Abstract
The Sucrose export defective1 (Sxd1) gene of maize was cloned and shown to encode a novel protein conserved between plants and cyanobacteria. The structure of the Sxd1 locus was determined in wild-type plants and two independent sxd1 alleles. Expression analysis demonstrated that the gene was transcribed in all green tissues, with highest levels in maturing leaf blades. In situ hybridization studies revealed high levels of Sxd1 mRNA in bundle sheath cells, with lower levels within the mesophyll. The SXD1 protein was localized to chloroplasts, in both bundle sheath and mesophyll cells. Levels of sucrose, glucose, and fructose were compared between wild-type and sxd1 plants. Mutant plants were fully capable of producing sucrose and accumulated all three sugars at concentrations above those measured in wild-type plants. Despite these increased sugar concentrations, photosynthetic gene expression was not significantly downregulated in affected areas of sxd1 leaf blades. These results are consistent with photosynthate being trapped within anthocyanin-accumulating regions of sxd1 leaves due to plasmodesmal occlusion at the bundle sheath–vascular parenchyma boundary of the minor veins. A model for SXD1 function is proposed in which the protein is involved in a chloroplast-to-nucleus signaling pathway necessary for proper late-stage differentiation of maize bundle sheath cells, including the developmentally regulated modification of plasmodesmata.
INTRODUCTION
The recessive maize mutant sucrose export defective1 (sxd1) represents the only known example of a plant in which plasmodesmal structure and/or function is compromised during leaf development. Affected regions of sxd1 leaves accumulate starch and anthocyanin, demonstrate vascular parenchyma (VP) cell death, and fail to efficiently export sucrose. In addition, minor veins in maturing sxd1 leaf blades exhibit specific occlusion of plasmodesmata between bundle sheath (BS) and VP cells (Russin et al., 1996). This phenotype suggests that loss of symplasmic continuity at the BS-VP boundary inhibits the delivery of sucrose to the phloem in sxd1 mutant leaves; the subsequent buildup of photosynthate would then result in starch accumulation and pigment synthesis (Russin et al., 1996; Stitt, 1996).
Recent work has supported such a model by demonstrating that symplasmic movement of the fluorescent tracer Lucifer Yellow (LYCH) is limited in sxd1 mutants (Botha et al., 2000). When the dye was injected into wild-type leaf blades, it was observed to move into the minor veins, but such movement was blocked in anthocyanin-accumulating regions of sxd1 leaf blades. Based on the presence of aggregates of aniline blue–staining material at the BS-VP interface, these authors concluded that the inhibition of LYCH movement involved the deposition of callose at plasmodesmata in the BS-VP cell walls. Thus, molecular characterization of SXD1, and subsequent analysis of its function within the developing maize leaf, could lead to the identification of a pathway involved in the alteration of plasmodesmal function.
In this report, we describe the cloning of Sxd1 and show that it encodes a novel chloroplast protein conserved between plants and cyanobacteria. Sugar analyses revealed that sxd1 plants produce abundant sucrose, indicating that SXD1 is not a direct regulator of photosynthate export from the chloroplast. Rather, this protein may play a role in regulating expression of nuclear-encoded genes, as suggested by a lack of photosynthetic gene repression in sxd1 leaves. Disruption of chloroplast-to-nucleus signaling in sxd1 mutants likely has pleiotropic effects on the developing leaf, resulting in the sealing of BS-VP plasmodesmata and subsequent VP cell death.
RESULTS
Cloning sxd1
Phenotypically wild-type individuals from families segregating sxd1 mutants were outcrossed to the lines B73 and R1Scm2. Individuals from the outcrosses were then self-crossed to obtain F2 families. For molecular analysis, wild-type plants were selected from nonsegregating families (to ensure that none was heterozygous for the sxd1 mutation) and compared with plants showing the sxd1 phenotype (homozygous mutants). The Amplification of Insertionally Mutagenized Sites (AIMS) (Frey et al., 1998) was used to identify genomic sequences adjacent to Mutator (Mu) elements present in sxd1 but absent in wild-type plants. AIMS products were identified that amplified from all sxd1, but not wild-type, templates (data not shown). These products were isolated from polyacrylamide gels, used as templates for re-amplification by polymerase chain reaction (PCR), and cloned.
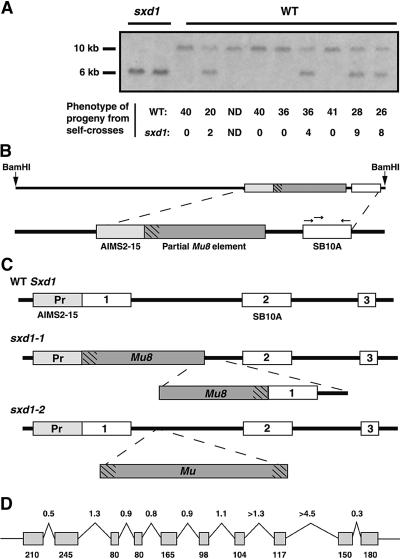
For analysis, the individual cloned PCR products were used as probes on DNA gel blots. A clone representing a portion of the sxd1 locus was expected to be a low copy number sequence with a restriction fragment length polymorphism (RFLP) between wild-type and sxd1 individuals. One of the five cloned AIMS products, the 275-bp AIMS2-15, met both of these criteria, because the probe recognized a single-copy sequence in the maize genome with an RFLP that cosegregated with the sxd1 phenotype. To ensure that the AIMS2-15 sequence was tightly linked to the sxd1 locus, we performed cosegregation analysis with >100 individual maize plants. The AIMS2-15 probe recognized a 6-kb BamHI fragment in all sxd1 mutant plants and a 10-kb BamHI fragment in all wild-type plants from nonsegregating families (data not shown). In a segregating family, the wild-type plants were either homozygous for the 10-kb band or heterozygous at the locus, and each of the heterozygotes segregated the sxd1 phenotype among its self-cross progeny (Figure 1A). Thus, the AIMS2-15 fragment seemed likely to represent a portion of a locus that was located adjacent to an sxd1-specific Mu element insertion.
Figure 1.
Structure of the Sxd1 Locus in Wild-Type and Mutant Plants.
(A) Segregation of an RFLP recognized by the AIMS2-15 fragment in a family carrying sxd1-1. Phenotypically wild-type (WT) plants were self-crossed to confirm that plants heterozygous for the RFLP segregated sxd1 mutants in the following generation.
(B) The SB10 genomic clone. The AIMS2-15 probe was used to isolate a 6-kb BamHI fragment from an sxd1 subgenomic library; this sequence was found adjacent to a partial Mu8 element. The region labeled SB10A showed homology to expressed sequence tags from Arabidopsis and rice. Arrows show the approximate position of primers used in reverse transcription (RT)–PCR experiments, and hatch marks represent the Mu TIR region.
(C) Structure of the 5′ end of the wild-type and mutant Sxd1 loci. White boxes represent exons, and grey boxes represent the AIMS2-15 sequence in the promoter region of the gene. PCR reactions using primers designed to the SB10A and AIMS2-15 regions were used to amplify the 5′ end of the Sxd1 locus from wild-type templates. The sxd1-1 allele carries a partial Mu8 element and a deletion of both the first exon and half of the first intron. The sxd1-2 allele, isolated by Trait Utility System for Corn (TUSC) screening, carries a Mu insertion in the first intron. Pr, promoter.
(D) Intron/exon structure of the Sxd1 locus. Numbers above the diagram represent lengths of introns (grey boxes) in kilobases, and numbers below indicate the sizes of exons (black lines) in base pairs.
A subgenomic library was constructed from an sxd1 individual to isolate the 6-kb BamHI fragment. Size-selected genomic DNA was used to create a library in the λ-ZAP vector. Three 6-kb clones, which hybridized to the AIMS2-15 probe, were isolated and shown to be identical by restriction mapping; one clone, designated SB10 (Figure 1B), was fully sequenced. Within this clone, the AIMS2-15 fragment was indeed found to lie adjacent to a Mu Terminal Inverted Repeat (TIR) sequence at one end of a Mu8 element. Interestingly, the Mu8 element was missing 570 bp of sequence, including the opposite TIR region. Portions of the SB10 clone were used to search the public databases for homologous sequences, and one short (240 bp) stretch of sequence, designated SB10A, showed homology to a rice expressed sequence tag and a putative Arabidopsis open reading frame.
Reverse transcription (RT)–PCR and RNA gel blot experiments were used to determine whether the SB10A sequence was part of a transcribed region of the genome. Primers were designed from the SB10A sequence to amplify the portion showing homology to putative genes (Figure 1B). With wild-type maize seedling poly(A)+ RNA templates, RT-PCR products of the expected size were amplified (data not shown), indicating that the SB10A sequence was part of an expressed sequence. The PCR products representing the SB10A region were then used as probes on RNA gel blots containing total RNA from leaves of both wild-type and sxd1 seedlings. The SB10A probe hybridized to a band of 1.6 kb in wild-type, but not sxd1, leaves (data not shown). Thus, the SB10A sequence was expressed in wild-type but absent in sxd1 leaves.
To obtain the full sequence encoded by the SB10 locus, we used primers designed against the SB10A region in 3′ and 5′ rapid amplification of cDNA ends (RACE) PCR experiments. The sequences of the 3′ and 5′ RACE products were combined to deduce the full-length 1.5-kb coding sequence of the putative Sxd1 cDNA (Figure 2). Next, primers were designed to the ends of the deduced sequence, and DNA was amplified by RT-PCR from wild-type seedling RNA. These products were cloned and sequenced to confirm that the 3′ and 5′ RACE products represented a single transcript.
Figure 2.
The Sxd1 cDNA with Its Deduced Amino Acid Sequence.
Gray triangles indicate the positions of introns, which divide the coding sequence into 10 exons, and asterisk indicates stop codon (GenBank accession number AF302187).
Identification of a Second Null Allele, sxd1-2
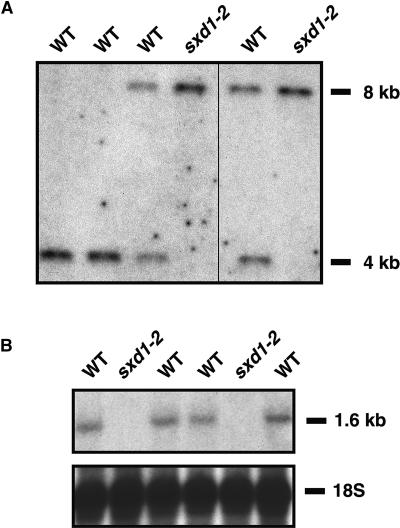
To confirm that the cloned sequence corresponded to the sxd1 phenotype, we next sought to obtain a second sxd1 allele by using the Pioneer HiBred Trait Utility System for Corn (TUSC). Primers were designed against the putative Sxd1 coding sequence and used in PCR reactions, along with primers specific for Mu TIR sequences, to screen Mu populations. From this screen, three families containing Mu element insertions in the first intron of the putative Sxd1 locus were identified. One family, PV03 206 C-03, segregated three out of 20 plants that accumulated anthocyanin in the same manner as sxd1 plants. DNA gel blot analysis of individuals within this family showed that only the anthocyanin-accumulating plants were homozygous for an RFLP at the 5′ end of the Sxd1 locus (Figure 3A). RNA gel blot analysis revealed that these plants did not produce Sxd1 mRNA in maturing leaves, whereas their wild-type siblings showed expression of Sxd1 (Figure 3B).
Figure 3.
The sxd1-2 Phenotype Is Associated with an RFLP at the Sxd1 Locus and Loss of mRNA Expression.
(A) DNA gel blot of genomic DNA from individuals in a family segregating the sxd1-2 phenotype. Both plants homozygous for the 8-kb fragment showed the sxd1 phenotype, whereas none of the plants carrying at least one copy of the 4-kb fragment expressed the phenotype.
(B) RNA gel blot of total RNA from leaf blades of wild-type and sxd1-2 plants. Levels of 18S rRNA show relative amounts loaded in each lane.
WT, wild type.
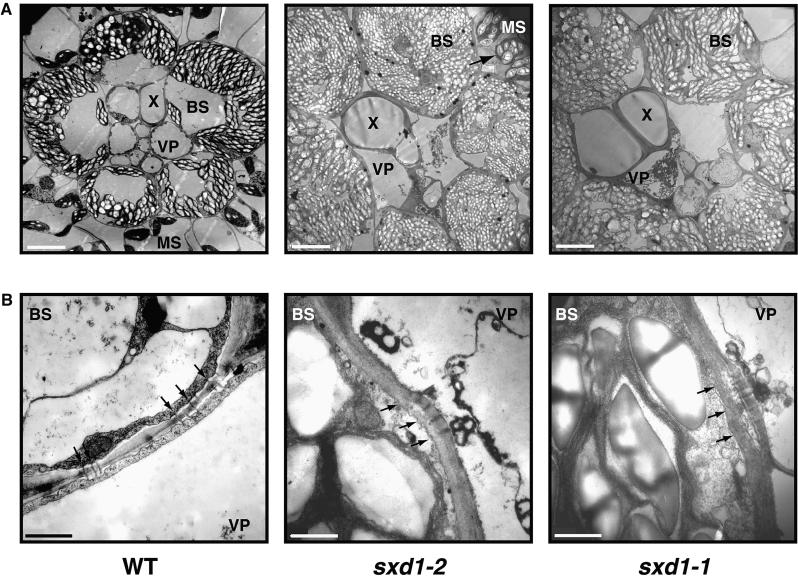
Heterozygous individuals from family PV03 206 C-03 were self-crossed to generate additional plant material for phenotypic analysis. One-quarter of plants from these self-crosses demonstrated a phenotype indistinguishable from that seen in the original sxd1 mutants (data not shown). Thus, the mutation in line PV03 206 C-03 was named sxd1-2, and the original sxd1 reference allele was renamed sxd1-1. In both sxd1-1 and sxd1-2 mutants, anthocyanin and starch accumulated in maturing leaf blades and were associated with the death of vascular parenchyma cells (Figure 4A). Furthermore, as shown in Figure 4B, occlusion of plasmodesmata was observed at the BS-VP interface of sxd1-1 and sxd1-2 mutants. Isolation of a second mutation at the same locus that eliminates expression of the gene and produces the same phenotype serves as confirmation that we have identified the Sxd1 locus.
Figure 4.
Comparison of Minor Vein Structure between Wild-Type and Two sxd1 Alleles.
(A) In contrast to wild-type (WT) plants, sxd1-1 and sxd1-2 BS cells accumulate starch granules to the point at which chloroplasts become distended, filling the cell volume. Plasmolysis of VP cells is evident in both mutants but absent from wild-type plants.  . MS, mesophyll cell; X, tracheary element.
. MS, mesophyll cell; X, tracheary element.
(B) In a manner similar to sxd1-1, sxd1-2 mutants show occlusion of plasmodesmata due to deposition of material at the BS-VP boundary, whereas in wild-type plants, plasmodesmata exhibit normal morphology. Black arrows mark the position of the BS plasma membrane. 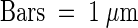 .
.
Structure of the Sxd1 Locus
To determine the structure of the wild-type Sxd1 locus, we conducted PCR experiments with both sxd1-1 and wild-type genomic DNA as templates. Primers designed from the AIMS2-15 and SB10A regions allowed amplification of the expected 1.5-kb fragment from the sxd1-1 template and a shorter 1.0-kb fragment from the wild-type template. The 1.5-kb fragment revealed the same sequence present in the SB10 genomic clone, confirming that the primers were specific for this region. In contrast, the fragment amplified from wild-type DNA contained sequence corresponding to the 5′ end of the cDNA sequence but lacking the partial Mu8 element. These results indicated that the sxd1-1 mutation involved a deletion of the 5′ end of the coding region and that the SB10A region is the second exon within the gene (Figure 1C). PCR products generated from sxd1-2 templates with the Mu TIR primer and each of two gene-specific primers were cloned and sequenced. This confirmed that a Mu element was present within the first intron of the sxd1-2 locus, as shown in Figure 1C. The lack of mRNA expression from this allele suggests that the Mu insertion may have disrupted a regulatory sequence within the first intron to produce a second null allele. Alternately, the Mu insertion may interfere with correct splicing of the gene, leading to early truncation.
The intron/exon structure of the Sxd1 gene was determined by PCR experiments using primer sets designed to amplify short segments of the cDNA. Introns were amplified from wild-type genomic DNA templates, and the intron/exon borders were sequenced. As shown in Figure 1D, the maize Sxd1 locus is >11 kb in length. The coding sequence is broken up into 10 exons by a series of large introns. By comparing Sxd1-hybridizing fragments seen on DNA gel blots (performed under both high- and low-stringency conditions) with the structure of the gene, it was determined that Sxd1 represents a single-copy sequence within the maize genome (data not shown).
The Sxd1 locus was mapped by utilizing a collection of recombinant inbred lines to compare the position of the Sxd1 sequence with those of previously mapped genes and markers (Burr et al., 1994). This analysis showed that Sxd1 lies near the centromere of chromosome 5, near the ris1 locus. This position corresponded well with B-A- and waxy-translocation mapping results, which indicated that the sxd1 phenotype mapped to the short arm of chromosome 5 (data not shown).
Sxd1 Encodes a Novel Protein
The Sxd1 cDNA and its deduced amino acid sequence are shown in Figure 2. Database searches revealed homology to only a small number of sequences, including expressed sequence tags from rice, wheat, tomato, soybean, and pine; an uncharacterized Arabidopsis gene on chromosome four; and a putative coding region from the unicellular photosynthetic bacteria Synechocystis. Alignment of the proteins from maize (ZmSXD1), Arabidopsis (AtSXD1) and Synechocystis (SySXD1) is shown in Figure 5. The N termini of the maize and Arabidopsis proteins were predicted to be plastid transit peptides with putative cleavage sites that corresponded to the N terminus of the Synechocystis protein.
Figure 5.
Alignment of the SXD1 Amino Acid Sequence with Homologs in Arabidopsis and Synechocystis.
Similar residues are outlined with boxes, and identical residues are shaded. Asterisks highlight conserved tryptophan residues. Arrow shows the predicted cleavage site for the putative transit peptide. The maize and Arabidopsis sequences are 62% identical, and the maize and Synechocystis sequences are 38% identical. ZmSXD1, Zea mays SXD1 protein; AtSXD1, Arabidopsis thaliana Sxd1, GenBank accession number AF302188; SySXD1, Synechocystis sp Sxd1, GenBank accession number BAA17775.
Neither the ZmSXD1 sequence nor the homologs found in the databases showed any similarity to characterized sequences, and no protein motifs could be identified from primary structure analysis. The predicted mature-sized ZmSXD1 and AtSXD1 should be hydrophilic proteins, with isoelectric points of 5.0 and 5.2, respectively; they also contain many tryptophan residues that are conserved in Synechocystis and spaced throughout the sequence (Figure 5). The Sxd1 gene appears to encode a novel protein present in plants and cyanobacteria and is unlikely to be a member of a gene family represented by multiple genes per organism.
Pattern of Sxd1 Expression
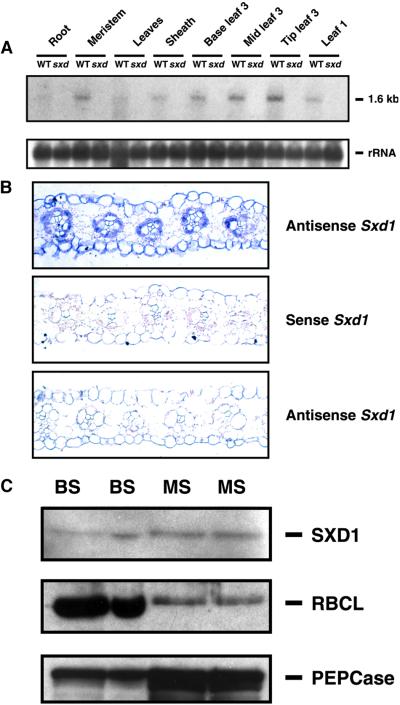
RNA gel blot analysis was conducted to determine the tissue specificity and timing of Sxd1 mRNA expression. Samples were collected from wild-type and sxd1-1 plants grown in the greenhouse until the fourth leaf had emerged. At this stage, the sxd1-1 plants showed anthocyanin accumulation in the first and second leaves and at the tip of the third leaf. As shown in Figure 6A, no Sxd1 mRNA was detected in any of the mutant samples. In wild-type plants, Sxd1 was present at low levels in all green tissues. Slight upregulation of Sxd1 expression was observed in the middle and tip of leaf three, corresponding to the parts of the leaf that were beginning to show a phenotype in the sxd1-1 plants.
Figure 6.
Pattern of Tissue and Cellular Expression of Sxd1 in Wild-Type and sxd1 Mutant Maize Plants.
(A) RNA gel blot showing levels of Sxd1 mRNA in wild-type (WT) and sxd1-1 tissues. Each lane contains 5 μg of total RNA collected from the indicated tissues. The lower panel shows 18S rRNA as a control for loading.
(B) In situ hydridization of maize leaf sections with Sxd1 probes. Transverse sections from a mature wild-type leaf hybridized with Sxd1 antisense (upper) and sense (middle) probes. The lower panel shows a section of a matched leaf from an sxd1 mutant plant hybridized with the Sxd1 antisense probe.
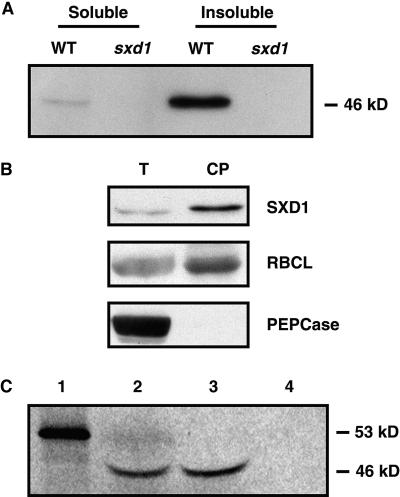
(C) Protein gel blots comparing protein expression in BS and mesophyll (MS) cells. Detection of SXD1, PEPCase, and RBCL is shown in two independent preparations of each cell type. PEPCase and RBCL distributions indicate the level of enrichment for each tissue, because PEPCase and RBCL are MS and BS specific, respectively.
In situ hybridization experiments were performed with mature leaves to determine whether Sxd1 expression was cell-type specific. In cross-sections of the mature leaf blade, signal was clearly detected in BS cells around both major and minor veins, and a weaker signal was detected in mesophyll cells (Figure 6B). No signal above background was detected on control slides hybridized with probe generated in the sense orientation or in samples from sxd1 mutants (Figure 6B). To confirm the expression of Sxd1 in mesophyll tissue, BS and mesophyll cells were isolated and analyzed at the protein level (Figure 6C). The large subunit of Rubisco, RBCL, was highly enriched in the BS samples, indicating that the mesophyll preparations contained low levels of BS contamination. The cytoplasmic protein phosphoenolpyruvate carboxylase (PEPCase), a marker for the mesophyll, was enriched in the mesophyll cell preparations, although some cross-contamination of the BS preparations with mesophyll cells was apparent. Levels of SXD1 were nearly uniform between the two preparations, indicating that, together with the in situ hybridization data, SXD1 is present in both BS and mesophyll cells.
SXD1 Is Targeted to Chloroplasts
Antibodies generated against SXD1 recognized a band at 46 kD on protein gel blots that was present in wild-type but not sxd1-1 leaf protein extracts (Figure 7A). Based on the deduced amino acid sequence, SXD1 was predicted to be 53 kD. However, the observed size was consistent with the predicted size after transit peptide cleavage. The majority of SXD1 was insoluble when leaf sections were ground, without detergent, in a glass homogenizer. This was also consistent with localization to chloroplasts, because most of the organelles would have remained intact and insoluble in this crude preparation.
Figure 7.
SXD1 Localizes To and Is Imported into Chloroplasts.
(A) Anti-SXD1 antiserum recognizes a 46-kD protein in wild-type (WT) but not sxd1 leaf protein extracts. Soluble proteins were extracted by homogenization without detergent. The insoluble fraction represents the proteins and organelles pelleted by centrifugation of the homogenized tissue and then solubilized in SDS PAGE sample buffer.
(B) Protein gel blots of total leaf proteins (T) versus chloroplast proteins (CP). The same samples were used for detection of SXD1, RBCL, and PEPCase. PEPCase is a cytoplasmic protein, and RBCL resides in the chloroplast stroma.
(C) Import of SXD1 into isolated pea chloroplasts. In vitro–translated 35S-labeled full-length SXD1 protein (lane 1) was incubated with pea chloroplasts under conditions favoring import. The chloroplasts were then untreated (lane 2) or treated with protease (lane 3) or with protease and Triton X-100 to disrupt the chloroplast envelope (lane 4).
To further analyze the subcellular localization of SXD1, we prepared intact chloroplasts from 10-day-old maize seedlings. Protein levels in these preparations were compared with those in total leaf protein extracts. Figure 7B depicts a protein gel blot showing that SXD1 was enriched in chloroplast preparations; RBCL, which localizes to the stromal region of the chloroplast, showed a similar enrichment, whereas PEPCase was detected only in the total protein extract, demonstrating that the isolated chloroplasts were free from cytosolic contamination.
In vitro import experiments were conducted to obtain additional proof that SXD1 is targeted to chloroplasts. The full-length Sxd1 coding sequence was used as a template for in vitro transcription/translation in the presence of 35S-labeled methionine. The 53-kD precursor protein was incubated with intact chloroplasts isolated from pea seedlings to test for import. After an incubation of 20 min, the reactions were treated with buffer only, with protease, or with protease and Triton X-100. As shown in Figure 7C, a labeled protein of 46 kD was detected in the chloroplasts and was protected from protease digestion. Collectively, these data indicate that the SXD1 precursor protein contains a functional transit peptide, is imported effectively into chloroplasts, and upon import is cleaved to the 46-kD size seen in maize leaf extracts.
sxd1 Mutants Accumulate High Sugar Levels
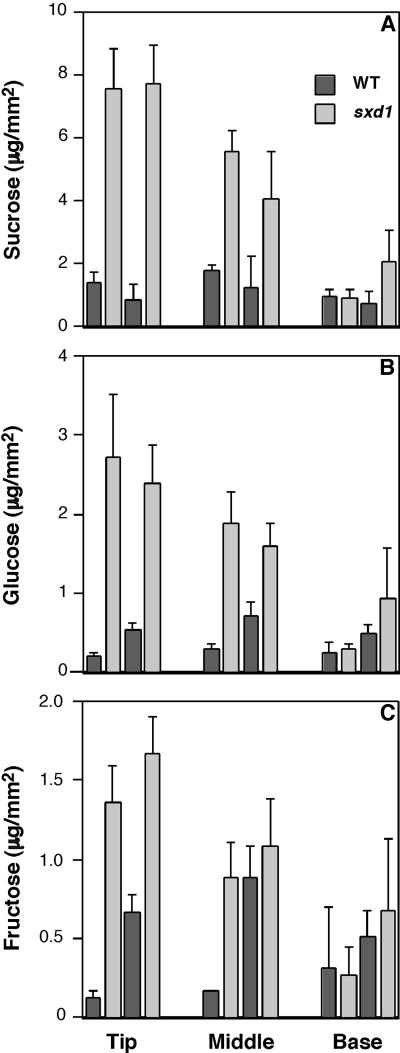
Russin et al. (1996) showed that sxd1 plants deposit large amounts of starch in bundle sheath cells and aberrantly accumulate starch in mesophyll cells (Figure 4). In addition, these mutants fail to effectively export sucrose from mature leaves. Given that SXD1 was localized to chloroplasts, the observed starch accumulation implicated a role for this protein in the regulation of photosynthate partitioning between the chloroplast and cytosol. If this were correct, then mature leaves of sxd1 mutants would be expected to contain reduced levels of sucrose during the photoperiod.
Sugar concentrations at the tip, middle, and base of leaf 3 from 16-day-old greenhouse-grown plants (with five or six expanded leaves) were compared between the wild-type and sxd1 genotypes. At this stage, the tips of the sxd1 leaf 3 showed strong anthocyanin accumulation, indicating the presence of high levels of starch and occluded plasmodesmata. The phenotype at the middle of the leaf was patchy, and at the base of leaf 3, no effects of the sxd1 mutation were apparent. Contrary to expectations, the sucrose levels were five to six times higher in the tips of sxd1 leaves than in the corresponding wild-type tissue (Figure 8A). Glucose and fructose also accumulated to higher levels in the tips of sxd1 leaves than in wild-type leaves (Figures 8B and 8C). The increase in sugar concentrations relative to wild-type levels correlated with the observed sxd1 phenotype. The differences were largest at the tip of leaf 3, intermediate in the middle, and insignificant at the base. Similar effects on the concentrations of all three sugars were observed when wild-type and sxd1 plants were grown under a lower light intensity in controlled environment conditions (data not shown). These results demonstrate that the loss of SXD1 function does not block the export of fixed carbon from the chloroplast.
Figure 8.
Analysis of Sugar Levels in Wild-Type and sxd1 Leaves.
Amounts of sucrose (A), glucose (B), and fructose (C) in the tip, middle, and base of the third leaf from wild-type (WT) and sxd1 plants. Leaves were sampled in the middle of a 14-hr photoperiod. Values from two independent experiments are shown, and each value represents the mean ±sd of five samples extracted from 15 4-mm-diameter leaf discs.
sxd1 Mutants Fail to Downregulate Photosynthetic Gene Expression
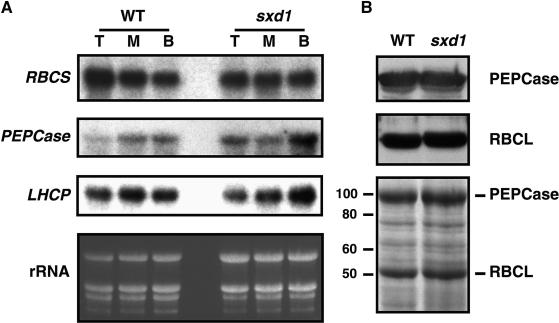
Accumulation of sugars in leaves, whether as a result of disruption of sucrose export (Jeannette et al., 2000) or of sugar feeding (Sheen, 1990; Krapp et al., 1993; Jang and Sheen, 1994; Ehness et al., 1997), has been shown to negatively regulate the expression of photosynthetic genes. Given the presence of elevated sugar levels in sxd1 leaves, we next examined whether the mutants responded to this accumulation by downregulating the expression of the small subunit of Rubisco (RBCS), the light harvesting complex protein (LHCP), and PEPCase. As for sugar analyses, the third leaf of 16-day-old plants was cut into sections representing the tip, middle, and base of the leaf, and total RNA was extracted. RNA gel blots revealed that expression levels for these genes were similar between the wild type and sxd1 in all three regions of the leaf blade (Figure 9A). This result was supported by analysis of protein levels in the tip of leaf 3. Here, the overall protein profiles did not demonstrate any major differences, and the two dominant photosynthetic proteins, RBCL and PEPCase, accumulated at the same level in wild-type and sxd1 tissues (Figure 9B).
Figure 9.
Expression of Photosynthetic Genes Is Not Downregulated in sxd1 Mutant Leaves.
(A) Total RNA was extracted from the tip (T), middle (M), and base (B) of the third leaf of 18-day-old wild-type (WT) and sxd1 seedlings. Leaf sections from three different plants were combined before extraction of RNA. The same blot was hybridized sequentially with probes specific for LHCP, RBCS, and PEPCase, and the levels of rRNA served as a control for loading.
(B) Protein gel blots comparing levels of photosynthetic proteins in the tip of leaf 3 from 18-day-old wild-type and sxd1 seedlings. The lower panel shows a Coomassie-stained gel showing the protein profiles in the samples used for protein gel blot analysis. Each sample consisted of total protein extract pooled from leaves excised from three plants. At left, molecular weight markers.
DISCUSSION
Sxd1 Encodes a Novel Chloroplast Protein
In this work, we have identified and characterized the Sxd1 gene of maize. A portion of the Sxd1 locus was isolated through the AIMS procedure (Frey et al., 1998), allowing the subsequent cloning of the full SXD1 coding sequence (Figure 2). Loss of Sxd1 expression was attributed to a partial Mu8 insertion and a deletion of the 5′ end of the gene in sxd1-1 mutants (Figure 1). This mutational event was associated with an RFLP that was tightly linked to the sxd1-1 phenotype and eliminated expression of a gene product normally found in tissues affected by the mutation. The identification of a second null allele, producing a phenotype indistinguishable from that of the original sxd1-1 mutants, confirmed that the isolated gene was indeed Sxd1. Interestingly, loss of expression in sxd1-2 resulted from insertion of a Mu element in the first intron. Because the presence of regulatory sequences within introns has been reported for several plant genes (Rowland and Strommer, 1985; Gallie and Young, 1994; Sieburth and Meyerowitz, 1997; Jeon et al., 2000), this finding suggests that the first intron of Sxd1 may contain elements necessary for transcription. Alternately, the Mu insertion could result in the improper splicing of the transcript. However, if this were case, the resulting mRNA must be either highly unstable or truncated close to the 5′ end, because no alternative forms of Sxd1 mRNA were detected in RNA gel blot analysis of the sxd1-2 mutant.
The Sxd1 cDNA represents a novel single-copy gene present in plants and photosynthetic bacteria but absent from yeast and invertebrates. Several lines of evidence strongly support the conclusion that SXD1 is targeted to, and functions within, chloroplasts. First, sequence analysis predicted the presence of a transit peptide with a cleavage site coincident with the N terminus of the Synechocystis SXD1 (Figure 5). Second, antibodies raised against SXD1 recognized a 46-kD protein in maize leaves (Figure 7), which corresponded to the predicted size of the mature protein after transit peptide cleavage. Third, SXD1 was enriched in chloroplast preparations (Figure 7). Finally, in vitro–translated SXD1 protein was imported into pea chloroplasts and subsequently cleaved to the expected size of 46 kD (Figure 7). Absence of detectable levels of the 53-kD full-length protein on protein gel blots and in import experiments indicated that the peptide was very efficiently imported into chloroplasts, suggesting no function for the protein in the cytosol.
Analysis of the SXD1 amino acid sequence revealed an absence of homology to any characterized motifs. However, the presence of numerous conserved tryptophan residues suggested that SXD1 might fall into the class of WD-repeat proteins. This family includes various regulatory proteins thought to function via protein–protein interactions (Neer et al., 1994). Members of this group are classified by the presence of four to eight repeats of a conserved motif 23 to 41 amino acids in length, often beginning with GH and ending with WD (Neer et al., 1994; Sondek et al., 1996). In SXD1, the spacing of tryptophan residues is irregular, conserved histidines cannot be found upstream of the tryptophans, and no repeating units were identified within the peptide. Thus, despite the presence of numerous tryptophan residues, SXD1 does not fit the characteristics of a WD-repeat protein. Future experiments will explore whether the tryptophan residues are essential to SXD1 function, and analyses of related sequences may show whether it is evolutionarily related to WD proteins.
Loss of SXD1 Indirectly Blocks Phloem Loading
Our finding that SXD1 localizes to chloroplasts precludes its function as a structural component of plasmodesmata, a cell wall peptide, or a plasma membrane protein involved in sucrose transport. Thus, the pleiotropic effects of the sxd1 mutation on sucrose export must be indirect and may initiate in mesophyll, BS, or VP cells. The accumulation of both sucrose and starch in sxd1 leaves established that photosynthate exchange occurs in these mutants, because carbon fixed in BS chloroplasts was available for sucrose synthesis in the cytosol of mesophyll cells (Lunn and Furbank, 1999). Hence, it is unlikely that SXD1 functions as a regulator of carbon partitioning. Rather, our results are consistent with earlier findings in which a block to phloem loading/translocation caused the buildup of starch, sugars, and anthocyanin (King et al., 1967; Neales and Incoll, 1968; Huber and Israel, 1982; von Schaewen et al., 1990; Heineke et al., 1992; Riesmeier et al., 1994; Geigenberger et al., 1996; Jeannette et al., 2000).
The failure of sxd1 leaves to export photosynthate suggests that sucrose is unable to reach the site of phloem loading. This model is consistent with recent results in which the fluorescent tracer LYCH was unable to diffuse into minor veins in affected regions of sxd1 leaves (Botha et al., 2000). If the primary cause of the symplasmic discontinuity observed in sxd1 plants was VP cell death, then one might invoke a model in which SXD1-mediated signals from plastids prevent activation of a cell death pathway. An alternative scenario would involve signaling between the chloroplast and nucleus in BS cells. A defect in such a communication pathway may affect final stages of BS differentiation during the sink-to-source transition, resulting in damage to the cells and subsequent deposition of callose at the plasmodesmata between BS and VP cells. Irrespective of where the blockage to phloem loading is initiated, our results provide additional evidence that sucrose must move symplasmically from the BS to the VP cells for access to the loading system.
A Model for SXD1 Involvement in Chloroplast-Nucleus Signaling
Mutations affecting both chloroplast function and cell differentiation provide evidence that signaling occurs between chloroplasts and the nucleus, although the mechanism(s) remains to be resolved. For instance, the late-stage differentiation of palisade mesophyll cells appears to require chloroplast function. The defective chloroplasts and leaves (dcl) mutation in tomato and the differentiation and greening (dag) mutation in Antirrhinum majus, both of which knock out the function of a nuclear-encoded protein targeted to plastids, prevent the differentiation of proplastids into chloroplasts. In leaf tissues in which this defect occurs, the palisade cells fail to elongate and assume their normal columnar shape (Chatterjee et al., 1996; Keddie et al., 1996). In maize, the Inhibitor of striate (Isr) gene product has been shown to play a role in cellular response to defective plastids. In plants carrying the striate2 mutation, leaves demonstrate stripes of plastid-defective tissue, and Isr inhibits development of these stripes in a dose-dependent manner. Recent work has shown that Isr encodes a protein likely to localize to plastids, which prevents the proliferation of leaf cells containing defective chloroplasts (Park et al., 2000).
Numerous experiments have shown that photooxida-tive damage to chloroplasts results in the downregulation of nuclear-encoded photosynthetic genes (Taylor, 1989; Goldschmidt-Clermont, 1998). These effects do not involve an inhibition of protein import into chloroplasts (Bolle et al., 1994). Furthermore, promoter–β-glucuronidase fusion experiments demonstrated the involvement of gene-specific cis regulatory sequences, some of which overlapped with sequences involved in light-mediated gene regulation (Bolle et al., 1994). In Arabidopsis, several mutations have been identified that uncouple chloroplast function from the expression of photosynthetic genes. These genomes un-coupled (gun) mutants retain high levels of RBCS and chlorophyll a/b binding (CAB) proteins expression in the absence of functional chloroplasts (Susek et al., 1993). Together, these results indicate that signals from plastids to the nucleus are necessary for normal regulation of photosynthetic gene expression. The gun mutations produce subtle alterations in plant development, and their recessive nature suggests that nonphotosynthetic plastids may send signals repressing transcription of these genes (Goldschmidt-Clermont, 1998).
In sxd1 leaves, photosynthetic gene expression remains high, despite the accumulation of high sugar levels (Figures 8 and 9). This observation, together with the effects of sxd1 loss of function on BS plasmodesmata and sugar export, leads us to hypothesize that sxd1 mutants may be defective in a chloroplast-to-nucleus signalling mechanism that is involved in sugar sensing. The mechanisms by which plant cells sense sugar levels are currently under investigation (Koch, 1996; Jang and Sheen, 1997; Smeekens and Rook, 1997; Gibson and Graham, 1999; Lalonde et al., 1999). Many photosynthetic genes are suppressed by high sugar levels via hexokinase-mediated signals (Jang et al., 1997) and encode proteins that are targeted to the chloroplast. Expression of these nuclear genes is coordinated by additional factors as well. For instance, their expression is repressed in nongreen tissues and enhanced in green tissues in a light-dependent and developmentally regulated manner (Taylor, 1989; Goldschmidt-Clermont, 1998). These aspects of gene regulation must involve some form of communication between plastids and the nucleus. Hence, it is possible that one of the pathways for hexokinase-mediated sugar sensing includes chloroplast factors, thereby allowing coordination of photosynthetic gene expression by the chloroplast. Future experiments will explore whether our observations on photosynthetic gene expression reflect a lack of repression in sxd1 mutants or a long-term response of maize leaves to maintained high sugar levels. If it can be conclusively determined that sxd1 mutants lack a photosynthetic gene repression mechanism, then SXD1 might represent a factor required in the chloroplasts to integrate hexokinase-mediated signals within the cell.
In maize, the sink-to-source transition is associated with late-stage differentiation of BS cell walls and plasmodesmata, which occurs as the tissue ceases import (Evert et al., 1996). Our results with SXD1 suggest that signals from the chloroplast may be necessary for proper gene regulation during BS differentiation. In plants lacking SXD1, perturbation of this pathway might result in mis-expression of nuclear genes, the deposition of callose at the BS-VP border, blockage of plasmodesmata, and the subsequent inhibition of photosynthate export.
METHODS
Plant Material
Maize plants (Zea mays) carrying sxd1-1 were grown in the field and outcrossed to the inbred lines B73 and R1Scm2. Outcross progeny were self-crossed to obtain F2 families that either segregated the sxd1 phenotype or contained only wild-type individuals. For amplification of insertionally mutagenized sites (AIMS) and cosegregation analyses, homozygous wild-type plants from nonsegregating families were compared with sxd1-1 mutants. For some experiments, plants were also grown under controlled environment (14-hr-light, 26°C/10-hr-dark, 20°C; 500 μmol m−2 sec−1 photosynthetically active radiation [PAR]; 70% humidity) or greenhouse conditions (14-hr photoperiod, noon levels of PAR ∼1000 μmol m−2 sec−1, 30°C).
Preparation of DNA, RNA, and DNA and RNA Gel Blots
DNA was prepared from leaf tissue using the urea extraction method (Chen and Dellaporta, 1994). For DNA gel blots, 5 μg of digested DNA was run on 0.8% agarose gels, transferred to nylon membranes (Hybond N+; Amersham, Piscataway, NJ), and hybridized at 65°C with 32P-labeled probes (Random Primer Extension System; DuPont–New England Nuclear, Boston, MA). Total RNA extracts were prepared from tissue ground under liquid N2, as described by Ruiz-Medrano et al. (1999). For RNA gel blots, 5 μg of total RNA was run on formaldehyde gels, transferred to nylon membranes, and hybridized to 32P-labeled probes at 65°C. Poly(A)+ RNA was prepared from total RNA using an Oligotex mRNA extraction kit (Qiagen, Chatsworth, CA), according to the manufacturer's protocols. Signal was detected by autoradiography on Kodak Biomax MS film.
Isolation of the SB10 Genomic Sequence
AIMS reactions were performed, using wild-type and sxd1-1 templates, as described (Frey et al., 1998), except 0.5 μL of unlabeled Mu Sel primer (10 μM) was included (in addition to the 0.5 μL of 32P-labeled Mu Sel) in the exponential polymerase chain reaction (PCR). Ultrapure dNTP mix (Clontech, Palo Alto, CA) was used in all reactions. Reamplified products were cloned into pCRII/TOPO (Invitrogen, Carlsbad, CA). For generation of a subgenomic library, sxd1-1 DNA was digested with BamHI, phenol/chloroform extracted, precipitated, and resuspended at 1 mg/mL. An aliquot of 200 μg was size-fractionated on a sucrose gradient (Weis, 1996), and an appropriate fraction was used to construct a library in λ-ZAP Express (Stratagene, La Jolla, CA), according to the manufacturer's protocols. Half of the primary library, 400,000 plaque-forming units, was plated and screened, and phagemids from three independent clones were excised according to the Stratagene protocol.
Sequencing and Analysis
Public databases were searched using BLASTP, BLASTN, FASTA, and PROSITE algorithms, and sequence alignments between pairs or groups of sequences were performed with EERIE or CLUSTALW algorithms (Baylor College of Medicine Search Launcher, Houston, TX). N-terminal chloroplast targeting sequences and cleavage sites were analyzed by the ChloroP algorithm (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark). The AtSXD1 sequence shown in Figure 5 represents the deduced amino acid sequence of a cDNA amplified from ecotype Landsberg erecta inflorescence tissue. This sequence differs from that entered in GenBank (accession number CAA18584) due to a misinterpretation of splice sites at the 5′ end of the gene in the original entry. The correct coding sequence has been submitted to GenBank under accession number AF302188.
PCR Procedures
cDNA was prepared from poly(A)+ RNA by reverse transcription (RT) at 42°C with SuperScript II RT enzyme (Gibco BRL, Rockland, MD) and oligo-dT-GAGA primer (5′-[GA]10ACTAGTCTCAG[T]18-3′). For RT-PCR reactions, 1 μL of the cDNA was used as template in PCR reactions using Advantage cDNA Polymerase Mix (Clontech). Primers SB-1 (5′-AGTTTCTGCTTTATGTACTCTGTTG-3′) and SB-3 (5′-CCAAAAATTGTTTGATTTCTCAGAGAACTGG-3′) were used to amplify the region corresponding to the rice expressed sequence tag, and primers SB-2 (5′-GTACCACTTTGATGGCACGGTTCC-3′) and SB-3 were used to amplify the region with similarity to the Arabidopsis sequence.
Experiments using 3′ rapid amplification of cDNA ends (RACE) were performed with the SB-2 and oligo dT-GAGA primers, 1 μL of cDNA, and Amplitaq DNA polymerase (PE Applied Biosystems, Foster City, CA). Amplification conditions were as follows: 94°C for 5 min; eight cycles of 94°C for 1 min, 68°C for 30 sec, minus 0.5°C per cycle, and 72°C for 1.3 min; 30 cycles of 94°C for 1 min, 64°C for 30 sec, and 72°C for 1.3 min; and finally, 72°C for 5 min. An aliqout (1 μL) from this first reaction was used as template in a second PCR with the nested SB-1 primer and oligo dT-GAGA, and the amplified fragments were cloned into pCRII/TOPO. Reactions for 5′ RACE were conducted with SB-3 and the nested primer SB-8 (5′-TATGTACTTATCGTCCGCTCCAAGG-3′) using the Gibco BRL 5′ RACE system, and products were cloned in pCRII/TOPO. To amplify the full-length SB10 cDNA, we used primers SB-13 (5′-ACAGAATTCTCTAGACATGAACCTCGCCGTCGCAGCCGC-3′) and SB-14 (5′-ACA-GAATTCTAGAGTTAGGCCTGGGGGCTTAAGAAATGGG-3′) designed from the 5′ and 3′ RACE fragments and used in RT-PCR reactions. For amplification of the 5′ end of the Sxd1 gene, the 215-4 primer (5′-CTAATTTTGACGCGCTCATTGGTCCCCAG-3′) was used with SB-3 to amplify products from BamHI-digested genomic DNA templates.
Identification of sxd1-2 by Trait Utility System for Corn Screening
The following primers, based on the Sxd1 cDNA sequence, were used by Pioneer HiBred to screen their collection of Mu insertion lines with terminal inverted repeat (TIR)–PCR: primers 35,149 (forward) 5′-CCTCGAGCTCCGTTTCCTCCTTTAC-3′ and 35,381 (reverse) 5′-TCATACCATCACGGAAGAGAGGGTTCTC-3′. Amplified fragments were sequenced, and plants containing potential inserts in the Sxd1 locus were grown under greenhouse conditions. Segregation of insertions was analyzed by DNA gel blotting.
In Situ Hybridization
Leaf tissues were excised from sxd1 leaf blades showing significant anthocyanin accumulation and matched leaves on wild-type seedlings. Tissue was processed, sectioned, and hybridized as described by Ruzin (1999), using probes generated against the Sxd1 cDNA.
Maize Protein Extraction and Protein Gel Blotting
Total protein was extracted by homogenizing leaf pieces in Hepes buffer (25 mM Hepes, pH 7.2, 150 mM NaCl, 10 μg/mL each leupeptin and aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride). Homogenate was filtered through miracloth, mixed with 6 × SDS-PAGE sample buffer (1 × SDS-PAGE sample buffer is 0.12 mg/mL bromophenol blue, 360 mM Tris, pH 6.8, 30% [v/v] glycerol, 10% [w/v] SDS, 600 mM DTT), boiled for 5 min, and cleared of insoluble material by centrifugation. For crude separation of soluble and insoluble proteins, the filtrate was centrifuged for 5 min at 16,000g before the addition of sample buffer. The supernatant was used as the soluble fraction, and the insoluble pellet was resuspended in 2 × sample buffer by boiling for 5 min. Proteins were separated on 10% polyacrylamide SDS–glycine gels and transferred to nitrocellulose membranes (Osmonics, Westborough, MA) by electroblotting. Protein gel blot detection was performed following standard protocols.
Preparation of mesophyll and BS cells was performed as described by Nelson (1994). For protein analysis, mesophyll cells were thawed in the presence of 2 × SDS-PAGE sample buffer, boiled for 5 min, and cleared of insoluble material by centrifugation. For BS protein extraction, vascular strands were ground under liquid nitrogen and then processed as described above for mesophyll samples.
For generation of antisera, the region of Sxd1 encoding residues 100 to 239 was subcloned to create an HIS tag fusion peptide in pRSET-B (Invitrogen). The peptide was overexpressed in Escherichia coli strain BL21(DE3), extracted from inclusion bodies, and purified on a Ni-NTA-Agarose column. Antisera were produced in rabbits by Strategic BioSolutions (Ramona, CA). A glutathione S-transferase:SXD1 fusion protein, made with the full-length Sxd1 cDNA in pGEX-KG (Guan and Dixon, 1991), was used to test the antisera for specificity and to affinity purify anti-SXD antibodies. Goat anti–rabbit IgG peroxidase conjugate (Sigma, St. Louis, MO) was used as a secondary antibody, and signal was detected with Renaissance chemiluminescence reagent (DuPont–New England Nuclear). Antisera against the large subunit of Rubisco (RBCL) and phosphoenolpyruvate carboxylase (PEPCase) were kindly provided by D. Schnell (Rutgers University, Newark, NJ) and W. Taylor (Commonwealth Scientific and Industrial Research Organization [CSIRO], Canberra, Australia), respectively.
Chloroplast Isolation and Import Assays
For comparison of total leaf versus chloroplast protein, chloroplasts were isolated as described by Musser and Theg (2000) from maize seedlings grown for 10 days under controlled environment conditions and then transferred to the dark for 24 hr. Intact chloroplasts were solubilized in SDS-PAGE sample buffer and used for protein gel blot analysis.
For import experiments, the Sxd1 cDNA was subcloned into pSP64 PolyA (Promega, Madison, WI) for in vitro translation. Transcription/translation reactions were performed in the presence of 35S-methionine with the TnT Coupled Reticulocyte Lysate System (Promega); 15 μL of 20 mM unlabeled methionine was added after translation to prevent further incorporation of label. Chloroplasts were isolated (Musser and Theg, 2000) from pea seedlings grown for 12 days in vermiculite under controlled environment conditions (8-hr- light/16-hr-dark, 250 μmol m−2 sec−1 PAR, 24°C, 60% humidity). In vitro–translated protein was then incubated for 20 min in the light with these chloroplasts (0.33 mg/mL chlorophyll) and 3 mM ATP in 60 μL 1 × import buffer (0.5 M K-Tricine, pH 8.0, 0.33 M sorbitol, 2 mM MgCl2) and then diluted in 600 μL import buffer containing 5 mM CaCl2. For protease treatment, the dilution contained 0.2 mg/mL thermolysin (Protease X; Sigma) ±0.1% (v/v) Triton X-100. After a 20-min incubation (0°C), EDTA was added to a final concentration of 50 mM, and chloroplasts were then pelleted by centrifugation. Proteins contained in the pellet were separated on SDS–polyacrylamide gels, and labeled proteins were detected by autoradiography.
Sugar Analysis
Leaf discs (4 mm diameter) were cut from the tip, middle, and base of leaf 3 of maize seedlings grown for 16 to 18 days under greenhouse conditions. Discs were immediately placed in 80% ethanol and heated to 100°C for 1 min and then extracted five times with 80% ethanol (70°C) and twice with water (70°C). Extracts were dried under vacuum, resuspended in 0.2 N H2SO4 and 1 mM EDTA, and cleaned with Sep-Pak Plus C18 cartridges (Waters Chromatography, Taunton, MA). Sugars were analyzed on an HPLC system consisting of a Hewlett Packard (HP) 1050 pump coupled to a Bio-Rad Refractive Index Monitor (model 1750 set at ×16 sensitivity) and an autosampler (HP 1050) operated by HP ChemStation software (version A.06.01). Isocratic separation in 8.5 mN H2SO4 was performed at 23°C over an Ion-300 (300 mm × 7.8 mm; 10 μm) column (Inter Action Chromatography, San Jose, CA) at a flow rate of 0.4 mL/min.
Acknowledgments
Thanks are due to Robert Meeley, Pioneer HiBred, for invaluable assistance and advice throughout the course of this work and especially with the TUSC screen, Steve Theg and members of his laboratory for assistance with chloroplast import assays, Betty Hess-Pierce for performing sugar analyses, David Weiss for helpful suggestions and assistance with photosynthetic gene expression experiments, and Jen Sheen for providing clones of maize RBCS, LHCP, and PEPCase. This work was supported by National Science Foundation (NSF) Grant No. IBN-9900539 (to W.J.L), NSF Research Training Grant No. 94-14106, and a University of California-Davis Jastro Shields Graduate Research Award (to L.M.P.).
References
- Bolle, C., Sopory, S., Lubberstedt, T., Klosgen, R.B., Herrmann, R.G., and Oelmuller, R. (1994). The role of plastids in the expression of nuclear genes for thylakoid proteins studied with chimeric β-glucoronidase gene fusions. Plant Physiol. 105 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha, C.E.J., Cross, R.H.M., van Bel, A.J.E., and Peter, C.I. (2000). Phloem loading in the sucrose-export-defective (SXD1) mutant maize is limited by callose deposition at plasmodesmata in bundle sheath–vascular parenchyma interface. Protoplasma 214 65–72. [Google Scholar]
- Burr, B., Burr, F.A., and Matz, E.C. (1994). Mapping genes with recombinant inbreds. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 249–254.
- Chatterjee, M., Sparvoli, S., Edmunds, C., Garosi, P., Findlay, K., and Martin, C. (1996). DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 15 4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chen, J., and Dellaporta, S. (1994). Urea-based plant DNA miniprep. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 526–527.
- Ehness, R., Ecker, M., Godt, D.E., and Roitsch, T. (1997). Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9 1825–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert, R.F., Russin, W.A., and Bosabalidis, A.M. (1996). Anatomical and ultrastructural changes associated with sink-to-source transition in developing maize leaves. Int. J. Plant Sci. 157 247–261. [Google Scholar]
- Frey, M., Stettner, C., and Gierl, A. (1998). A general method for gene isolation in tagging approaches: Amplification of insertion mutagenised sites (AIMS). Plant J. 13 717–721. [Google Scholar]
- Gallie, D.R., and Young, T.E. (1994). The regulation of gene expression in transformed maize aleurone and endosperm protoplasts. Plant Physiol. 106 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger, P., Lerchl, J., Stitt, M., and Sonnewald, U. (1996). Phloem-specific expression of pyrophosphatase inhibits long-distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ. 19 43–55. [Google Scholar]
- Gibson, S., and Graham, I. (1999). Another player joins the complex field of sugar-regulated gene expression in plants. Proc. Natl. Acad. Sci. USA 96 4746–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177 115–180. [DOI] [PubMed] [Google Scholar]
- Guan, K., and Dixon, J.E. (1991). Eukaryotic proteins expressed in E. coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192 262–267. [DOI] [PubMed] [Google Scholar]
- Heineke, D., Sonnewald, U., Büssis, D., Günter, G., Leidreiter, K., Wilke, I., Raschke, K., Willmitzer, L., and Heldt, H.W. (1992). Apoplastic expression of yeast-derived invertase in potato. Effects on photosynthesis, leaf solute composition, water relations, and tuber composition. Plant Physiol. 100 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, S.C., and Israel, D.W. (1982). Biochemical basis for partitioning of photosynthetically fixed carbon between starch and sucrose in soybean (Glycine max Merr.) leaves. Plant Physiol. 69 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.C., and Sheen, J. (1994). Sugar sensing in higher plants. Plant Cell 6 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.C., and Sheen, J. (1997). Sugar sensing in higher plants. Trends Plant Sci. 2 208–214. [Google Scholar]
- Jang, J.C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannette, E., Reyss, A., Gregory, N., Gantet, P., and Prioul, J.L. (2000). Carbohydrate metabolism in a heat-girdled maize source leaf. Plant Cell Environ. 23 61–69. [Google Scholar]
- Jeon, J.S., Lee, S., Jung, K.H., Jun, S.H., Kim, C., and An, G. (2000). Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 123 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie, J.S., Carroll, B., Jones, J.D.G., and Gruissem, W. (1996). The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 15 4208–4217. [PMC free article] [PubMed] [Google Scholar]
- King, R.W., Wardlaw, I.F., and Evans, L.T. (1967). Effect of assimilate utilization on photosynthetic rate in wheat. Planta 77 261–276. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 509–540. [DOI] [PubMed] [Google Scholar]
- Krapp, A., Hofmann, B., Schafer, C., and Stitt, M. (1993). Regulation of the expression of rbcS and other photosynthetic genes by carbohydrates: A mechanism for the ‘sink regulation’ of photosynthesis? Plant J. 3 817–828. [Google Scholar]
- Lalonde, S., Boles, E., Hellmann, H., Barker, L., Patrick, J.W., Frommer, W.B., and Ward, J.M. (1999). The dual function of sugar carriers: Transport and sugar sensing. Plant Cell 11 707–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn, J.E., and Furbank, R.T. (1999). Sucrose biosynthesis in C4 plants. New Phytol. 143 221–237. [Google Scholar]
- Musser, S.M., and Theg, S.M. (2000). Characterization of the early steps of OE17 precursor transport by the thylakoid DpH/Tat machinery. Eur. J. Biochem. 267 2588–2598. [DOI] [PubMed] [Google Scholar]
- Neales, T.F., and Incoll, L.D. (1968). The control of leaf photosynthesis rate by the level of assimilate concentration in the leaf: A review of the hypothesis. Bot. Rev. 34 107–125. [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371 297–300. [DOI] [PubMed] [Google Scholar]
- Nelson, T. (1994). Preparation of DNA and RNA from leaves: Expanded blades and separated bundle sheath and mesophyll cells. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 542–543.
- Park, S.H., Park, S.H., Chin, H.G., Cho, M.J., Martienssen, R.A., and Han, C. (2000). Inhibitor of striate conditionally suppresses cell proliferation in variegated maize. Genes Dev. 14 1005–1016. [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J.W., Willmitzer, L., and Frommer, W.B. (1994). Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 13 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, L.J., and Strommer, J.N. (1985). Insertion of an unstable element in an interveneing sequence of maize Adh1 affects transcription but not processing. Proc. Natl. Acad. Sci. USA 82 2875–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Russin, W.A., Evert, R.F., Vanderveer, P.J., Sharkey, T.D., and Briggs, S.P. (1996). Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell 8 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy (New York: Oxford University Press).
- Sheen, J. (1990). Metabolic repression of transcription in higher plants. Plant Cell 2 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth, L.E., and Meyerowitz, E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens, S., and Rook, F. (1997). Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol. 115 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek, J., Bohm, A., Lambright, D.G., Hamm, H., and Sigler, P.B. (1996). Crystal structure of a GA protein βγ dimer at 2.1Å resolution. Nature 379 369–374. [DOI] [PubMed] [Google Scholar]
- Stitt, M. (1996). Plasmodesmata play an essential role in sucrose export from leaves: A step toward an integration of metabolic biochemistry and cell biology. Plant Cell 8 565–571. [Google Scholar]
- Susek, R.E., Ausubel, F.M., and Chory, J. (1993). Signal-transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799. [DOI] [PubMed] [Google Scholar]
- Taylor, W.C. (1989). Regulatory interactions between nuclear and plastid genomes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40 211–233. [Google Scholar]
- von Schaewen, A., Stitt, M., Schmidt, R., Sonnewald, U., and Willmitzer, L. (1990). Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 9 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, J.H. (1996). Size fractionation using sucrose gradients. In Current Protocols in Molecular Biology. 1. F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, K. Struhl, eds (New York: John Wiley), pp. 5.3.2–5.3.8. [DOI] [PubMed]