Abstract
The role of inositol 1,4,5-trisphosphate (Ins[1,4,5]P3) in transducing the abscisic acid (ABA) signal during seed germination and in the stress responses of mature plants is poorly understood. We have considered the contributions of the phospholipase C1 (encoded by AtPLC1) and an Ins(1,4,5)P3 5-phosphatase (encoded by AtIP5PII) to ABA signaling by using a modified version of the glucocorticoid-inducible system to regulate transgene expression. In the presence of the dexamethasone (Dex) inducer, transgenic lines expressing the AtPLC1 antisense and AtIP5PII sense transgenes showed no inhibition of germination and growth by ABA, whereas in the absence of the inducer they were sensitive. In the presence of Dex, these lines accumulated lower Ins(1,4,5)P3 levels upon ABA treatment compared with that of the control transgenic lines. RNA gel blot analysis revealed a decrease in the induction of the ABA-responsive genes RD29a, KIN2, and RD22 but not COR47 in the Dex-induced transgenic plants. In transgenic lines expressing the inducible AtPLC1 sense transgene, an increase in AtPLC1 expression was not sufficient to activate the expression of ABA-responsive genes in vegetative tissues. In vitro experiments demonstrated the induced PLC1 expression when extracts were assayed in the presence of calcium, but no increase in Ins(1,4,5)P3 levels in vivo was detected, suggesting that the PLC1 enzyme was latent. Our results indicate that although an increase in PLC1 activity and increased Ins(1,4,5)P3 levels are necessary for maximal gene induction by ABA, overexpression of AtPLC1 itself is not sufficient to trigger the expression of ABA-responsive genes. We propose that AtPLC1 plays a role in secondary ABA responses.
INTRODUCTION
The phytohormone abscisic acid (ABA) regulates seed maturation and maintains seed dormancy. In adult plants, ABA acts as a negative regulator of growth by controlling the adaptive responses of many physiological and developmental processes to environmental stresses such as cold, drought, and high salinity (Giraudat, 1995; reviewed in Merlot and Giraudat, 1997). At the physiological level, this stress hormone is responsible for decreasing the turgor of the guard cells that surround the stomatal pore, resulting in a reduction of pore size and an increase in water retention (MacRobbie, 1998). These adaptive developmental responses are supported in large part by changes in the transcription of a set of genes triggered by ABA (Shinozaki and Yamaguchi-Shinozaki, 2000). Transduction of the ABA signal for gene expression (Wu et al., 1997) and for stomatal closure (Gilroy et al., 1990; McAinsh et al., 1990; Schroeder and Hagiwara, 1990) has been shown to require an increase in the cytosolic Ca2+ concentration.
Ca2+ is a major component in many signaling pathways in animal and plant systems, and it is thought that information is encoded in the period and amplitudes of Ca2+ waves (reviewed in Berridge, 1993; Clapham, 1995). In animal cells, there are two major mechanisms for the release of Ca2+ from internal stores. The first is the inositol 1,4,5-trisphosphate (Ins[1,4,5]P3)–dependent mobilization of Ca2+ by activation of the Ins(1,4,5)P3 receptor, and the second is the cyclic ADP ribose (cADPR)–mediated release of stored Ca2+ via the ryanodine receptor (Lee and Aarhus, 1991; Galione, 1993; Lee et al., 1995). Ca2+ also can activate the ryanodine receptor to release Ca2+ from internal stores by a process called Ca2+-induced Ca2+ release (Clapham, 1995). Several studies in animal cells suggest that cADPR, Ins(1,4,5)P3, and Ca2+ may participate in the regulation of Ca2+ oscillations by determining the onset, persistence, and intensity of a signal (Clapham, 1995; Cancela et al., 1998, 2000; Trewavas, 1999). In the case of plant cells, ABA treatment has been shown to cause a transient increase in the levels of cADPR (Wu et al., 1997) and Ins(1,4,5)P3 (Lee et al., 1996). However, the relative importance of the two compounds in mediating calcium release and ABA responses has not been addressed.
We have shown previously that cADPR can activate the expression of two ABA-responsive fusion genes, RD29a-GUS and KIN2-GUS, in single cells of tomato and that this activation is mediated by calcium (Wu et al., 1997). Moreover, in Arabidopsis plants, cADPR levels increased after ABA treatment and before changes in transcription of ABA-responsive genes. Using the same single-cell assay, we showed that Ins(1,4,5)P3 also can activate the expression of RD29a-GUS and KIN2-GUS (Wu et al., 1997) and that this activation is sensitive to heparin, a specific inhibitor of Ins(1,4,5)P3-gated channels. In contrast, ABA induction of these two promoters is insensitive to heparin (Wu et al., 1997), suggesting that Ins(1,4,5)P3 is not absolutely required for the induction of these two ABA-responsive genes. This observation is consistent with the observation that transcript levels of the Arabidopsis (AtPLC1) gene are low or undetectable in untreated plants and are induced upon ABA treatment (Hirayama et al., 1995). Together, these results suggest that cADPR mediates the primary ABA response and that AtPLC1 and Ins(1,4,5)P3 may be involved as a secondary signal to potentiate the primary ABA response.
To expand our knowledge of the role of Ins(1,4,5)P3 in the transduction of the ABA signal during germination and in the stress response of mature plants, we have used a modified version of the glucocorticoid-inducible system for regulated gene expression in plants (Aoyama and Chua, 1997). We have investigated the contributions of phospholipase C1 (encoded by AtPLC1) as well as an Ins(1,4,5)P3 5-phosphatase (encoded by AtIP5PII) to ABA signaling. Transgenic plants expressing these inducible genes, in either the sense or the antisense orientation, were generated and analyzed.
RESULTS
We have used a modified version of the two-component glucocorticoid-inducible system (Aoyama and Chua, 1997) for the present work. Figure 1A shows the T-DNA region of pTA211, in which two series of constructs were made containing the AtPLC1 and AtIP5PII coding sequences in either the sense or the antisense orientation. pTA211 encodes a glucocorticoid-regulated transcription factor, GVG (for Gal4-VP16-glucocorticoid binding domain; Aoyama and Chua, 1997). The GVG fusion protein is composed of a Gal4 DNA binding domain, a transactivating domain from the herpes simplex virus VP16 protein, and the hormone binding domain of the rat glucocorticoid receptor (Aoyama and Chua, 1997). GVG is expressed from the constitutive G10-90 promoter (Ishige et al., 1999), which has higher expression levels than does the cauliflower mosaic virus 35S promoter used in the previous pTA7001 version. The GVG fusion protein mediates dexamethasone (Dex)-inducible transcription of promoters containing Gal4 upstream activation sequence (or UAS) (Aoyama and Chua, 1997; McNellis et al., 1998).
Figure 1.
Structures of the Transgenic Constructs and Alignment of the Ins(1,4,5)P3 5-Phosphatase Amino Acid Sequences.
(A) Structure of the pTA211 plasmid with the left and right T-DNA borders. AtPLC1 and AtIP5PII cDNAs were cloned in both the sense and antisense orientations into the multiple cloning site of the binary plasmid pTA211, which is a modification of the original pTA7001 vector (Aoyama and Chua, 1997) (see Methods). RB, right T-DNA border; G1090, G10-90 promoter (Ishige et al., 1999); GVG, glucocorticoid-inducible chimeric transcription factor; E9, pea rbcS-E9 polyadenylation sequence; NOS, nopaline synthetase promoter; HPT, hygromycin phosphotransferase coding sequence; NOST, nopaline synthetase polyadenylation sequence; 6XUAS-46, GVG-regulated promoter; MCS, multiple cloning site; 3AT, rbcS-3A polyadenylation sequence; LB, left T-DNA border.
(B) Alignment of the conserved amino acid sequences of the human Ins(1,4,5)P3 5-phosphatase type II (HuI5P2; Jefferson and Majerus, 1995) and Arabidopsis AtIP5PI and AtIP5PII. Identical amino acids are shaded in black. Alignment was performed using Clustal methods with the DNASTAR (Madison, WI) program.
Arabidopsis cDNAs encoding AtPLC1 (GenBank accession number D38544) and Ins(1,4,5)P3 5-phosphatase (AtIP5PII cDNA; GenBank accession number AF289634) were obtained from Dr. K. Shinozaki (Tsukuba Life Science Center of Plant Molecular Biology, Ibaraki, Japan) and Dr. W. Gruissem (Swiss Federal Institute of Technology, Zurich, Switzerland), respectively. Arabidopsis AtPLC1 has been previously characterized by Hirayama et al. (1995). Analysis of the putative coding sequence of AtIP5PII indicates an extensive homology (57%) with the human Ins(1,4,5)P3 5-phosphatase type II (Jefferson and Majerus, 1995) in the catalytic domain (Figure 1B). We expressed Arabidopsis AtIP5PII in Escherichia coli, purified it as a His-tagged protein, and assayed the purified protein for 5-phosphatase activity. The AtIP5PII protein had Ins(1,4,5)P3 and Ins(1,3,4,5)P4 5-phosphatase activities 100-fold greater than those of a control protein (glutathione S-transferase; Table 1). No 5-phosphatase activity was found by using phosphatidylinositol 4, 5 bisphosphate as a substrate.
Table 1.
Inositol 5-Phosphatase Activity of the AtIP5PII Gene Producta
AtIP5PII was expressed in E. coli as a His-tagged protein. Values shown are nanomoles hydrolyzed per 50 ng of protein.
PIP2, phosphatidylinositol 4, 5 bisphosphate.
GST, glutathione S-transferase.
Six to eight independent transgenic lines were produced for each construct. All lines were selected for hygromycin resistance, and several T3 homozygous lines were selected for further studies. As controls, a number of transgenic lines were generated containing the pTA211 vector alone. In this work, we focused on the analysis of transgenic plants containing the AtPLC1 sense and antisense transgenes and the AtIP5PII sense transgene.
Recently, the pTA7001 Dex-inducible transcription system has been found to cause growth defects in certain Arabidopsis transgenic lines (Kang et al., 1999; T. Aoyama and N.-H. Chua, unpublished data) and to induce defense-related genes such as PDF1.2. Most of the transgenic plants (ecotype Columbia or C24) carrying the G10-90 GVG transgene from the new pTA211 vector did not display any growth defects or unusual phenotypes upon Dex induction. In the experiments of Kang et al. (1999), plants were grown in liquid medium with constant agitation, whereas in our experiments, plants were grown on plates and induced for 1 to 24 hr by using a hydroponics system. We found that GVG transcript levels did not vary significantly between transgenic lines and did not correlate with morphological defects in some transgenic plants. Induction of PDF1.2 was observed rarely in the transgenic lines (1 of 15) and did not correlate with Dex induction. However, we found that improper handling of plants induced PDF1.2 transcript expression (data not shown).
When treated with Dex, transgenic lines carrying the pTA211 vector alone did not display any induction or repression in the presence of ABA of >20 genes (ABI1, ABI2, COR47, COR15a, COR15b, CDP kinase, CDP synthase, 4-5IP kinase, IP5PI, IP5PII, KIN1, KIN2, PLC1, PLC2, RAB18, RD22, RD29a, RAC1, PDF1.2, PR1, and genes encoding ζ-crystallin) (data not shown).
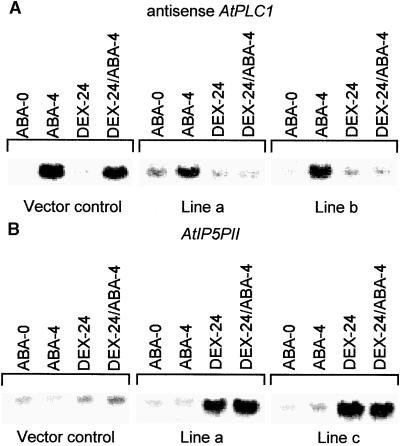
Dex Induction of the AtPLC1 Antisense and AtIP5PII Sense Transcripts in Transgenic Plants
Transgenic lines containing the AtPLC1 antisense transgene, the AtIP5PII sense transgene, and the pTA211 vector alone were analyzed by RNA gel blotting after treatment with either Dex or ABA or both. The vector control line and the uninduced transgenic lines had low levels of AtPLC1 transcript in the absence of ABA (Figure 2A, lane ABA-0). After a 4-hr exposure to ABA, however, these lines showed a fivefold induction of the endogenous AtPLC1 transcript (Figure 2A, lane ABA-4). Although a 20-hr exposure to Dex did not affect the basal expression of AtPLC1, Dex-treated transgenic AtPLC1 antisense lines did not show an increase in AtPLC1 transcript levels after a subsequent 4-hr incubation with ABA (Figure 2A, lanes DEX-24 and DEX-24/ABA-4). Under our experimental conditions, the AtPLC1 probe hybridized specifically with the AtPLC1 but not the AtPLC2 transcript.
Figure 2.
RNA Gel Blot Analysis of Transgenic Lines Expressing AtIP5PII Sense and AtPLC1 Antisense Transgenes.
For each construct, two independent lines were selected, grown, and treated as described in Methods.
(A) Induction of the AtPLC1 transcript in two AtPLC1 antisense lines and a vector control transgenic line. ABA-0, plants collected at 0 hr in the ABA medium; ABA-4, plants incubated with ABA (50 μM) for 4 hr; DEX-24, plants incubated with Dex (30 μM) for 24 hr; DEX-24/ABA-4, plants incubated with Dex (30 μM) for 24 hr followed by an additional 4 hr with ABA (50 μM). For details, see Methods.
(B) Induction of the AtIP5PII transcript in two AtIP5PII sense transgenic lines and a control transgenic line containing the pTA211 vector. Lanes are the same as in (A).
Two transgenic lines expressing AtIP5PII in the sense orientation displayed the normal AtIP5PII expression profile, as did a vector control line in the absence of Dex (Figure 2B). In all of these plants, the AtIP5PII transcript level was not affected by ABA. However, a 24-hr incubation with Dex resulted in a significant (sixfold) increase in AtIP5PII transgene expression levels, which were not affected significantly by ABA treatment (Figure 2B, lanes DEX-24 and DEX-24/ABA-4). Under our experimental conditions, the AtIP5PII probe hybridized specifically with the AtIP5PII but not the AtIP5PI transcript. These results indicate that antisense AtPLC1 transgenic plants showed greatly diminished AtPLC1 expression in response to ABA. Moreover, expression of the transgenic AtIP5PII transcript can be induced by Dex.
Effect of ABA on Transgenic Seed Germination
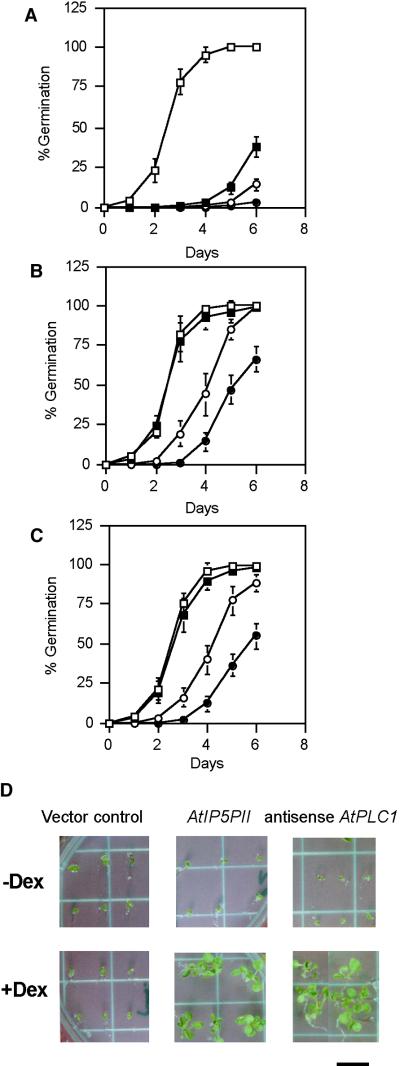
In addition to mediating stress responses in adult plants, ABA also inhibits seed germination. Screening for mutants insensitive to ABA in a germination assay has yielded several mutants altered in components of the ABA signal transduction pathway (Giraudat, 1995). To determine whether the transgenic plants display altered ABA signaling, we tested their ABA sensitivity during germination. Seed from transgenic lines containing an AtIP5PII sense construct and an AtPLC1 antisense construct were germinated on 2 μM ABA in the presence and absence of Dex (Table 2). Seed from transgenic lines carrying the AtPLC1 antisense transgene displayed 93% germination in the presence of Dex, whereas only 6% of the AtPLC1 antisense plants germinated in the absence of Dex (Table 2). In comparison, 2 to 3% germination was observed in vector control lines and in transgenic lines carrying an inducible AtPLC1 sense transgene with or without Dex (Table 2). As expected, the ABA-insensitive mutant abi1 was unaffected by the presence of Dex and germinated at a frequency of 95% (Table 2). Similarly, 95% of seed of transgenic lines containing the AtIP5PII sense transgene germinated in the presence of the Dex; in the absence of inducer, however, germination was inhibited by ABA (Table 2). No effects on germination were observed in several antisense AtIP5PII lines (Table 2). There was no significant difference in the germination frequencies of the different transgenic lines in the presence or absence of Dex without ABA (data not shown). At higher concentrations of ABA (2 to 10 μM), the AtIP5PII sense and AtPLC1 antisense lines in the presence of Dex were also insensitive to ABA with a 40 to 90% germination frequency (see Figures 3A to 3C).
Table 2.
Germination Frequency of Transgenic Lines in the Presence of ABAa
|
AtPLC1
|
AtIP5PII
|
|||||
|---|---|---|---|---|---|---|
| Vector Control | Antisense | Sense | Antisense | Sense | abi1-1 | |
| +Dex | 1.9 (0.6) | 93.3 (5.2) | 2.4 (1.0) | 1.8 (0.7) | 94.9 (4.1) | 95.9 (8.1) |
| −Dex | 2.5 (0.9) | 6.5 (1.5) | 1.5 (0.5) | 2.9 (1.0) | 2.1 (0.8) | 93.2 (6.5) |
Germination was measured in the presence of 2 μm ABA (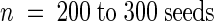 ). Values shown are % germination, expressed as average value (standard deviation).
). Values shown are % germination, expressed as average value (standard deviation).
Figure 3.
Effects of ABA on the Germination and Growth of Transgenic Plants.
(A) to (C) Seeds from transgenic lines carrying the pTA211 vector (A), from transgenic lines carrying the AtPLC1 antisense gene (B), and from transgenic lines carrying the AtIP5PII sense gene (C) were incubated on plates containing 0 μM (open squares), 2.5 μM (closed squares), 5 μM (open circles), and 10 μM (closed circles) ABA under normal light conditions (see Methods) for 1 to 6 days in the presence of Dex (30 μM). Germination frequency was measured by scoring for radicle emergence. Average values ±sd are shown for three independent lines of each construct. Approximately 100 to 150 seeds of each line were plated in duplicates.
(D) Transgenic plants were grown in ABA (2 μM) for 18 days in the absence (−) or presence (+) of Dex (30 μM). Representative plants of three lines for each transgenic construct tested and a vector control transgenic line are shown. 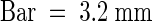 .
.
We next examined the effects of ABA on growth in the AtIP5PII sense, AtPLC1 antisense, and vector control transgenic lines after 16 days of growth under normal conditions in the presence and absence of Dex. A marked difference was observed in the growth of AtIP5PII sense and AtPLC1 antisense lines. In the presence of Dex, the plants grew normally, but almost no growth was detected in the absence of the chemical inducer (Figure 3D). These results suggest that ABA inhibition of germination and seedling growth can be overcome in lines expressing AtPLC1 antisense or AtIP5PII sense transgenes.
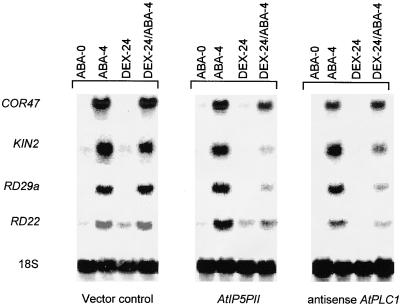
Analysis of ABA-Responsive Gene Expression in Transgenic Plants Carrying AtPLC1 Antisense and AtIP5PII Sense Transgenes
The ABA insensitivity of transgenic plants containing either the AtPLC1 antisense or the AtIP5PII sense transgene prompted us to analyze the expression of ABA-responsive genes in these plants. Induction of RD29a, KIN2, and RD22 (Giraudat, 1995) by ABA was normal in vector control transgenic lines and AtPLC1 antisense and AtIP5PII sense transgenic lines not treated with Dex (Figure 4). However, in the presence of Dex, although the accumulation of these transcripts was still responsive to ABA, it was threefold to sevenfold lower in the AtPLC1 antisense and AtIP5PII sense transgenic lines than in the vector control lines (Figure 4). In contrast, COR47 expression was unaffected by Dex in either the AtPLC1 antisense and AtIP5PII sense transgenic lines or the vector control transgenic lines. These results show that in the presence of Dex, induction of the ABA-responsive genes RD29a, KIN2, and RD22 was diminished in transgenic lines containing either the AtPLC1 antisense transgene or the AtIP5PII sense transgene. Analysis of the AtIP5PII antisense transgenic lines revealed no effects on the expression of ABA-responsive genes (data not shown). Together, our results suggest that a decrease in AtPLC1 levels or an increase in AtIP5PII levels interferes with normal ABA signaling, leading to attenuated gene expression.
Figure 4.
Induction of ABA-Responsive Genes in Transgenic Plants Expressing the AtPLC1 Antisense and AtIP5PII Sense Transgenes.
Two independent lines for each construct were grown on plates for 2 weeks, and the seedlings were transferred to a hydroponic medium (Aoyama and Chua, 1997). After 2 days, the medium was replaced with fresh medium containing either Dex (30 μM) or ABA (50 μM) or both. Total RNA was isolated at various times (0, 4, and 24 hr), and RNA gel blots were hybridized with RD29a, KIN2, RD22, and COR47 probes and 18S rDNA. The vector control line contained the pTA211 plasmid alone. For details, see Methods. ABA-0, plants collected at 0 hr in the ABA medium; ABA-4, plants incubated with ABA (50 μM) for 4 hr; DEX-24, plants incubated with Dex (30 μM) for 24 hr; DEX-24/ABA-4, plants incubated with Dex (30 μM) for 24 hr followed by an additional 4 hr with ABA (50 μM).
Analysis of Ins(1,4,5)P3 Levels in Transgenic Plants in Response to ABA
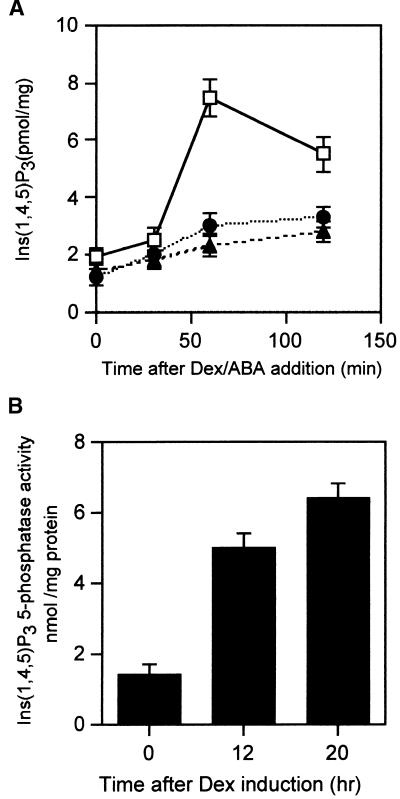
To determine whether expression of the AtPLC1 antisense gene and the AtIP5PII sense genes led to the expected changes in Ins(1,4,5)P3 levels, we measured the latter in transgenic lines treated with ABA in the presence and absence of Dex. Treatment of the vector control transgenic line with 30 μM ABA resulted in a 3.5-fold increase in Ins(1,4,5)P3 levels after 1 hr (Figure 5A). Although a small increase in the levels of Ins(1,4,5)P3 was detected in transgenic plants expressing the AtPLC1 antisense and AtIP5PII sense transgenes, the levels were significantly lower than the levels in vector control plants. The reduced accumulation of Ins(1,4,5)P3 in ABA-treated transgenic lines expressing the AtPLC1 antisense transgene likely was due to a decrease in PLC1 activity in vivo.
Figure 5.
Analysis of Ins(1,4,5)P3 Levels and Ins(1,4,5)P3 5-Phosphatase Activity in Transgenic Lines.
(A) Changes in Ins(1,4,5)P3 levels in transgenic lines expressing the AtPLC1 antisense and AtIP5PII sense transgenes in response to ABA. Fourteen-day-old transgenic seedlings were transferred to a hydroponic system (Aoyama and Chua, 1997) for 2 days before the addition of fresh medium containing Dex (30 μM) or ABA (50 μM) or both. Plants were collected at 0, 30, 60, and 120 min after ABA addition, and the amount of Ins(1,4,5)P3 was determined as described in Methods. Vector control transgenic plants contained the pTA211 plasmid alone. Open squares, vector control line with ABA (50 μM); closed circles, AtPLC1 antisense line with Dex (30 μM) and ABA (50 μM); closed triangles, AtIP5PII sense line with Dex (30 μM) and ABA (50 μM). Mean values ±sd are provided for three independent induction experiments.
(B) Ins(1,4,5)P3 5-phosphatase activity in transgenic lines expressing the Dex-responsive AtIP5PII sense transgene. Fourteen-day-old transgenic seedlings were transferred to a hydroponic system (Aoyama and Chua, 1997) for 2 days before the addition of fresh medium containing Dex (30 μM). Plants were collected from different lines at 0, 14, or 20 hr after Dex addition, and Ins(1,4,5)P3 was assayed as described in Methods. Vector control lines displayed only background levels upon the addition of Dex (0-hr time point; data not shown). Each bar represents the average value of three independent induction experiments with standard deviations.
We analyzed the Ins(1,4,5)P3 5-phosphatase activity in total extracts prepared from AtIP5PII sense transgenic lines after Dex induction. A fourfold to fivefold increase in phosphatase activity was found after 20 hr of induction with Dex (Figure 5B), suggesting that an active AtIP5PII was produced. The increase of the Ins(1,4,5)P3 5-phosphatase would result in the hydrolysis of Ins(1,4,5)P3 that accumulated in response to ABA, thus leading to lower steady state levels.
Analysis of ABA-Responsive Gene Expression in Transgenic Plants Carrying an Inducible AtPLC1 Sense Transgene
To further examine the contribution of PLC1 to ABA signaling, we analyzed the transgenic lines containing the AtPLC1 sense transgene and the pTA211 vector alone by RNA gel blotting after treatment with Dex. AtPLC1 transgenic transcript was detected after 3 hr of Dex induction and reached a maximal level at 6 hr, whereas no transcript was detected in the vector control line (Figure 6A). Biochemical experiments showed that 3 to 6 hr after the induction, there was a threefold to fourfold increase in phospholipase C activity when the extracts were assayed in the presence of calcium in vitro (Figure 6C). However, almost no increase in Ins(1,4,5)P3 levels in vivo was detected in these transgenic plants (Figure 6D). As expected, a vector control transgenic line showed no increase in PLC1 activities or Ins(1,4,5)P3 levels after Dex treatment. Notwithstanding the increase in Dex-induced PLC1 expression, the transgenic lines did not express any of the ABA-responsive genes (KIN2, RD29a, or RD22) even after 20 hr of induction (Figure 6A). Moreover, the induction of KIN2 and RD29a by ABA was normal in the presence or absence of Dex in the AtPLC1 sense transgenic lines (Figure 6B).
Figure 6.
Molecular and Biochemical Analysis of Transgenic Lines Expressing the Dex-Inducible AtPLC1 Sense Transgene.
(A) Dex induction of the AtPLC1 transgenic transcript in AtPLC1 sense lines and in a vector control transgenic line. Plants were collected at 3, 6, and 20 hr after treatment with Dex (30 μM), and the expression of PLC1, KIN2, RD29a, and RD22 probes and 18S rDNA was examined by RNA gel blot hybridization.
(B) Transgenic lines containing the AtPLC1 sense transgene were grown on plates for 2 weeks, and the seedlings were transferred to a hydroponic medium (Aoyama and Chua, 1997). After 2 days, the medium was replaced with fresh medium containing either Dex (30 μM) or ABA (50 μM) or both. Total RNA was isolated at various times (0, 4, and 24 hr), and the gel blots were hybridized with RD29a and KIN2 probes and 18S rDNA. Vector control transgenic lines contained the pTA211 plasmid alone. For details, see Methods. ABA-0, plants collected at 0 hr in the ABA medium; ABA-4, plants incubated with ABA (50 μM) for 4 hr; DEX-24, plants incubated with Dex (30 μM) for 24 hr; DEX-24/ABA-4, plants incubated with Dex (30 μM) for 24 hr followed by an additional 4 hr with ABA (50 μM).
(C) and (D) Seedlings of a transgenic line containing the inducible AtPLC1 sense transgene were grown and treated with Dex as described in (B), except that plants were collected at 0, 3, and 6 hr after treatment. Total protein extracts were assayed for PLC1 enzyme activity (C), and the amount of Ins(1,4,5)P3 was determined (D). Each bar represents the average value of three independent induction experiments with standard deviations. For details, see Methods. Solid bars, transgenic line with the inducible AtPLC1 sense gene; open bars, vector control transgenic line. PIP2, phosphatidylinositol 4, 5 bisphosphate.
A vector control line showed similar levels of ABA-induced expression of KIN2 and RD29a. In addition, the transgenic lines were neither insensitive (Table 2) nor hypersensitive (data not shown) to ABA during germination. Together, these results suggest that in the absence of ABA, an increase in AtPLC1 expression is not sufficient to cause an increase in Ins(1,4,5)P3 levels and to induce the expression of ABA-responsive genes in vegetative tissues. It is likely that the expressed AtPLC1 enzyme is latent because it requires a calcium signal for activation in vivo. Furthermore, the increase in AtPLC1 apparently did not interfere with the induction of KIN2 and RD29a by ABA.
DISCUSSION
The Dex-inducible system (Aoyama and Chua, 1997) can be used for the regulated expression of transgenes in plants and is a useful tool to probe the function of proteins in plant signaling pathways. We have used a modified version of this system to generate a series of Arabidopsis transgenic lines carrying cDNAs encoding either the phospholipase C1 (AtPLC1) transgene or the Ins(1,4,5)P3 5-phosphatase (AtIP5PII) transgene in the sense and antisense orientations. The germination, biochemical, and molecular phenotypes of the transgenic lines reported here are gene specific and completely dependent on whether the cDNA is expressed in the sense or the antisense orientation.
Decreased Expression of AtPLC1 and Increased Expression of AtIP5PII Reduce Ins(1,4,5)P3 Levels in ABA-Treated Plants
RNA gel blot analysis showed that ABA induction of the endogenous AtPLC1 in the transgenic lines carrying the AtPLC1 antisense transgene was decreased markedly only after Dex-induced expression. The Dex induction of the AtPLC1 antisense transgene did not affect AtPLC2 expression levels, indicating specificity (data not shown). Upon Dex treatment, the Ins(1,4,5)P3 levels in AtPLC1 antisense transgenic lines showed only a small increase in response to ABA (Figure 4A). These results demonstrate a decrease in ABA-induced PLC1 enzymatic activity as well as in Ins(1,4,5)P3 levels in these transgenic lines.
In the AtIP5PII sense transgenic lines, the Dex-induced increase in AtIP5PII transcript levels was accompanied by a corresponding increase in specific Ins(1,4,5)P3 5-phosphatase activity. When treated with ABA, these plants showed only a small increase in Ins(1,4,5)P3 levels compared with that of vector control transgenic lines (Figure 5A). These results suggest that the increase in the Ins(1,4,5)P3 5-phosphatase activity in AtIP5PII sense transgenic lines most likely is responsible for the reduced Ins(1,4,5)P3 levels.
AtPLC1 Expression Is Necessary but Not Sufficient for the Maximal Induction of ABA-Responsive Genes
Analysis of ABA-responsive gene expression in Dex-induced adult transgenic lines carrying the AtIP5PII sense and AtPLC1 antisense genes revealed lower levels of KIN2, RD29a, and RD22 induction by ABA, whereas COR47 was induced normally. In addition, we found that the abi1-1 mutant was impaired in the ABA induction of these three genes but not COR47 (J.-P. Sanchez and N.-H. Chua, unpublished results), suggesting that the action of the two transgenes is specific to the ABI1-mediated ABA signaling pathway. Our results indicate that increases in the PLC1 and Ins(1,4,5)P3 levels are required for maximal induction of a subset of genes by ABA.
Transgenic lines containing the AtPLC1 sense transgene displayed induction of the AtPLC1 transgenic transcript only upon the addition of Dex, but no induction in the expression of KIN2, RD29a, and RD22 was observed in the Dex-treated lines. Therefore, in the absence of ABA, the expression of AtPLC1 itself is not sufficient to trigger expression of the three ABA-responsive genes. At different times after Dex induction, extracts prepared from the transgenic plants showed an increase in PLC1 activity when assayed in vitro in the presence of calcium, demonstrating the expected induction of the enzyme. However, there was no difference in the Ins(1,4,5)P3 levels between AtPLC1 sense and vector control transgenic lines. These results can be explained if the expressed PLC1 is latent and requires an increase in cytosolic Ca2+ for activation.
We also examined the induction of the AtPLC1 transgene in the presence of Dex and ABA. We found that in the AtPLC1 sense transgenic lines, the induction of KIN2 and RD29a by ABA was similar to that in a vector control line and was not affected by Dex. These results suggest that AtPLC1 expression apparently does not interfere with the ABA induction of KIN2 and RD29a.
We propose that the Ca2+ signal required for activation of the expressed latent PLC1 is induced by cADPR, which shows a rapid and transitory increase upon ABA treatment and likely mediates the primary ABA response (Wu et al., 1997). This notion is consistent with the observation that the expression of AtPLC1 itself is induced by ABA (Hirayama et al., 1995). The subsequent activation of AtPLC1 and increase in Ins(1,4,5)P3 levels are needed as secondary signals to amplify the primary response and to maximize the induced gene expression. In previous single-cell experiments, the induction of RD29a-GUS and KIN2-GUS by cADPR was observed, and this was likely to be the primary ABA response. In pancreatic acinar cells treated with the hormone cholecystokinin, mobilization of calcium from the endoplasmic reticulum and amplification of the calcium signal are mediated by cADPR and nicotinic acid adenine dinucleotide phosphate, and Ins(1,4,5)P3 participates only as a terminal amplifier of the calcium wave signal (Cancela et al., 2000).
AtPLC1-Dependent Events Are Required for Sensitivity of Germination and Seedling Growth to ABA
To date, limited information is available concerning the molecular events that take place during Arabidopsis seed germination other than the fact that ABA and gibberellic acid (GA) have opposing effects. The germination of Arabidopsis seed is sensitive to ABA, suggesting that an ABA signaling pathway operates in seed to block this process. This observation has been used to identify ABA signaling components through the isolation and characterization of mutants insensitive to ABA. In principle, the inducible expression of antisense and sense transcripts encoding a positive regulatory component of the ABA signaling pathway should phenocopy loss-of-function and gain-of-function mutations, respectively, in the pathway. A well-known regulator of the ABA signaling pathway is ABI1, a protein phosphatase 2C. A dominant mutation (abi1-1) in this phosphatase can render plants insensitive to ABA (Giraudat, 1995). We have observed that transgenic plants expressing abi1-1 can become insensitive to ABA, whereas transgenic plants expressing ABI1 have the same sensitivity as wild-type plants (Y. Wu, J.-P. Sanchez, and N.-H. Chua, unpublished results).
Transgenic seeds were germinated in 2 μM ABA in the presence or absence of Dex to examine the effect of regulated transgene expression on ABA signaling pathways that operate during germination. In the case of the AtPLC1 transgenic plants, only lines expressing the antisense transgene were able to germinate in the presence of Dex (93%), whereas in the absence of Dex almost no germination was observed. In the absence of Dex, 3% germination was observed in the AtIP5PII sense transgenic lines, compared with the 95% germination frequency observed in the presence of the inducer. No ABA insensitivity was seen in lines expressing AtIP5PII in the antisense orientation.
Figure 3D shows that growth of AtIP5PII sense and AtPLC1 antisense transgenic plants in the presence of ABA (without Dex) was arrested for >18 days, whereas in the presence of Dex they were able to grow. Furthermore, after 2 to 3 weeks of incubation in the presence of Dex, AtIP5PII sense and AtPLC1 antisense transgenic lines showed no growth inhibition when challenged subsequently with ABA. Mild inhibition was observed if the plants were first germinated (4 to 5 days) in the absence of ABA and subsequently transferred to medium containing 2 μM ABA (previously induced for 24 hr in Dex), suggesting that the major effect occurs during germination (data not shown). These results also suggest that the effects exerted by ABA during germination are mediated by PLC1 and increased Ins(1,4,5)P3 levels. Similar experiments were performed with transgenic plants harboring the antisense AtPLC2 transgene, which encodes a phospholipase C2 (Hirayama et al., 1997). Seed of these transgenic lines, however, displayed no ABA insensitivity during germination, suggesting that the effect seen with AtPLC1 is specific to the PLC1 gene product (data not shown).
Analysis of AtPLC1 sense transgenic seed germinated in 2 μM ABA in the presence or absence of Dex to examine the effect of regulated transgene expression on the ABA signaling pathway revealed neither a hypersensitivity nor an insensitivity to ABA (hypersensitivity, data not shown; see Table 2 for insensitivity).
During Arabidopsis seed germination, it is possible that the ABA signal is mediated by a Ca2+-dependent pathway acting in parallel with the GA signal. Studies of barley aleurone cells suggest that GA stimulates Ca2+ production in an Ins(1,4,5)P3-independent manner and that ABA inhibits this response (reviewed in Lovegrove and Hooley, 2000). Furthermore, ABA has been postulated to mediate the repression of an α-amylase through a Ca2+-dependent pathway (Chen, 1997; Lovegrove and Hooley, 2000). How these ABA signaling events in barley aleurone cells relate to the Arabidopsis system remains to be determined.
METHODS
Reagents
AtPLC1 (GenBank accession number D38544) and AtPLC2 (GenBank accession number D50804) cDNAs were kindly provided by Dr. K. Shinozaki. AtIP5PI (GenBank accession number AF289633) and AtIP5PII (GenBank accession number AF289634) cDNA were provided by Dr. W. Gruissem. KIN2 and RD29a cDNAs were obtained as expressed sequence tag (EST) clones (Arabidopsis Information Management System [AIMS] accession numbers 103216T7 and ATT53026; Arabidopsis Biological Resource Center [ABRC], Ohio State University, Columbus). COR47 and RD22 were amplified by polymerase chain reaction (PCR) from a cDNA library provided by Dr. Qi Xie (Institute of Molecular Agrobiology, Singapore), with primers flanking the coding sequence. The 18S rDNA probe was obtained from Dr. Taku Takahashi (Department of Biology, Hokkaido University, Sapporo, Japan). The authenticity of the probes was verified by sequencing.
Cloning of AtIP5PI and AtIP5PII cDNAs
An EST database search with sequences from the human inositol 1,4,5-trisphosphate (Ins[1,4,5]P3) 5-phosphatase revealed two different groups of Arabidopsis thaliana ESTs that contain the highly conserved domain for Ins(1,4,5)P3 5-phosphatase. The EST used for the isolation of the AtIP5PII full-length clone was AIMS accession number ATT0437 obtained from ABRC. A cDNA fragment of 1.2 kb was excised with EcoRI and XhoI and used as a probe for cDNA library screening. A similar procedure was used to isolate a full-length cDNA of AtIP5PI by use of the EST clone AIMS accession number ATT04703. The EST was used as a probe to screen an Arabidopsis cDNA library CD4-15 (ABRC) to obtain full-length cDNAs for AtIP5PI and AtIP5PII.
Transformation Constructs
A modified version of the binary transformation plasmid pTA7001 containing the complete two-component Gal4-VP16-glucocorticoid binding domain (GVG) (Aoyama and Chua, 1997) was used. In contrast to pTA7001, which was derived from pBI101, the new plasmid, pTA211 (P. Spielhofer and N.-H. Chua, unpublished results), is derived from a pPZP vector (Hajdukiewicz et al., 1994); in addition, the cauliflower mosaic virus 35S promoter for transcription of the GVG fusion gene was replaced by the G10-90 promoter (Ishige et al., 1999). The pTA211 plasmid was digested with XhoI and SpeI (New England BioLabs, Beverly, MA). The coding region of the AtIP5PII gene was engineered by PCR to have a XhoI restriction site at the 5′ end and a SpeI site at the 3′ end. The sequences of the forward and reverse primers were 5′-GGCTCGAGATGGTATTCGCGGCCGCGT-TTGTACTCTCC-3′ and 5′-GCACTAGTTCAAAAAGAAGGTTCAGGA-TGAACAGCAGAA-3′, respectively. For the AtIP5PII antisense construct, the same gene-specific sequences were used as primers, except that the restriction sites were switched. The AtPLC1 antisense construct was engineered by PCR to have a XhoI site at the 3′ end and a SpeI site at the 5′ end by using PCR amplification. The sequences of the forward and reverse primers were 5′-GGCTCG-AGCTAACGAGGCTCCAAGACAAACCGCATGAGC-3′ and 5′-GGA-CTAGTATGATATGTTGTGTAAGAAACTTCAAGGTGA-3′, respectively. For the AtPLC1 sense construct, the same primers were used, except that the restriction sites were switched. The PCR-amplified DNA fragment was digested with XhoI and SpeI, gel purified using the Qiaquick Gel extraction protocol (Qiagen, Valencia, CA), and ligated into the pTA211 plasmid by use of T4 DNA ligase (New England BioLabs). A clone for each construct was verified by restriction analysis. All constructs were introduced into Agrobacterium tumefaciens strain ABI (Aoyama and Chua, 1997).
Plant Transformation
Arabidopsis plants (ecotypes C24 and Landsberg) were transformed with Agrobacterium by use of the vacuum infiltration procedure (Bent et al., 1994). The C24 line contains a transgene consisting of the ABA-responsive KIN2 promoter fused to the coding sequence of firefly luciferase (Foster and Chua, 1999). Seed collected from the vacuum-infiltrated plants were surface sterilized by treating them with a solution of 1.5% sodium hypochlorite and 0.01% Tween 20 (Sigma) for 10 min and washed three times with sterile water. The sterilized seed were then resuspended in 0.1% agarose and sown in Petri dishes containing medium A (full-strength Murashige and Skoog [1962] salts, pH 5.7, and 1% sucrose solidified with 0.8% Bactoagar [Gibco BRL, Grand Island, NY]) and 20 μg/mL hygromycin B (Sigma). The plated seed were vernalized for 4 days and then transferred to a growth chamber maintained at 22°C under long-day conditions (16 hr of light/8 hr of dark). Transgenic T1 seedlings were selected on a plate containing hygromycin (20 mg/mL), and after 2 to 3 weeks of growth, the presence of the transgene was confirmed by PCR analysis.
Dexamethasone Treatments
ABA (Sigma) was dissolved in 100% methanol to make a 100 mM stock solution, and dexamethasone (Dex; Sigma) was dissolved in DMSO to make a 100 mM stock solution. Both solutions were stored at −20°C. To monitor transgene expression, we surface sterilized transgenic seed and sowed them in Petri dishes as described above. After vernalization at 4°C for 4 days, the plates were incubated for 2 weeks in a growth chamber maintained at 22°C under long-day conditions (16 hr of light/8 hr of dark). Seedlings were removed from the plates and grown for 2 days in a hydroponic system containing liquid medium A (full-strength Murashige and Skoog salts, pH 5.7, and 1% sucrose; Gibco BRL). Fresh medium containing either Dex (30 μM) or ABA (50 μM) or both was added, and plants were removed at the designated times and then washed and frozen in liquid nitrogen. For the vector control transgenic lines, identical conditions were used, and the experiments were performed in parallel.
RNA Analysis
Total RNA was isolated from seedlings and adult plants using the Qiagen RNA purification kit. RNA gel blot analysis was performed according to Ausubel et al. (1994). Each lane contained 10 μg of total RNA. KIN2, RD29a, COR47, RD22, and 18S rDNA fragments were obtained by PCR amplification with Pfu polymerase (Pyrococcus furiosus; Stratagene, La Jolla,CA) as described above. For the AtPLC1 and AtIP5PII genes, we used the coding region or the 3′ noncoding sequence as a probe. Fragments were purified using the Qiaquick Gel extraction protocol (Qiagen). All DNA fragments were labeled with 32P-dCTP and 32P-dATP by random priming (Amersham, Arlington Heights, IL). Hybridization signals were quantified using the PhosphorImager STORM system (Molecular Dynamics, Sunnyvale, CA), and the data were analyzed with the Image Quant program (version 1.1).
Quantification of Ins(1,4,5)P3
Twenty-day-old plants treated with ABA (50 μM) and Dex (30 μM) as described above were collected and frozen rapidly in liquid nitrogen. Frozen tissue (including stem, roots, and leaves) were ground to a fine powder with a porcelain mortar and pestle precooled in liquid nitrogen. The frozen powder was resuspended in 0.2 volume of ice-cold 20% perchloric acid and incubated on ice for 20 min. Extracts were sonicated for 10 sec to disrupt any aggregates, and samples were purified as described by the manufacturer of the Ins(1,4,5)P3 assay system kit (Amersham) except that we used a Partisil 5 SAX column (Whatman) (Shears, 1997). Ins(1,4,5)P3, Ins(1,4)P2, and Ins(1,3,4,5)P4 standards were prepared according to the manufacturer's specifications (Sigma).
Inositol Polyphosphate 5-Phosphatase Purification and Assay
A full-length cDNA of AtIP5PII was cloned in the vector pET28a (Novagen, Madison, WI) and transformed into Escherichia coli BL21-Codon Plus (DE3) (Stratagene, La Jolla, CA). The pET28a vector without an insert served as a control. E. coli cells were grown at 37°C overnight. A 0.5-mL overnight culture was used to inoculate 100 mL of 1 × Luria-Bertani medium, and the culture was induced at 37°C for 2 hr with a final concentration of 0.1 mM isopropyl-β-d-thiogalactopyranoside (Sigma). The culture was grown for another 2 hr at 24°C. Cells were harvested by centrifugation, and the pellets were resuspended in ice-cold bacterial lysis buffer (50 mM phosphate buffer, pH 7.4, 100 mM NaCl, and 100 μg/mL lysozyme). Bacterial cells were lysed by sonication, and the cell lysate was centrifuged at 15,000g for 30 min at 4°C. The supernatant was collected, and the recombinant protein was purified as recommended by the manufacturer (Histrap kit; Amersham Pharmacia Biotech) except that a chromatography system (Perfusion Chromatography System; Biocad-Sprint, Cambridge, MA) was used with the column. The protein was incubated at 37°C for 30 min with 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 2 mM MgCl2, and 3H-labeled Ins(1,4,5)P3 or Ins(1,3,4,5)P4 (10 μM 3H, 0.01 μCi) in 50 μL. The reaction was initiated by the addition of the substrate and terminated by the addition of 1 mL of distilled water followed by immediate heating at 100°C for 3 min (Connolly et al., 1985). After cooling, the samples were applied to a Sep-Pak Accell Plus QMA cartridge (Waters; Millipore, Milford, MA) and eluted (Maslanski and Busa, 1990). One-milliliter fractions were collected, and the radioactivity was determined by scintillation counting. Samples were then analyzed by HPLC using a Partisil 5 SAX column (250 × 4.6 mm) according to the procedure described by Shears (1997).
Phospholipase C Assay
Twenty-day-old plants were treated with Dex (30 μM) as described above, collected, and frozen rapidly in liquid nitrogen. Extracts were prepared as described (Deutscher, 1990), and phosphatidylinositol 4, 5 bisphosphate hydrolysis was assayed according to Hirayama et al. (1995).
Acknowledgments
We thank Dr. K. Shinozaki for the cDNA clones encoding AtPLC1 and AtPLC2, Dr. W. Gruissem for the cDNA clones encoding AtIP5PI and AtIP5PII, and Dr. Peter Hare for critical reading of the manuscript. This work was supported by Department of Energy Grant No. DOE94ER20143 to N.-H.C.
References
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kinston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: John Wiley).
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. [DOI] [PubMed] [Google Scholar]
- Berridge, M.J. (1993). Inositol triphosphate and calcium signaling. Nature 361 315–325. [DOI] [PubMed] [Google Scholar]
- Cancela, J.M., Mogami, H., Tepikin, A.V., and Petersen, O.H. (1998). Intracellular glucose switches between cyclic ADP-ribose and inositol trisphosphate triggering of cytosolic Ca2+ spiking. Curr. Biol. 16 865–868. [DOI] [PubMed] [Google Scholar]
- Cancela, J.M., Gerasimenko, O., Gerasimenko, J., Tepikin, A., and Petersen, O. (2000). Two different but converging messenger pathways to intracellular Ca2+ release: The roles of nicotinic acid adenine dinucleotide phosphate, cyclic ADP-ribose and inositol trisphosphate. EMBO J. 19 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. (1997). Cloning of a Ca-ATPase gene and the role of cytosolic calcium in the gibberellin signalling pathway in aleurone cells. Plant J. 11 363–371. [DOI] [PubMed] [Google Scholar]
- Clapham, D. (1995). Calcium signaling. Cell 80 259–268. [DOI] [PubMed] [Google Scholar]
- Connolly, T.M., Bross, T.E., and Majerus, P.W. (1985). Isolation of a phosphomonoesterase from human platelets that specifically hydrolyzes the 5-phosphate of inositol 1,4,5-trisphosphate. J. Biol. Chem. 260 7868–7874. [PubMed] [Google Scholar]
- Deutscher, M.P. (1990). Methods in Enzymology, Vol. 182: Guide for Protein Purification. (San Diego, CA: Academic Press).
- Foster, R., and Chua, N.-H. (1999). An Arabidopsis mutant with deregulated ABA gene expression: Implications for negative regulator function. Plant J. 17 363–372. [DOI] [PubMed] [Google Scholar]
- Galione, A. (1993). Cyclic ADP-ribose: A new way to control calcium. Science 259 325–326. [DOI] [PubMed] [Google Scholar]
- Gilroy, S., Read, N., and Trewavas, A. (1990). Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature 343 769–771. [DOI] [PubMed] [Google Scholar]
- Giraudat, J. (1995). Abscisic acid signaling. Curr. Opin. Cell Biol. 7 232–238. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Ohto, C., Mizoguchi, T., and Shinozaki, K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92 3903–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T., Mitsukawa, N., Mizoguchi, T., and Shinozaki, K. (1997). At-PLC2, a gene encoding a phosphatidylinositol-specific phospholipase, is constitutively expressed in vegetative and floral tissue in Arabidopsis thaliana. Plant Mol. Biol. 34 175–180. [DOI] [PubMed] [Google Scholar]
- Ishige, F., Takaishi, M., Foster, R., Chua, N.-H., and Oeda, K. (1999). A G-box motif (GCCACGTGCC) tetramer confers high levels of constitutive expression in dicot and monocot. Plant J. 18 443–448. [Google Scholar]
- Jefferson, A.B., and Majerus, P.W. (1995). Properties of type II inositol polyphosphate 5-phosphatase. J. Biol. Chem. 270 9370–9377. [DOI] [PubMed] [Google Scholar]
- Kang, H.G., Fang, Y., and Singh, K.B. (1999). A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 20 127–133. [DOI] [PubMed] [Google Scholar]
- Lee, H.C., and Aarhus, R. (1991). ADP-ribosyl cyclase: An enzyme that cyclizes NAD+ into a calcium-mobilizing metabolite. Cell Regul. 2 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.C., Aarhus, R., and Graeff, R.M. (1995). Sensitization of calcium-induced calcium release by cyclic ADP-ribose and calmodulin. J. Biol. Chem. 270 9060–9066. [DOI] [PubMed] [Google Scholar]
- Lee, Y.C., Suh, S.L., Assmann, S., Kelleher, J., and Crain, C. (1996). Abscisic acid–induced phosphoinositide turnover in guard cells protoplasm of Vicia faba. Plant Physiol. 110 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovegrove, A., and Hooley, R. (2000). Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 5 102–110. [DOI] [PubMed] [Google Scholar]
- MacRobbie, E.A. (1998). Signal transduction and ion channels in guard cells. Philos. Trans. R. Soc. Lond. Biol. Sci. 353 1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslanski, J., and Busa, W.B. (1990). A sensitive and specific mass assay for myo-inositol and inositol phosphates. In Methods in Phosphoinositide Research, R.F. Irvine, ed (New York: Raven Press), pp. 112–122.
- McAinsh, M., Browlee, C., and Hetherinton, A. (1990). ABA-induced elevation of guard cells cytosolic Ca2+ precedes stomatal closure. Nature 343 186–188. [Google Scholar]
- McNellis, T.W., Mudgett, M.B., Li, K., Aoyama, T., Horvath, D., Chua, N.-H., and Staskawicz, B.J. (1998). Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J. 14 247–257. [DOI] [PubMed] [Google Scholar]
- Merlot, S., and Giraudat, J. (1997). Genetic analysis of abscisic acid signal transduction. Plant Physiol. 114 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Schroeder, J., and Hagiwara, S. (1990). Repetitive increases in cytosolic Ca2+ of guard cells by abscisic acid activation of nonselective Ca2+ permeable channels. Proc. Natl. Acad. Sci. USA 87 9305–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears, H.B. (1997). Measurement of inositol phosphate turnover in intact cells and cell-free system. In Signalling by Inositides: A Practical Approach, H.B. Shears, ed; B.D. Hames, series ed (New York: IRL Press/Oxford University Press), pp. 47–51.
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signalling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Trewavas, A. (1999). Le calcium, C'est la vie: Calcium makes waves. Plant Physiol. 120 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Kuzma, J., Marechal, E., Graeff, R., Lee, H.C., Foster, R., and Chua, N.-H. (1997). Abscisic acid signaling through cyclic ADP-ribose in plants. Science 278 2126–2130. [DOI] [PubMed] [Google Scholar]