Abstract
A mutant screen was conducted in Arabidopsis that was based on deregulated expression of auxin-responsive transgenes. Two different tightly regulated (i.e., very low expression in the absence of auxin treatment and very high expression after exogenous auxin treatment) auxin-responsive promoters were used to drive the expression of both a β-glucuronidase (GUS) reporter gene and a hygromycin phosphotransferase (HPH)–selectable marker gene. This screen yielded several mutants, and five of the mutations (axe1-1 to axe1-5) mapped to the same locus on chromosome 5. A map-based cloning approach was used to locate the axe1 mutations in an Arabidopsis RPD3-like histone deacetylase gene, referred to as HDA6. The axe1 mutant plants displayed increased expression of the GUS and HPH transgenes in the absence of auxin treatment and increased auxin-inducible expression of the transgenes compared with nonmutant control plants. None of a variety of endogenous, natural auxin-inducible genes in the mutant plants were upregulated like the transgenes, however. Results of treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine suggest that the axe1 mutations affect transgene silencing; however, histone deacetylase inhibitors had no affect on transgene silencing in mutant or control plants. The specific effect of AtHDA6 mutations on the auxin-responsive transgenes implicates this RPD3-like histone deacetylase as playing a role in transgene silencing. Furthermore, the effect of AtHDA6 on transgene silencing may be independent of its histone deacetylase activity.
INTRODUCTION
Acetylation and deacetylation of histones are known to play important roles in the regulation of gene expression in eukaryotes (reviewed in Meyer, 2000). Histone acetyltransferases (reviewed in Grant and Berger, 1999; Strahl and Allis, 2000) and histone deacetylases (HDACs; reviewed in Johnson and Turner, 1999; Knoepfler and Eisenman, 1999; Ng and Bird, 2000) have been identified in large, multisubunit complexes that target these enzymes to specific sites in nuclear DNA. In the nucleosomes of tightly coiled, condensed chromatin, positively charged lysine residues in the N-terminal tails of core histones interact with DNA and other chromosomal proteins. Acetylation of the lysine residues by histone acetyltransferases disrupts these interactions, resulting in relaxation of the chromatin structure, which allows gene expression to occur (Meyer, 2000). Conversely, the deacetylation of lysine residues in histones by HDACs results in the condensation of chromatin structure and the repression of gene expression (Meyer, 2000).
HDAC genes and proteins have been isolated and characterized from a variety of animals, fungi, and plants. There are two subgroups of HDACs found in all eukaryotes. One subgroup is most closely related to the yeast RPD3 HDAC, and the other is more closely related to yeast HDA1 (Johnson and Turner, 1999). These HDAC proteins contain a core of conserved amino acids, and mutations within this core result in the loss of HDAC activity and the loss of transcriptional repression (Hassig et al., 1997; Kadosh and Struhl, 1998). In addition to these two subgroups of HDACs, plants contain a family of HDACs (HD2) that appear to be unique and unrelated to yeast RPD3 (Lusser et al., 1997; Kölle et al., 1999; Lechner et al., 2000; Wu et al., 2000). HDACs generally are found as part of multiprotein complexes that contain transcriptional repressors, corepressors, and a variety of other proteins (Ng and Bird, 2000). In some cases, HDAC complexes contain ATP-dependent chromatin-remodeling proteins and proteins that bind to methylated DNA (Ahringer, 2000).
Several studies indicate a connection between DNA methylation and histone deacetylation. The mammalian methyl-CpG binding protein MeCP2 recruits the SIN3–HDAC complex to methylated DNA to mediate transcriptional repression (Nan et al., 1998), whereas a related protein, MBD2, directs the NuRD–HDAC complex to methylated DNA (Zhang et al., 1999). In several cases, gene silencing has been reported to be relieved by treatment with either HDAC inhibitors or an inhibitor of DNA methylation (Chen and Pikaard, 1997; Pikaart et al., 1998; Selker, 1998). The data suggest that DNA methylation results in histone deacetylation and gene silencing, but one report also suggests that HDACs may control DNA methylation, because methylation was reduced in Neurospora cells treated with an HDAC inhibitor (Selker, 1998). Two other findings that link chromatin remodeling factors with DNA methylation and transcriptional silencing are the recent cloning of the ddm1 and mom loci in Arabidopsis (Jeddeloh et al., 1999; Amedeo et al., 2000). ddm1 mutants have 70% less genomic cytosine methylation than do wild-type plants, and the DDM1 gene encodes a predicted protein with a high degree of similarity to the SWI2/ SNF2 family of chromatin remodeling factors (Jeddeloh et al., 1999). Mutations in the MOM gene release the transcriptional silencing of methylated genes without affecting their methylation patterns. The MOM gene encodes a nuclear protein that contains a region related to part of the ATPase region of the SWI2/SNF2 family of proteins (Amedeo et al., 2000).
Until recently, little attention has been given to the roles that plant HDACs may play in transcriptional repression or gene silencing. Two Arabidopsis genes with similarity to maize HD2 (AtHD2A and AtHD2B) have been identified, and their gene products have been characterized as repressors (Wu et al., 2000). Very recently, a homolog of yeast RPD3 (AtHD1) was cloned from Arabidopsis (Tian and Chen, 2001). Hyperacetylation of histone H4 and a range of developmental abnormalities were observed in plants expressing an antisense AtHD1 construct (Tian and Chen, 2001). We have isolated mutations (five alleles) in the HDA6 gene of Arabidopsis, which also is closely related to yeast RPD3. These alleles were identified in a screen designed to isolate mutants with increased expression of transgenes and natural genes regulated by the plant hormone auxin. Our results with the AtHDA6 mutant plants suggest that this HDAC, in particular, plays a role in the silencing of transgenes.
RESULTS
Development of a Screen for Mutants with Increased Expression of Auxin-Regulated Transgenes
A screen for Arabidopsis mutants with increased expression of auxin-regulated transgenes was performed, which involved the use of a selectable marker gene encoding hygromycin resistance (hygromycin phosphotransferase [HPH]; Gritz and Davies, 1983) and the β-glucuronidase (GUS) reporter gene (Jefferson, 1987). Promoters containing auxin-responsive elements (AuxREs) were ligated upstream of each of these genes, and the constructs were used to transform Arabidopsis plants.
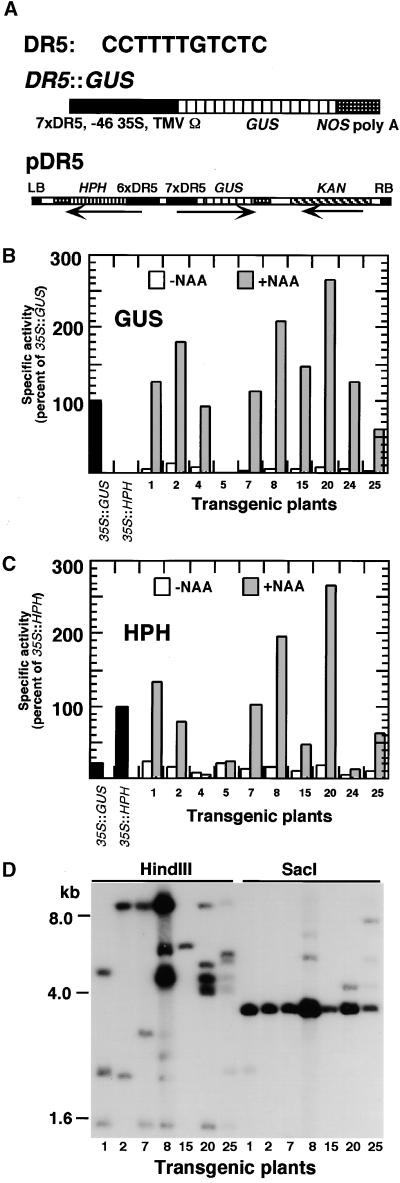
Diagrams of the T-DNA regions of the binary Ti vectors pDR5 and p2xD0 are shown in Figures 1A and 2A, respectively. In pDR5, the HPH and GUS genes were controlled by promoters containing multimers of the minimal, synthetic AuxRE DR5 (Figure 1A, top; Ulmasov et al., 1997). The GUS construct (Figure 1A, middle) contained seven copies of DR5 ligated upstream of a minimal cauliflower mosaic virus 35S promoter and the tobacco mosaic virus Ω sequence. The HPH construct was similar, but it had six copies of DR5. Both genes were terminated by the nopaline synthase polyadenylation sequence. The genes were transferred to the binary Ti vector pBIN19 (Bevan, 1984) to create pDR5 (Figure 1A, bottom). This vector also carried a kanamycin resistance gene for the selection of transgenic plants.
Figure 1.
The pDR5 Construct and Expression in Transgenic Plants.
(A) Top: Sequence of the pDR5 AuxRE. Middle: The DR5::GUS gene, which has seven copies of DR5 (7xDR5) ligated upstream of the minimal 35S promoter (−46 35S) and the tobacco mosaic virus Ω sequence (TMV Ω). The β-glucuronidase coding sequence (Escherichia coli uidA gene, referred to as GUS) was terminated by the nopaline synthase polyadenylation sequence (NOS poly A). Bottom: T-DNA region of the binary vector pDR5 containing DR5::GUS (GUS), DR5::HPH (HPH), and a gene encoding kanamycin resistance (KAN). DR5::HPH was similar to DR5::GUS but was driven by six copies of DR5 and contained the coding region for the HPH gene. Arrows indicate the direction of transcription of each gene. LB, left T-DNA border; RB, right T-DNA border.
(B) GUS-specific activities in seedlings from individual T2 lines transformed with pDR5. Six-day-old seedlings were treated with water (white bars) or water supplemented with 50 μM NAA (gray bars) for 24 hr and assayed for GUS activity fluorimetrically. Data for control lines transformed with 35S::GUS or 35S::HPH also are shown (black bars). Data are means of duplicate assays and are expressed as percentages of the activity in the 35S::GUS line.
(C) HPH-specific activities in seedlings from individual T2 lines transformed with pDR5. Seedlings were treated as described for (B). Data are expressed as percentages of the activity in the 35S::HPH line.
(D) DNA gel blot analysis of selected transgenic lines. Each lane contained ∼20 μg of DNA isolated from T2 seedlings of lines 1, 2, 7, 8, 15, 20, or 25 digested with the restriction endonuclease HindIII or SacI as indicated. The blot was probed with a radiolabeled HPH cDNA fragment.
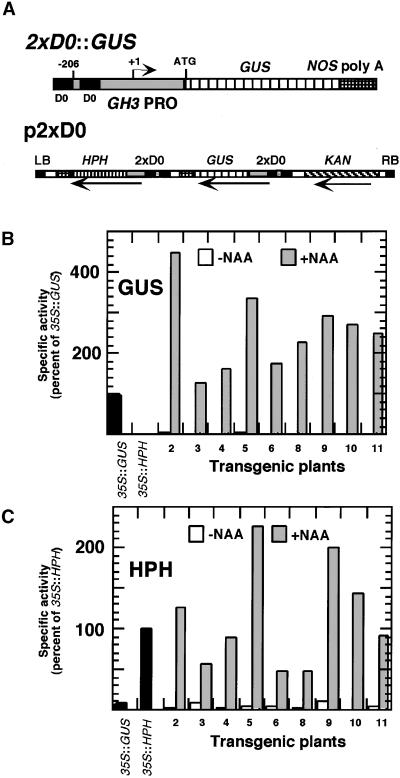
Figure 2.
The p2xD0 Construct and Expression in Transgenic Plants.
(A) Top: The 2xD0::GUS gene. The promoter was derived from the soybean GH3 gene, deleted to a position 260 bp upstream of the transcription start site. A second copy of the D0 element, present in the promoter, was ligated upstream of the first. The GUS gene was terminated by the nopaline synthase polyadenylation (NOS poly A) sequence. Bottom: T-DNA region of the binary vector p2xD0, which is similar to that of pDR5 (see legend to Figure 1), but with the GUS and HPH genes driven by the 2xD0 promoter rather than the DR5 promoter. LB, left T-DNA border; RB, right T-DNA border.
(B) GUS-specific activities in seedlings of T2 lines transformed with p2xD0. Seedlings were treated and data are presented as described for Figure 1B.
(C) HPH-specific activities in seedlings of T2 lines transformed with p2xD0. Seedlings were treated and data are presented as described for Figure 1C.
Arabidopsis (ecotype Columbia [Col]) plants were transformed with pDR5, and independent T2 lines were tested for GUS and HPH activities in seedlings (Figures 1B and 1C). In most lines, water-treated seedlings exhibited very low levels of GUS and HPH activities; however, after treatment with the auxin 1-naphthaleneacetic acid (NAA; 50 μM for 24 hr), activities were increased up to 100-fold. We reported previously GUS histochemical staining results that indicated that exogenous application of auxin induced this promoter within most, if not all, organs of 7-day-old Arabidopsis seedlings (Ulmasov et al., 1997). Segregation of kanamycin resistance was used to identify lines that were transformed at single loci. These plants were analyzed using a DNA gel blot to identify lines with intact T-DNA inserts and to estimate the T-DNA copy numbers at the transgenic loci. The blot contained DNA from T2 seedlings of the seven lines digested with either HindIII or SacI and probed with an HPH cDNA fragment. SacI cut within the T-DNA and had a predicted HPH-hybridizing fragment of 3.4 kb (Figure 1D). All transgenic lines contained hybridizing SacI fragments of this size. The HPH gene in pDR5 is flanked by a HindIII restriction site and the left T-DNA border, so that after HindIII digestion, hybridizing bands will vary in size depending on the location of the T-DNA inserts in the plant genome. The number and intensity of hybridizing bands give an indication of the number of T-DNA inserts in each line. Most lines contained multiple inserts, and the two lines with the most inserts, lines 8 and 20, also showed the highest expression of the GUS and HPH transgenes. Only line 15 appeared to contain a single T-DNA insert. The two lines (8 and 20) with the greatest increases in auxin-regulated gene expression and the highest expression after auxin treatment were chosen for mutagenesis.
In addition to the synthetic DR5 promoter construct, a more natural auxin-responsive promoter (2xD0), which was derived from the soybean GH3 gene (Hagen et al., 1991; Liu et al., 1994; Ulmasov et al., 1995), was constructed. The GH3 promoter was deleted to a position 206 bp upstream of the transcription start site. A 71-bp fragment from the same promoter, containing the D0 AuxRE (positions −110 to −181 with respect to the transcription start site), was ligated upstream of the deleted promoter to create a duplication of the D0 element (Figure 2A, top). The binary vector p2xD0 (Figure 2A, bottom) was similar to DR5, but with the 2xD0 promoter in place of the DR5 promoters. Transgenic plants carrying this construct showed strong auxin-inducible expression of both the GUS and HPH transgenes (Figures 2B and 2C). GUS histochemical staining of 1- to 2-week-old seedlings indicated that the 2xD0 promoter, like the DR5 promoter, was induced by 1 to 50 μM NAA in most, if not all, organs (data not shown). Lines 2 and 10 were chosen for mutagenesis. On the basis of DNA gel blot analysis, line 2 contained multiple T-DNA inserts and line 10 contained a single insert (data not shown).
Segregation of kanamycin resistance was used to identify homozygous T3 seed batches. Approximately 106,000 (24,000 pDR5 line 8; 44,000 pDR5 line 20; 19,000 p2xD0 line 2; and 19,000 pDR5 line 10) homozygous seed were mutagenized with ethyl methanesulfonate and grown in soil to obtain M2 seed. The M2 seed were germinated on medium containing hygromycin. For each transgenic line, a concentration of hygromycin was identified that was just high enough to kill transgenic plants. The concentration varied for different lines because they had varying levels of background HPH gene expression (i.e., expression in the absence of exogenous auxin). Plants derived from pDR5 lines 8 and 20 and p2xD0 lines 2 and 10 were treated with hygromycin concentrations of 30, 100, 25, and 20 mg/L, respectively. Approximately 665,000 M2 seedlings (360,000 from pDR5 and 305,000 from p2xD0) were tested for increased hygromycin resistance. Resistant plants (120 from pDR5 and 70 from p2xD0) were transferred to soil, and M3 seed were collected. The seed were tested for inheritance of the hygromycin resistance phenotype and for increased GUS activity.
Eight mutant lines were identified that had heritable, quantifiable increases in both HPH and GUS activities. These were backcrossed with wild-type Col plants and outcrossed with Landsberg erecta (Ler) plants for mapping. Segregation of hygromycin resistance among progeny from these crosses indicated that all eight mutations were recessive (data not shown). Preliminary mapping data indicated that five of the mutations were located near the bottom of chromosome 5, and complementation crosses between these lines indicated that they were allelic (data not shown). GUS and HPH activity data for these five mutants (axe1-1 to axe1-5, for auxin gene expression mutants) are shown in Figures 3A and 3B. axe1-4 was derived from p2xD0 line 2, whereas axe1-1, axe1-2, axe1-3, and axe1-5 were derived from pDR5 line 8. All five lines showed at least twofold higher expression of the transgenes compared with nonmutant transgenic controls.
Figure 3.
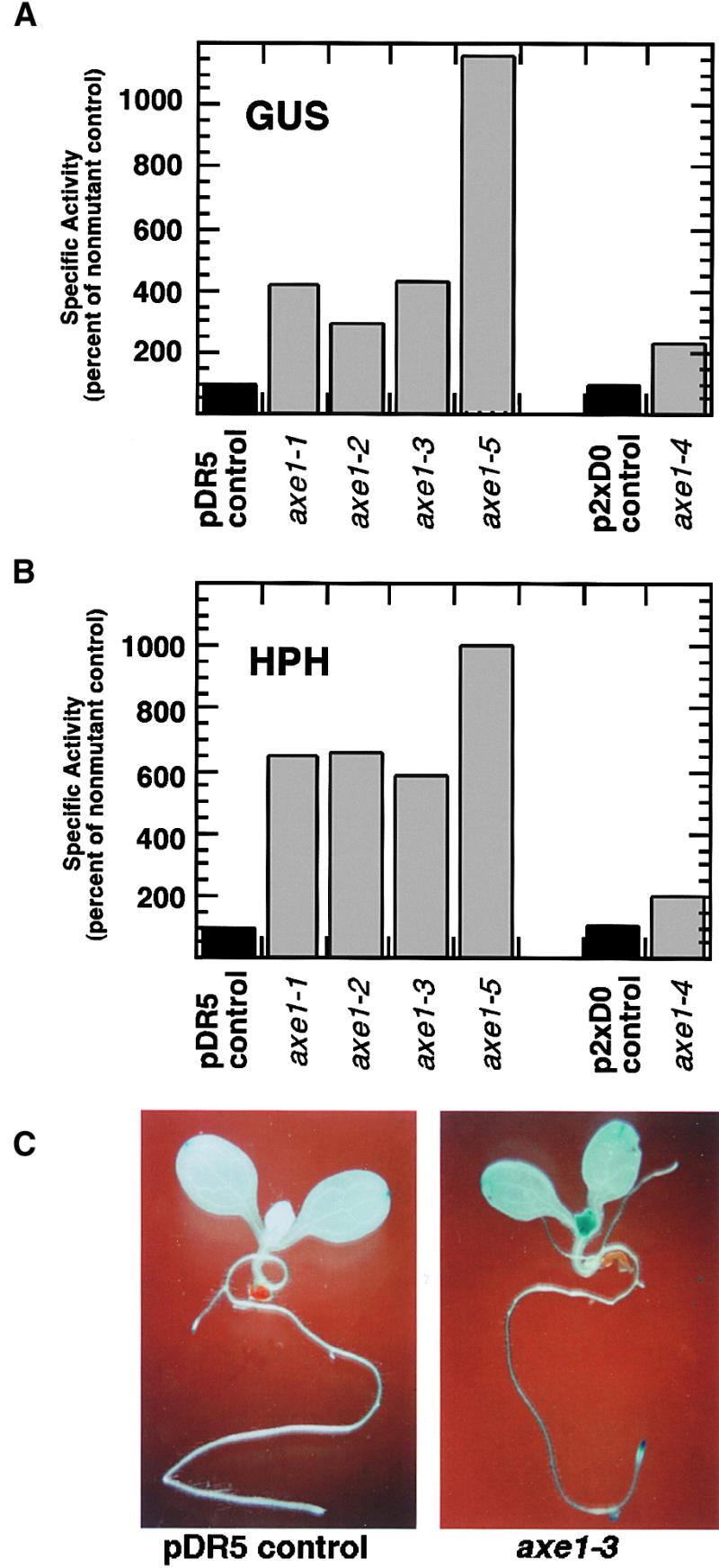
GUS and HPH Expression in axe1 Mutants.
(A) GUS-specific activities in 6-day-old M3 seedlings (gray bars) are expressed as percentages of nonmutant, transgenic controls (black bars). axe1-1, axe1-2, axe1-3, and axe1-5 were derived from pDR5 transgenic line 8, and axe1-4 was derived from p2xD0 line 2. Data are means of duplicate assays.
(B) HPH-specific activities in mutant seedlings are presented as described for (A).
(C) Histochemical staining for GUS activity in 8-day-old axe1-3 (right) and control (left) seedlings.
Seedlings from each mutant line were analyzed by histochemical staining for GUS activity. Nonmutant pDR5 and p2xD0 seedlings showed low levels of GUS expression in root tips, young leaves, and cotyledon tips (Ulmasov et al., 1997; data not shown). The five axe1 mutants displayed patterns of expression that were similar to those of nonmutant controls, but with levels of expression increased throughout the plants. Typical results are shown in Figure 3C. The nonmutant pDR5 seedling in this case showed detectable GUS expression only in the tips of the cotyledons. The axe1-3 seedling showed low levels of GUS staining throughout the plant, and higher expression in young leaves, cotyledon tips, and root tips. This pattern of expression was dissimilar to that produced by auxin treatment of wild-type plants, which resulted in very high levels of expression throughout the plant, with greatest expression in the roots (Ulmasov et al., 1997).
The axe1 Mutations Occur within a Putative HDAC Gene
A population of 803 mutant F2 plants from the Ler × axe1-3 cross was used for fine-mapping of the axe1 locus. The mutation was flanked by the cleaved amplified polymorphic sequence (CAPS) markers LFY3 and g2368 (Konieczny and Ausubel, 1993; http://www.arabidopsis.org/aboutcaps.html). Fifty-seven plants with recombinations between these markers were identified. Sequence data from the Arabidopsis sequence database were used to develop new simple sequence length polymorphism markers between LFY3 and g2368 (Bell and Ecker, 1994; see Methods for details). By this method, a 55-kb region of the P1 clone MDC12 (GenBank accession number AB008265) was identified that contained the mutation. This region contained 16 putative genes. One of these, MDC12.7, was chosen for sequencing because it encoded a homolog of the yeast HDAC RPD3, which is known to be a transcriptional repressor. Polymerase chain reaction (PCR) primers were designed to amplify sections of MDC12.7 covering the entire coding region. These were used to amplify the gene from each of the axe1 mutants, and the products were sequenced. Amplification and sequencing were repeated three times to confirm the results. In each mutant, a single base change (G to A or C to T) within the coding region of the gene was identified. The locations of the mutations are shown in Figure 4. In axe1-1, axe1-2, and axe1-3, the base changes resulted in missense mutations in the predicted amino acid sequence. In axe1-4 and axe1-5, the base changes occurred at intron splice sites.
Figure 4.
Amino Acid Sequence Alignment of RPD3-Like HDACs from Arabidopsis, Yeast RPD3, and Human HDAC1.
The sequences of Arabidopsis HDA6 (At6), HDA1 (At1), HDA7 (At7), and HDA9 (At9), S. cerevisiae RPD3 (Sc), and human HDAC1 (Hs) are compared. Identical amino acids are boxed in black, and similar amino acids are boxed in gray. Amino acid substitutions for axe1-1 (G127R), axe1-2 (G284D), and axe1-3 (A294V) are indicated as R, D, and V, respectively. Splice site mutations in axe1-4 and axe1-5 are indicated as //. Conserved H residues that are likely to be important for deacetylase activity are indicated by asterisks. Numbers to the right indicate amino acid residues from the initiator methionine.
The predicted MDC12.7 protein was highly similar to RPD3 from yeast (Saccharomyces cerevisiae) and the RPD3 homolog HDAC1 from humans; it is referred to as AtHDA6, in line with the designation given by the Plant Chromatin Database (http://ag.arizona.edu/chromatin/chromatin.html). Three other Arabidopsis proteins (HDA1 [referred to as AtHD1 in Tian and Chen, 2001], HDA7, and HDA9) also are similar to yeast RPD3 (The Arabidopsis Genome Initiative, 2000). Figure 4 shows an alignment of AtHDA6 with the three Arabidopsis HDA6-related proteins, yeast RPD3, and human HDAC1. Each of the missense mutations in AtHDA6 occurs within a conserved region of the gene; however, the mutations lie outside of the conserved core (Figure 4, asterisks), which is thought to be required for deacetylase activity (Hassig et al., 1997; Kadosh and Struhl, 1998).
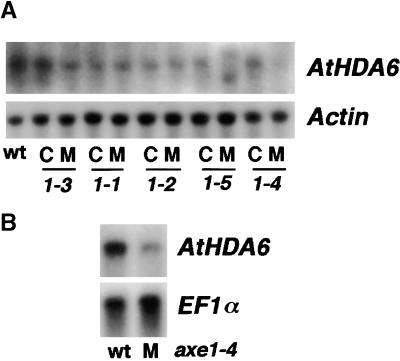
RNA gel blots were used to examine the expression of the AtHDA6 gene in the axe1 mutants. The mutants were backcrossed twice, and F3 lines from the second backcrosses were identified as either mutant or homozygous wild type at the axe1 locus. The wild-type F3 families were used as internal controls for each mutant line. Figure 5A shows the levels of AtHDA6 message in total RNA extracted from seedlings of mutants and controls. The abundance of AtHDA6 message in seedlings with any of the three missense mutations (axe1-1, axe1-2, and axe1-3) was similar to that found in wild-type seedlings. In the axe1-5 splice site mutant, AtHDA6 message of normal length was not detected, but two AtHDA6-hybridizing messages with altered lengths, one shorter and one longer than the wild type, were detected. In the axe1-4 splice site mutant, AtHDA6 message of normal length was detected, but the abundance was reduced. To more carefully assess the relative abundance of AtHDA6 mRNA in wild-type and axe1-4 mutant seedlings, we used polyadenylated RNA for hybridization. The blots shown in Figure 5B clearly show a reduction of AtHDA6 mRNA abundance in the axe1-4 seedlings.
Figure 5.
RNA Gel Blot Analysis of AtHDA6 Expression in axe1 Mutants.
(A) Analysis of total RNA (20 μg) from 8-day-old seedlings of wild-type Col (wt), mutants (M), and internal controls (C) for each mutant line (axe1-3, axe1-1, axe1-2, axe1-5, and axe1-4). The blot was probed with both a radiolabeled PCR product from the AtHDA6 gene and a radiolabeled Actin-2 cDNA, which was used as a loading control.
(B) Analysis of polyadenylated RNA (400 ng) from seedlings of wild-type Col (wt) and axe1-4 mutants (M). The blot was probed with both the AtHDA6 PCR product and a cDNA from EF1α, which was used as a loading control.
The axe1 Mutants Show No Alterations in Expression of Endogenous Auxin Response Genes
The axe1 mutants had no obvious alterations in morphology, size, or growth rate (data not shown). They were tested for alterations in root gravitropic responses and for altered root elongation rates in the presence of exogenous auxin, because these responses are altered in some previously characterized auxin-related mutants (Leyser et al., 1993, 1996; Timpte et al., 1994; Hobbie and Estelle, 1995; Bennett et al., 1996; Hobbie et al., 2000). The axe1 mutants showed normal gravitropic and root growth responses (data not shown).
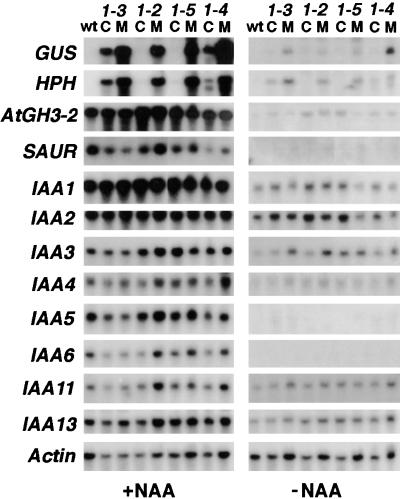
RNA gel blots were used to examine the expression of a number of genes that are rapidly upregulated by auxin (e.g., GH3, SAUR, and Aux/IAA; reviewed in Abel and Theologis, 1996; Guilfoyle et al., 1998) in mutant and control seedlings. As described for Figure 5, we used F3 seed batches from the second backcrosses for RNA isolation and gel blotting. These batches were all homozygous for the transgenes and either homozygous mutant or homozygous wild type at the axe1 locus. Untreated seedlings and seedlings treated with auxin (50 μM NAA for 2 hr) were examined. The blots shown in Figure 6 were probed with DNA fragments derived from HPH, GUS, an Arabidopsis GH3 homolog that we refer to as AtGH3-2, SAUR-AC1 (Gil et al., 1994), and eight Aux/IAA genes (Abel et al., 1995). Each mutant line showed greater expression of the GUS and HPH transgenes, compared with its internal control, in both untreated seedlings and seedlings treated with auxin; however, there were no clear alterations in expression of the endogenous auxin-regulated genes. The small differences observed between mutants and controls for some lines (e.g., expression of SAUR-AC1, IAA6, and IAA11 in auxin-treated axe1-2 seedlings) were not consistent with other lines and were most likely due to slight differences in gel loading (see data for Actin-2 control). On the autoradiographs shown in Figure 6, expression of some of the genes in untreated seedlings was not detected. After longer exposures, however, expression of all of the genes except SAUR-AC1 could be detected (data not shown). In total, the RNA gel blot data indicated that although expression of the auxin-responsive transgenes was upregulated in axe1 mutant seedlings, the endogenous auxin-responsive transgenes were expressed at similar levels in wild-type and mutant seedlings that were exposed or not exposed to exogenous auxin.
Figure 6.
RNA Gel Blot Analysis of Auxin-Regulated Genes in axe1 Mutants.
Analysis of total RNA (20 μg) from 8-day-old seedlings that were treated with either water (−NAA) or water containing 50 μM NAA (+NAA) for 2 hr. Seedlings from wild-type Col (wt), mutants (M), and internal controls (C) for each mutant line (axe1-3, axe1-2, axe1-5, and axe1-4) were analyzed. The blots were probed with cDNAs from GUS, HPH, an Arabidopsis GH3 homolog (AtGH3-2), the SAUR-AC1 gene (SAUR), and eight Aux/IAA genes (IAA1, -2, -3, -4, -5, -6, -11, and -13). The Actin-2 gene was used as a loading control.
To examine global gene expression patterns in axe1 mutants compared with nonmutant controls, we submitted RNA samples from axe1-4 mutant and control seedlings to the Arabidopsis Functional Genomics Consortium (AFGC) Microarray Facility at Michigan State University (see http://afgc.stanford.edu/). Results from this experiment are available at the Stanford Microarray Database (see data for Guilfoyle at http://genome-www4.Stanford.EDU/MicroArray/SMD/). Results from the microarray analysis are consistent with the RNA gel blot data presented in Figure 6 in that no members of the GH3, SAUR, or Aux/IAA auxin response gene families showed significantly altered expression in the axe1-4 mutant seedlings.
Effects of Methylation and HDAC Inhibitors on Transgene Expression in axe1 Mutants
Among the internal control lines shown in Figure 6, there was variation in expression of the transgenes, suggesting that transgene silencing may have affected the different F3 batches to varying degrees. The mutant plants were backcrossed four times, and new generations were always selected on medium containing hygromycin. Each generation retained the ability to grow at a level of hygromycin that killed the original T3 seed used for mutagenesis; however, a reduction in GUS expression levels has been observed in newer generations of mutants and their nonmutant siblings, with different mutant lines being affected to different degrees (data not shown). These observations, along with the results shown in Figure 6, suggest the possibility that the AtHDA6 mutations may actually affect the level of transgene silencing rather than auxin signaling.
The plant lines used for our mutant screen contained multiple copies of the transgenes at a single locus (Figure 1D; data not shown), and repetitive DNA is known to result in methylation and transgene silencing (Flavell, 1994; Park et al., 1996; Vaucheret et al., 1998). Because histone deacetylation appears to be connected with DNA methylation in the repression and silencing of genes (Chen and Pikaard, 1997; Nan et al., 1998; Pikaart et al., 1998; Selker, 1998; Bird and Wolffe, 1999; Zhang et al., 1999) and our mutations were found in an HDAC gene, the effects of DNA methylation and HDAC inhibitors on derepression of the transgenes were examined. To determine if demethylation might increase the expression of DR5::GUS in mutant and control plants, we tested an inhibitor of cytosine methylation, 5-aza-2′-deoxycytidine (aza-dC; Jones, 1985; Chen and Pikaard, 1997). Mutant and control Arabidopsis seedlings were germinated on medium containing 0, 1, 5, 10, or 20 mg/L aza-dC. Four days after germination, the seedlings were analyzed for GUS activity by using a histochemical stain. All of the aza-dC treatments resulted in higher expression of the GUS transgene, with similar levels of induction from concentrations of 5, 10, or 20 mg/L (data not shown). These treatments also severely inhibited plant growth; however, plants that were grown on 5 mg/L aza-dC could be rescued after transfer to medium lacking aza-dC. Therefore, a concentration of 5 mg/L aza-dC was used to grow plants for quantitative GUS assays.
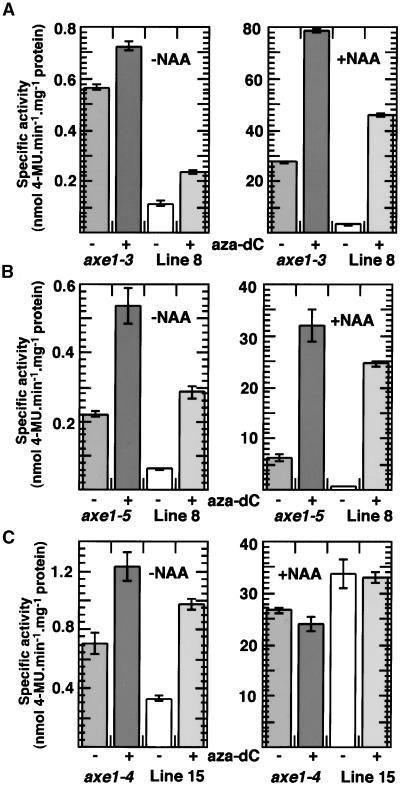
Figures 7A and 7B show the expression of DR5::GUS in axe1-3 and axe1-5 mutants after growth on unmodified medium or medium containing aza-dC. In each case, the mutants were compared with their respective internal controls as described for Figures 5 and 6. Both of these mutants (and their controls) were derived from pDR5 line 8. The 4-day-old seedlings were treated with potassium phosphate buffer or buffer plus NAA (10 μM, 24 hr) before the protein extracts were prepared. Both mutants showed greater expression than their controls with and without auxin treatment. After growth on aza-dC, the mutants still showed greater expression than the controls; however, the differences in expression levels between mutants and controls were reduced. These results suggested that transgene expression in both mutant and control plants was partially silenced due to methylation, because growth on aza-dC resulted in higher expression. The large increases in expression after auxin treatment suggest that methylation strongly inhibits auxin induction of the transgenes. The results also are consistent with the possibility that the AtHDA6 mutations resulted in increased expression of the transgenes through some effect involving DNA methylation, because treatment with aza-dC reduced the differences between mutants and controls. The differences were not eliminated, however. This finding may be due to the incomplete removal of methyl groups after aza-dC treatment or may indicate that some function other than methylation-induced silencing (i.e., histone deacetylation or intrinsic gene repression by AtHDA6) is affected in the axe1 mutants.
Figure 7.
GUS Expression in Mutant and Control Seedlings Grown with and without aza-dC.
(A) Specific activities in seedlings of axe1-3 mutants and pDR5 line 8 controls that had been grown for 4 days on unsupplemented growth medium (−) or medium containing 5 mg/L aza-dC (+) and then treated with potassium phosphate buffer (−NAA) or buffer containing 10 μM NAA (+NAA) for 2 hr. Note that the −NAA and +NAA histograms have different scales for specific activities (expressed as nmol 4-methylumbelliferone [4-MU]·min−1·mg−1 protein). Data are means of triplicate assays, and error bars indicate standard deviations.
(B) Specific activities in seedlings of axe1-5 mutants and pDR5 line 8 controls. The seedlings were treated and the data are presented as described for (A).
(C) Specific activities in seedlings of axe1-4 mutants in the pDR5 line 15 background and pDR5 line 15 controls. Seedlings were treated and the data are presented as described for (A).
Because single transgene copies are less susceptible to silencing than multiple copies (Flavell, 1994; Matzke and Matzke, 1995), transgene expression was examined in an axe1 mutant containing a single T-DNA insert. First, an axe1-4 (second backcrossed F3) line that was homozygous at the axe1 locus and heterozygous at the transgenic (p2xD0) locus was identified. Then, F4 progeny were selected that were mutant but nontransformed (i.e., GUS negative). These were crossed with pDR5 line 15 plants, which contained a single T-DNA insert (Figure 1D). F3 seed batches from this cross that were homozygous at both the axe1-4 and transgenic loci were identified. These plants are more resistant to hygromycin than are the nonmutant pDR5 line 15 plants (data not shown). GUS expression data for these axe1-4 pDR5 line 15 plants, compared with homozygous nonmutant line 15 controls, are shown in Figure 7C. The seedlings were grown in the presence or absence of aza-dC and treated with or without NAA as described for Figures 7A and 7B. In the pDR5 line 15 background, the differences in expression between the mutants and controls were less pronounced than in backgrounds containing multiple transgenes. Plants grown on aza-dC and not treated with auxin showed greater expression than plants grown in the absence of aza-dC, suggesting that these single-copy transgenic lines also may be partially affected by methylation-induced silencing. In contrast, the auxin-treated plants showed similar expression levels regardless of whether they were grown on aza-dC, indicating that the plants may have been less silenced than the lines containing multiple inserts. Mutant plants grown in the absence of aza-dC and not treated with auxin showed approximately twofold greater GUS expression than did the controls, but after growth on aza-dC, the difference between mutants and controls was greatly reduced. After auxin treatment, the control plants had slightly higher GUS activity than did the mutants, and this difference was not affected by growth on aza-dC. These results are consistent with the possibility that the AtHDA6 mutations resulted in increased expression of the transgenes through some effect involving DNA methylation.
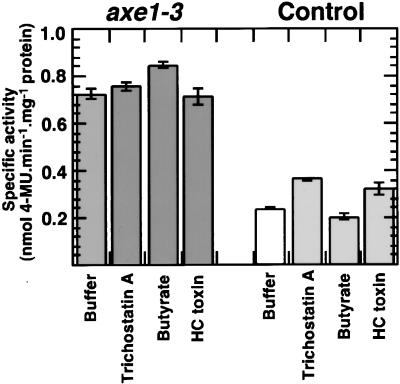
The effects of HDAC inhibitors on transgene expression in mutant and control plants also were investigated. Three different inhibitors were tested: sodium butyrate (Kruh, 1982; Cuisset et al., 1997) at concentrations ranging from 0.1 to 100 mM; trichostatin A (Yoshida et al., 1990, 1995) at concentrations of 1, 10, and 100 μM; and HC toxin (Brosch et al., 1995) at 2.5 μM. Several different transgenic pDR5 and p2xD0 lines, along with axe1 mutant and control seedlings (4 to 6 days old), were treated with potassium phosphate buffer or buffer containing these inhibitors for 24 hr and then tested for GUS activity by histochemical staining. Seedlings treated with HDAC inhibitors displayed GUS histochemical staining that was indistinguishable from that shown by seedlings that were not treated with the inhibitors, in contrast with seedlings that showed a strong increase in GUS histochemical staining after aza-dC treatment (data not shown). Seedlings also were treated with NAA (0, 0.1, or 1 μM, 24 hr) after sodium butyrate treatment (0, 0.1, or 1 mM, 24 hr). Based on histochemical staining, the sodium butyrate did not affect auxin-inducible GUS expression (data not shown). In two recent reports, silenced genes were shown to be derepressed by treatment with DNA methylation inhibitors followed by trichostatin A (Cameron et al., 1999; Lorincz et al., 2000). Therefore, the expression of DR5::GUS was tested in mutant and control plants that were first grown on aza-dC and treated subsequently with each of the three HDAC inhibitors. Results for axe1-3 mutants and their internal controls are shown in Figure 8. The quantitative GUS assay results are presented in this case because previous treatment with aza-dC resulted in a substantial gain in GUS expression compared with untreated controls. The combination of aza-dC plus HDAC inhibitors had little or no effect on GUS expression in mutants and controls beyond the effect seen with aza-dC treatment alone. The HDAC inhibitor experiments suggest that the derepression of transgene expression in axe1 plants may be related not to alterations in AtHDA6 deacetylase activity but to some other function of AtHDA6 (e.g., corepressor or repressor recruitment to an HDAC complex).
Figure 8.
GUS Expression in axe1-3 Mutant and Control Seedlings Treated with HDAC Inhibitors.
Seedlings of mutant axe1-3 (dark gray bars) and pDR5 line 8 (white and light gray bars) were grown for 4 days on medium supplemented with 5 mg/L aza-dC and then treated for 24 hr with potassium phosphate buffer (Buffer) or buffer containing 1 mM trichostatin A, 1 mM sodium butyrate (Butyrate), or 2.5 mM HC toxin. Specific activity data (expressed as nmol 4-methylumbelliferone [4-MU]· min−1·mg−1 protein) are means of triplicate assays, and error bars indicate standard deviations.
DISCUSSION
The Plant Chromatin Database (http://ag.arizona.edu/chromatin/atgenes.html), using sequence data supplied by The Arabidopsis Genome Initiative (2000), lists 15 HDAC genes in Arabidopsis, four of which are highly similar in amino acid sequence to the yeast RPD3 HDAC (Figure 4). Little is known about how the HDAC gene products function in Arabidopsis, but on the basis of studies with animal and fungal HDACs, they would be expected to play roles in deacetylating histones and transcription factors, repressing transcription, and silencing genes (Struhl, 1998; Ng and Bird, 2000). Antisense suppression of Arabidopsis HDA1 resulted in hyperacetylation of histone H4 and ectopic expression of at least one tissue-specific gene, indicating that this HDAC does play a role similar to that of RPD3-like HDACs in animals and fungi (Tian and Chen, 2001). The RPD3-like HDACs in Arabidopsis (i.e., HDA1, HDA6, HDA7, and HDA9) are strikingly similar throughout their amino acid sequences, with the exception of their C-terminal regions (Figure 4). Whether these related HDACs play redundant roles in regulating transcription or have distinct functions remains to be determined. Our identification of five independent mutations found exclusively in AtHDA6 suggests that the function of this HDAC may be different from those of other Arabidopsis HDACs. It is possible, however, that AtHDA6 was targeted in our mutant screen because it might be the predominant RPD3-like HDAC expressed in Arabidopsis seedlings. Expressed sequence tags available from the Arabidopsis database suggest that both AtHDA1 and AtHDA6 are expressed under a variety of conditions. On the other hand, no expressed sequence tags have been identified for AtHDA7 or AtHDA9.
Although the axe1 mutations affected the expression of the auxin-responsive transgenes, they apparently have no affect on the expression of natural auxin response genes in Arabidopsis. This conclusion is based on RNA gel blot analysis of 10 auxin-responsive genes, including members of the GH3, SAUR, and Aux/IAA gene families, and is supported by microarray analysis. The microarray analysis was performed with mRNA prepared from wild-type and axe1-4 mutant seedlings. Because the axe1-4 mutant plants display reduced amounts of AtHDA6 mRNA but are not depleted entirely of AtHDA6 mRNA, the microarray results must be interpreted with caution; however, this analysis did reveal several genes, not known to be auxin responsive, that were differentially expressed in wild-type and mutant plants (these data are accessible at the Stanford Microarray Database; see data for Guilfoyle at http://genome-www4.Stanford.EDU/MicroArray/SMD/).
The observation that axe1 mutations affected only the auxin-responsive transgenes and not natural auxin response genes raised the possibility that the mutants were isolated due to some effect involving transgene silencing rather than auxin signaling. Transgene silencing is a well-known phenomenon and is related mechanistically to other forms of epigenetic gene silencing (reviewed in Vaucheret et al., 1998; Grant, 1999; Wolffe and Matzke, 1999; Matzke et al., 2000). Two separate mechanisms for silencing are recognized, one referred to as transcriptional gene silencing (TGS) and the other referred to as post-transcriptional gene silencing (PTGS). In TGS, the promoters of transgenes become heavily methylated, and this corresponds to a reduction in the rate of transcription of the transgenes. In PTGS, transcription is not affected, but the levels of transgene-encoded mRNA are greatly reduced. PTGS correlates with an increase in methylation of the transcribed regions of the transgenes. The mechanisms by which de novo methylation of the transgenes occurs are not well understood; however, in the case of TGS, interactions between DNA repeats, and particularly inverted repeats, are implicated because these are more likely to become heavily methylated and silenced (Matzke et al., 2000). The strong correlation between promoter hypermethylation and gene silencing suggests that promoter methylation represses transcription.
The transgenic lines used for mutagenesis in this study contained repetitive DNA resulting from multiple transgene inserts at single loci. Furthermore, the T-DNA of the pDR5 construct contained an inverted repeat of ∼260 bp within its auxin-responsive promoters (i.e., seven copies of the DR5 AuxRE in one direction drove expression of the GUS gene and six copies in the opposite direction drove expression of the HPH gene; Figure 1A). The p2xD0 construct contained a direct repeat of the 2xD0 promoter (i.e., one of the repeats drove expression of the GUS gene and the second repeat drove expression of the HPH gene; Figure 2A), and within each 2xD0, there was a 71-bp direct repeat of the D0 AuxRE. The AuxRE repeats within the promoters were required for high-level auxin-inducible expression of the transgenes, and a repeat of the auxin-inducible promoters (i.e., pDR5 and p2xD0) was required to drive expression of the GUS reporter gene and the HPH-selectable marker gene. The repeated sequences in the auxin-responsive promoters might be good targets for methylation and gene silencing, based on previous studies (Matzke et al., 2000). This is supported by the silencing effects of the GUS gene that were observed through successive generations in both wild-type and mutant plants and by the partial reversibility of the silencing with aza-dC. The reduced silencing observed with the HPH gene was probably the result of selective pressure brought about by germinating each successive generation of mutant seedlings on hygromycin at a concentration that killed wild-type seedlings.
Recent results in animal systems indicate that repression of gene expression by methylation can involve the recruitment of HDACs by methyl-CpG binding proteins such as MeCP2 and that this repression is sensitive to deacetylase inhibitors such as trichostatin A (Jones et al., 1998; Nan et al., 1998). Several studies have indicated that gene silencing can be reversed or partially reversed either by treatment with the DNA methylation inhibitors aza-dC or 5′-azacytidine or by treatment with HDAC inhibitors such as trichostatin A, trapoxin, and butyrate (Chen and Pikaard, 1997; Chen et al., 1997; Pikaart et al., 1998; Selker, 1998). In these cases, it is thought that the acetylation of histones and the demethylation of promoter DNA somehow lead to derepression of the silenced gene. In other cases, silencing cannot be reversed by treatment with HDAC inhibitors alone but only by treatment with DNA methylation inhibitors followed by HDAC inhibitors (Cameron et al., 1999; Lorincz et al., 2000). This type of silencing suggests that HDAC activity may be the primary effector of repression with genes that have a low density of methylation but that an HDAC-independent mechanism operates in the repression of highly methylated genes. In other cases, treatment with DNA methylation inhibitors is effective in activating silenced or repressed genes, whereas HDAC inhibitors are ineffective even if applied after the methylation inhibitors (Chen et al., 2001).
Treatment with the DNA methylation inhibitor aza-dC is at least partially effective in reversing the silencing of the auxin-responsive transgenes in wild-type or axe1 mutant plants, but treatment with HDAC inhibitors is ineffective whether alone or in combination with aza-dC. This finding presents an intriguing situation, because repression of the transgenes is partially relieved by AtHDA6 mutations. One possible explanation for our results is that silencing of the transgenes may involve one or more HDACs, including AtHDA6, that are resistant to HDAC inhibitors. A trichostatin A–insensitive HDAC (i.e., HOS3) has been observed in yeast, but this is not an RPD3-like HDAC (Carmen et al., 1999). A second possibility is that the turnover of acetyl groups on the acetylated histones is too slow or the acetylase activity is too weak to be affected during HDAC inhibitor treatment. Another possibility is that AtHDA6 or an AtHDA6 complex has an affect on gene silencing that is independent of deacetylase activity.
Genetic studies on yeast RPD3, which is related to AtHDA6, have shown that although deacetylation is an important part of RPD3 repressive action, it is not the only mechanism by which transcription is repressed by this HDAC (Kadosh and Struhl, 1998). The results with yeast RPD3 suggest that HDACs may have functions beyond deacetylation that contribute to the repression of transcription. Some of this repression may be intrinsic to HDACs, and some may result from specific repressors associated with HDAC complexes (Kadosh and Struhl, 1997; Kingston and Narlikar, 1999). If this is the case, HDAC inhibitors may have only mild to moderate effects on derepressing silenced natural genes or transgenes. The missense axe1-1, axe1-2, and axe1-3 mutations in the AtHDA6 gene lie outside of a conserved core domain that has been shown to be required for HDAC activity (Kadosh and Struhl, 1998) and do not correspond to residues involved in contacting trichostatin A (Finnin et al., 1999). Additional experiments are required to determine if the axe1 mutations are defective in deacetylase activity or have activities similar to wild-type AtHDA6.
Note that several studies indicate a correlation between auxin treatment and DNA methylation levels in plant cell cultures. In carrot cell cultures, increases in exogenously added auxin levels resulted in increases in DNA methylation (LoSchiavo et al., 1989; Arnholdt-Schmitt, 1993; Arnholdt-Schmitt et al., 1991). Removal of auxin from the culture medium resulted in embryogenesis and an initial reduction in DNA methylation, followed by an increase in methylation during late embryogenesis (LoSchiavo et al., 1989). At this stage, it is difficult to see a connection between these observations and our results with the Arabidopsis AtHDA6 mutants. The transgenes used in this study were partially activated by aza-dC and activated to high levels of expression by auxin. These results suggest that the transcriptional mechanisms affected in the axe1 mutants are different from the pathways affecting methylation levels in the auxin-treated carrot cell cultures.
Future studies are required to determine the role that AtHDA6 plays in transgene silencing and what role it might play in regulating the expression of natural genes in Arabidopsis. It will be important to determine if AtHDA6 is part of an HDAC multisubunit complex and, if so, what components are in the complex. It may be that AtHDA6 will be found in a complex with other proteins that are known to play a role in gene silencing, including DDM1 (Jeddeloh et al., 1999) and MOM1 (Amedeo et al., 2000).
METHODS
Plant Growth Conditions
For all experiments involving seedlings, Arabidopsis thaliana seed were surface sterilized and plated on medium containing half-strength Murashige and Skoog (1962) salts, 1% sucrose, Gamborg's B5 vitamins (Sigma), and 0.6% agar, pH 5.7. For some experiments, this medium was supplemented with kanamycin, hygromycin B, or 5-aza-2′-deoxycytidine (aza-dC) as described in Results. The plates were left at 4°C in the dark for 2 days and then transferred to a growth chamber and incubated at 22°C under continuous light. Plants treated with 1-naphthaleneacetic acid (NAA) or histone deacetylase (HDAC) inhibitors and untreated controls were removed from the growth medium and placed in either water or potassium phosphate buffer (20 mM, pH 6), supplemented as described in Results, and incubated with gentle shaking. Plants used for crossing or mapping were transferred to soil after ∼2 weeks and grown in a chamber with a 16-hr-light/8-hr-dark cycle at 22°C.
Vector Construction, Plant Transformation, and Mutagenesis
The vectors pDR5 (Ulmasov et al., 1997) and p2xD0 were constructed using the binary Ti vector pBIN19 (Bevan, 1984) as described in Results. The −46 cauliflower mosaic virus 35S promoter, tobacco mosaic virus Ω sequence, β-glucuronidase (GUS) open reading frame, and nopaline synthase polyadenylation sequence were derived from pAGUS1-TN2 (Skuzeski et al., 1990). For the hygromycin phosphotransferase (HPH) constructs, the GUS open reading frame was replaced by that of the hygromycin B phosphotransferase gene (Gritz and Davies, 1983). The binary vectors were transferred to the Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986) and used to transform Arabidopsis ecotype Columbia (Col) plants by vacuum infiltration (Bechtold et al., 1993). Ethyl methanesulfonate mutagenesis was performed as described by Chory et al. (1989). To screen for mutants, we germinated progeny of plants grown from ethyl methanesulfonate–treated seed (M2 generation) on medium supplemented with hygromycin B. The concentration of hygromycin depended on the transgenic line (see Results). Plants that survived this treatment were transferred to soil.
Assays for GUS and HPH Activity
Fluorimetric and histochemical GUS assays were performed as described by Jefferson (1987) and Hagen et al. (1991). HPH activity assays were performed as described by Haas and Dowding (1975). Specifically, 6-day-old seedlings (15 to 20 per transgenic line) were homogenized in 0.3 mL of assay buffer (0.067 M Tris-HCl base, 0.042 M MgCl2, 0.4 M NH4Cl, and 1.7 mM DTT; pH adjusted to 7.1 with maleic acid). Undissolved material was removed by centrifugation. Reactions were prepared on ice using 10 μL of assay buffer, 10 μL of sample, and 2 μL of hygromycin B solution (1 mg/mL). To start each reaction, we added 10 μL of ATP solution (75 μM ATP and 100 μCi/mL γ-32P-ATP, pH 7.2), and the reaction was incubated at 37°C for 20 min. Reactions were ended by pipetting 20 μL onto 1-cm squares of cellulose phosphate paper (Whatman P81). The paper squares were washed three times in hot water (70 to 80°C) and then dried, and the radioactivity bound to each piece was measured in a scintillation counter. Sample protein concentrations were assayed using a protein assay kit (Bio-Rad), and specific activities (cpm/mg protein) were calculated.
Mapping and Sequencing of the axe1 Locus
The axe1 locus was mapped using F2 progeny from crosses between mutant Col and wild-type Landsberg erecta (Ler) plants. DNA was extracted from inflorescences of the mutant (hygromycin resistant) F2 plants. Bulked segregant restriction fragment length polymorphism analysis (Michelmore et al., 1991) and cleaved amplified polymorphic sequence (CAPS) mapping (Konieczny and Ausubel, 1993) were used to place the mutant locus on chromosome 5 between the CAPS markers LFY3 and g2368. P1 clones of sequences between these markers were downloaded from the Kazusa laboratory Web site (http://www.kazusa.or.jp/kaos/) and scanned for simple sequence repeats (i.e., repeats of A, AT, CA, CAG, CAT, CTT, or GA). Polymerase chain reaction (PCR) primers were designed that would amplify short sequences (100 to 200 bp) containing these repeats. Eighteen primer pairs were tested, and six of these gave polymorphic PCR products from Col and Ler DNA with differences that could be distinguished using agarose (4 or 5%) gels. Two of these primer pairs amplified sequences from within the MDC12 P1 clone, and these sequences, which are separated by ∼55 kb, were found to flank the mutant locus. Of the annotated genes located within this 55-kb region, an HDAC gene was sequenced. PCR primers were designed to amplify the entire coding region of the gene in three fragments, each ∼600 to 700 bp long. The gene was amplified from DNA extracted from each of the five axe1 lines, and the PCR products were sequenced using an ABI 377 sequencer with ABI BigDye Terminator chemistry (Applied Biosystems Inc., Foster City, CA).
DNA and RNA Gel Blot Analysis
DNA was extracted from 8-day-old seedlings, and DNA gel blots were prepared as described by Bernatzky (1988). Total RNA was extracted from 8-day-old seedlings, and RNA gel blots were prepared as described by Ulmasov et al. (1999). Probes for RNA gel blotting were either purified PCR or cDNA fragments labeled with 32P by random priming. The Arabidopsis GH3 homolog (AGH3-2) was cloned by homology with the soybean GH3 gene and will be described in detail elsewhere. The SAUR-AC1 cDNA was obtained by PCR from genomic DNA (Col). Aux/IAA cDNA clones and the Actin-2 cDNA clone (GenBank accession number ATU37281) were provided by Dr. Athanasios Theologis (U.S. Department of Agriculture Plant Gene Expression Center, Albany, CA) and Dr. John Walker (University of Missouri, Columbia), respectively. The EF1α cDNA was obtained from the Arabidopsis Functional Genomics Consortium (AFGC) Microarray Facility.
Microarray Analysis
Plants used for microarray analysis were germinated and grown on nonselective medium for 8 days. Approximately 400 mutant and control seedlings were used for RNA extraction. Total RNA was isolated using the RNA Wiz kit (Ambion, Austin, TX). Polyadenylated RNA samples were prepared, and quality control experiments were performed according to instructions from the AFGC Microarray Facility (http://afgc.stanford.edu/).
Acknowledgments
We thank Dr. Mannie Liscum for his valuable advice on approaches to mapping the axe1 locus and Drs. Athanasios Theologis and John Walker for providing cDNA clones. We are also grateful to Jennifer Kelley, Joel Baumgart, Matt Heebner, Eric Tomko, and Xumin Feng for providing technical assistance. This work was supported by grants from the United States Department of Agriculture (Grant No. CSREES 98-35304-6674) and the National Science Foundation (Grant No. NSF POWRE 98108541) to J.M., National Science Foundation Grant Nos. IBN 9303956, MCB 9604208, and MCB 0080096 to T.J.G. and G.H., the University of Missouri Research Board, and the University of Missouri F21C Program.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251 533–549. [DOI] [PubMed] [Google Scholar]
- Ahringer, J. (2000). NuRD and SIN3. Trends Genet. 16 351–356. [DOI] [PubMed] [Google Scholar]
- Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405 203–206. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Arnholdt-Schmitt, B. (1993). Rapid changes in amplification and methylation pattern of genomic DNA in cultured carrot root explants (Daucus carota L.). Theor. Appl. Genet. 85 793–800. [DOI] [PubMed] [Google Scholar]
- Arnholdt-Schmitt, B., Holzapfel, B., Schillinger, A., and Neumann, K.-H. (1991). Variable methylation and differential replication of genomic DNA in cultured carrot root explants during growth induction as influenced by hormonal treatments. Theor. Appl. Genet. 82 283–288. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Ser. III Sci. Vie 316 1194–1199. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273 948–950. [DOI] [PubMed] [Google Scholar]
- Bernatzky, R. (1988). Restriction fragment length polymorphism. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–18.
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A.P., and Wolffe, A.P. (1999). Methylation-induced repression: Belts, braces and chromatin. Cell 99 451–454. [DOI] [PubMed] [Google Scholar]
- Brosch, G., Ransom, R., Lechner, T., Walton, J.D., and Loidl, P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, E.E., Bachman, K.E., Myöhänen, S., Herman, J.G., and Baylin, S.B. (1999). Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21 103–107. [DOI] [PubMed] [Google Scholar]
- Carmen, A.A., Griffin, P.R., Calaycay, J.R., Rundlett, S.E., Suka, Y., and Grunstein, M. (1999). Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96 12356–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Yang, M.C., and Yang, T.P. (2001). Evidence that silencing of the HPRT promoter by DNA methylation is mediated by critical CpG sites. J. Biol. Chem. 276 320–328. [DOI] [PubMed] [Google Scholar]
- Chen, W.Y., Bailey, E.C., McCune, S.L., Dong, J.Y., and Townes, T.M. (1997). Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylases. Proc. Natl. Acad. Sci. USA 94 5798–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J., and Pikaard, C.S. (1997). Epigenetic silencing of RNA polymerase 1 transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 991–999. [DOI] [PubMed] [Google Scholar]
- Cuisset, L., Tichonicky, L., Jaffray, P., and Delpech, M. (1997). The effects of sodium butyrate on transcription are mediated through activation of a protein phosphatase. J. Biol. Chem. 272 24148–24153. [DOI] [PubMed] [Google Scholar]
- Finnin, M.S., Donigian, J.R., Cohen, A., Richon, V.M., Rifkind, R.A., Marks, P.A., Breslow, R., and Pavletich, N.P. (1999). Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401 188–193. [DOI] [PubMed] [Google Scholar]
- Flavell, R.B. (1994). Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc. Natl. Acad. Sci. USA 91 3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, P., Liu, Y., Orbovic, V., Verkamp, E., Poff, K.L., and Green, P.J. (1994). Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P.A., and Berger, S.L. (1999). Histone acetyltransferase complexes. Semin. Cell Dev. Biol. 10 169–177. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. (1999). Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell 96 303–306. [DOI] [PubMed] [Google Scholar]
- Gritz, L., and Davies, J. (1983). Plasmid-encoded hygromycin B resistance: The sequence of the hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25 179–188. [DOI] [PubMed] [Google Scholar]
- Guilfoyle, T., Hagen, G., Ulmasov, T., and Murfett, J. (1998). How does auxin turn on genes? Plant Physiol. 118 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, M.J., and Dowding, J.E. (1975). Aminoglycoside-modifying enzymes. Methods Enzymol. 43 611–628. [DOI] [PubMed] [Google Scholar]
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17 567–579. [DOI] [PubMed] [Google Scholar]
- Hassig, C.A., Fleischer, T.C., Billin, A.N., Schreiber, S.L., and Ayer, D.E. (1997). Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89 341–347. [DOI] [PubMed] [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7 211–220. [DOI] [PubMed] [Google Scholar]
- Hobbie, L., McGovern, M., Hurwitz, L.R., Pierro, A., Liu, N.Y., Bandyopadhyay, A., and Estelle, M. (2000). The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127 23–32. [DOI] [PubMed] [Google Scholar]
- Jeddeloh, J.A., Stokes, T.L., and Richards, E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22 94–97. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying for chimeric genes in plants: The GUS fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Johnson, C.A., and Turner, B.M. (1999). Histone deacetylases: Complex transducers of nuclear signals. Semin. Cell Dev. Biol. 10 179–188. [DOI] [PubMed] [Google Scholar]
- Jones, P.A. (1985). Altering gene expression with 5-azacytidine. Cell 40 485–486. [DOI] [PubMed] [Google Scholar]
- Jones, P.L., Veenstra, G.J., Wade, P.A., Vermaak, D., Kass, S.U., Landsberger, N., Strouboulis, J., and Wolffe, A.P. (1998). Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19 187–191. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and Struhl, K. (1997). Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89 365–371. [DOI] [PubMed] [Google Scholar]
- Kadosh, D., and Struhl, K. (1998). Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston, R.E., and Narlikar, G.J. (1999). ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13 2339–2352. [DOI] [PubMed] [Google Scholar]
- Knoepfler, P.S., and Eisenman, R.N. (1999). Sin meets NuRD and other tails of repression. Cell 99 447–450. [DOI] [PubMed] [Google Scholar]
- Kölle, D., Brosch, G., Lechner, T., Pipal, A., Helliger, W., Taplick, J., and Loidl, P. (1999). Different types of maize histone deacetylases are distinguished by a highly complex substrate and site specificity. Biochemistry 38 6769–6773. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of a TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Konieczny, A., and Ausubel, F. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Kruh, J. (1982). Effects of sodium butyrate, a new pharmacological agent, on cells in culture. Mol. Cell. Biochem. 42 65–82. [DOI] [PubMed] [Google Scholar]
- Lechner, T., Lusser, A., Pipal, A., Brosch, G., Loidl, A., Goralik-Schramel, M., Sendra, R., Wegener, S., Walton, J.D., and Loidl, P. (2000). RPD3-type histone deacetylases in maize embryos. Biochemistry 39 1683–1692. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin activating enzyme E1. Nature 364 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T.J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz, M.C., Schübeler, D., Goeke, S.C., Walters, M., Groudine, M., and Martin, D.I.K. (2000). Dynamic analysis of proviral induction and de novo methylation: Implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 20 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoSchiavo, F., Pitto, L., Giuliano, G., Torti, G., Nuti-Ronchi, V., Marazziti, D., Vergara, R., Orselli, S., and Terzi, M. (1989). DNA methylation of embryogenic carrot cell cultures and its variations as caused by mutation, differentiation, hormones and hypomethylating drugs. Theor. Appl. Genet. 77 325–331. [DOI] [PubMed] [Google Scholar]
- Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277 88–91. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., and Matzke, A.J.M. (1995). How and why do plants inactivate homologous (trans)genes? Plant Physiol. 107 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M.A., Mette, M.F., and Matzke, A.J.M. (2000). Transgene silencing by the host genome defense: Implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43 401–415. [DOI] [PubMed] [Google Scholar]
- Meyer, P. (2000). Transcriptional transgene silencing and chromatin components. Plant Mol. Biol. 43 221–234. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., Paran, I., and Kesseli, R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–479. [Google Scholar]
- Nan, X., Ng, H.-H., Johnson, C.A., Laherty, C.D., Turner, B.M., Eisenman, R.N., and Bird, A. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393 386–389. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., and Bird, A. (2000). Histone deacetylases: Silencers for hire. Trends Biochem. Sci. 25 121–126. [DOI] [PubMed] [Google Scholar]
- Park, Y.-D., Papp, I., Moscone, E.A., Iglesias, V.A., Vaucheret, H., Matzke, A.J.M., and Matzke, M.A. (1996). Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J. 9 183–194. [DOI] [PubMed] [Google Scholar]
- Pikaart, M.J., Recillas-Targa, F., and Felsenfeld, G. (1998). Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker, E.U. (1998). Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95 9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuzeski, J.M., Nichols, L.M., and Gesteland, R.F. (1990). Analysis of leaky viral translation termination codons in vivo by transient expression of improved β-glucuronidase vectors. Plant Mol. Biol. 15 65–79. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403 41–45. [DOI] [PubMed] [Google Scholar]
- Struhl, K. (1998). Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12 599–606. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpte, C., Wilson, A.K., and Estelle, M. (1994). The axr2–1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics 138 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z.-B., Hagen, G., and Guilfoyle, T.J. (1995). Composite structure of auxin response elements. Plant Cell 7 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19 309–319. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.-B., Mourrain, P., Palauqui, J.-C., and Vernhettes, S. (1998). Transgene-induced silencing in plants. Plant J. 16 651–659. [DOI] [PubMed] [Google Scholar]
- Wolffe, A.P., and Matzke, M.A. (1999). Epigenetics: Regulation through repression. Science 286 481–486. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22 19–27. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265 17174–17179. [PubMed] [Google Scholar]
- Yoshida, M., Horinouchi, S., and Beppu, T. (1995). Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17 423–430. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Ng, H.-H., Erdjument-Bromage, H., Tempst, P., Bird, A., and Reinberg, D. (1999). Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]