Abstract
The time of flowering in Arabidopsis is controlled by multiple endogenous and environmental signals. Some of these signals promote the onset of flowering, whereas others repress it. We describe here the isolation and characterization of two allelic mutations that cause early flowering and define a new locus, EARLY BOLTING IN SHORT DAYS (EBS). Acceleration of flowering time in the ebs mutants is especially conspicuous under short-day photoperiods and results from a reduction of the adult vegetative phase of the plants. In addition to the early flowering phenotype, ebs mutants show a reduction in seed dormancy, plant size, and fertility. Double mutant analysis with gibberellin-deficient mutants indicates that both the early-flowering and the precocious-germination phenotypes require gibberellin biosynthesis. Analysis of the genetic interactions among ebs and several mutations causing late flowering shows that the ft mutant phenotype is epistatic over the early flowering of ebs mutants, suggesting that the precocious flowering of ebs requires the FT gene product. Finally, the ebs mutation causes an increase in the level of expression of the floral homeotic genes APETALA3 (AP3), PISTILLATA (PI), and AGAMOUS (AG) and partially rescues the mutant floral phenotype of leafy-6 (lfy-6) mutants. These results suggest that EBS participates as a negative regulator in developmental processes such as germination, flowering induction, and flower organ specification.
INTRODUCTION
The reproductive success of plants depends on initiation of flowering occurring under the most favorable conditions. Plants have developed mechanisms to sense environmental conditions as well as their developmental and nutritional status and to integrate this information to regulate their flowering time. Flowering in Arabidopsis is promoted by low nonfreezing temperatures (vernalization) and long photoperiods, and delayed by short photoperiods (Koornneef et al., 1998). Before the induction of flowering, Arabidopsis plants grow vegetatively as rosettes that result from the repetitive production of leaves from lateral primordia initiated at the flanks of the apical meristem. Two developmental phases, juvenile and adult, have been distinguished during rosette growth on the basis of leaf morphology, trichome distribution, and acquisition of meristem competence to flower (Telfer et al., 1997). As a result of floral induction, leaf production is inhibited, lateral primordia develop into flowers, and the main stem elongates to give rise to an inflorescence. Analyses of Arabidopsis mutants have allowed the identification of many genes involved in the regulation of flowering time. Physiological, genetic, and molecular analyses of flowering time mutants have shown that flowering is promoted or inhibited by several pathways, some of which are dependent on the environment (reviewed by Koornneef et al., 1998; Levy and Dean, 1998; Piñeiro and Coupland, 1998; Simpson et al., 1999).
Three floral-promotion pathways have been proposed in Arabidopsis: the long-day (LD) pathway, the autonomous pathway, and a gibberellin-dependent pathway. Genes in the LD pathway, such as FHA, CONSTANS (CO), GIGANTEA (GI), LATE ELONGATED HYPOCOTYL, FWA, and FT, have been identified by mutations that delay flowering specifically under LD (Koornneef et al., 1998). Among them, the differential interactions of ft and fwa with mutations that affect flower meristem identity suggest that FT and FWA participate in the final steps of the LD pathway (Ruiz-García et al., 1997; Nilsson et al., 1998; Kardailsky et al., 1999; Kobayashi et al., 1999). The autonomous pathway has been defined on the basis of mutants delayed in flowering under both LD and short days (SD), which are responsive to vernalization, and include genes such as FCA, FPA, FVE, FY, and LUMINIDEPENDENS (Koornneef et al., 1998). Finally, flowering of Arabidopsis under noninductive SD conditions is absolutely dependent on gibberellin biosynthesis. This is demonstrated by the inability to flower under SD of strong gibberellic acid (GA)–deficient mutants such as ga1-3 (Wilson et al., 1992) and the early SD flowering phenotype of spindly (spy) mutants, which have a constitutively activated GA signal transduction pathway (Jacobsen and Olszewski, 1993).
The repression of flowering in Arabidopsis has not been analyzed so intensively, and only a few genetic interactions among late- and early-flowering mutants have been described (reviewed by Hicks et al., 1996a; Koornneef et al., 1998; Levy and Dean, 1998). Some of the genes involved in repression of the floral transition act independently of environmental factors. Among them, HASTY is required early in development to regulate the competence to flower of the shoot apical meristem (Telfer and Poethig, 1998). Once plants have reached the adult vegetative phase and are competent to flower, several regulatory systems prevent floral initiation until the appropriate developmental stage is reached and inductive environmental conditions are present. Genes such as TERMINAL FLOWER 1 seem to function to repress reproductive development irrespective of photoperiodic conditions, allowing the plant to reach further vegetative development before flowering (Alvarez et al., 1992). Under noninductive photoperiods (SD), flowering inhibition depends largely on genes involved in light perception (phytochrome B and related phytochromes) and signal transduction (Hicks et al., 1996a). Mutants defective in phytochromes, such as long hypocotyl 1 (hy1), hy2, and hy3 (=phytochromeB) (Koornneef et al., 1980), or in phytochrome signal transduction, such as phytochrome-signaling early-flowering 1 (pef1), pef2, and pef3 (Ahmad and Cashmore, 1996), show reduced sensitivity to photoperiodic inhibition of flowering. Furthermore, early flowering 3 shows almost complete photoperiod insensitivity (Hicks et al., 1996b). Finally, in vernalization-requiring genotypes, flowering repression under regular growing temperatures is provided by dominant alleles at loci such as FRIGIDA (Levy and Dean, 1998) and FLOWERING LOCUS C (FLC; Michaels and Amasino, 1999; Sheldon et al., 1999). The abundance of the FLC transcript seems to be negatively controlled by both vernalization and the activity of the autonomous flowering promotion pathway (Michaels and Amasino, 1999; Sheldon et al., 1999).
Ultimately, the promotion and repression pathways regulate the initiation of flowering by modulating the expression of floral meristem identity genes such as LEAFY (LFY) and APETALA1 (AP1) (Simon et al., 1996; Kardailsky et al., 1999; Kobayashi et al., 1999; Blázquez and Weigel, 2000). Mutations in any of these genes produce flowers with shootlike characteristics, supporting their role in the specification of floral fate (Irish and Sussex, 1990; Schultz and Haughn, 1991). Furthermore, LFY has been identified as an upstream regulator of AP1, APETALA3 (AP3), and AGAMOUS (AG), which are responsible for the A, B, and C functions, respectively, in the specification of flower organ identity (Parcy et al., 1998; Busch et al., 1999; Wagner et al., 1999).
In a screening for early-flowering mutants under SD, we have identified two allelic mutations that define a new locus of Arabidopsis, EARLY BOLTING IN SHORT DAYS (EBS), that is involved in flowering repression. These mutants show a regular juvenile phase but bolt early once the adult vegetative phase has been reached. Moreover, ebs mutants show additional phenotypic defects, including reduced dormancy of the seed. The construction and characterization of double mutants show that both the early-flowering and germination phenotypes of ebs mutants require gibberellin biosynthesis, whereas the early-flowering phenotype also requires FT function. Moreover, a partial rescue of petal and stamen development was observed in the ebs-1 lfy-6 double mutant. We discuss the role of EBS in flowering repression and other developmental processes.
RESULTS
Isolation of Mutant Alleles of the EBS Locus
Early-flowering mutants in the Landsberg erecta (Ler) background were selected under SD from one ethyl methanesulfonate (EMS)–mutagenized M2 population and from progeny families derived from self-fertilizing plants carrying mobilized Dissociation (Ds) elements (Long et al., 1997). Two mutants, one derived from each population, showed a similar early-flowering phenotype and were studied further. In both mutants, a recessive mutation at a single locus was responsible for an early-bolting phenotype, particularly under SD but also under LD. Phenotypic similarities observed in both mutant plants suggested that the mutations might be allelic, and this was confirmed by a complementation test. The locus was named EBS for EARLY BOLTING IN SHORT DAYS, and the isolated alleles were named ebs-1 and ebs-2 for the EMS- and transposon-induced alleles, respectively (this locus has been designated SPEEDY in previous reviews [Levy and Dean, 1998; Simpson et al., 1999]). Both alleles were backcrossed to Ler twice before further analyses.
Plants homozygous for the ebs-1 allele were crossed to Columbia to determine the map position of the mutation relative to molecular markers. ebs-1 was located on the lower arm of chromosome 4, specifically at 2.29 ± 0.46 centimorgan (cM) south from marker g3883 and 3.08 ± 0.02 cM north from RPS2. No other early-flowering mutation had been mapped to this location, indicating that EBS could be a new locus regulating flowering time. The ebs-2 line carried a single Ds element, and genetic analysis showed that the Ds was tightly linked to the mutation (<0.4 cM). However, molecular analyses of the transposon-induced allele indicated that the Ds insertion was associated with a chromosomal rearrangement, probably a deletion or an inversion (see Methods). This precluded the direct identification of the EBS gene but suggested that ebs-2 probably is a null allele.
Mutations at the EBS Locus Cause Early Flowering and Have Pleiotropic Effects on Shoot, Leaf, and Flower Development
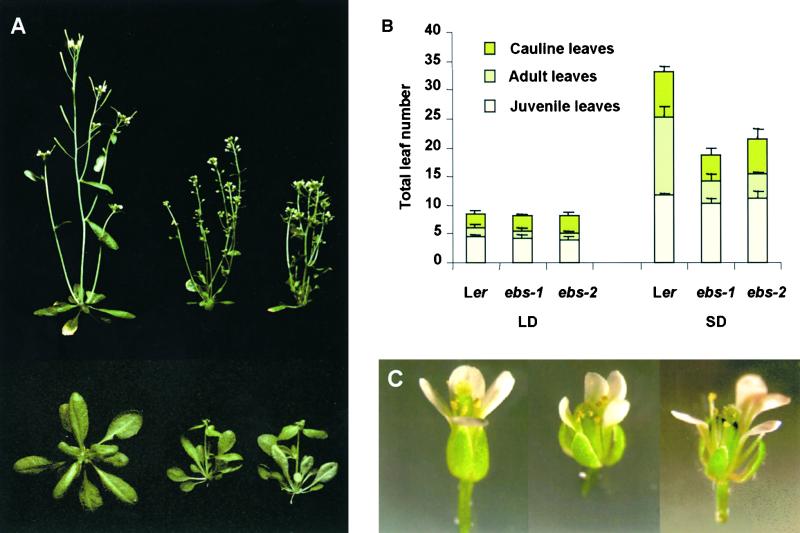
Plants homozygous for each of the mutant alleles were grown under inductive (LD) and noninductive (SD) photoperiods and compared with Ler wild-type plants (Figures 1A and 1B). Under LD, mutant plants flowered slightly earlier than did the wild type (20 versus 25 days) and with fewer leaves (eight versus nine). However, the early-flowering phenotype was much more conspicuous under SD. Under short photoperiods, both mutant alleles flowered after 35 days with 18 to 20 leaves, whereas wild-type plants took more than 50 days to flower and produced more than 30 leaves. Thus, the ebs mutations cause premature flowering under both LD and SD, although the mutant alleles retain a photoperiodic response. To determine whether the ebs mutations shortened a specific developmental phase or all of the phases, we analyzed the presence of trichomes in the abaxial surface of the leaves, which has been used as a criterion to distinguish juvenile and adult rosette leaves (Telfer et al., 1997). As shown in Figure 1B, the ebs mutations seemed to shorten specifically the adult vegetative phase, because fewer leaves of this class were produced in both mutant plants under LD or SD. This effect was particularly conspicuous when plants were grown under SD; under this condition, both mutant alleles produced ∼10 adult leaves fewer than did the wild type. The fact that both mutations caused a very similar phenotype indicates that the EMS allele ebs-1 is as strong as the presumed null allele.
Figure 1.
Phenotype of ebs Mutants.
(A) Ler (left), ebs-1 (middle), and ebs-2 (right) 5-week-old plants grown under LD (top) or SD (bottom).
(B) Average number of juvenile, adult, or cauline leaves for Ler and ebs mutants grown under LD or SD photoperiods. Bars indicate ±se.
(C) Flowers from Ler (left), ebs-1 (middle), and ebs-2 (right).
Mutant plants also showed other pleiotropic effects in shoot development. Mutant leaves were generally narrower and smaller than were wild-type leaves. The stems, inflorescences, and flower pedicels were shorter, and the plants showed a semidwarf phenotype (Figure 1A). Mutant flowers were smaller and slightly asymmetric. Flower organs also showed some developmental defects: petals were generally narrower, stamens produced a reduced amount of pollen, and carpels often showed fusion abnormalities (Figure 1C). Occasionally, mutant plants grown under higher-intensity illumination formed terminal flowers (data not shown).
Early Flowering of ebs Mutants under SD Requires Gibberellin Biosynthesis
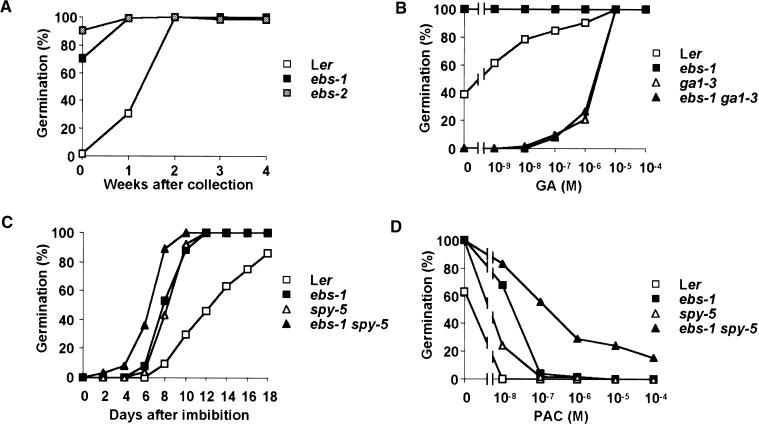
Flowering under short photoperiods in Arabidopsis is strongly dependent on gibberellin biosynthesis (Wilson et al., 1992). Because ebs mutants are early flowering under SD, we tested whether this phenotype was dependent on gibberellin biosynthesis or signal transduction. Double mutants carrying ebs-1 and mutations affecting GA biosynthesis (ga1-3, ga2-1) or response (spy-5) were constructed. Both ga1-3 and ga2-1 mutations affect early steps in GA biosynthesis (Sun and Kamiya, 1994; Yamaguchi et al., 1998) and delay flowering under LD and SD (Wilson et al., 1992). Furthermore, the ga1-3 mutation completely prevents flowering under SD (Wilson et al., 1992). The double mutants ebs-1 ga1-3 and ebs-1 ga2-1 showed the same flowering time phenotype as the single GA-deficient mutants under both LD and SD (Figures 2A and 2B). In both cases, the phenotype of these double mutants was very similar to the phenotype of the single GA-deficient mutants (Figure 2C). The GA deficiency mutations, therefore, are epistatic over the ebs phenotype, indicating that GA biosynthesis is required for early flowering of ebs mutants.
Figure 2.
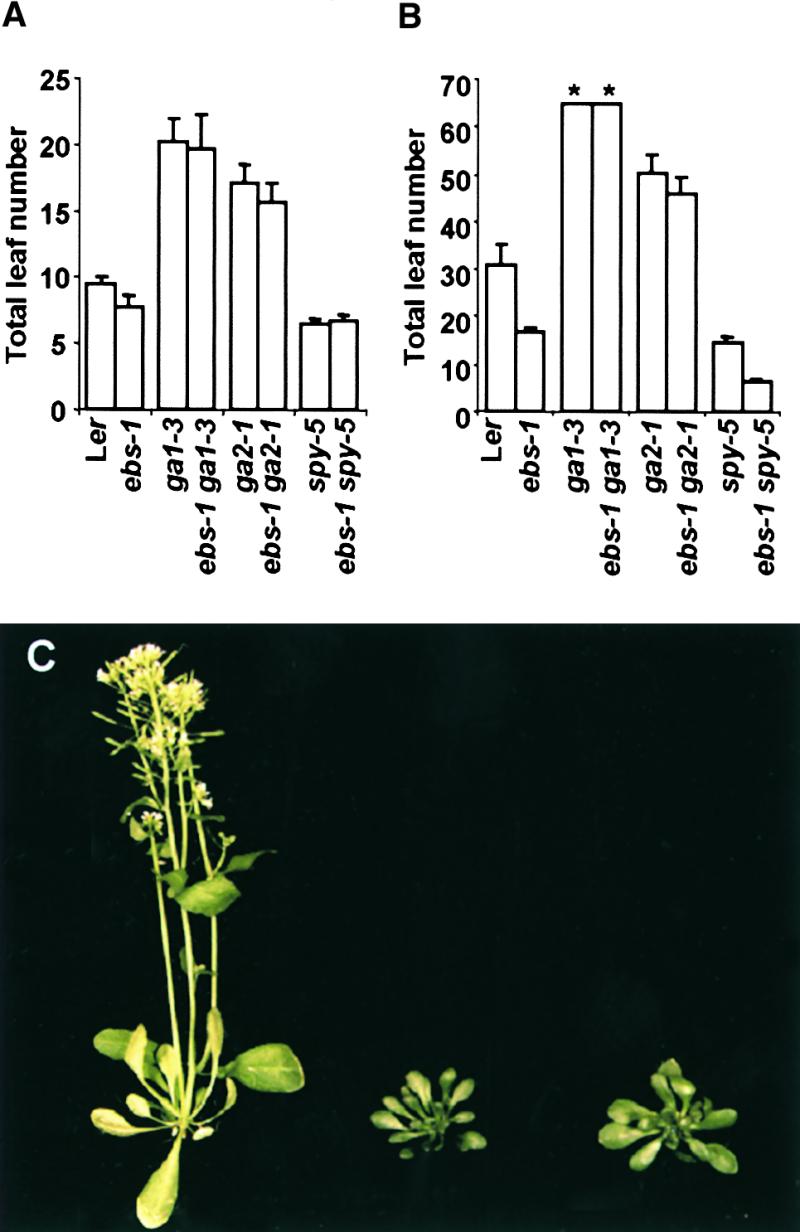
Effect of the ebs Mutation on Total Leaf Number of ga1-3, ga2-1, and spy-5 Mutants.
(A) Total leaf number under LD.
(B) Total leaf number under SD. Asterisks indicate that plants were unable to flower after 3 months of growth under SD. During this time, they produced 65 leaves.
(C) Phenotype of ebs-1 (left), ebs-1 ga1-3 (middle), and ga1-3 (right) 5-week-old plants grown under LD.
Error bars in (A) and (B) indicate ±se.
The spy mutation alters GA signal transduction, producing a constitutive GA response even in the absence of gibberellins (Jacobsen and Olszewski, 1993). The flowering time phenotype of spy mutants is similar to that of ebs: they flower earlier than does the wild type under both LD and SD, but they are sensitive to photoperiod, showing a delay in flowering time under SD. The double mutant ebs-1 spy-5 was similar to spy-5 in flowering time and slightly earlier than ebs under LD (Figure 2A). Furthermore, this double mutant was daylength insensitive, flowering with the same number of leaves under both LD and SD (Figures 2A and 2B). These mutations, therefore, have an additive effect on flowering under SD.
ebs Mutant Seed Shows Reduced Dormancy
The phenotype of the ebs-1 ga1-3 double mutant demonstrated that GA biosynthesis is required for the early-flowering phenotype of ebs mutants, suggesting that ebs might affect flowering time by enhancing GA biosynthesis or response. GA also is required for germination (Koornneef and van der Veen, 1980); therefore, we tested whether ebs had an effect on germination. Seeds of Ler, ebs-1, and ebs-2 were stored for different periods of time after harvest, and their ability to germinate was scored 14 days after sowing. In contrast to the wild type, ebs-1 and ebs-2 mutant seeds showed almost no dormancy response, and a much higher percentage of mutant than wild-type seeds germinated when sown immediately after harvest (Figure 3A). These freshly harvested mutant seeds also were able to germinate in complete darkness, a condition that prevents germination of fresh wild-type seeds (data not shown).
Figure 3.
Reduction in the Dormancy of ebs Mutant Seeds and Its Effect on the Germination of Double Mutants with ga1-3 and spy-5.
Germination was scored after 2 weeks of incubation except in (C), where germination was scored as indicated.
(A) Germination of Ler, ebs-1, and ebs-2 seeds after different weeks of storage.
(B) Germination rates of Ler, ebs-1, ga1-3, and ebs-1 ga1-3 freshly harvested seeds in the presence of different GA concentrations.
(C) Time course of germination of Ler, ebs-1, spy5, and ebs-1 spy-5 freshly harvested seeds.
(D) Germination rates of Ler, ebs-1, spy5, and ebs-1 spy-5 freshly harvested seeds in the presence of different concentrations of PAC.
Mutations affecting GA biosynthesis, such as ga1-3, can completely abolish the ability of seeds to germinate unless GA is added exogenously (Koornneef and van der Veen, 1980). Because the early flowering phenotype of ebs mutants requires GA, we tested whether the reduced seed dormancy of these mutants also requires GA. Freshly harvested seeds of Ler wild type, ebs-1, ga1-3, and the double mutant ebs-1 ga1-3 were sown with increasing concentrations of GA, and germination was scored after 14 days. As shown in Figure 3B, seeds of the double mutant ebs-1 ga1-3 germinated only in the presence of concentrations of GA very similar to those required for the germination of ga1-3. Therefore, as observed for the early-flowering phenotype, the premature germination of ebs mutants also requires GA biosynthesis.
Phenotypic analysis of the double mutant ebs-1 spy-5 showed that both mutations act additively in the control of flowering time in SD. Because both the ebs and spy mutations show reduced dormancy and increased resistance to paclobutrazol (PAC), an inhibitor of GA biosynthesis, we tested the effect of combining both mutations on the germination of freshly harvested seeds and on the resistance of these seeds to PAC. Seeds of the double mutant ebs-1 spy-5 showed a further reduction in dormancy and started germinating earlier than any of the single mutants (Figure 3C). Furthermore, the double mutant also showed increased resistance to PAC compared with either single mutant (Figure 3D), suggesting that the ebs and spy mutations also have additive effects in reducing dormancy.
FT Is Required for the Early Flowering of ebs Mutants
To test the interaction between EBS and the pathways proposed to promote flowering in Arabidopsis, we made double mutants carrying ebs and mutations in representative genes for each of the pathways that promote flowering. Double mutants in which ebs was combined with mutations that affect the autonomous pathway (fve-2 or fpa-1) showed an intermediate phenotype measured as the total number of leaves produced before flowering when grown under LD or SD (Figures 4A and 4B). The time of bolting was also intermediate in these double mutants (data not shown). All of the pleiotropic effects caused by the ebs mutations on the morphology of the plant, such as the reduced elongation of inflorescence stems and the smaller size of leaves and flowers, also were present in these double mutants (data not shown). These results demonstrate a lack of interaction between these mutations and suggest that EBS acts in a pathway that is parallel to the pathway represented by the genes FVE and FPA.
Figure 4.
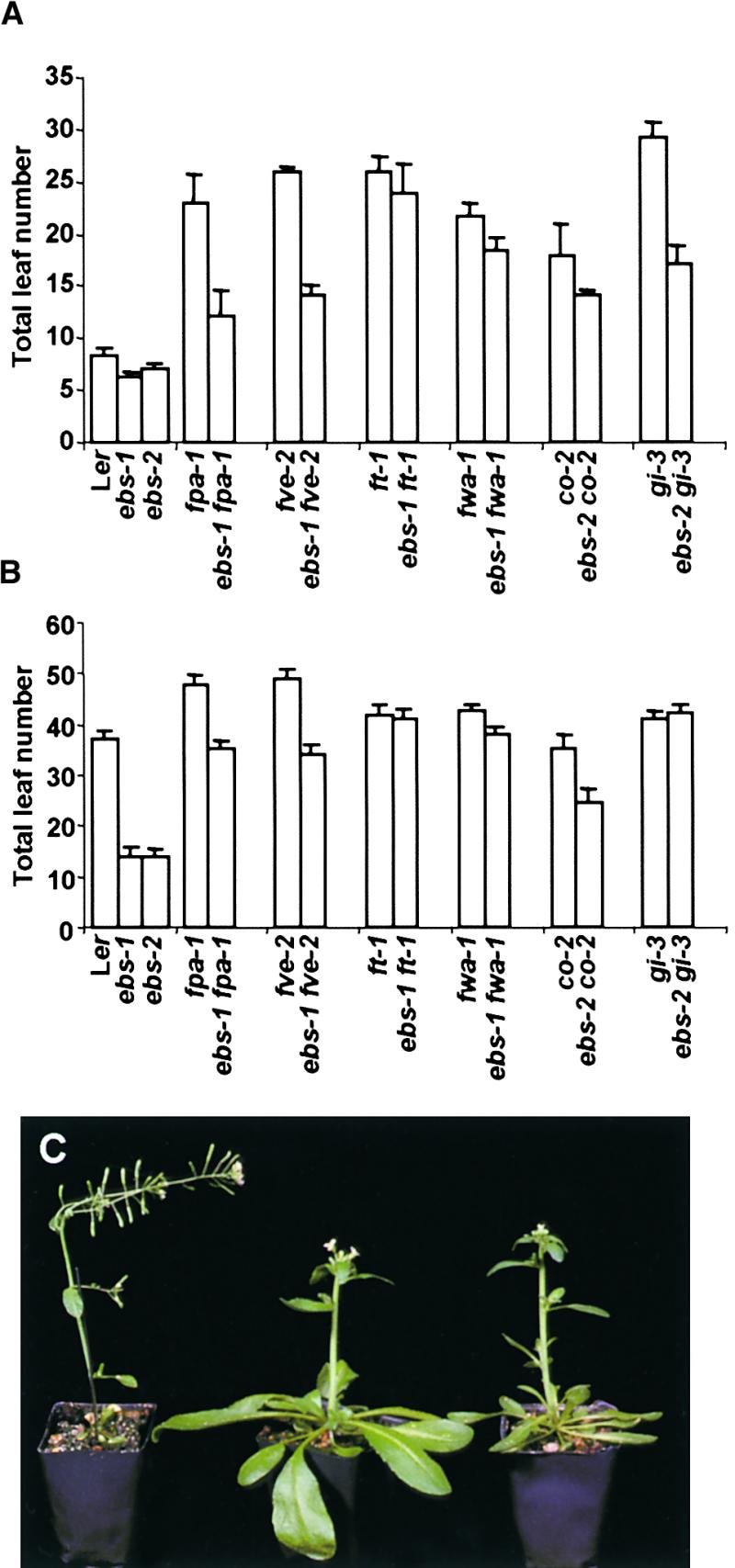
Effect of the ebs Mutation on Total Leaf Number of Late- Flowering Mutants Affecting the LD Pathway and the Autonomous Pathway.
(A) Total leaf number under LD.
(B) Total leaf number under SD.
(C) Phenotype of ebs-1 (left), ft-1 (middle), and ebs-1 ft-1 (right) 5-week-old plants grown under LD.
Error bars in (A) and (B) indicate ±se.
Double mutants carrying ebs and different mutations affecting the LD pathway showed flowering time phenotypes that differed depending on the late-flowering mutant used. The double mutants ebs-2 co-2 and ebs-2 gi-3 showed an intermediate flowering time under LD (Figure 4A). Under SD, the double mutant ebs-2 co-2 also exhibited an intermediate phenotype in terms of both total number of leaves (Figure 4B) and bolting time (data not shown). However, the double mutant ebs-2 gi-3 bolted 2 weeks earlier than did the late flowering parent, although with a number of rosette and cauline leaves similar to that of gi-3 (ebs-2 gi-3, 30.0 ± 1.2 rosette and 12.4 ± 0.7 cauline leaves; gi-3, 31.2 ± 1.7 rosette and 10.0 ± 0.5 cauline leaves; Figure 4B). These results indicate that under SD, the rate of leaf production is higher in ebs-2 gi-3 than in the gi-3 parent. Moreover, the double mutant ebs-2 gi-3 formed a large number of coflorescences not subtended by leaves (19.7 on average) before the development of the first flower, suggesting a delay in the establishment of floral meristem identity. These results, together with a number of other pleiotropic effects displayed by gi mutants, such as elongated hypocotyls and higher starch levels (Araki and Komeda, 1993; Eimert et al., 1995), suggest a unique role for GI in the LD promotion pathway. In conclusion, the phenotypes of the double mutants with co and gi indicate a lack of interaction between EBS and CO and GI loci in the regulation of bolting time. However, the phenotypic differences observed between these double mutants support the existence of different roles for CO and GI within the LD pathway and an SD requirement of the GI function for the initiation of flowers in ebs mutants.
Within the LD pathway, ft and fwa behave differently from the other mutants when combined with the flower meristem identity mutation lfy, and they may participate in later steps of this pathway (Ruiz-García et al., 1997; Nilsson et al., 1998). Consequently, we analyzed the possible interaction between ebs and these two mutations. The ebs-1 ft-1 and ebs-1 fwa-1 double mutants showed a different phenotype than did the other combinations of ebs with late-flowering mutants. When grown under both LD and SD, the ebs-1 ft-1 and ebs-1 fwa-1 double mutants showed a late-flowering phenotype similar to that of each single late parent, measured either as number of leaves or time of bolting, indicating that the ft and fwa mutant phenotypes are epistatic to ebs with respect to flowering time (Figures 4A and 4B). These double mutants also showed the pleiotropic phenotypes caused by the ebs mutation (Figure 4C). These results indicate that the early-flowering phenotype of ebs mutants requires the FT function. Considering the dominant nature of the fwa-1 mutation, this result could be interpreted as FWA having a negative effect on the induction of flowering downstream of the EBS function. Exactly the same results were obtained when these double mutants were generated with the ebs-2 allele (data not shown).
ebs Partially Corrects the lfy Phenotype
FT seems to be required together with LFY in the determination of flower meristem identity (Ruiz-García et al., 1997; Nilsson et al., 1998). Because FT was required for the early flowering of ebs mutants, we tested whether LFY also was required for the early-flowering phenotype of ebs. A double mutant carrying ebs-1 and the strong mutant allele lfy-6 was constructed. The lfy-6 mutant is slightly delayed in flowering time under LD with respect to wild-type plants; however, the double mutant ebs-1 lfy-6 flowered at the same time and with a similar number of rosette leaves as did the ebs mutants (ebs-1, 5.7; lfy-6, 8.7; ebs-1 lfy-6, 5.5, on average) and showed the characteristic ebs phenotype in rosettes and inflorescences. The absence of the LFY function prevents the development of petals and stamens in the flowerlike structures produced by lfy-6 (Weigel et al., 1992).
Interestingly, ebs-1 lfy-6 formed a variable number of petaloid and staminoid organs with variable degrees of differentiation as well as mosaic organs intermediate between petals and stamens (Figure 5). This partial rescue of the floral organ phenotype of lfy-6 mutants was observed in all ebs-1 lfy-6 double mutant plants, under both LD and SD. To quantify the phenomenon, 102 flowers from 10 LD-grown ebs-1 lfy-6 double mutant plants were tested for the presence of floral organs normally absent in lfy-6 flowerlike structures. Petals or petaloid organs were present in 90% of the ebs-1 lfy-6 flowerlike structures analyzed. These structures contained an average of 2.8 ± 1.8 petals or petaloid organs. The occurrence of stamens or staminoid organs was less frequent: up to 25% of the flowerlike structures analyzed showed an average of 1.2 ± 0.6 stamens or staminoid organs. However, pollen was observed only occasionally in the flowers of the ebs-1 lfy-6 double mutant. These transformations were observed only rarely in the lfy-6 progenitor, in which less than 1% of flowers contained petaloid structures and no staminoid structures were observed in 10 plants tested (Figure 5A). In conclusion, the early-flowering ebs phenotype does not require LFY function, and ebs mutations are able to partially rescue the specification of floral organ identity in the second and third whorls of lfy-6 flowers.
Figure 5.

Partial Rescue of the lfy-6 Floral Phenotype by the ebs Mutation.
(A) Inflorescence of a lfy-6 mutant plant grown under LD.
(B) Inflorescence of an ebs-1 lfy-6 double mutant plant grown under LD.
(C) Inflorescence of an ebs-1 lfy-6 double mutant plant grown under SD.
(D) Flower from an SD-grown ebs-1 lfy-6 plant showing the partial rescue of petals and stamens.
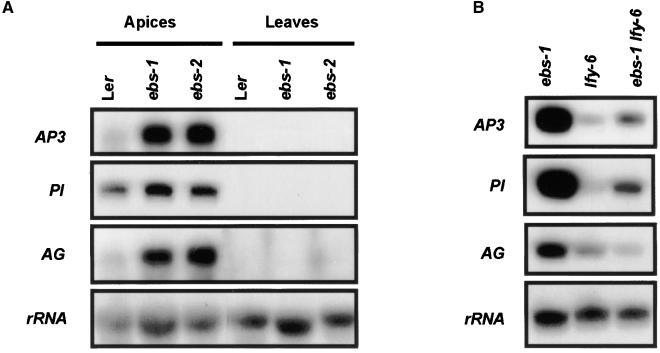
The overexpression of AP3 under the control of the 35S cauliflower mosaic virus (CaMV) promoter can partially rescue the development of petals and stamens in flowers of the lfy-6 mutant (Jack et al., 1994). Because ebs-1 lfy-6 mutants are able to develop second and third whorl organs in older flowers, AP3 and/or PI expression could be enhanced by ebs mutations. To test this hypothesis, we performed RNA gel blot experiments using total RNA from reproductive apices and leaves of wild-type Ler plants and ebs mutants. As shown in Figure 6A, AP3 and PI mRNA were found at higher levels in the apices of ebs mutants than in those of wild-type plants, whereas neither transcript was detected in leaves. The level of AG mRNA, which also is involved in the specification of third whorl organs, also was higher in apices of ebs mutants than in apices of Ler wild-type plants (Figure 6A). Furthermore, the expression of AP3 and PI was enhanced in the double mutant ebs-1 lfy-6 compared with lfy-6 (Figure 6B). In contrast, the level of AG mRNA was not altered significantly in the ebs-1 lfy-6 double mutant compared with the lfy-6 single mutant, indicating a differential effect of the ebs mutation on the regulation of AP3/PI and AG in the lfy-6 mutant background. The observed increase in the expression of AP3 and PI caused by the ebs mutation seems to be enough to promote the development of petals and stamens in the absence of LFY product, suggesting a role for EBS in the regulation of AP3 and PI expression.
Figure 6.
Expression of AP3, PI, and AG in ebs-1 and the ebs-1 lfy-6 Double Mutant.
Total RNA was isolated from reproductive apices or leaves of LD-grown plants, and 20 μg was loaded in each lane. Blots were probed with radiolabeled AP3, PI, and AG cDNAs and then reprobed with rDNA as a loading control.
(A) Steady state levels of AP3, PI, and AG mRNA in apices and leaves of Ler and ebs mutants.
(B) Steady state levels of AP3, PI, and AG mRNA in apices of ebs-1, lfy-6, and ebs-1 lfy-6 plants.
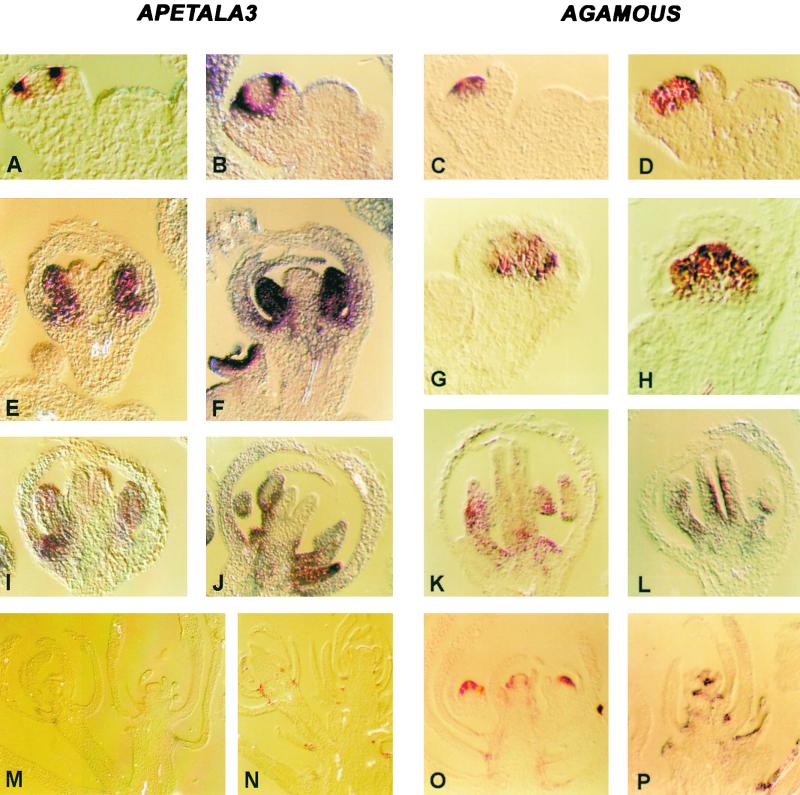
The lack of homeotic transformations in the organs of ebs flowers suggests that AP3 and AG messages are not expressed ectopically during flower development. To test this possibility, we performed RNA in situ hybridization with AP3 and AG probes on flower meristems and flowers of Ler and ebs-1 plants. As shown in Figure 7, the temporal and spatial patterns of AP3 and AG expression were very similar in wild-type and ebs mutant plants. AP3 mRNA was first detected in early stage 3 flowers, localized in whorls 2 and 3, in both Ler and ebs (Figures 7A and 7B). Later in flower development (stages 6 and 9), AP3 transcript was detected on developing stamens and petals, with a similar pattern in Ler (Figures 7E and 7I) and ebs (Figures 7F and 7J). A greater hybridization signal was always detected on floral meristems of the ebs mutant, consistent with the results obtained by RNA gel blot hybridization. The pattern of expression of AG also was the same in Ler and ebs: during stages 3 and 5, AG mRNA was restricted to the central region of the floral meristem (Ler, Figures 7C and 7G; ebs, Figures 7D and 7H). Later in development, during stage 9, AG mRNA was located in developing carpels and stamens in both Ler (Figure 7K) and ebs (Figure 7L).
Figure 7.
Pattern of Expression of AP3 and AG in Floral Meristems of ebs and Inflorescences of the ebs-1 lfy-6 Double Mutant.
The expression of AP3 and AG was analyzed by in situ hybridization on longitudinal sections of apical buds from plants grown under LD.
(A) Ler stage 3 flower probed with AP3.
(B) ebs-1 stage 3 flower probed with AP3.
(C) Ler stage 3 flower probed with AG.
(D) ebs-1 stage 3 flower probed with AG.
(E) Ler stage 6 flower probed with AP3.
(F) ebs-1 stage 6 flower probed with AP3.
(G) Ler stage 5 flower probed with AG.
(H) ebs-1 stage 5 flower probed with AG.
(I) Ler stage 9 flower probed with AP3.
(J) ebs-1 stage 9 flower probed with AP3.
(K) Ler stage 9 flower probed with AG.
(L) ebs-1 stage 9 flower probed with AG.
(M) lfy-6 mutant inflorescence probed with AP3.
(N) ebs-1 lfy-6 double mutant inflorescence probed with AP3.
(O) lfy-6 mutant inflorescence probed with AG.
(P) ebs-1 lfy-6 double mutant inflorescence probed with AG.
To determine whether the partial rescue of petal and stamen development in the ebs-1 lfy-6 double mutant could be caused by the observed increase in AP3 expression, we performed RNA in situ hybridizations on inflorescences of lfy-6 and ebs-1 lfy-6 plants. As expected, AP3 expression was reduced markedly in lfy-6 plants compared with wild-type plants, although with low frequency a weak hybridization signal could be observed in the axils of the bracts and on the basal region of the flowerlike structures of lfy-6 mutants (Figure 7M). When the inflorescences of the ebs-1 lfy-6 double mutant were analyzed, the localization of the AP3 transcript was similar to that observed in the lfy-6 single mutant, although the hybridization signal was higher and detected more frequently in the axils of the bracts (Figure 7N). The pattern of expression of AG was similar in the inflorescences of lfy-6 and ebs-1 lfy-6 (Figures 7O and 7P). In conclusion, the observed increase in AP3 and AG expression in the ebs mutant background is not the result of ectopic expression of these homeotic genes, suggesting a role for EBS in their regulation in those cells in which they are normally expressed.
DISCUSSION
EBS, a New Locus Required for the Repression of Flowering in Arabidopsis
The early-flowering phenotype of ebs mutants and the map position of EBS indicate that it is a new locus involved in the regulation of flowering time. Two aspects of the mutant phenotypes strongly support a role for EBS as a flowering repressor under noninductive photoperiods. First, the reduction in flowering time caused by the ebs mutations is not the result of a general acceleration of the development of the plant; rather, it results specifically from a reduction in the duration of the adult vegetative phase (Figure 1). During this phase, the apical meristem is already competent to initiate reproductive development once the environmental conditions are adequate (Telfer et al., 1997). This observation places the role of the EBS locus in the negative regulation of flowering time once the shoot apical meristem is competent to flower. Second, the epistatic relationships revealed by the late-flowering phenotype of the ebs ft double mutant suggest a specific role for EBS in the repression of FT, a gene required (along with LFY) to promote the initiation of flowering in Arabidopsis (Ruiz-García et al., 1997).
FT has been shown to induce flowering under SD when it is expressed constitutively from a CaMV 35S promoter (Kardailsky et al., 1999; Kobayashi et al., 1999). The pleiotropic phenotype shown by ebs mutants, which include reduced seed dormancy, reduced plant size, altered flower morphology, and reduced fertility, suggests the involvement of EBS in other developmental processes in addition to the repression of flowering. However, we cannot completely exclude the possibility that EBS might have a very early role in development that could affect later developmental stages. Most of the early-flowering mutants already characterized in Arabidopsis also show pleiotropic defects (Koornneef et al., 1998). On the one hand, early-flowering mutants such as elongated (Halliday et al., 1996), early flowering 1 (Scott et al., 1999), and early flowering in short days (Soppe et al., 1999) are affected in either dormancy or plant size. On the other hand, mutants defective in light or gibberellin response also can show an early-flowering phenotype. This is the case with mutants defective in phytochrome biosynthesis, such as hy1 (Parks and Quail, 1991) and phyB (Somers et al., 1991), or in signal transduction, such as pef (Ahmad and Cashmore, 1996), constitutively photomorphogenic 1 (Deng and Quail, 1992), and de-etiolated 1 (Pepper et al., 1994), which show early-flowering phenotypes together with alterations in organ and plant size and chlorophyll content. Some of the mutants altered in GA-mediated signal transduction, such as spy (Jacobsen and Olszewski, 1993), also show early flowering together with reduced dormancy and increased elongation of the plant.
GA-deficient mutants are impaired in germination and flowering under noninductive photoperiods (Koornneef and van der Veen, 1980; Wilson et al., 1992). The reduced dormancy of ebs mutants and their early bolting under SD would suggest that EBS could act as a repressor of GA biosynthesis or could participate as a negative regulator in GA-mediated signal transduction. However, several aspects of the mutant and double mutant phenotypes are not consistent with these hypotheses. The gibberellin requirement shown by ebs mutants for both early flowering and premature germination is not in agreement with EBS negatively regulating GA-mediated signal transduction, because a GA-independent phenotype like that observed in spy mutants (Jacobsen and Olszewski, 1993) would have been expected. On the other hand, the semidwarf phenotype shown by ebs mutants is not consistent with the phenotype of Arabidopsis transgenic plants that overproduce GA (Hedden and Phillips, 2000) or the phenocopies obtained in Arabidopsis by exogenous GA treatments (Chandler and Dean, 1994). Thus, the phenotypes of ebs and the double mutants ebs-1 ga1-3 and ebs-1 ga2-1 do not support a role for EBS in the regulation of GA biosynthesis or signaling pathways. However, EBS could participate as a repressor in at least two developmental processes (germination and flowering under SD) that also are regulated by gibberellins.
EBS Mediates the Repression of Flowering through FT
Analyses of double mutants carrying ebs and late-flowering mutations indicate a specific interaction of EBS with FT and FWA genes, which have been shown to act downstream of other genes participating in the photoperiod-dependent pathway (Kardailsky et al., 1999; Kobayashi et al., 1999). These results suggest that EBS could act as a direct or indirect repressor of FT expression under noninductive photoperiods. This is consistent with recent reports demonstrating that the overexpression of FT is enough to promote early flowering under SD (Kardailsky et al., 1999; Kobayashi et al., 1999). Furthermore, considering the dominant nature of the fwa mutations, the late-flowering phenotype displayed by the double mutant ebs-1 fwa-1 indicates that FWA could act as a repressor downstream of EBS. This hypothesis is in agreement with the late-flowering phenotype of fwa 35S::FT plants, which suggests that FWA represses flowering downstream of FT (Kardailsky et al., 1999; Kobayashi et al., 1999). Two reports have suggested a transcriptional regulation of FT by CO. First, there is a direct correlation between CO expression and the FT transcript level (Kobayashi et al., 1999; Samach et al., 2000). Second, ft mutations partially suppress the early-flowering phenotype of transgenic Arabidopsis plants expressing a 35S::CO construct (Onouchi et al., 2000). However, the intermediate-flowering time phenotype of ebs-2 co-2 suggests that EBS and CO might regulate flowering through FT independently.
The gibberellin requirement for flowering under noninductive conditions in Arabidopsis is clearly established (Wilson et al., 1992), and these hormones have been proposed to function in regulating LFY expression (Blázquez et al., 1998; Blázquez and Weigel, 2000). We have shown here that GA biosynthesis is absolutely required for the early-flowering phenotype of ebs mutants under SD and that this early-flowering phenotype does not require the LFY function. These observations, together with the FT requirement for the early flowering of ebs, suggest that GA also could have a regulatory role on either FT or CO, or downstream of them, a hypothesis that can be tested in future experiments.
Thus, EBS could participate in the repression of flowering under SD through the repression of FT. Upon removal of EBS repression, probably as a consequence of inductive photoperiods, FT activation still would be dependent on gibberellins and CO transcriptional activation. This model of interaction could be extended to other developmental processes such as germination, in which EBS-mediated repression and GA-mediated activation could act on common targets with additional specific transcriptional activators. This hypothesis is consistent with the additive phenotype shown by the double mutant ebs-1 spy-5 in terms of both germination and flowering initiation under noninductive conditions. Furthermore, the lack of a photoperiodic response of these double mutants suggests that SD inhibition of flower induction depends largely on both EBS-mediated flowering repression and GA-mediated flowering activation. The roles of photoreceptors and photoperiod in the regulation of these two pathways remain to be elucidated.
EBS Involvement in the Regulation of Floral Organ Identity Genes
Partial rescue of the differentiation of petals and stamens in the ebs-1 lfy-6 double mutant suggests increased activation of AP3 and PI in the ebs background. In fact, higher steady state transcript levels for AP3, PI, and AG were confirmed by RNA gel blot experiments in apices of the ebs mutants. Contrary to what has been reported for mutants such as clf (Goodrich et al., 1997), in situ hybridization experiments indicate that both AG and AP3 are not expressed ectopically in the ebs mutants. These results suggest a direct or indirect repression effect of EBS on AG and AP3 expression, independent of the positional regulation provided by additional factors. The expression of AP3 and PI in the ebs-1 lfy-6 background suggests that these genes can be activated in the absence of LFY product. However, the enhancement of AG expression in ebs mutants might be dependent on LFY, because the levels of AG transcript are similar in lfy-6 and the ebs-1 lfy-6 double mutant. Because the ft mutation is epistatic over the ebs mutation for the initiation of flowering, it is possible that the EBS effect on AP3, PI, and AG expression could be mediated by FT. Alternately, EBS could participate in the repression of those genes independently of FT. Our genetic analysis does not discriminate between these two hypotheses. However, the fact that 35S::FT does not correct the lfy-6 flower phenotype (D. Weigel, personal communication) supports the second possibility. Additional experiments are under way to elucidate the FT requirement in AP3, PI, and AG overexpression. Whether gibberellins also are required for complete activation of these homeotic genes remains to be shown.
In conclusion, we have identified a new locus of Arabidopsis whose product participates as a negative regulator in several developmental processes during the life cycle of the plant, from germination to flower development. We also show that in at least two of these processes, which are positively regulated by gibberellins, biosynthesis of these hormones is required for the observation of the ebs mutant phenotype. Finally, in two of these processes, flower induction and flower development, we have identified genes such as FT, AP3, PI, and AG that could be direct or indirect targets of the EBS function. Confirmation of these hypotheses and further research on the molecular role of the EBS gene product require its molecular cloning and characterization, a task that is currently under way.
METHODS
Plant Material
The Arabidopsis thaliana mutant lines used in this work are all in the ecotype Landsberg and carry the erecta mutation. Seed stocks were obtained from the Arabidopsis Biological Resource Center of Ohio State University (Columbus) or the Nottingham Arabidopsis Centre (UK). The monogenic mutants fve-2, fpa-1, ft-1, fwa-1, co-2, and gi-3 were described by Koornneef et al. (1991), ga1-3 and ga2-1 were described by Koornneef and van der Veen (1980), spy-5 was described by Jacobsen and Olszewski (1993), and lfy-6 was described by Weigel et al. (1992). Because ga1-3 and ga2-1 mutants require gibberellin treatment for germination, seed carrying these mutations were incubated with 100 μM gibberellic acid (GA) during 2 days in darkness and were rinsed thoroughly with water before sowing.
The ebs-1 mutant allele was isolated from ethyl methanesulfonate (EMS)–mutagenized seed, whereas the ebs-2 mutant allele was identified in a mutant screen of a population carrying Ds elements. The ebs-2 mutant line contains a single transposed Ds insertion that is tightly linked to the ebs mutation. In an attempt to identify the Ds insertion site on ebs-2, an inverse polymerase chain reaction fragment on the 3′ end of the Ds element was generated and sequenced. However, the flanking sequence predicted to be adjacent to the 5′ end of the transposon was not present in ebs-2, suggesting a chromosomal rearrangement generated upon insertion of the Ds. We confirmed that both mutations were allelic by their failure to complement the early flowering phenotype in F1 plants derived from crosses between them. In addition, all plants from the F2 generation exhibited early-flowering and a similar pleiotropic phenotype when grown under short day (SD) photoperiods.
Growth Conditions
Plants were grown in plastic pots containing a mixture of substrate and vermiculite (3:1). Controlled environmental conditions were provided in growth chambers at 18°C and 80% RH. Plants were illuminated with cool-white fluorescent lights. Long day (LD) conditions consisted of 16 hr of light followed by 8 hr of darkness; SD conditions consisted of 8 hr of light followed by 16 hr of darkness. For germination experiments, sterilized seed were sown aseptically in 9-cm Petri dishes on 0.8% (w/v) agar containing Murashige and Skoog (1962) mineral salts supplemented with 1% sucrose. Germination tests were performed subsequently under the LD conditions used for plant growth or in total darkness. For GA or paclobutrazol (PAC) sensitivity tests, sterilized seed were sown on plates in the presence of the GA or PAC concentrations indicated in Figures 4C and 4D, respectively. Unless mentioned otherwise, germination was scored after 2 weeks of incubation.
Flowering Time Analysis
The total number of leaves was recorded as an adequate measurement of flowering time. Total leaf number was scored as the number of leaves in the rosette (excluding cotyledons) plus the number of leaves in the inflorescence at the time of opening of the first flower. The appearance of abaxial trichomes was monitored using a magnifying glass. Adult and juvenile leaves were scored independently. Rosette leaves lacking abaxial trichomes were considered juvenile leaves (Telfer et al., 1997).
Genetic Analysis
The ebs-1 mutation was mapped relative to cleaved-amplified polymorphic sequence (Bell and Ecker, 1994) and simple sequence length polymorphism molecular markers (Konieczny and Ausubel, 1993). Recombination fractions were used to calculate the map distances using the Kosambi mapping function (Kosambi, 1944). Double mutants were constructed by crossing the monogenic ebs-1 or ebs-2 mutant with lines carrying the mutations fve-2, fpa-1, ft-1, fwa-1, co-2, gi-3, ga1-3, ga2-1, spy-5, and lfy-6. Double mutants were isolated from selfed F2 progeny that showed the ebs phenotype and that segregated for the second mutation. The genotypes of double mutants were confirmed by crosses with the monogenic parental lines.
Expression Analysis
Total RNA was isolated from both leaves and apices using the FastRNA Kit-GREEN (BIO101; Vista, California) according to the instructions of the manufacturer. Total RNA was electrophoresed in 1.5% formaldehyde agarose gels (Sambrook et al., 1989) and transferred to Hybond N+ membranes (Amersham). The AP3 probe was an EcoRI–BglII fragment from pD793 plasmid that contains the cDNA of the AP3 gene (Jack et al. 1992). The PI probe was a NsiI–XhoI fragment from plasmid pNX that contains the cDNA of the PI gene (Goto and Meyerowitz, 1994). The AG probe was a ScaI–EcoRI fragment from the pCIT565 plasmid that contains the cDNA of the AG gene (Yanofsky et al., 1990). As a loading control, we used a 300-bp fragment of the cauliflower 18S rDNA gene. For in situ hybridization, apical buds were fixed and embedded by standard methods. In situ hybridization was performed essentially as described by Huijser et al. (1992). Immunological detection was performed according to the DIG Nucleic Acid Detection kit (Boehringer Mannheim). Antisense probes for the AP3 messenger were made using T7 RNA polymerase and pD793 plasmid linearized with BglII, which cuts once, 216 bp from the 5′ end of the cDNA, just past the 3′ end of the MADS box (Jack et al. 1992). For the AG probe, plasmid pCIT565 was linearized with HindIII, and labeled RNA was made using SP6 polymerase (Drews et al., 1991).
Acknowledgments
We thank Dr. Maarten Koornneef (Laboratory of Genetics, University of Wageningen, The Netherlands) and the Nottingham Arabidopsis Stock Centre for supplying seed stocks, and Dr. Detlef Weigel (Salk Institute, La Jolla, CA) for sharing unpublished results. We also thank Gemma Bravo for her technical assistance. This work was supported by Grant No. BIO4-CT97-2340 from the European Union and Grant No. AGF98-0206 from Comisión Interministerial de Ciencia y Tecnología, Spain. Support for research activity at Centro Nacional de Biotecnología is provided through a specific agreement with Consejo Superior de Investigaciones Científicas–Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA). C.G-M. was supported by an INIA (Spain) predoctoral fellowship. M.P. was supported by the European Molecular Biology Organization and the Biotechnology and Biological Science Research Council. J.M.F.-Z. was funded by a predoctoral fellowship from Dirección General de Investigación Científica y Técnica (Spain).
References
- Ahmad, M., and Cashmore, A.R. (1996). The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 10 1103–1110. [DOI] [PubMed] [Google Scholar]
- Alvarez, J., Guli, C.L., Yu, X.-H., and Smyth, D.R. (1992). TERMINAL FLOWER, a gene affecting inflorescence development in Arabidopsis thaliana. Plant J. 2 103–116. [Google Scholar]
- Araki, T., and Komeda, Y. (1993). Analysis of the role of the late-flowering locus, GI, in the flowering of Arabidopsis thaliana. Plant J. 3 231–239. [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404 889–892. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Green, R., Nilsson, O., Sussman, M.R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, M.A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285 585–587. [DOI] [PubMed] [Google Scholar]
- Chandler, J., and Dean, C. (1994). Factors influencing the vernalization response and flowering time of late flowering mutants of Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 45 1279–1288. [Google Scholar]
- Deng, X.-W., and Quail, P.H. (1992). Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 2 83–85. [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowitz, E.M. (1991). Negative regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. [Google Scholar]
- Eimert, K., Wang, S.-M., Lue, W.-l., and Chen, J. (1995). Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. Plant Cell 7 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A polycomb group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44–51. [DOI] [PubMed] [Google Scholar]
- Goto, K., and Meyerowitz, E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. [DOI] [PubMed] [Google Scholar]
- Halliday, K., Devlin, P.F., Whitelam, G.C., Hanhart, C., and Koornneef, M. (1996). The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J. 9 305–312. [DOI] [PubMed] [Google Scholar]
- Hedden, P., and Phillips, A.L. (2000). Manipulation of hormone biosynthetic genes in transgenic plants. Curr. Opin. Biotechnol. 11 130–136. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Sundås, A., and Meeks-Wagner, D.R. (1996. a). Arabidopsis early-flowering mutants reveal multiple levels of regulation in the vegetative to floral transition. Semin. Cell Dev. Biol. 7 409–418. [Google Scholar]
- Hicks, K.A., Millar, A.J., Carré, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996. b). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Huijser, P., Klein, J., Lönnig, W.E., Meijer, H., Saedler, H., and Sommer, H. (1992). Bracteomania, an inflorescence anomaly, is caused by loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 11 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack, T., Brockman, L.L., and Meyerowitz, E.M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. [DOI] [PubMed] [Google Scholar]
- Jack, T., Fox, G.L., and Meyerowitz, E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76 703–716. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1993). Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100 147–160. [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late-flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Peeters, A.J., and Soppe, W. (1998). Genetic control of flowering time in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 345–370. [DOI] [PubMed] [Google Scholar]
- Kosambi, D.D. (1944). The estimation of map distance from recombination values. Ann. Eugen. 12 172–175. [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, D., Goodrich, J., Wilson, K., Sundberg, E., Martin, M., Puangsomlee, P., and Coupland, G. (1997). Ds elements on all five Arabidopsis chromosomes and assessment of their utility for transposon tagging. Plant J. 11 145–148. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nilsson, O., Lee, I., Blázquez, M.A., and Weigel, D. (1998). Flowering time genes modulate the response to LEAFY activity. Genetics 150 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi, H., Igeño, M.I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395 561–566. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, A.E., Delaney, T., Washburn, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78 109–116. [DOI] [PubMed] [Google Scholar]
- Piñeiro, M., and Coupland, G. (1998). The control of flowering time and floral identity in Arabidopsis. Plant Physiol. 117 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-García, L., Madueño, F., Wilkinson, M., Haughn, G., Salinas, J., and Martínez-Zapater, J.M. (1997). Different roles of flowering time genes in the activation of floral initiation genes in Arabidopsis. Plant Cell 9 1921–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schultz, E.A., and Haughn, G.W. (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D.B., Jin, W., Ledford, H.K., Jung, H.-S., and Honma, M.A. (1999). EAF1 regulates vegetative-phase change and flowering time in Arabidopsis. Plant Physiol. 120 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R., Igeño, M.I., and Coupland, G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384 59–62. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., Gendall, A.R., and Dean, C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 99 519–550. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Sharrock, R.A., Tepperman, J.M., and Quail, P.H. (1991). The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3 1263–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe, W.J.J., Bentsink, L., and Koornneef, M. (1999). The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126 4763–4770. [DOI] [PubMed] [Google Scholar]
- Sun, T.-p., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1998). HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125 1889–1898. [DOI] [PubMed] [Google Scholar]
- Telfer, A., Bollman, K.M., and Poethig, R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W.M., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Wilson, R.N., Heckman, J.W., and Somerville, C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S., Smith, M.W., Brown, R.G.S., Kamiya, Y., and Sun, T.-p. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky, M.F., Ma, H., Bowman, J.L., Drews, G.N., Feldmann, K.A., and Meyerowitz, E.M. (1990). The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]