 seedlings were subjected to sonication and centrifugation to isolate the soluble and membrane fractions and then subjected to immunoblot analysis. Mt, purified mitochondria; Inf, insoluble fraction; Sf, soluble fraction.
Assuming that the translation of the RF2 protein begins at the first methionine codon in the cDNA, its predicted molecular mass would be 59.4 kD. However, the RF2 protein contains a predicted mitochondrial targeting sequence (MARRAASSLVSRCLLARAPAGAPPAAPSAPRRTVPADGM-HRLLPGVLQRFS) whose predicted cleavage site is Leu42 or Ser51 (underlined). If the predicted mitochondrial targeting sequence is cleaved at one of these sites, the molecular mass of the mitochondrial RF2 protein would be 55.3 or 54.1 kD, respectively. On the basis of SDS-PAGE analysis, the RF2 protein isolated from mitochondria was estimated to be 53.2 kD (data not shown). These data demonstrate that the RF2 protein is translated as a precursor that is processed after import into the matrix space. These data are consistent with either Leu42 or Ser51 being the cleavage site, because SDS-PAGE analysis cannot provide the level of resolution required to determine the actual cleavage site. Hence, it would be necessary to conduct N-terminal sequencing to determine the actual cleavage site.
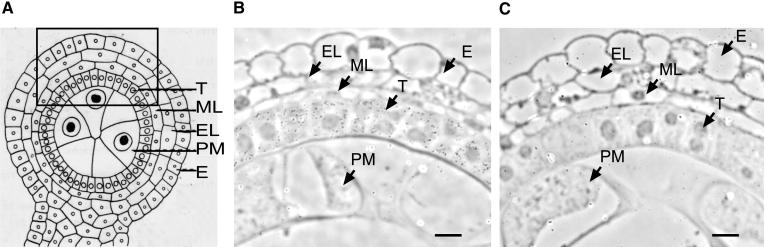
Immunolocalization experiments revealed that accumulation of the RF2 protein is substantially greater in the cells of the tapetum than in other cell layers of the anther wall during the premeiosis stage (Figure 3). This result is consistent with the massive accumulation of mitochondria that occurs in the tapetal cells between the premeiotic and tetrad stages of microsporogenesis (Warmke and Lee, 1978). This result also suggests that during the premeiosis stage, the amount of RF2 protein per mitochondrion may be higher in tapetal cells than in other cells. This hypothesis is based on the observation that little RF2 signal was observed in pollen mother cells, even though the number of mitochondria per unit volume in pollen mother cells is approximately twofold greater than that in tapetal cells during this stage (Warmke and Lee, 1978).
Figure 3.
Immunolocalization of the RF2 Protein.
(A) Diagram of a cross-section of one locule of an anther (from Kiesselbach, 1980). The approximate position of the areas shown in (B) and (C) is indicated by the box.
(B) Cross-section of a T cytoplasm anther of the inbred line Ky21 that is homozygous for the Rf2-Ky21 allele incubated with affinity-purified rabbit anti-RF2 antibodies, followed by gold-labeled goat anti-rabbit IgG antibodies and silver enhancement.
(C) Cross-section of an anther from T cytoplasm rf2-m8904/rf2-m8904 treated as described for (B).
E, epidermis; EL, endothecium; ML, middle layer; PM, microspore pollen mother cell; T, tapetum. 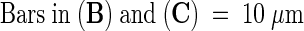 .
.
Recombinant RF2 Exhibits ALDH Activity
The predicted RF2 protein exhibits a high level of sequence similarity to ALDHs; however, the His-tagged recombinant RF2 protein produced by pLB333 did not exhibit ALDH activity (data not shown). This construct included a His tag, an S tag, and the predicted maize mitochondrial targeting sequence, any of which might interfere with ALDH activity. Hence, the rf2 cDNA was cloned into another expression vector, pET-17b, which does not include a His tag or an S tag, in such a way that the putative maize mitochondrial targeting sequence was removed and only two vector-derived amino acids were added in the N terminus of the RF2 protein. The resulting plasmid, pMAP11, was transformed into the E. coli strain BL21(DE3). Expression of RF2 was induced by isopropylthio-β-galactoside (IPTG) and examined via immunoblot analysis. Although most of the recombinant RF2 protein produced by these cells accumulated in inclusion bodies, the soluble fraction of the crude cellular extract accumulated levels of RF2 protein detectable via immunoblot analyses (data not shown).
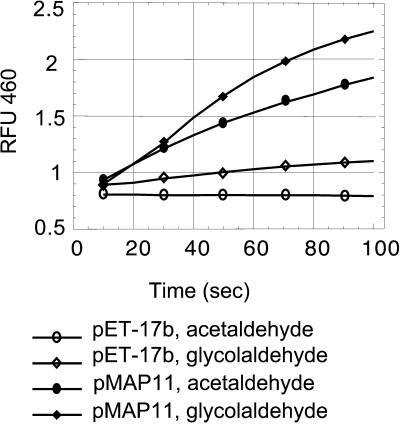
This soluble fraction was subjected to assays for ALDH activity in which the reduction of NAD+ to NADH was monitored with a fluorescence spectrophotometer at 460 nm. Acetaldehye and glycolaldehyde were selected for these initial enzyme assays because acetaldehyde is a substrate in the fermentation pathway, which we have hypothesized may be involved in fertility restoration (Cui et al., 1996). Glycolaldehyde is structurally similar to lactaldehyde, which is not commercially available. Lactaldehyde is the in vivo substrate for the ALDH that is mutated in the JA111(DE3) strain of E. coli. When acetaldehyde was used as a substrate, ALDH activity was detected in extracts from BL21(DE3)pMAP11 (Figure 4). Control assays using extracts from BL21(DE3)pET-17b showed no ALDH activity (Figure 4). When glycolaldehyde was used as a substrate, ALDH activity was detected in extracts from both BL21(DE3)pMAP11 and BL21(DE3)pET-17b cells. This finding indicates that the BL21(DE3) strain contains endogenous glycolaldehyde dehydrogenase activity. However, the level of glycolaldehyde dehydrogenase activity was substantially greater in extracts from BL21(DE3)pMAP11 cells (Figure 4). These data demonstrate that the RF2 protein has both acetaldehyde and glycolaldehyde dehydrogenase activities.
Figure 4.
ALDH Assay of E. coli–Expressed Recombinant RF2.
Crude extracts of E. coli expressing the RF2 protein were assayed for ALDH activity. RFU 460, relative fluorescence units at 460 nm.
The rf2 Gene Can Complement an E. coli ALDH Mutant
The E. coli strain 3 (Caballero et al., 1983) can grow on media in which 1,2-propanediol is the sole carbon source. To use this carbon source, strain 3 must first oxidize 1,2-propanediol to l-lactaldehyde, which is then further oxidized to l-lactate by an ALDH. l-Lactate is subsequently converted to pyruvate, which can enter central metabolism (Boronat and Aguilar, 1979). E. coli strain JA111 differs from strain 3 in that it carries a mutation in the ald gene that encodes an ALDH capable of oxidizing l-lactaldehyde to l-lactate. Hence, unlike strain 3, strain JA111 is unable to grow on media in which 1,2-propanediol is the sole carbon source (Hidalgo et al., 1991).
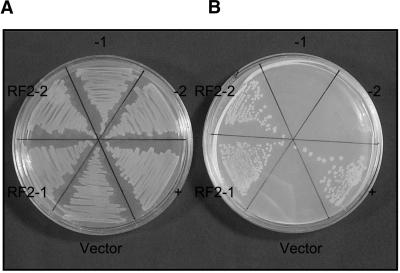
To determine whether rf2 can complement the ald mutant, JA111 was first lysogenized with λDE3, which carries the T7 RNA polymerase gene. This was done because rf2 expression is under the control of the T7 promoter in pMAP11. The resulting strain JA111(DE3) exhibited the same inability to grow on media in which 1,2-propanediol is the sole carbon source as JA111 (Figure 5). Subsequently, the plasmids pMAP11, pET-17b, and pALD9 (a plasmid that carries the E. coli ald gene and that can complement the ald-deficient mutant of JA111) were transformed into JA111(DE3). The ability of JA111(DE3)pMAP11 to express RF2 was tested using immunoblot analysis and ALDH activity assays (data not shown). All of the strains grew normally on glucose media, but only JA111(DE3)pALD9 and JA111(DE3)pMAP11 were able to grow on 1,2-propanediol media (Figure 5). This result indicates that the recombinant RF2 protein exhibits l-lactaldehyde dehydrogenase activity.
Figure 5.
The rf2 Gene Can Complement an E. coli ald-Deficient Mutant.
(A) Basal medium plus glucose.
(B) Basal medium plus 1,2-propanediol.
−1, JA111; −2, JA111(DE3); (+), JA111(DE3)pALD9; Vector, JA111 (DE3)pET-17b; RF2-1, JA111(DE3)pMAP11-1; RF2-2, JA111(DE3) pMAP11-2.
It should be noted that the complementation analyses described above were performed in the absence of IPTG induction. This was because E. coli strain JA111(DE3)pMAP11 failed to grow on 1,2-propanediol media when induced with all tested levels of IPTG (0.5, 1.0, and 1.5 mM). In contrast, E. coli cells that expressed the endogenous ald gene from the pALD9 construct grew on 1,2-propanediol media under all tested levels of IPTG. Both cells could grow on glucose media containing various levels of IPTG. The reason for this discrepancy is not known.
ALDH Activity in Maize Mitochondrial Extracts
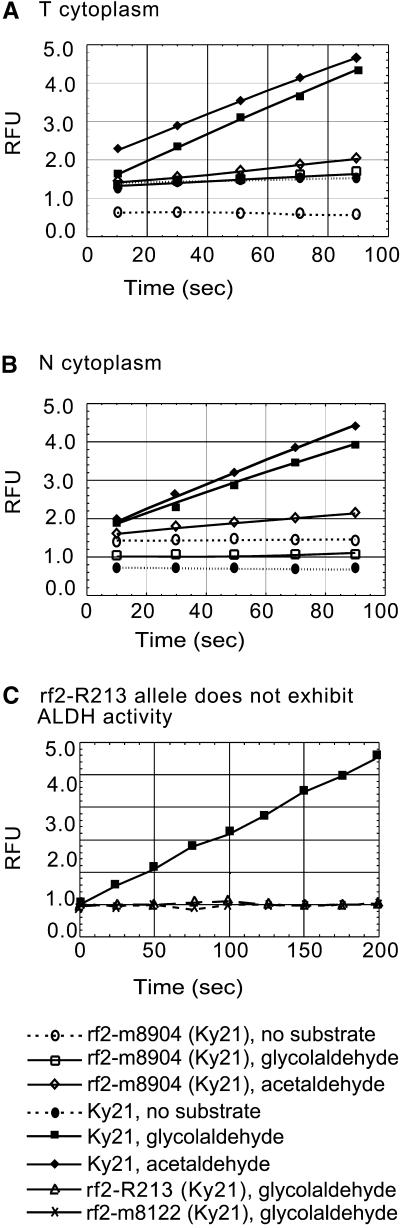
To compare the enzyme activity of recombinant RF2 protein to that of the native RF2 protein, we performed ALDH assays on protein extracts from maize mitochondria prepared from immature unpollinated ears of the inbred line Ky21 (which is homozygous for the functional Rf2-Ky21 allele) and a near-isogenic line homozygous for rf2-m8904. When acetaldehyde was used as a substrate, mtALDH activity was approximately four- to fivefold greater in wild-type Ky21 plants than in mutant plants. When glycolaldehyde was used as a substrate, wild-type plants exhibited 16- to 34-fold greater levels of ALDH activity than did mutants (Table 2 and Figures 6A and 6B). These results demonstrate that like the recombinant RF2 protein, the native RF2 protein exhibits acetaldehyde and glycolaldehyde dehydrogenase activities. The levels of ALDH activity were not significantly different in mitochondrial extracts from N and T cytoplasm plants (Table 2), which is consistent with the fact that the accumulation of RF2 protein does not differ between these genotypes (Figure 1). ALDH assays of submitochondrial fractions revealed that ALDH activity is confined to the mitochondrial matrix (data not shown), which is consistent with the results of immunoblot analyses (Figure 2D).
Table 2.
Maize mtALDH Assaya
| T Cytoplasm
|
N Cytoplasm
|
|||
|---|---|---|---|---|
| Substrate | Rf2b | rf2-mc | Rf2b | rf2-mc |
| Acetaldehyde (17 μM) |
7.03 ± 0.58 | 1.43 ± 0.08 | 7.23 ± 0.15 | 1.87 ± 0.25 |
| Glycolaldehyde (20 μM) |
7.73 ± 0.29 | 0.23 ± 0.13 | 6.43 ± 0.29 | 0.40 ± 0.17 |
Reaction rates are expressed as change in relative fluorescence units at 460 nm per minute per milligram of mitochondrial protein. Data represent the average ±sd of three measurements.
Inbred line Ky21.
rf2-m8904/rf2-m8904 backcrossed into Ky21 for three generations.
Figure 6.
Mitochondrial ALDH Assay.
ALDH activity was measured in extracts from mitochondria purified from unpollinated ears produced by T cytoplasm (A) or N cytoplasm (B) plants or from etiolated seedlings (C) of the indicated genotypes. Results from one typical experiment are shown. Data from (A) and (B) are summarized in Table 2. RFU, relative fluorescence units.
Molecular Lesion in the rf2-R213 Allele
The inbred line R213 carries a mutant allele of rf2 (rf2-R213) that is not capable of restoring male fertility to T cytoplasm maize. Surprisingly, this inbred line accumulates RF2 protein (Figure 1B) of the same size and at the same level as does the inbred line Ky21, which carries a functional Rf2 allele. In addition, mitochondrial extracts from R213 exhibit a small amount of ALDH activity (data not shown). In contrast, a stock that is homozygous for rf2-R213 but near isogenic with Ky21 accumulates rf2 mRNA (Cui et al., 1996) and RF2 protein (data not shown) but does not exhibit ALDH activity (Figure 6C). Hence, it appears that the rf2-R213 allele does not code for a functional ALDH.
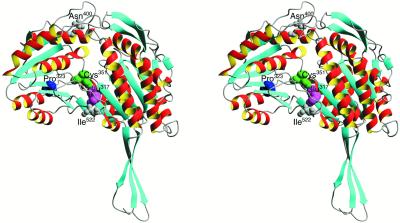
To identify the molecular lesion in the rf2-R213 allele, we amplified the coding region of this allele via reverse transcription–polymerase chain reaction (PCR) and sequenced. Sequence analysis revealed that the rf2-R213 allele (GenBank accession number AF269064) has four SNPs relative to the wild-type rf2-B73 allele (GenBank accession number U43082). These polymorphisms introduce three amino acid substitutions (one SNP is a silent third position substitution). The RF2 protein structure was predicted with SWISS-MODEL (http://www.expasy.ch/swissmod; Guex and Peitsch, 1997), using as a template bovine mtALDH (Protein Data Bank number 1AG8), which exhibits 62% identity and 70% similarity to RF2. The Asn400-to-Lys and the Ile522-to-Met substitutions in the rf2-R213 allele are both located at the periphery of the RF2 protein structure (Figure 7), whereas the Pro323-to-Ser substitution is located in the substrate binding pocket (Liu et al., 1997; Steinmetz et al., 1997) and in the conserved catalytic Glu317 domain predicted by the Motifs function of the Genetics Computer Group (Madison, WI) package. The Asn400-to-Lys substitution mimics the functional bovine mtALDH, and the Ile522- to-Met substitution is conservative. In contrast, the mutation of Pro323-to-Ser is likely to greatly affect the protein structure. Hence, this substitution is most likely to be responsible for the loss of ALDH activity in the rf2-R213 allele.
Figure 7.
Structure of the RF2 Protein.
This structure was predicted by SWISS-MODEL (Guex and Peitsch, 1997) using bovine mtALDH (Protein Data Bank number 1AG8) as a template. The stereo images were prepared using MOLMOL (Koradi et al., 1996). Residues Glu317, Pro323, Cys351, Asn400, and Ile522 in the RF2 sequence are equivalent to amino acid residues Glu267, Pro273, Cys301, Lys350, and Ser472 in Protein Data Bank number 1AG8, respectively.
The rf2 Gene Is Required for Normal Anther Development in N Cytoplasm Maize
Most maize inbred lines carry functional Rf2 alleles, even though they have never been exposed to T cytoplasm. This suggests that the RF2 protein has an important physiological role other than restoring male fertility to plants that carry T cytoplasm (Schnable and Wise, 1994). To characterize this physiological role of the rf2 gene, the effects on anther development of two independent transposon insertion mutant alleles, rf2-m8904 and rf2-m9323, were characterized. These characterizations were performed on plants homozygous for these two alleles that were derived from three and four generations, respectively, of backcrossing to the inbred line N cytoplasm Ky21. The tassels of the backcrossed rf2-m plants exert anthers and shed functional pollen. However, various degrees of anther arrest (Figure 8) were observed in plants homozygous for these two alleles compared with the near-isogenic N cytoplasm Ky21 plants (Figure 8A). Arrested anthers were smaller than those at the same developmental stage in wild-type N cytoplasm Ky21 plants (data not shown) and failed to shed pollen. Arrested anthers were distributed evenly within affected tassels.
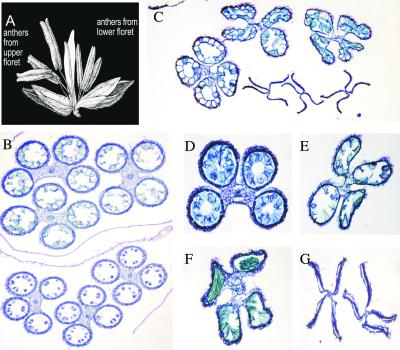
Figure 8.
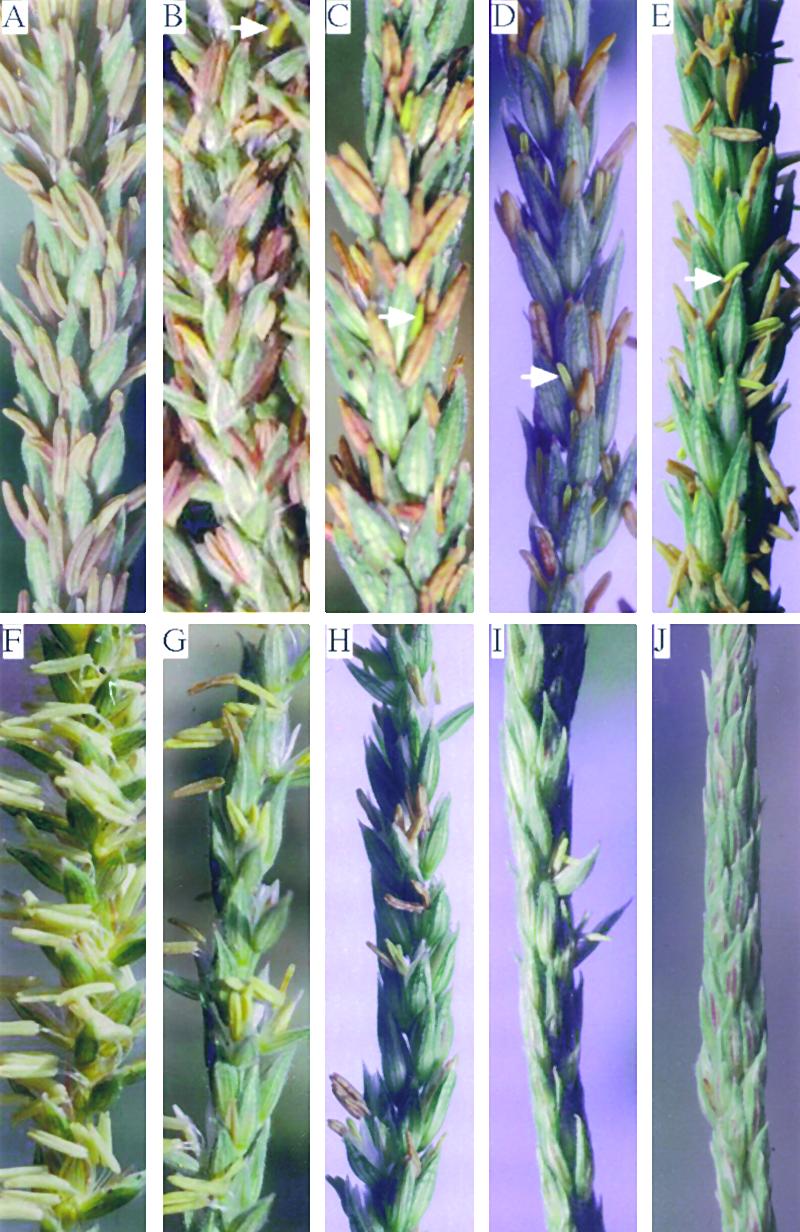
Anther Arrest in rf2 Plants Carrying N or T Cytoplasm.
(A) Normal anther development on a tassel branch from N cytoplasm Ky21.
(B) to (E) Various degrees of anther arrest on tassel branches from different plants in a segregating population resulting from the cross (N) rf2-m9323/rf2-m9323 × rf2-m9323/Rf2-Ky21. Arrows identify arrested anthers.
(F) Normal anther development on a tassel branch from a fully suppressed T cytoplasm plant homozygous for rf2-m9390.
(G) to (I) Various degrees of anther arrest on tassel branches from partially suppressed T cytoplasm plants homozygous for rf2-m9437.
(J) Completely sterile tassel branch from an unsuppressed T cytoplasm plant homozygous for rf2-m9437.
A maize spikelet consists of an upper and a lower floret, each of which contains three anthers. Anthers in the lower floret progress through development ∼3 days after those in the upper floret of the same spikelet (Hsu and Peterson, 1991). When sections of spikelets from N cytoplasm rf2-m plants were observed microscopically, various defects were observed, from slightly affected microspores to completely degenerated microspores and shriveled locules. As expected, on the basis of the gross examination of spikelets, these defects are observed mainly in the anthers from lower florets (Figure 9).
Figure 9.
Microscopic Observation of Anther Arrest.
(A) Illustration of a normal maize spikelet.
(B) A spikelet from an N cytoplasm Ky21 plant.
(C) A spikelet from an N cytoplasm plant homozygous for rf2-m9323 and near-isogenic with Ky21.
(D) and (E) Anthers from the lower florets of N cytoplasm plants homozygous for rf2-m8904.
(F) and (G) Anthers from the lower florets of N cytoplasm plants homozygous for rf2-m9323.
To demonstrate that the anther arrest phenotype is caused by loss of Rf2 function, plants from a segregating population, resulting from the test cross (N) wx rf2-m8904/wx rf2-m8904 × wx rf2-m8904/Wx Rf2-Ky21, were observed for anther arrest. Plants were categorized into five classes according to the severity of anther arrest (Figure 8). Among the progeny of this test cross, the anther arrest phenotype is well correlated with the waxy kernel phenotype, which serves as a marker for rf2 (data not shown). To further characterize the relationship between anther arrest and the rf2 mutation, some plants from the N, ‘N’, A, and AA categories (see Methods for a description of the categories) were genotyped at the rf2 locus via restriction fragment length polymorphism (RFLP) analysis, using the rf2 cDNA as a probe (Table 3, row No. 98 6503-06). The ambiguous category ‘A’ (Figure 8C) was not genotyped. All 38 plants from the N and ‘N’ categories were heterozygous rf2-m8904/Rf2-Ky21, and all 23 plants from the A and AA categories were homozygous for the rf2-m8904 allele. Similar results were obtained from a population segregating for the other tested rf2 mutant allele, rf2-m9323. However, in this case, two exceptions were observed. Both exceptional plants were homozygous for rf2-m9323 but did not exhibit anther arrest (Table 3, row No. 98 6509-10). These results indicate that either (1) the rf2 mutation is the cause of the anther arrest phenotype and the two exceptions in the population segregating for rf2-m9323 represent instances of incomplete penetrance or indicate the presence of a suppressing factor, or (2) the genetic factor that causes anther arrest is not rf2 but is closely linked to rf2.
Table 3.
Correlation between Anther Arrest and rf2 Genotype in Progeny from the Test Cross (N) rf2-m/rf2-m × rf2-m/Rf2
| Phenotypeb
|
|||
|---|---|---|---|
| Row No. | Genotypea | N + ‘N’ | A + AA |
| 98 6503-06 | rf2-m8904/rf2-m8904 | 0 | 23 |
| rf2-m8904/Rf2-Ky21 | 38 | 0 | |
| 98 6509-10 | rf2-m9323/rf2-m9323 | 2 | 17 |
| rf2-m9323/Rf2-Ky21 | 30 | 0 | |
| 99 6655-56 | rf2-m8904/rf2-R213 | 2 | 10 |
| rf2-m8904/Rf2′1036W103 | 9 | 0 | |
| 99 6657-58 | rf2-m8904/rf2-R213 | 2 | 12 |
| rf2-m8904/Rf2′1036W110 | 13 | 0 | |
Genotypes were established via DNA gel blot analyses, using the partial rf2 cDNA as a probe.
Anther phenotypes were scored in the morning during the period of pollen shedding. Examples of each phenotype are shown in Figures 8A to 8E.
Two independent revertant (Rf2′) alleles derived from the excision of the nonautonomous transposon insertion from the rf2-m8904 allele were used to distinguish between these two possibilities. Both of these revertant alleles, Rf2′1036W103 and Rf2′1036W110, confer male fertility to plants that carry T cytoplasm. Because these two revertant alleles would be expected to have exactly the same haplotype in the vicinity of the rf2 locus as does the rf2-m8904 mutant from which they arose, these alleles represent useful reagents for determining whether the anther arrest phenotype is controlled by rf2 or a closely linked gene.
Observation of 29 progeny from the cross (N) rf2-m8904/rf2-m8904 × Rf2′1036W103/Rf2′1036W103 revealed that all exhibited normal anther development. This revertant allele also was used to generate a segregating population from the test cross (N) wx rf2-m8904/wx rf2-m8904 × Wx rf2-R213/wx Rf2′1036W103. All of the nine resulting progeny that carried the revertant allele were normal, and 10 of 12 of the progeny that carried rf2-m8904/rf2-R213 exhibited the anther arrest phenotype (Table 3, row No. 99 6655-56). Similar results were obtained with the Rf2′1036W110 revertant allele (Table 3, row No. 99 6657-58). Hence, these data demonstrate that Rf2 function is required to prevent anther arrest in N cytoplasm plants.
Partial ALDH Activity Causes Anther Arrest in T Cytoplasm
Because the lack of rf2-encoded mtALDH activity causes complete male sterility in T cytoplasm plants but only partial anther arrest in N cytoplasm plants, we hypothesized that restoration of fertility to T cytoplasm plants requires more RF2 activity than does the prevention of anther arrest in N cytoplasm plants. To test this hypothesis, suppressible rf2 Mu insertion alleles rf2-m9437, rf2-m9390, and rf2-m8110 were used to observe the phenotype associated with low, but not null, levels of RF2. Mu suppression is the loss of a Mu-induced allele's capacity to condition a mutant phenotype in the absence of excisional loss of the associated Mu transposons (Martienssen et al., 1989). Many maize Mu-induced mutant alleles, such as hcf106::Mu1, exhibit Mu suppression (Martienssen et al., 1989, 1990; Barkan and Martienssen, 1991). Mu suppression occurs in the absence of high activity of the autonomous Mu transposon MuDR (Lowe et al., 1992; Greene et al., 1994; Martienssen and Baron, 1994).
The suppressible rf2 alleles fail to condition male sterility in T cytoplasm plants in the absence of MuDR (X. Cui, A.P. Hsia, D.A. Ashlock, R.P. Wise, and P.S. Schnable, submitted manuscript). Various degrees of Mu suppression can be obtained from these suppressible alleles when Mu activity is at intermediate levels. Plants that carry partially suppressed rf2-m alleles exert various numbers of anthers (from a few to almost all), some of which shed pollen (Figures 8G to 8I). Analysis of individual spikelets revealed that exerted anthers are always derived from the upper florets, whereas the anthers in most lower florets are arrested in their development. The rf2-mediated anther arrest that occurs in these partially suppressed T cytoplasm plants closely resembles the arrest that occurs in N cytoplasm plants completely lacking RF2 activity.
DISCUSSION
CMS systems play important roles in the production of hybrid seed. In addition, they serve as useful models for the study of mitochondrial mutations and nucleus–mitochondria interactions (Mackenzie et al., 1994). Despite the considerable effort that has been directed toward understanding CMS systems, the rf2 gene remains the only nuclear restorer of CMS to be cloned from any species. As such, it represents a unique resource for the study of the molecular mechanisms associated with fertility restoration.
CMS systems often are associated with the presence of novel mitochondrial open reading frames (Schnable and Wise, 1998). In at least some cases, it is known that the gene products associated with these open reading frames cause male sterility (Gengenbach et al., 1981; Kemble et al., 1982; Umbeck and Gengenbach, 1983; Fauron et al., 1987; Wise et al., 1987b; He et al., 1996). Hence, nuclear genes that disrupt the normal accumulation of these gene products can function as nuclear restorers. Examples of this type of nuclear restorer include the Fr gene, which is capable of directing the loss of the CMS-associated pvs sequence from the Phaseolus vulgaris mitochondrial genome (Mackenzie and Chase, 1990), and the rf1 gene, which reduces the accumulation of the cms-T–associated URF13 protein (Dewey et al., 1987; Kennell and Pring, 1989). In contrast, even though the rf2 gene is a nuclear restorer of cms-T, it does not affect the accumulation of URF13. Hence, the analysis of the rf2 gene has defined another general mechanism by which nuclear genes can function as restorers. Instead of affecting the accumulation of CMS-associated gene products, “compensatory restorers,” such as rf2, provide “work around” solutions that compensate for the metabolic dysfunctions caused by CMS-associated gene products.
The rf2 Gene Encodes an mtALDH
The first two steps toward defining the mechanism by which rf2 compensates for the as-yet-undefined metabolic disruptions induced by URF13 were to determine the enzymatic function and the localization of the RF2 protein. The sequence-based prediction that the rf2 gene encodes an ALDH (Cui et al., 1996) was tested via three independent approaches. First, it was established that recombinant RF2 protein has acetaldehyde and glycolaldehyde dehydrogenase activities. Second, the rf2 gene was shown to complement an E. coli mutant that requires l-lactaldehyde dehydrogenase activity. Third, protein extracts of mitochondria purified from wild-type maize plants were found to contain more acetaldehyde and glycolaldehyde dehydrogenase activities than similar extracts from rf2 mutant plants. Hence, we conclude that the rf2 gene encodes an ALDH.
The accumulation of the RF2 protein to high levels in the tapetal layer of anthers is consistent with the observation that premature degeneration of the tapetum occurs in unrestored cms-T plants (Warmke and Lee, 1977). Subcellular and suborganellar fractionation experiments established that the RF2 protein accumulates in the mitochondrial matrix and thereby have confirmed and extended the sequence-based predictions of Cui et al. (1996).
mtALDH Activity Is Required for Fertility Restoration of cms-T Maize
The results described above establish that the rf2 gene encodes an mtALDH. However, it is formally possible that some feature of the RF2 protein, other than its ALDH activity, is responsible for its ability to function as a restorer. For example, the ALDH-derived ω-crystallin lacks ALDH activity and functions as a structural protein in the lens of squid and octopus (Zinovieva et al., 1993). Analysis of a spontaneous rf2 mutant allele provided data to test this hypothesis. Plants homozygous for rf2-R213 accumulate mRNA (Cui et al., 1996) and protein (Figure 1B) of the same sizes and in the same amounts as do nonmutant plants. However, the protein encoded by this allele does not exhibit ALDH activity (Figure 6). Sequence analysis of rf2-R213 defined a single amino acid substitution, Pro323 to Ser, that is likely to be responsible for the loss of ALDH activity. Hence, these data strongly support the view that the rf2 gene functions as a restorer of fertility by virtue of its ability to encode ALDH activity.
Metabolic Role of rf2-Encoded mtALDH
One of the challenges of the postgenomic era is how to assign enzymatic functions to thousands of newly identified proteins. In many instances, it is possible to obtain clues regarding the functions of these proteins on the basis of sequence similarities and high throughput functional assays (Martzen et al., 1999). However, once an enzymatic function has been established, there remains the even more difficult “post–post-genomic” challenge of defining the precise metabolic roles of the enzymatic functions within an organism. This challenge exists because orthologous proteins can have different metabolic functions among organisms and the same protein can have different functions within an organism.
Because mtALDHs typically have many potential substrates (Klyosov, 1996), the task of determining the specific aldehyde(s) that must be oxidized during fertility restoration is particularly challenging. Although the three-dimensional structure of the RF2 protein has been predicted via modeling using the known structure of a mammalian class II mtALDH as a template (Steinmetz et al., 1997), this information does not provide information regarding the metabolically important substrate(s) of this enzyme. Biochemical approaches to defining this substrate are complicated by the fact that mutants of the rf2 gene exert their effects on male fertility (at least in T cytoplasm maize) in only a single internal cell layer of the anther (i.e., the tapetum).
One function of mtALDHs in mammals is the detoxification of ethanol-derived acetaldehyde. It has been established that enzymes involved in fermentation (i.e., pyruvate decarboxylase [EC 4.1.1.1], alcohol dehydrogenase [EC 1.1.1.1], and aldehyde dehydrogenase [EC 1.2.1.3]) are expressed during microspore development (reviewed in Tadege et al., 1999); this study established that the RF2 protein exhibits acetaldehyde dehydrogenase activity. Hence, we are currently using a genetic approach to test the hypothesis that the metabolically significant substrate of RF2 is acetaldehyde.
However, diverse aldehydes are produced via numerous metabolic pathways, including the peroxidation of membrane lipids that can result from oxidative stress (Siu and Draper, 1982); glycine biosynthesis (Davies, 1959); the catabolism of threonine (Styrvold et al., 1986), phenylalanine (Ferrandez et al., 1997), and tyrosine (Ferrandez et al., 1997); vitamin A metabolism (McCaffery and Drager, 1993); and indole-3-acetyl acetate biosynthesis (Marumo, 1986). Other biogenic aldehydes and retinaldehydes also are possible substrates of RF2. In mammalian systems, trace amounts of such products are capable of affecting gene action (Sladek et al., 1989; Lindahl, 1992). Hence, in parallel with testing specific hypotheses, we are also conducting genetic screens for suppressors and enhancers of rf2 mutants with the expectation that the nature of the genes identified via these screens will provide clues regarding the specific biochemical pathway(s) in which the rf2-encoded mtALDH functions during fertility restoration.
The rf2 Gene Has a Function Independent of Its Role in Restoration
Most of the examined maize inbred lines (15 of 17) carry functional alleles of the rf2 gene (Wise et al., 1999). The two exceptions, R213 and Wf9, carry the same mutant rf2 allele, because R213 was derived from Wf9 (D. Duvick, personal communication). Because most inbred lines have never (or have only recently) been exposed to T cytoplasm, it has been hypothesized that the rf2 gene must have been selected during evolution for a function other than fertility restoration (Schnable and Wise, 1994). According to this hypothesis, the rf2 gene was recruited only recently to serve as a nuclear restorer. However, the nature of this other (presumed) function was not predicted, because rf2 was not known to confer any phenotype in maize that does not carry T cytoplasm.
The detailed analyses reported here establish that the rf2 gene plays a significant developmental role, even in plants that do not carry T cytoplasm. Specifically, this gene is required for normal anther development in plants that carry N cytoplasm. The recruitment of the rf2 gene to serve as a nuclear restorer demonstrates the ability of plant genomes to respond to novel metabolic disturbances.
Because the rf2 gene has been recruited only recently to serve its new function as a restorer of CMS, it is not surprising that the levels of rf2 mRNA (Cui et al., 1996), RF2 protein, and rf2-encoded ALDH activity are not higher in T cytoplasm than in N cytoplasm maize (Figures 1 and 6). However, this latter observation also indicates that rf2-encoded levels of ALDH activity are high enough in N cytoplasm plants to accommodate the enzymatic function(s) required for restoration of T cytoplasm plants. This is true even though an analysis of suppressible rf2 alleles suggests that higher levels of rf2 expression are required for fertility in T cytoplasm maize than for anther development in N cytoplasm maize. However, it should be noted that although rf2 mutants affect anther development in plants that carry either N or T cytoplasm, the metabolic role that the RF2 protein plays during the restoration of cms-T may differ from the role that it plays during anther development in N cytoplasm maize.
Although the upper floret develops several days earlier than does the lower floret from the same spikelet (Hsu and Peterson, 1991), the two florets do not exhibit visually detectable differences in their developmental programs. However, because this study revealed that rf2 mutations preferentially affect the lower florets in plants that carry either T or N cytoplasm, we hypothesize that even at the same developmental stage, upper and lower florets will exhibit different patterns of gene expression.
METHODS
Maize Strains and rf2 Alleles
The inbred maize (Zea mays) line Ky21 is homozygous for functional alleles of the rf1 (Rf1-Ky21) and rf2 (Rf2-Ky21) loci. The inbred line R213 is fixed for Rf1 and rf2. Because R213 was derived from a cross between Ky21 and Wf9 (rf1-Wf9, rf2-Wf9) (D. Duvick, personal communication), the Rf1 allele carried by R213 must be Rf1-Ky21. Similarly, the rf2 allele in R213 must be identical to rf2-Wf9. However, to be consistent with previous nomenclature, this allele is referred to as rf2-R213. A Texas (T) cytoplasm version of Ky21 was generated by backcrossing a T cytoplasm version of the inbred line R213 to Ky21 for seven generations. Restriction fragment length polymorphism (RFLP) analysis established that the resulting line is homozygous for Rf1-Ky21 and Rf2-Ky21. The T cytoplasm versions of Ky21 homozygous for the rf2-m or rf2-R213 mutant alleles were generated by backcrossing each allele to Ky21 for at least three generations. All lines used in this study carry the Rf1-Ky21 allele.
The rf2-m alleles were derived from Mutator or Spm/En transposon tagging experiments (Wise and Schnable, 1994). The progenitor alleles of rf2-m8122, rf2-m9323, rf2-m9390, and rf2-m8110 have been described (Cui et al., 1996). On the basis of sequence comparisons, the progenitor allele of rf2-m9437 (GenBank accession number AF318138) is Rf2-B79 (GenBank accession number AF318135). The progenitor allele of rf2-m8904 (GenBank accession numbers AF318130, AF318131, and AF318132) has not yet been identified. However, it is not Rf2-B79, Rf2-Q66 (GenBank accession number AF318133), Rf2-Q67 (GenBank accession number AF318134), or Rf2-B77 (GenBank accession numbers AF318136 and AF318137). When originally isolated, the rf2-m8904 allele conferred a stable male-sterile phenotype. However, it appears to contain a transposon insertion. This conclusion is based on the observation that the allele can produce functional revertant alleles (Rf2′) that contain an RFLP relative to rf2-m8904 (see below).
Genetic analyses established that the rf2-m8904 allele does not contain an autonomous Spm/En transposon insertion (Schnable and Wise, 1994). However, some crosses (e.g., 95 1902-21/2945) between stocks that carried rf2-m8904 and related stocks derived from the Spm/En tagging population yielded progeny that produced tassels with male-fertile sectors, presumably because they were chimeric for revertant Rf2′ alleles. Pollen from revertant sectors was used to pollinate T cytoplasm R213 plants. Some of the resulting progeny were male fertile and self-pollinated. Plants that were homozygous for two revertant alleles, Rf2′1036W103 (GenBank accession numbers AF318139, AF318140, and AF318141) and Rf2′1036W110 (GenBank accession numbers AF318142, AF318143, and AF318144), were identified by RFLP analysis. The 1.2-kb partial rf2 cDNA probe (Cui et al., 1996) detected two distinct HindIII fragments of ∼9 and 10 kb in DNA from plants homozygous for rf2-m8904 but only a single 10-kb fragment in DNA from plants homozygous for the revertant alleles. In contrast, the six introns (844 bp) of these alleles that have been sequenced are identical to the corresponding introns of rf2-m8904, even though 28 SNPs were identified between these alleles and the Rf2-B73 allele (GenBank accession number AF215823). Hence, it appears that Rf2′1036W103 and Rf2′1036W110 arose from rf2-m8904 via the excision of a nonautonomous transposon that was activated by the introduction of an autonomous transposon via the cross described above.
Genotype and Phenotype Determinations
Immature ears or young leaves were collected from plants to be genotyped. DNA was extracted from ears according to the procedure of Dellaporta et al. (1983). Ten micrograms of DNA was digested with KpnI at 37°C for 3 hr and separated on an 0.8% agarose gel. DNA gel blot analyses (Sambrook et al., 1989) were conducted on these DNA samples using as a probe the full-length rf2 cDNA isolated from prf27311 (Cui et al., 1996) labeled with 32P.
Male fertility and anther arrest phenotypes were scored in the morning for several days during the period of pollen shed. Plants were scored for male fertility according to the scale of Schnable and Wise (1994). The degrees of anther arrest were categorized into five classes (N, ‘N’, ‘A’, A, and AA). The N category is equivalent to the phenotype displayed by Ky21 (Figure 8A); ‘N’ category plants have very few arrested anthers (Figure 8B); ‘A’ category plants have some arrested anthers, but fewer than half of the lower florets contain arrested anthers (Figure 8C); most of the lower florets in A category plants contain arrested anthers (Figure 8D); and most anthers in most of the lower florets in AA category plants are arrested (Figure 8E).
Expression of the RF2 Protein in Escherichia coli
Polymerase chain reaction (PCR) was performed using primers rf2-B2 (5′-AAGATCTGATGCACAGGCTGTTGCCA-3′) and rf2-xq (5′-CCAACTTTCCAGGCATACATCA-3′) to amplify a portion of the rf2 cDNA (nucleotides 368 to 918) from plasmid prf27311 that contains the 2.2-kb full-length rf2 cDNA (Cui et al., 1996). The 5′ primer (rf2-B2) was synthesized such that it contained an extra BglII site; the 3′ primer (rf2-xq) was downstream of an internal NdeI site. A three-fragment ligation was conducted using the 367-bp BglII-NdeI PCR product, the 1493-bp NdeI-KpnI fragment from prf27311, and the BglII-KpnI fragment of the expression vector pET-30b (Novagen, Madison, WI). The resulting plasmid was termed pLB333. Plasmid pMAP11 was generated in a similar fashion except that the 5′ primer (rf2-lp1, 5′-GTCTAGAACCGCAGCAGCAGTAGAGG-3′) carried an NheI site, the PCR product included nucleotides 407 to 918 from prf27311, and the cloning vector was NheI-KpnI–digested pET-17b.
Both plasmids were transformed into the E. coli strain BL21(DE3) for expression. Transformed cells were incubated at 37°C overnight, transferred to 200 mL of fresh medium (1.6% tryptone, 1.0% yeast extract, and 0.5% NaCl) the next morning, and incubated in a 37°C shaker. Once the OD600 reached 0.8 to 1.0, 1 mM isopropylthio-β-galactoside (IPTG) was added and incubation was continued for another 3 hr or until the OD600 reached 1.5.
Antibody Preparation and Purification
RF2 protein that had been expressed from pLB333 as a fusion protein containing six histidines (His tag) and the S tag was purified by Ni2+ affinity chromatography under denaturing conditions using the protocol described in the sixth edition of the Novagen pET system manual. The eluted fraction was dialyzed sequentially against TEN buffer (100 mM Tris, pH 8.0, 10 mM EDTA, and 50 mM NaCl) containing 3, 1, or 0.5 M urea. Finally, this fraction was dialyzed twice against TEN buffer without urea. The dialysis tubing containing RF2 protein was embedded in ground sucrose overnight, and then its contents were passed through a Sephadex G-75 column equilibrated with 50 mM Tris, pH 8.0, and 0.1 M NaCl. Aliquots of fractions of the flow through were subjected to SDS-PAGE and then transferred to nitrocellulose membranes. The RF2-containing fractions could be recognized because they contained detectable levels of S tag peptide, which was detected according to the manufacturer's instructions (Novagen). Fractions that contained the S tag (and hence the RF2 protein) were pooled. SDS-PAGE analysis of the pooled fractions revealed only a single Coomassie Brilliant Blue R 250–stainable protein of approximately the size expected for the recombinant RF2 protein (i.e., 57 kD).
The recombinant RF2 protein was concentrated to 1 mg/mL by using a Millipore (Bedford, MA) spin column (Centriplus 30) and then used as an antigen to inject rabbits. Two weeks after the second injection, antisera were collected and used directly or further purified according to Dumbroff and Gepstein (1993) with the following modifications. Maize tassels with the genotype rf2-m8904/rf2-m8904 Ky21 were ground in TBS buffer (25 mM Tris, pH 7.5, and 150 mM NaCl) supplemented with 1 mM phenylmethylsulfonyl fluoride (Sigma), 5 mM EDTA, and 1 μg/mL E-64 (Sigma), filtered through four layers of cheesecloth, and spun at 12,000g for 10 min. An 8 × 8-cm2 piece of nitrocellulose membrane was incubated at room temperature on a shaker for 3 hr in the resulting supernatant, which contained 100 mg of protein extracted from the rf2-m8904/rf2-m8904 tassels. The membrane was washed in TBS four times and rinsed in PBS (0.8% NaCl, 0.02% KCl, 0.14% Na2HPO4, and 0.024% KH2PO4, pH 7.2). The bound protein was fixed to the membrane by treatment with 0.2% glutaraldehyde in PBS for 1 hr and subsequently washed sequentially in PBS and TBS. One milliliter of antiserum was added to 10 mL of TBS and incubated with the membrane overnight at 4°C. The solution was collected and loaded onto a protein A–agarose column and washed with 10 volumes of TBS. Antibodies were eluted from the column using 0.1 M glycine, pH 3.0, neutralized with 1.0 M Tris, pH 8.0, passed through a PBS-equilibrated Sephadex G-25 spin column, divided into aliquots, and stored at −20°C.
Immunolocalization
Maize anthers at the premeiotic stage were dissected and fixed at 4°C for 6 hr in 2% glutaraldehyde, 3% paraformaldehyde, and 0.1 M sodium cacodylate, pH 7.0. The fixed anthers were dehydrated in 25, 50, 75, 80, 90, 95, and 100% (twice) ethanol for 20 min each and then infiltrated with ethanol/LR White (Electron Microscopy Sciences, Fort Washington, PA) in ratios of 1:3, 1:1, and 3:1, and LR White (twice), for 30 min each (modified from Parthasarathy, 1994). The embedded anthers were left in a 60°C oven overnight and then cross-sectioned into 1-μm-thick sections. These sections were mounted on polylysine-treated slides, blocked with 5% dry milk in TBS, and incubated with purified anti-RF2 polyclonal antibodies (final concentration, 36.5 μg/mL) at 37°C for 3 hr. Slides were washed with TBS three times for 10 min each and then incubated with 10-nm gold-labeled goat anti-rabbit IgG secondary antibodies (1:3000; Sigma) at 37°C for 3 hr. The gold particles were enhanced using a silver-enhancing kit (model SE-100; Sigma) and were viewed and photographed with a phase contrast light microscope.
Light Microscopy
Maize spikelets and anthers at various stages were fixed in 50% ethanol, 5% acetic acid, and 3.7% formaldehyde at room temperature for 24 hr. Fixed tissues were dehydrated, infiltrated, and embedded in paraffin (Sylvester and Ruzin, 1994). Cross-sections of 10 to 20 μm were deparaffined and stained with toluidine blue for observation via bright-field microscopy.
Sequencing
DNA sequencing was performed at the Iowa State University DNA Sequencing and Synthesis Facility on an automated sequencer (model ABI 373A; Applied Biosystems, Foster City, CA). Sequence assembling and analyses were performed using the Wisconsin GCG software package, version 10.0-Unix, from the Genetics Computer Group. Polymorphisms detected between rf2-R213 cDNA and wild-type Rf2-B73 were confirmed by sequencing polymorphic regions obtained by PCR amplification of the rf2-R213 allele from genomic DNA of the inbred line R213.
E. coli Complementation
E. coli strain JA111 was lysogenized with a recombinant λDE3 phage (Novagen) according to the manufacturer's instructions. The resulting strain was designated JA111(DE3) and carried a T7 RNA polymerase gene under the control of the lacUV5 promoter. Plasmid pALD9 carries an E. coli aldehyde dehydrogenase that complements the E. coli ald-deficient mutant JA111 (Hidalgo et al., 1991). Plasmids pALD9, pMAP11, and pET-17b were each transformed into JA111(DE3) by electroporation. Two pMAP11 transformants (pMAP11-1 and pMAP11-2) and one pET-17b transformant were selected for further analysis. Each E. coli culture was streaked in duplicate on basal medium (Boronat and Aguilar, 1979) supplemented with either 40 mM glucose or 40 mM 1,2-propanediol as the sole carbon source and incubated at 37°C. Cells grown on glucose were scored after 24 hr of incubation, whereas cells grown on 1,2-propanediol were scored after 3 days.
Cell Fractionation and Mitochondria Preparation
Mitochondria to be used in the subcellular localization experiment were purified from 4-day-old etiolated Ky21 seedlings via Percoll gradient centrifugation (Jackson and Moore, 1979). Chloroplasts were prepared from 7-day-old green maize Ky21 seedlings according to Leegood and Walker (1983). Chlorophyll (Leegood and Walker, 1983), catalase (Vigil, 1983), alcohol dehydrogenase (Freeling and Schwartz, 1973; Quail, 1979), and cytochrome c oxidase (Errede et al., 1967) were used as markers in chloroplastic, microsomal, cytosolic, and mitochondrial fractions, respectively. For the catalase assay, fractions were passed through a Sephadex G-15 spin column equilibrated with 0.1 M phosphate buffer, pH 7.0, before the enzyme assay.
Mitochondria for other purposes were prepared from either etiolated maize seedlings or unpollinated ears according to the method of Payne et al. (1980) and then further purified by sucrose cushion centrifugation (Jackson and Moore, 1979). The proteinase protection assay of mitochondria was conducted according to Huang et al. (1990), except that papain was used instead of proteinase K.
To isolate the membrane and soluble fractions from mitochondria, we resuspended purified mitochondria in 20 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.5, and 1 mM DTT to yield a 0.2 mg/mL protein solution and sonicated three times for 10 sec each using a Fisher model 60 sonicator set at 15 W. The solution was then spun at 100,000g for 2 hr. The supernatant contained the soluble mitochondrial proteins. The pellet was resuspended and washed once with 20 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.5, and 1 mM DTT and centrifuged as described above. The resulting pellet was collected and resuspended in the buffer described above.
Preparation of Protein Extracts, SDS-PAGE, and Immunoblot Analysis
E. coli protein was prepared according to the sixth edition of the Novagen pET system manual. Total maize protein was prepared by grinding tissue in 2 to 4 volumes of 0.1 M Tris, pH 8.0, 0.1 M NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 0.1% Triton X-100, and 1% insoluble polyvinylpyrrolidone and then centrifuging the resulting material at 14,000g for 20 min. The supernatant was subjected to SDS-PAGE according to Sambrook et al. (1989). Separated proteins were transferred to a nitrocellulose membrane using a semidry electrophoretic transfer cell (Bio-Rad, Hercules, CA) at 20 V for 2 hr. Membranes were blocked using 3% BSA in TBS and incubated with purified anti-RF2 antibodies at room temperature for 3 hr, washed with TBS and 0.05% Tween 20 three times for 10 min each, and then incubated with anti-rabbit IgG alkaline phosphatase secondary antibody (1:30,000 dilution; Sigma) for 1 hr. The result was visualized by staining with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Sigma).
Aldehyde Dehydrogenase Assays
For E. coli aldehyde dehydrogenase (ALDH) assays (modified from op den Camp and Kuhlemeier, 1997), cells were washed with water and resuspended in 0.1 volume of the original cell cultures of 0.1 M Hepes, pH 7.4, 1 mM EDTA, 2 mM DTT, and 0.1% Triton X-100, treated with lysozyme (0.1 mg/mL) on ice for 20 min, and sonicated three times for 10 sec each using a Fisher model 60 sonicator at maximum output. Between sonication treatments, the cellular extract was cooled in an ice-water bath. The lysate was centrifuged at 14,000g for 20 min. The resulting supernatant was used as a crude enzyme extract.
For maize mitochondrial ALDH assays, mitochondria were prepared from maize seedlings or unpollinated ears according to Payne et al. (1980). The resulting mitochondrial pellet was resuspended in 0.1 M Hepes, pH 7.4, 1 mM EDTA, 2 mM DTT, and 0.1% Triton X-100, sonicated, and centrifuged as described above. The resulting supernatant served as a crude enzyme extract.
For each ALDH assay, 200 μg of E. coli protein extract or 180 μg of mitochondrial protein extract was added to a reaction mixture containing 1.5 mM NAD (Sigma) and 0.1 M sodium pyrophosphate buffer, pH 8.5, and the total volume was adjusted to 1.0 mL with water. After 30 sec of prereaction, aldehyde (17 μM acetaldehyde or 20 μM glycolaldehyde) was added to the mixture, which was excited at 360 nm, and the emission fluorescence of NADH was recorded at 460 nm at 10-sec intervals for up to 2 min on a model F-2000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). ALDH activity was expressed as nanomoles of NADH production per minute per milligram of protein.
Reverse Transcription–PCR of rf2-R213
RNA from immature tassels (still in the whorl) from the inbred line R213 was extracted according to Dean et al. (1985). First-strand DNA synthesis was conducted from 5 μg of RNA using a 3′ rapid amplification of cDNA ends kit (Gibco BRL, Rockville, MD) and rf2-specific primers (rf2-xq, 5′-CCAACTTTCCAGGCATACATCA-3′; and RF2C2, 5′-CCAGGCTAGGGCAAATCTTAT-3′). The resulting DNA was used as a template in subsequent PCR reactions with various rf2 primer combinations that covered the entire rf2 coding sequence. Both strands of the PCR products were sequenced completely.
Acknowledgments
We thank Juan Aguilar (Department of Biochemistry, School of Pharmacy, University of Barcelona, Spain) for the gift of the ald-deficient E. coli strain JA111 and the plasmid pALD9, and Robert Thornburg (Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University) for providing access to the fluorescence spectrophotometer. We also thank Miwa Kojima for illustrating the maize spike-let. This research was supported in part by competitive grants from the United States Department of Agriculture National Research Initiative program to P.S.S. (Nos. 9801805 and 0001478) and from the Human Frontiers in Science Program (No. RG0067) to Cris Kuhlemeier (Institute of Plant Physiology, University of Berne, Switzerland) and P.S.S. This is journal paper No. 18706 of the Iowa Agriculture and Home Economics Experiment Station (Ames, IA), project No. 3554, supported by Hatch Act and State of Iowa funds.
References
- Asker, H., and Davies, D.D. (1985). Mitochondrial aldehyde dehydrogenase from plants. Phytochemistry 24 689–693. [Google Scholar]
- Barkan, A., and Martienssen, R.A. (1991). Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc. Natl. Acad. Sci. USA 88 3502–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat, A., and Aguilar, J. (1979). Rhamnose-induced propanediol oxidoreductase in Escherichia coli: Purification, properties, and comparison with the fucose-induced enzyme. J. Bacteriol. 140 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero, E., Baldoma, L., Ros, J., Boronat, A., and Aguilar, J. (1983). Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J. Biol. Chem. 258 7788–7792. [PubMed] [Google Scholar]
- Comstock, J.C., and Scheffer, R.P. (1973). Role of host-selective toxin in colonization of corn leaves by Helminthosporium carbonum. Phytopathology 63 24–29. [Google Scholar]
- Cui, X., Wise, R.P., and Schnable, P.S. (1996). The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272 1334–1336. [DOI] [PubMed] [Google Scholar]
- Davies, D.D. (1959). The purification and properties of glycolaldehyde dehydrogenase. J. Exp. Bot. 11 289–295. [Google Scholar]
- Dean, C., Van Den Elzen, P., Tamaki, S., Dunsmuir, P., and Bedbrook, J. (1985). Differential expression of the eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J. 4 3055–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version 2. Plant Mol. Biol. Rep. 1 19–22. [Google Scholar]
- Dewey, R.E., Levings III, C.S., and Timothy, D.H. (1986). Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell 44 439–449. [DOI] [PubMed] [Google Scholar]
- Dewey, R.E., Timothy, D.H., and Levings III, C.S. (1987). A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc. Natl. Acad. Sci. USA 84 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbroff, E.B., and Gepstein, S. (1993). Immunological methods for assessing protein expression in plants. In Methods in Plant Molecular Biology and Biotechnology, B.R. Glick and J.E. Thompson, eds (Boca Raton, FL: CRC Press), pp. 207–241.
- Errede, B., Kamen, M.D., and Hatefi, Y. (1967). Preparation and properties of complex IV (ferrocytochrome c:oxidoreductase EC 1.9.3.1). Methods Enzymol. 10 41–48. [DOI] [PubMed] [Google Scholar]
- Fauron, C.M.-R., Abbott, A.G., Brettell, R.I.S., and Gesteland, R.F. (1987). Maize mitochondrial DNA rearrangements between the normal type, the Texas male sterile cytoplasm, and a fertile revertant cms-T regenerated plant. Curr. Genet. 11 339–346. [Google Scholar]
- Ferrandez, A., Prieto, M.A., Garcia, J.L., and Diaz, E. (1997). Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 406 23–27. [DOI] [PubMed] [Google Scholar]
- Forde, B.G., and Leaver, C.J. (1980). Nuclear and cytoplasmic genes controlling synthesis of variant mitochondrial polypeptides in male sterile maize. Proc. Natl. Acad. Sci. USA 77 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling, M., and Schwartz, D. (1973). Genetic relationships between the multiple alcohol dehydrogenases of maize. Biochem. Genet. 8 27–36. [DOI] [PubMed] [Google Scholar]
- Gengenbach, B.G., Connelly, J.A., Pring, D.R., and Conde, M.F. (1981). Mitochondrial DNA variation in maize plants regenerated during tissue culture selection. Theor. Appl. Genet. 59 161–167. [DOI] [PubMed] [Google Scholar]
- Greene, B., Walko, R., and Hake, S. (1994). Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hack, E., Lin, C., Yang, H., and Horner, H.T. (1991). T-URF13 protein from mitochondria of Texas male-sterile maize (Zea mays L.). Plant Physiol. 95 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S., Abad, A.R., Gelvin, S.B., and Mackenzie, S.A. (1996). A cytoplasmic male sterility–associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proc. Natl. Acad. Sci. USA 93 11763–11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo, E., Chen, Y.-M., Lin, E.C.C., and Aguilar, J. (1991). Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J. Bacteriol. 173 6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker, A.L., Smith, D.R., Jim, S.R., and Beckett, J.B. (1970). Reaction of corn seedlings with male-sterile cytoplasm to Helminthosporium maydis. Plant Dis. Rep. 54 708–712. [Google Scholar]
- Hsu, S.U., and Peterson, P.A. (1991). The upper and lower florets of spikelets in maize. J. Genet. Breed. 45 215–222. [Google Scholar]
- Huang, J., Lee, S.-H., Lin, C., Medici, R., Hack, E., and Myers, A.M. (1990). Expression in yeast of the T-URF13 protein from Texas male-sterile maize mitochondria confers sensitivity to methomyl and to Texas-cytoplasm–specific fungal toxins. EMBO J. 9 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C., and Moore, A.L. (1979). Isolation of intact higher-plant mitochondria. In Plant Organelles, Methodological Surveys (B): Biochemistry, Vol. 9, E. Reid, ed (Chichester, UK: Ellis Horwood), pp. 1–12.
- Kemble, R.J., Flavell, R.B., and Brettell, R.I.S. (1982). Mitochondrial DNA analysis of fertile and sterile maize plants derived from tissue culture with the Texas male sterile cytoplasm. Theor. Appl. Genet. 62 213–217. [DOI] [PubMed] [Google Scholar]
- Kennell, J.C., and Pring, D.R. (1989). Initiation and processing of atp6, T-urf13 and ORF221 transcripts from mitochondria of T cytoplasm maize. Mol. Gen. Genet. 216 16–24. [Google Scholar]
- Kiesselbach, T.A. (1980). The Structure and Reproduction of Corn. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Klyosov, A.A. (1996). Kinetics and specificity of human liver aldehyde dehydrogenase toward aliphatic, aromatic, and fused polycyclic aldehydes. Biochemistry 35 4457–4467. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. (1996). MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Korth, K.L., Kaspi, C.I., Seidow, J.N., and Levings III, C.S. (1991). URF13, a maize mitochondrial pore-forming protein, is oligomeric and has a mixed orientation in Escherichia coli plasma membranes. Proc. Natl. Acad. Sci. USA 88 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood, R.C., and Walker, D.A. (1983). Chloroplast. In Isolation of Membranes and Organelles from Plant Cells, J.L. Hall and A.L. Moore, eds (London: Academic Press), pp. 185–210.
- Levings III, C.S. (1990). The Texas cytoplasm of maize: Cytoplasmic male sterility and disease susceptibility. Science 250 942–947. [DOI] [PubMed] [Google Scholar]
- Levings III, C.S. (1993). Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell 5 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings III, C.S., and Siedow, J.N. (1992). Molecular basis of disease susceptibility in the Texas cytoplasm of maize. Plant Mol. Biol. 19 135–147. [DOI] [PubMed] [Google Scholar]
- Lindahl, R. (1992). Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 27 283–335. [DOI] [PubMed] [Google Scholar]
- Lindahl, R., and Petersen, D.R. (1991). Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem. Pharmacol. 41 1583–1587. [DOI] [PubMed] [Google Scholar]
- Liu, Z.J., Sun, Y.J., Rose, J., Chung, Y.J., Hsiao, C.D., Chang, W.R., Kuo, I., Perozich, J., Lindahl, R., Hempel, J., and Wang, B.C. (1997). The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 4 317–326. [DOI] [PubMed] [Google Scholar]
- Lowe, B., Mathern, J., and Hake, S. (1992). Active Mutator elements suppress the Knotted phenotype and increase recombination at the Kn1–0 tandem duplication. Genetics 132 813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S., and Chase, C.D. (1990). Fertility restoration is associated with loss of a portion of the mitochondrial genome in cytoplasmic male-sterile common bean. Plant Cell 2 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S., Shichuan, H., and Lyznik, A. (1994). The elusive plant mitochondrion as a genetic system. Plant Physiol. 105 775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R., and Baron, A. (1994). Coordinate suppression of mutations caused by Robertson's Mutator transposons in maize. Genetics 136 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R.A., Barkan, A., Freeling, M., and Taylor, W.C. (1989). Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson's Mutator. EMBO J. 8 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R., Barkan, A., Taylor, W.C., and Freeling, M. (1990). Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 4 331–343. [DOI] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., and Phizicky, E.M. (1999). A biochemical genomics approach for identifying genes by the activity of their products. Science 286 1153–1155. [DOI] [PubMed] [Google Scholar]
- Marumo, S. (1986). Auxins. In Chemistry of Plant Hormones, N. Takahashi, ed (Boca Raton, FL: CRC Press), pp. 9–56.
- McCaffery, P., and Drager, U.C. (1993). Retinoic acid synthesis in the developing retina. Adv. Exp. Med. Biol. 328 181–190. [DOI] [PubMed] [Google Scholar]
- Nakai, K. (1991). Predicting various targeting signals in amino acid sequences. Bull. Inst. Chem. Res. Kyoto Univ. 69 269–291. [Google Scholar]
- op den Camp, R.G., and Kuhlemeier, C. (1997). Aldehyde dehydrogenase in tobacco pollen. Plant Mol. Biol. 35 355–365. [DOI] [PubMed] [Google Scholar]
- Osakovskii, V.L., Ponomarev, A.G., Bubyakina, V.V., Tatarinova, T.D., Kononova, S.K., Li, N.G., Khokhlachev, A.V., Kovalenko, V.A., Pashkov, V.I., and Yakunina, N.B. (1992). Aldehyde dehydrogenase of mung bean (Phaseolus aureus) root mitochondria: Characterization and preparation of monospecific polyclonal antibodies. Biokhimiya 57 418–429. [Google Scholar]
- Parthasarathy, M.V. (1994). Transmission electron microscopy: Chemical fixation, freezing methods, and immunolocalization. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 118–134.
- Payne, G., Kono, Y., and Daly, J.M. (1980). A comparison of purified host specific toxin from Helminthosporium maydis, race T, and its acetate derivative on oxidation by mitochondria from susceptible and resistant plants. Plant Physiol. 65 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozich, J., Nicholas, H., Wang, B.C., Lindahl, R., and Hempel, J. (1999). Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 8 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring, D.R., and Lonsdale, D.M. (1989). Cytoplasmic male sterility and maternal inheritance of disease susceptibility in maize. Annu. Rev. Phytopathol. 27 483–502. [Google Scholar]
- Quail, P.H. (1979). Plant cell fractionation. Annu. Rev. Plant Physiol. 30 425–484. [Google Scholar]
- Rajagopal, R. (1971). Metabolism of indole-3-acetaldehyde. III. Some characteristics of the aldehyde oxidase of Avena coleoptile. Physiol. Plant. 24 272–281. [Google Scholar]
- Rocha, V., and Ting, I.P. (1970). Preparation of cellular plant organelles from spinach leaves. Arch. Biochem. Biophys. 140 398–407. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schnable, P.S., and Wise, R.P. (1994). Recovery of heritable, transposon-induced, mutant alleles of the rf2 nuclear restorer of T-cytoplasm maize. Genetics 136 1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P.S., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3 175–180. [Google Scholar]
- Siu, G.M., and Draper, H.H. (1982). Metabolism of malonaldehyde in vivo and in vitro. Lipids 17 349–355. [DOI] [PubMed] [Google Scholar]
- Sladek, N.E., Manthey, C.L., Maki, P.A., Zhang, Z., and Landkamer, G.J. (1989). Xenobiotic oxidation catalyzed by aldehyde dehydrogenases. Drug. Metab. Rev. 20 697–720. [DOI] [PubMed] [Google Scholar]
- Steinmetz, C.G., Xie, P., Weiner, H., and Hurley, T.D. (1997). Structure of mitochondrial aldehyde dehydrogenase: The genetic component of ethanol aversion. Structure 5 701–711. [DOI] [PubMed] [Google Scholar]
- Styrvold, O.B., Falkenberg, P., Landfald, B., Eshoo, M.W., Bjornsen, T., and Strom, A.R. (1986). Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester, A.W., and Ruzin, S.E. (1994). Light microscopy. I. Dissection and microtechnique. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 89–92.
- Tadege, M., Dupuis, I., and Kuhlemeier, C. (1999). Ethanolic fermentation: New function for an old pathway. Trends Plant Sci. 4 320–325. [DOI] [PubMed] [Google Scholar]
- Ullstrup, A.J. (1972). The impacts of the southern corn leaf blight epidemics of 1970–1971. Helminthosporium turcicum. Annu. Rev. Phytopathol. 10 37–50. [Google Scholar]
- Umbeck, P.F., and Gengenbach, B.G. (1983). Reversion of male-sterile T-cytoplasm maize to male fertility in tissue culture. Crop Sci. 23 584–588. [Google Scholar]
- Vasiliou, V., Bairoch, A., Tipton, K.F., and Nebert, D.W. (1999). Eukaryotic aldehyde dehydrogenase (ALDH) genes: Human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9 421–434. [PubMed] [Google Scholar]
- Vigil, E.L. (1983). Microbodies. In Isolation of Membranes and Or-ganelles from Plant Cells, J.L. Hall and A.L. Moore, eds (London: Academic Press), pp. 211–236.
- Vojtechova, M., Hanson, A.D., and Munoz-Clares, R.A. (1997). Betaine-aldehyde dehydrogenase from amaranth leaves efficiently catalyzes the NAD-dependent oxidation of dimethylsulfoniopropionaldehyde to dimethylsulfoniopropionate. Arch. Biochem. Biophys. 337 81–88. [DOI] [PubMed] [Google Scholar]
- Wang, X., Mann, C.J., Bai, Y., Ni, L., and Weiner, H. (1998). Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J. Bacteriol. 180 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke, H.E., and Lee, S.L.J. (1977). Mitochondrial degeneration in Texas cytoplasmic male-sterile corn anthers. J. Hered. 68 213–222. [Google Scholar]
- Warmke, H.E., and Lee, S.L.J. (1978). Pollen abortion in T cytoplasmic male-sterile corn (Zea mays): A suggested mechanism. Science 200 561–563. [DOI] [PubMed] [Google Scholar]
- Wightman, F., and Cohen, D. (1968). Intermediary steps in the enzymatic conversion of tryptophan to IAA in cell-free systems from mung bean seedlings. In Biochemistry and Physiology of Plant Growth Substances: Proceedings of the 6th International Conference on Plant Growth Substances, F. Wightman and G. Setterfield, eds (Ottawa, Canada: Runge Press), p. 273.
- Wise, R.P., and Schnable, P.S. (1994). Mapping complementary genes in maize: Positioning the rf1 and rf2 nuclear-fertility restorer loci of Texas (T) cytoplasm relative to RFLP and visible markers. Theor. Appl. Genet. 88 785–795. [DOI] [PubMed] [Google Scholar]
- Wise, R.P., Fliss, A.E., Pring, D.R., and Gengenbach, B.G. (1987. a). urf13-T of T-cytoplasm maize mitochondria encodes a 13 kD polypeptide. Plant Mol. Biol. 9 121–126. [DOI] [PubMed] [Google Scholar]
- Wise, R.P., Pring, D.R., and Gengenbach, B.G. (1987. b). Mutation to male fertility and toxin insensitivity in Texas (T)-cytoplasm maize is associated with a frame shift in a mitochondrial open reading frame. Proc. Natl. Acad. Sci. USA 84 2858–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, R.P., Bronson, C.R., Schnable, P.S., and Horner, H.T. (1999). The genetics, pathology, and molecular biology of T-cytoplasm male sterility in maize. Adv. Agron. 65 79–130. [Google Scholar]
- Yoder, O.C. (1973). A selective toxin produced by Phyllosticta maydis. Phytopathology 63 1361–1366. [Google Scholar]
- Yoshida, A., Rzhetsky, A., Hsu, L.C., and Chang, C. (1998). Human aldehyde dehydrogenase gene family. Eur. J. Biochem. 251 549–557. [DOI] [PubMed] [Google Scholar]
- Zinovieva, R.D., Tomarev, S.I., and Piatigorsky, J. (1993). Aldehyde dehydrogenase–derived omega-crystallins of squid and octopus: Specialization for lens expression. J. Biol. Chem. 268 11449–11455. [PubMed] [Google Scholar]