Abstract
Many aspects of plant development are regulated by photoreceptor function and the circadian clock. Loss-of-function mutations in the Arabidopsis EARLY FLOWERING 3 (ELF3) and PHYTOCHROME B (PHYB) genes cause early flowering and influence the activity of circadian clock–regulated processes. We demonstrate here that the relative abundance of the ELF3 protein, which is a novel nucleus-localized protein, displays circadian regulation that follows the pattern of circadian accumulation of ELF3 transcript. Furthermore, the ELF3 protein interacts with PHYB in the yeast two-hybrid assay and in vitro. Genetic analyses show that ELF3 requires PHYB function in early morphogenesis but not for the regulation of flowering time. This suggests that ELF3 is a component of a PHYB signaling complex that controls early events in plant development but that ELF3 and PHYB control flowering via independent signal transduction pathways.
INTRODUCTION
Light regulation of shoot development in higher plants appears to involve a complex interaction of several classes of photoreceptors with factors that function in downstream signaling pathways. The complexity of this system lies in part in the number of functionally redundant photoreceptors and in the potential for the downstream signal transduction pathways either to directly influence the light-regulated output response or to function via the input of signals to the circadian clock. These signaling pathways eventually influence a number of major aspects of shoot morphogenesis, including cell elongation, photosynthesis, and the transition to flowering.
In Arabidopsis, there are five red/far-red light–absorbing phytochromes (phytochrome A [PHYA], PHYB, PHYC, PHYD, and PHYE) and at least two blue/UV-A light–absorbing cryptochromes (CRY1 and CRY2) (Chory, 1997; Cashmore et al., 1999). These two types of photoreceptors control many aspects of photomorphogenesis, and their functions overlap in controlling physiological responses (Cashmore et al., 1999). The signal transduction network that is responsive to these photoreceptors is beginning to be elucidated, and several factors involved in phytochrome signal transduction have been defined (Whitelam et al., 1993; Ahmad and Cashmore, 1996; Wagner et al., 1997; Genoud et al., 1998; Ni et al., 1998, 1999; Choi et al., 1999; Fankhauser et al., 1999; Halliday et al., 1999; Hoecker et al., 1999; Hudson et al., 1999; Smith, 1999). PHYA and PHYB differ in their roles in plant development, perhaps because of differences in their structural activity—with PHYA being light labile and PHYB being light stable—and in part because of the differential association of downstream signaling pathways activated by these two photoreceptors (Quail et al., 1995). These distinctions have led to the commonly held view that PHYA is a primary sensor of continuous far-red light, whereas PHYB primarily mediates the responses to continuous red light (Chory, 1997).
In plants, the circadian clock regulates many aspects of development, including hypocotyl elongation and the photoperiodic induction of flowering (Kreps and Kay, 1997). Recently, several circadian clock–associated Arabidopsis genes (e.g., CCA1, LHY, FKF1, and ZTL) have been implicated in the control of plant photomorphogenesis, including the control of hypocotyl elongation and flowering time (Schaffer et al., 1998; Wang and Tobin, 1998; Nelson et al., 2000; Somers et al., 2000). In addition, the photoreceptors also regulate the entrainment of the endogenous circadian oscillator to light/dark cycles and to modulating circadian clock function (Millar et al., 1995; Somers et al., 1998). However, the relationship between these clock-associated genes and the photoreceptors that condition the clock to recognize light/dark cycles is unclear.
Previous work has shown that Arabidopsis elf3 mutants are defective in light perception or light-mediated signal transduction; they also are defective in circadian rhythm responses when grown in constant light but not when grown in constant dark (Hicks et al., 1996; Zagotta et al., 1996). Recently, Reed et al. (2000) reported a possible independent action of EARLY FLOWERING3 (ELF3) and PHYB to control hypocotyl elongation and flowering time. In this study, our goal was to elucidate ELF3 function in signal transduction pathways that relate ELF3 to photoreceptor function and the regulation of the circadian clock. We show that ELF3 is a component of a PHYB signaling complex that controls hypocotyl elongation, whereas ELF3 and phytochrome control flowering via independent signal transduction pathways. This leaves open the possibility suggested by Zagotta et al. (1996) that ELF3 interacts with cryptochrome or with other potential blue light receptors such as FKF1 and ZTL to regulate the photoperiodic induction of flowering in Arabidopsis.
RESULTS
ELF3 Encodes a Novel Nuclear Protein
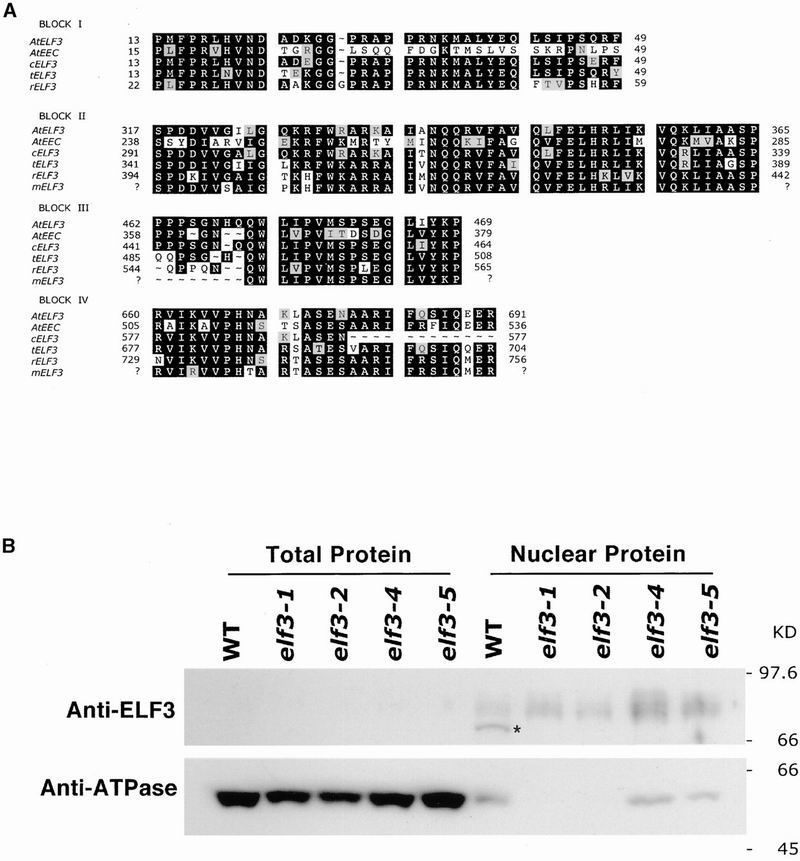
Cloning of ELF3 revealed that this gene encodes a novel 695–amino acid protein without any significant homology to proteins of known function (Hicks et al., 2001). A recent report indicates that the ELF3 gene is transcribed in nematode feeding structures that form in Arabidopsis roots upon infection by sedentary nematodes (Puzio et al., 1999), but the relationship of this transcriptional activity to the role of ELF3 in flowering and circadian clock function is not apparent. The predicted ELF3 protein has glutamine/threonine–rich sequences in the C-terminal region and a nuclear localization signal within the glutamine/threonine–rich region, suggesting that ELF3 may function as a transcription factor (Peterson et al., 1990; Lee et al., 1994). Database searches have identified putative ELF3 homologs in several plant species as well as a very similar duplicate gene, named ESSENCE OF ELF3 CONSENSUS (EEC), in Arabidopsis. Analysis of these sequences, shown in Figure 1A, revealed several highly conserved domains in the related proteins, suggesting that ELF3 function may be conserved between dicots and monocots.
Figure 1.
Sequence Comparison of ELF3 Homologs and Localization of the ELF3 Protein to the Nucleus.
(A) Multiple sequence alignment shows four highly conserved regions within ELF3 and putative ELF3 homologs from Arabidopsis (AtEEC) and other plant species (Cardamine oligosperma [cELF3], tomato [tELF3], rice [rELF3], and maize [mELF3]). Protein designations are given at left. Amino acid residues are numbered at both left and right. Residues shaded in black indicate identity of at least three ELF3/ELF3–related sequences in the alignment; light-shaded residues indicate similarity to consensus. Nucleotide sequences from C. oligosperma were obtained by sequencing polymerase chain reaction products using degenerate oligonucleotides to the Arabidopsis ELF3 gene and genomic DNA or cDNA prepared from C. oligosperma seedlings. Sequences were aligned and analyzed using CLUSTAL W (Thompson et al., 1994) and PrettyBox (Genetics Computer Group, Madison, WI). GenBank accession numbers for ELF3 and putative ELF3 homologs are as follows: AtELF3 (AC004747), AtEEC (AB023045), tELF3 (AW093790, AI894513, AI488927, AI486934, and AI894398), rELF3 (AP000399), and mELF3 (AI637184).
(B) Total protein and protein from isolated nuclei were prepared from either wild-type (WT) or elf3 mutant lines of Arabidopsis seedlings, electrophoresed, and transferred to a nitrocellulose membrane. Both total protein and nuclear protein immunoblots were probed with ELF3 polyclonal antibody and β-ATPase antibody.
The cellular localization of the ELF3 protein was determined using isolated nuclear and total protein fractions from wild-type and elf3 mutant seedlings. As shown in Figure 1B, total protein from wild-type seedlings revealed no binding of the ELF3 antibody, whereas the nuclear protein fraction showed strong binding of antibody to a protein of an approximate molecular weight of 77 kD as predicted for ELF3. Immunoblots of these same protein fractions with the antibody against the chloroplast-localized β-subunit of the ATPase complex demonstrated that the purified nuclear fractions have only minor cytoplasmic protein contamination. All elf3 mutants tested lacked protein detectable by the ELF3 antibody. Thus, as predicted by sequence analysis (Hicks et al., 2001), all elf3 mutant alleles tested are likely to be null alleles.
Light and the Circadian Clock Regulate ELF3 Protein Accumulation in Nuclei
In related studies, it was determined that the level of ELF3 transcript cycles with a circadian periodicity that is not affected by the relative lengths of the light/dark periods, suggesting that ELF3 activity is regulated, at least in part, by a circadian clock (Hicks et al., 2001; Covington et al., 2001). Immunoblot analysis, shown in Figure 2A, demonstrated that the ELF3 protein accumulates in the nucleus in a periodic manner when seedlings are grown in a 12-hr-light/12-hr-dark photoperiod. Interestingly, ELF3 protein accumulation reaches a maximum level just before the onset of the dark phase of the 24-hr-light/dark cycle. This pattern of protein oscillation is nearly identical to that observed for the ELF3 transcript, suggesting that transcriptional regulation of the ELF3 gene is a key mechanism controlling ELF3 function in photomorphogenesis. To determine whether ELF3 protein accumulation is regulated in a circadian manner, we examined the level of ELF3 protein after a 48-hr period before which plants were moved from 12/12 to either constant light (LL) or constant dark (DD). After this 48-hr acclimation period and during the subsequent 48-hr period in either LL or DD conditions, plants were sampled for levels of ELF3 protein. The accumulation of ELF3 in nuclear extracts increased after shifting plants to LL (Figure 2B) and has a circadian component, with the maximum occurring in the subjective night of the subsequent LL condition (Figure 2C), as demonstrated for the ELF3 transcript (Hicks et al., 2001; Covington et al., 2001). The absolute level of the ELF3 protein decreases when the light condition shifts from 12/12 to DD (Figure 2D), making it difficult to determine if the circadian regulation of ELF3 continues in the subsequent DD condition (data not shown). The rapid decrease in the amount of ELF3 protein in DD may indicate that ELF3 has little, if any, role in regulating circadian clock function in the dark. This hypothesis is consistent with the previous observation that elf3 null mutant seedlings displayed nearly normal circadian clock output function when grown in DD (Hicks et al., 1996).
Figure 2.
Accumulation of the ELF3 Protein Is Regulated by Light and the Circadian Clock.
Wild-type seedlings were germinated and grown for 14 days in a 12-hr-light/12-hr-dark photoperiod (12/12). Tissue was then collected every 4 hr (A). Seedlings were also transferred to LL or DD for 48 hr for the collection of LL and DD tissues ([B] and [D], respectively). ELF3 protein was detected with ELF3 antibody, and 0.5% Ponceau S (Pon; Sigma) was used to stain the transferred membrane before probing with ELF3 antibody.
(A) Accumulation of the ELF3 protein oscillates in 12/12.
(B) Accumulation of the ELF3 protein increases in LL and keeps oscillating in the nucleus.
(C) Repeat experiments of ELF3 protein accumulation in LL. Wild-type seedlings were germinated and grown for 14 days in 12/12 and transferred to LL for 48 hr before the collection of tissue every 4 hr during the subsequent 48 hr. The level of ELF3 protein was determined by measuring anti-ELF3 signal relative to the Ponceau staining intensity of the transferred membrane before probing with ELF3 antibody. The results of two independent experiments are shown, with ELF3 protein levels normalized to the highest level detected in each experiment.
(D) Accumulation of the ELF3 protein decreases rapidly when plants are transferred to DD.
ELF3 Interacts with PHYB in Vitro
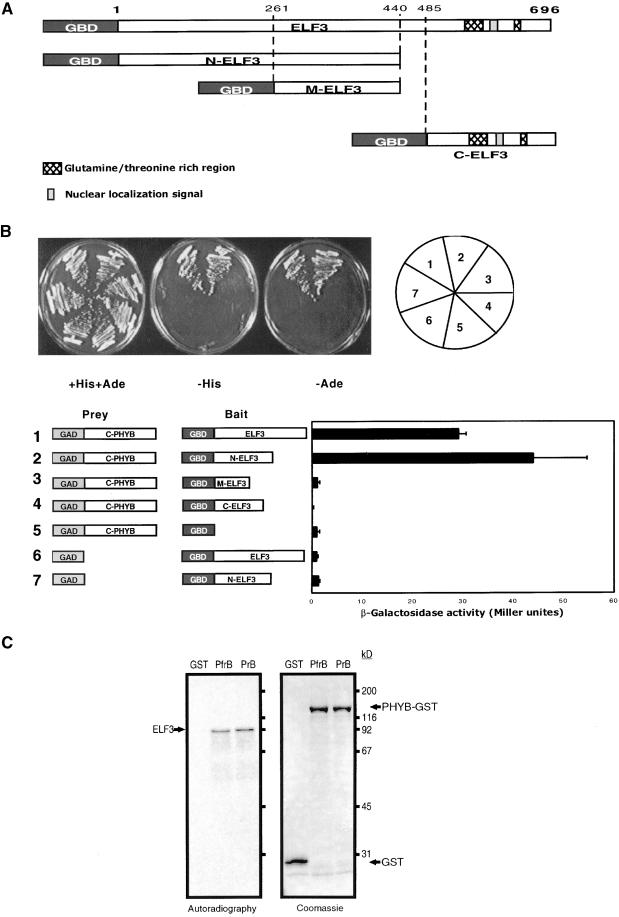
To investigate possible mechanisms by which ELF3 functions in photomorphogenesis, we conducted a yeast two-hybrid screen to identify proteins that interact with ELF3. Using four independent ELF3 DNA fragments as bait clones, shown in Figure 3A, we found that the screen identified several novel proteins capable of interacting specifically with ELF3 and two proteins known to function in light signal transduction: ELF3 and PHYB. A clone encoding the C-terminal domain of ELF3 was recovered and found to interact strongly with the ELF3 bait protein, suggesting that ELF3 may form a dimer. Also, a clone encoding the C-terminal domain of PHYB was isolated, and the interaction between ELF3 and PHYB was further characterized. Yeast two-hybrid experiments showed that ELF3 interacts only with the C-terminal region of PHYB and that it is the N-terminal region of ELF3 that facilitates this interaction (Figure 3B). Similar experiments with cloned regions of the PHYA gene failed to reveal any interaction between ELF3 and PHYA (data not shown), suggesting that ELF3 interacts specifically with PHYB. The interaction between ELF3 and PHYB was confirmed using full-length PHYB protein purified from yeast and in vitro transcribed and translated ELF3. ELF3 binds both the red and far-red light–absorbing forms of PHYB, even though the two spectral forms have different protein conformations (Figure 3C). Our observation that the ELF3 protein accumulates in nuclei in the light (Figure 2C) along with the recently reported light-dependent nuclear import of PHYB (Sakamoto and Nagatani, 1996; Kircher et al., 1999) suggests that a PHYB–ELF3 interaction in vivo could regulate aspects of PHYB-mediated photomorphogenesis.
Figure 3.
ELF3 and PHYB Proteins Interact in the Yeast Two-Hybrid System and in Vitro.
(A) Diagram of the GAL4 DNA binding domain (GBD):ELF3, ELF3 N-terminal domain (N-ELF3), ELF3 intermediate domain (M-ELF3), and ELF3 C-terminal domain (C-ELF3) fusions used as bait to test for protein–protein interactions in the yeast two-hybrid system.
(B) Yeast two-hybrid results with various ELF3 bait constructs and the C-terminal PHYB (C-PHYB) prey plasmid isolated in the initial yeast two-hybrid screen performed with ELF3. Different combinations of plasmids are represented by numbers 1 to 7. Yeast colony growth is shown under different selection media, and the quantitation of ELF3–PHYB interactions is shown by the production of β-galactosidase. GAD, Gal 4 activation domain; GBD, Gal 4 DNA binding domain. Error bars indicate ±se.
(C) In vitro interaction of ELF3 and full-length PHYB in the far-red light–absorbing form (PfrB) and in the red light–absorbing form (PrB). PHYB was expressed as a glutathione S-transferase (GST) fusion protein (PHYB-GST). Autoradiography of the 35S-labeled ELF3 protein and Coomassie blue staining of total protein are shown on the same gel.
ELF3 Is Involved in PHYB-Mediated Red Light Responses but Not in Flowering
To further analyze ELF3 function in phytochrome-mediated early photomorphogenesis, we analyzed elf3-1 single mutants and double mutants of elf3-1 with the phytochrome mutations phyA-211 and phyB-9. As shown in Figure 4A, consistent with previous reports (Neff and Chory, 1998; Reed et al., 2000), the phyA-211 mutation confers an altered far-red response, whereas the phyB-9 mutation renders seedlings defective in red light response. The elf3-1 single mutant seedlings displayed increased hypocotyl elongation in red light but not in far-red light, similar to that observed in phyB-9 mutants, supporting the hypothesis that ELF3 is involved in at least one PHYB signal transduction pathway regulating aspects of seedling development. The mean hypocotyl length of elf3-1 phyB-9 double mutants in red light was similar to that of elf3-1 single mutants and phyB-9 single mutants, indicating that PHYB and ELF3 function in the same pathway with regard to hypocotyl development. However, the mean hypocotyl length of elf3-1 phyB-9 double mutants in white light was much longer than that of either the elf3-1 or phyB-9 single mutants, suggesting that the elf3-1 mutation renders seedlings less sensitive to other wavelengths of light, perhaps via the action of blue light photoreceptors. Under blue and green wavelengths of light, hypocotyl elongation in elf3-1 mutant seedlings is less responsive than it is in wild-type seedlings (Zagotta et al., 1996), indicating that ELF3 is at least partially responsive to these wavelengths of light.
Figure 4.
Genetic Interaction of elf3-1 with phyB and phyA Mutations.
(A) Hypocotyl length of seedlings grown under dark, white light, red light, and far-red light conditions.
(B) Flowering time for wild-type (Columbia ecotype [Col]) and mutant plants grown under long-day (18-hr-light/6-hr-dark; 200 μmol m−2 sec−1 white fluorescent light) or short-day (9-hr-light/15-hr-dark; 150 μmol m−2 sec−1 cool-white fluorescent light) growth conditions. Flowering time was determined by the number of rosette leaves produced by a plant with a 1-cm inflorescence height.
Error bars indicate ±se.
In contrast to the elf3-1 phyB-9 double mutant, elf3-1 phyA-211 double mutant seedlings produced elongated hypocotyls in both red and far-red light (Figure 4A). Thus, PHYA and ELF3 appear to function in separate signal transduction pathways to control aspects of seedling development.
When flowering time of the elf3-1, phyA-211, and phyB-9 single and double mutants was examined in long-day and short-day photoperiods, a somewhat complicated picture was revealed, and this discrepancy most likely is due to the number of developmental pathways that impinge on the vegetative-to-floral transition (Koornneef et al., 1995; Jackson and Thomas 1997; Koornneef and Peeters, 1997; Levy and Dean, 1998). Previous studies (Neff and Chory, 1998; Reed et al., 2000) and our observations (Figure 4B) show that phyB mutants flower slightly earlier than do the wild-type, whereas phyA mutants flower slightly later than do the wild- type, and that both types of mutants remain sensitive to photoperiod. Two notable results in this analysis were the partial suppression of the early flowering phenotype of elf3-1 by the phyA-211 mutation and the complete suppression of the photoperiodic response of the phyB-9 mutant by the introduction of the elf3-1 mutation (Figure 4B). The elf3-1 phyB-9 double mutants also flowered earlier than did elf3-1 in both long-day and short-day photoperiods, and this additive effect is consistent with the observation of Reed et al. (2000), which suggests that ELF3 may function independently of PHYB in the regulation of flowering. PHYA is considered to be an essential component of the photoperiodic pathway in Arabidopsis and other long-day plants (Koornneef et al., 1995; Jackson and Thomas, 1997; Koornneef and Peeters, 1997; Levy and Dean, 1998). Interestingly, the elf3-1 phyA-211 double mutants retained sensitivity to photoperiod and initiated flowering later than did elf3-1 single mutants in short-day photoperiods, suggesting that PHYA also controls flowering independently of ELF3.
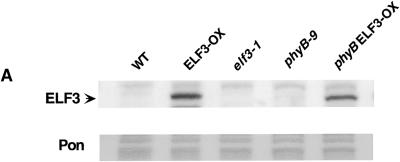
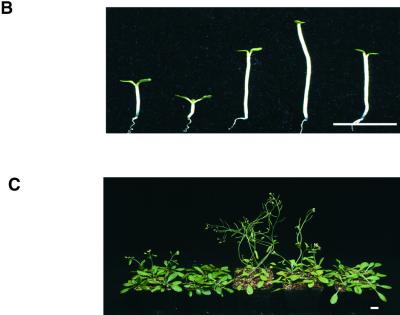
To further assess ELF3 function in PHYB and phytochrome signal transduction, we generated transgenic Arabidopsis lines that constitutively overexpressed the ELF3 protein (ELF3-OX; Figure 5A) and evaluated these overexpression lines, phyB-9, phyB-9 ELF3-OX double mutants, a phytochrome chromophore mutant (hy2-200; Parks and Quail, 1991; Zagotta et al., 1996), and hy2-200 ELF3-OX double mutants for hypocotyl elongation and flowering time (Table 1). When grown in monochromatic red light, the ELF3-OX seedlings displayed extremely short hypocotyls, which is indicative of enhanced sensitivity to red light. The phyB-9 ELF3-OX seedlings showed reduced responsiveness to red light compared with that of the ELF3-OX seedlings, and their responsiveness was similar to that of the phyB-9 single mutants for this phenotype. These findings support the hypothesis that ELF3 regulates hypocotyl elongation in response to red light mainly through the activity of PHYB (Figure 5B and Table 1).
Figure 5.
Genetic Interactions of the ELF3 Overexpression (ELF3-OX) Line with the phyB Mutation.
(A) Protein gel blot of the total protein of young seedlings with ELF3 antibody, showing that both ELF3-OX and phyB ELF3-OX plants overexpress the ELF3 protein. Pon, Ponceau S; WT, wild type.
(B) Hypocotyl length of wild-type, ELF3-OX, elf3-1 mutant, phyB mutant, and phyB ELF3-OX double mutant seedlings (shown left to right) grown under constant red light. Bar = 1 cm.
(C) Flowering time for wild-type, ELF3-OX, elf3-1 mutant, phyB mutant, and phyB ELF3-OX double mutant plants (shown left to right) grown in long-day conditions. Bar = 1 cm.
Table 1.
Hypocotyl and Flowering Characteristics of Wild-Type, Mutant, and ELF3-OX Plantsa
| Flowering Timec
|
Flowering Timed
|
||||
|---|---|---|---|---|---|
| Genotype | Hypocotyl Lengthb | LD | SD | LD | SD |
| Col-0 | 5.6 ± 0.55 (21) | 10.8 ± 1.36 (20) | 64.6 ± 5.10 (10) | 29.0 ± 2.02 (20) | 102.4 ± 6.41 (10) |
| ELF3-OX | 2.9 ± 0.52 (27) | 42.5 ± 4.42 (16) | 57.0 ± 1.37 (47) | 60.6 ± 7.53 (16) | 96.9 ± 0.92 (47) |
| elf3-1 | 12.4 ± 0.94 (27) | 5.15 ± 0.73 (20) | 6.6 ± 2.95 (17) | 20.8 ± 1.26 (20) | 47.1 ± 6.59 (17) |
| phyB-9 | 14.6 ± 0.86 (20) | 7.17 ± 1.34 (18) | NA | 25.8 ± 1.98 (18) | NA |
| phyB-9 ELF3-OX | 10.0 ± 0.70 (19) | 44.1 ± 5.21 (27) | NA | 64.4 ± 9.58 (27) | NA |
| hy2-200 | 14.24 ± 0.85 (15) | 2.77 ± 0.80 (13) | NA | 20.00 ± 4.19 (13) | NA |
| hy2-200 ELF3-OX | 10.08 ± 0.65 (16) | 35.39 ± 11.38 (18) | NA | 61.17 ± 5.35 (18) | NA |
Arabidopsis seedlings overexpressing ELF3 have a reduced sensitivity to red light in hypocotyl elongation, and they flower late in cool-white light long-day conditions. Mean hypocotyl length (in millimeters) and flowering time ±se are indicated. The number of plants measured for each character and genotype is indicated in parentheses.
In red light.
Number of rosette leaves on plants with a 1-cm inflorescence.
Days from sowing to 1-cm inflorescence.
The ELF3-OX plants initiate flowering much later than do the wild-type in long-day photoperiods and at the same time as do the wild-type in short-day photoperiods (Table 1). The late flowering phenotype in long-day photoperiods, caused by the overexpression of ELF3, suppresses the early flowering phenotype of the phyB-9 mutation. In fact, ELF3-OX is epistatic to the phyB-9 mutation with regard to flowering time in long-day photoperiods (Figure 5C). On the basis of these findings and the results of the elf3-1 phyB-9 double mutant (Figure 4B), it appears that ELF3 can influence the timing of the floral transition independently of PHYB function.
The hy2-200 ELF3-OX plants showed a phenotype similar to that of phyB-9 ELF3-OX in flowering time and red light responses, indicating that ELF3 regulates the timing of floral transition independently of phytochrome function. This hypothesis is consistent with previous observations regarding the flowering time of elf3-1 hy2-200 double mutants (Zagotta et al., 1996).
DISCUSSION
ELF3 Encodes a New Clock-Associated Protein
A number of photoreceptors, DNA binding proteins, and proteins possessing PER-ARNT-SIM (PAS)–related domains have been identified as being involved in circadian clock function and regulation in eukaryotes (Dunlap, 1999; Somers et al., 2000). Previously, it was shown that the ELF3 gene product likely functions in a light-input pathway to the circadian oscillator (Hicks et al., 1996). Our results on ELF3 protein oscillation in various photoperiodic conditions further demonstrate that ELF3 is closely associated with circadian clock function. However, sequence analysis of the ELF3 protein reveals no significant homology with proteins of known function. ELF3 has one Arabidopsis homolog (EEC), and the sequence alignment of ELF3 with EEC and with homologs in other plant species demonstrates that ELF3 has four highly conserved domains (Figure 1A), indicating that ELF3 encodes the first member of a newly defined gene family. Protein blot analysis showed that the ELF3 protein localized to nuclei, which suggests that ELF3 may function directly to regulate gene expression.
ELF3 Has a Functional Role in PHYB Signaling in Early Morphogenesis
The defect in red light–dependent inhibition of hypocotyl elongation in elf3 mutant plants and the hypersensitivity of red light–dependent inhibition of hypocotyl elongation in ELF3-overexpressing plants establish that ELF3 functions in PHYB signaling in early morphogenesis. The suppression of the hypersensitive red light–dependent inhibition of hypocotyl elongation of ELF3 overexpression by a phyB mutation further suggests that there is a functional interaction between ELF3 and PHYB signaling.
Zagotta et al. (1996) found that elf3 mutant seedlings were less responsive to all wavelengths of light than were wild-type seedlings in light-mediated inhibition of hypocotyl elongation. We found that elf3 has little effect on far-red light responses, indicating that ELF3 may not be involved in PHYA signaling. Dowson-Day and Millar (1999) also suggested that circadian dysfunction may cause aberrant hypocotyl elongation patterns in elf3 mutants. Our genetic data demonstrate that there are additive effects on hypocotyl elongation in both elf3-1 phyB-9 double mutants and phyB-9 ELF-OX double mutants under different light conditions. Similar additive effects on hypocotyl elongation were observed in elf3-1 35S::PHYB double mutants (Reed et al., 2000), suggesting that both ELF3 and PHYB act redundantly with other proteins or photoreceptors to control hypocotyl elongation.
ELF3 Interacts Directly with PHYB
The in vitro binding of ELF3 to PHYB indicates that the interaction exhibits a significant degree of molecular specificity. Observations using the yeast two-hybrid system demonstrate that ELF3 interacts with itself in the C-terminal region of the protein. Thus, ELF3 may form a homodimer to regulate its own function. The PHYB C-terminal domain, which we initially isolated from the screen of a two-hybrid library by using ELF3 as the bait, interacts specifically with the ELF3 N-terminal region. We did not attempt to use the yeast two-hybrid system to monitor ELF3 interaction with the full-length PHYB because the GAL:full-length phytochrome fusion product was unable to enter the yeast nucleus (Ni et al., 1998). However, given that the C-terminal domain of phytochrome was reported to play a critical role in signal transfer to downstream components (Quail et al., 1995; Wagner et al., 1996), ELF3 binding to the PHYB C-terminal domain may be involved in signaling in vivo.
In the yeast two-hybrid system, ELF3 interacts specifically with PHYB but not with the same C-terminal region of PHYA (data not shown). This result is consistent with the genetic data showing that ELF3 has no functional role in far-red light responses. Together with the genetic evidence for ELF3 involvement with PHYB signaling, this finding supports the conclusion that ELF3 is likely to be a direct modulator of signal information from PHYB through a physical interaction with PHYB.
Functions of ELF3 in the Intracellular Path of PHYB Signaling
Two other proteins, PIF3 and PKS1, were recently reported as interacting with PHYB and regulating PHYB signal transduction through one of several steps: translocation of PfrPHYB to the nucleus; binding of PfrPHYB to the constitutively nuclear protein PIF3; and interaction of the PfrPHYB-PIF3 complex directly or indirectly with target genes (Ni et al., 1998, 1999; Fankhauser et al., 1999; Smith, 1999; Martinez-Garcia, et al., 2000). PKS1 negatively regulates phytochrome signaling and was suggested to be important for retaining phytochrome in the cytoplasm (Fankhauser et al., 1999). Our data indicate that ELF3 is another nuclear factor required for PHYB function. Given the evidence showing the genetic redundancy of PHYB and ELF3 functions in early morphogenesis discussed above, it is possible that ELF3 and PIF3 act redundantly in regulating PHYB downstream signaling.
ELF3 may function in PHYB signaling by interacting directly with DNA regulatory sequences, although there is not an obvious DNA binding domain in the ELF3 protein. Alternately, the PHYB-ELF3 complex may regulate gene expression by a PHYB-mediated modification of the ELF3 protein, such as phosphorylation, that allows ELF3 to interact with other proteins to control gene transcription. In fact, several putative Arabidopsis transcription factors believed to interact with ELF3 were isolated in the yeast two-hybrid assay (X.L. Liu and D.R. Wagner, unpublished data), and these are candidates for additional factors in a PHYB-ELF3 signal transduction pathway.
The PHYB-ELF3 Complex May Modulate Circadian Light Responses
The PHYB–ELF3 interaction appears to control red light inhibition of hypocotyl elongation in Arabidopsis. Previously, elf3 mutants were shown to be defective in circadian rhythm responses under LL, and the ELF3 gene product likely functions in a light-input pathway to the circadian oscillator (Hicks et al., 1996). PHYB has been reported to affect the input of signals to the circadian clock (Millar et al., 1995; Somers et al., 1998), suggesting that the PHYB-ELF3 complex may modulate circadian light responses. Strong support for the functional importance of ELF3 and PHYB interaction gating light response to the circadian clock was provided by studies showing that both loss-of-function mutations and gain-of-function overexpression of ELF3 have strong effects on red light, causing arrythmicity of cab2-luc transcription (Hicks et al., 1996; Covington et al., 2001).
ELF3 Controls Flowering Independently of Phytochrome
Photoreceptors play a critical role in the photoperiodic induction of flowering (Jackson and Thomas, 1997). Interestingly, our genetic analysis showed that PHYB function is not required for the delayed flowering phenotype of ELF3-OX plants in long-day conditions, suggesting that ELF3 can regulate the initiation of flowering independently of PHYB photoreceptor function. This raises the question of whether other photoreceptors are able to regulate flowering via an interaction with ELF3. Results presented here indicate that PHYA is not a likely candidate for such a photoreceptor. The suppression of the hy2 flowering-time phenotype by ELF3 overexpression suggests that other phytochromes are also not likely candidates for such photoreceptors. However, given the conservation of PHYB, PHYC, PHYD, and PHYE sequences, and the fact that mutations in either PHYD or PHYE alter flowering time (Devlin et al., 1998, 1999), these two phytochromes appear to be the most obvious candidates for testing this hypothesis. In addition, elf3 mutations are epistatic to cry1 and cry2 mutations with respect to floral initiation (Zagotta et al., 1996; Mockler et al., 1999), and it is possible that ELF3 controls the photoperiodic induction of flowering directly via a cryptochrome-mediated pathway. Finally, the circadian clock has been implicated in the regulation of the photoperiodic induction of flowering (Kreps and Kay, 1997). Several circadian clock–associated Arabidopsis genes (e.g., CCA1, LHY, FKF1, and ZTL) have been shown to control flowering time (Schaffer et al., 1998; Wang and Tobin, 1998; Nelson et al., 2000; Somers et al., 2000). Thus, further analysis of additional photomorphogenic and circadian clock pathways is necessary to determine whether ELF3 regulates the induction of flowering via interactions with any of these photoreceptors and/or via regulation of the circadian clock.
METHODS
Plant Materials and Growth Conditions
All mutations used in this study were in the Columbia ecotype (Col-0) background of Arabidopsis thaliana. The genotypes of the various mutant combinations were confirmed by the following polymerase chain reactions. The elf3-1 mutation was identified using the primers 5′-TGTTGGTCAGTCTTCTCCGA-3′ and 5′-TCCCTACTGTCATTCAAGGG-3′, followed by digestion with HincII. The phyA211 mutation was identified by amplifying genomic DNA with primers 5′-GTC-ACAAGATCTGATCATGGC-3′, 5′-AACAACCGAAGGGCTGAATC-3′, 5′-TTATCCACAGGGTTACAGGG-3′, and 5′-GCATTCTCCTTGCATCATCC-3′, followed by resolution of 1243- and 1136-bp fragments for the wild type and a 1243-bp fragment for the phyA211 mutant. The dCAPS marker used to identify the phyB-9 mutant and seedling growth and light conditions were as described by Neff and Chory (1998).
Plants overexpressing ELF3 were obtained by in planta transformation of wild-type Col-0 with a pBI121 vector (Clontech, Palo Alto, CA) containing the 35S promoter and ELF3 cDNA and genomic clone. ELF3 cDNA was amplified with primers 5′-CCGGATCCA-TGAAGAGAGGGAAAGATGAGGA-3′ (A1) and 5′-CCCGGGTCGACA-GGCTTAGAGGAGTCATAGCGTT-3′ (A6) and cloned into BamHI and SmaI sites of pBI121, and the ELF3 cDNA in pBI121 was then replaced by a 4.0-kb SacI fragment of ELF3 genomic clone. Homozygous lines with a single T-DNA insertion of the ELF3 genomic clone were selected for analysis and crosses. ELF3 overexpression was confirmed by immunoblots of total protein with ELF3 antibody.
Generation of ELF3 Antibodies
An internal fragment of the ELF3 cDNA encoding 180 amino acids of the ELF3 protein was amplified with primers 5′-GGATCCATGGCA-ACGGAAAATCATTCACAA-3′ and 5′-GTCGACTTAGTAGTTGGATTG-TTGATGATGACCT-3′ and cloned in BamHI and SalI sites of the pET-30a(+) vector (Novagen, Madison, WI). The His-ELF3M180 fusion protein was overexpressed in Escherichia coli and affinity purified using nickel–nitrilotriacetic acid agarose (Qiagen, Valencia, CA). The purified His-ELF3M180 protein was used to immunize rabbits in which to raise polyclonal antibodies. An His-ELF3M180 protein affinity column was used to purify the ELF3 antibodies.
Isolation of Nuclei and Nuclear Proteins
The method of nuclei isolation was as follows: 5 to 10 g of 2-week-old seedlings was homogenized in 30 mL of extraction buffer (50 mM Mes, pH 8.5, 25 mM NaCl, 5 mM MgCl2, 30% glycerol, 5% sucrose, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mg/mL chymostatin [Sigma], 5 mg/mL leupeptin [Sigma], 5 mg/mL antipain [Sigma], 0.5% Triton X-100, 15 mM spermine, and 0.5 mM spermidine) with a Polytron PT 2100 (Brinkmann, Westbury, NY). The crude homogenate was filtered through eight layers of Miracloth, and the nuclei were pelleted by centrifugation at 4°C and 3400g for 20 min. The nuclei pellet was then washed four times with 5 mL of extraction buffer (without Triton X-100) at 4°C and pelleted at 2200g for 10 min, 1700g for 10 min, 1700g for 8 min, and 1700g for 6 min, respectively. The isolated nuclei were resuspended in 500 μL of homogenization buffer (40 mM 2-mercaptoethanol, 10% sucrose, 100 mM Tris HCl, pH 7.2, 5 mM EDTA, 2 mM PMSF, 5 mg/mL chymostatin, 5 mg/mL leupeptin, and 5 mg/mL antipain) for determination of protein concentration.
Immunodetection of ELF3
Total protein was extracted from young rosette leaves of 20-day-old seedlings grown in long-day conditions. One hundred milligrams of tissue was ground with 500 μL of homogenization buffer.
SDS sample buffer (5×) was added in the nuclear or total protein extract, boiled for 3 min, and fractionated by 10% (w/v) SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and detected with ELF3 antibody according to protocols described by Ausubel et al. (1988).
Yeast Two-Hybrid System
A bait coding for the full-length ELF3 was constructed by cloning ELF3 cDNA amplified by primers A1 and A6 into BamHI and SalI sites of pGBDU-C1 (James et al., 1996). The bait was used to screen an Arabidopsis λACT cDNA expression library for the yeast two-hybrid system (Kim et al., 1997). Positive clones were tested by means of bait plasmid loss for specificity.
Protein in Vitro Binding Analysis
In vitro transcription/translation of ELF3 was performed using the TNT Coupled Wheat Germ Extract system (Promega, Madison, WI), and in vitro binding of ELF3 to PHYB–glutathione S-transferase was as described by Fankhauser et al. (1999).
Acknowledgments
We thank K. Kallio, E. Last, and J. Remington for technical assistance; D. Rivers, D. Mitchell, and G. Sprague for providing yeast two-hybrid vectors; the Arabidopsis Biological Resource Center (Columbus, OH) for providing seed, a two-hybrid cDNA library, and clones; and members of the Wagner laboratory for helpful discussions. In addition, we thank Daniel Yarbrough for his work leading to the purification of the ELF3 protein fragment for antibody production and Holly Johnson for sequence analysis of the ELF3 ortholog from Cardamine oligosperma. This research was supported by National Science Foundation Grant MCB-9808208 and United States–Israel Binational Agricultural Research and Development Agency Grant US-2964-97 to D.R.W. and by a National Institutes of Health Grant to J.C., who is an Associate Investigator of the Howard Hughes Medical Institute. M.F.C. was supported by National Institutes of Health training Grant GM07413. C.F. was a fellow of the Swiss National Science Foundation and the Human Frontier Science Program.
References
- Ahmad, M., and Cashmore, A.R. (1996). The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J. 10, 1103–1110. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1988). Current Protocols in Molecular Biology. (New York: John Wiley), pp. 10.8.1–10.8.5.
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.R., and Song, P.S. (1999). Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Chory, J. (1997). Light modulation of vegetative development. Plant Cell 9, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington, M.F., Panda, S., Liu, X.L., Strayer, C.A., Wagner, D.R., and Kay, S.A. (2001). ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., Patel, S.R., and Whitelam, G.C. (1998). Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10, 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., Robson, P.R., Patel, S.R., Goosey, L., Sharrock, R.A., and Whitelam, G.C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyle elongation patterns in Arabidopsis. Plant J. 17, 63–71. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular basis for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Millar, A.J., Nishizawa, N., Kay, S.A., Schafer, E., Nagatani, A., and Chua, N.H. (1998). An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10, 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday, K.J., Hudson, M., Ni, M., Qin, M., and Quail, P.H. (1999). poc1: An Arabidopsis mutant perturbed in phytochrome signaling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 96, 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carre, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274, 790–792. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Albertson, T.M., and Wagner, D.R. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13, 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S., and Thomas, B. (1997). Photoreceptors and signals in the photoperiodic control of development. Plant Cell Environ. 20, 790–795. [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., and Peeters, A.J.M. (1997). Floral transition mutants in Arabidopsis. Plant Cell Environ. 20, 779–784. [Google Scholar]
- Koornneef, M., Hanhart, C., van Leonen-Martinet, P., and Blankestijn de Vries, H. (1995). The effect of daylength on the transition to flowering in phytochrome-deficient, late flowering and double mutants of Arabidopsis thaliana. Physiol. Plant. 95, 260–266. [Google Scholar]
- Kreps, J.A., and Kay, S.A. (1997). Coordination of plant metabolism and development by the circadian clock. Plant Cell 9, 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Aukerman, M.J., Gore, S.L., Lohman, K.N., Michaels, S.D., Weaver, L.M., John, M.C., Feldmann, K.A., and Amasino, R.M. (1994). Isolation of LUMINIDEPENDENS: A gene involved in the control of flowering time in Arabidopsis. Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Straume, M., Chory, J., Chua, N.H., and Kay, S.A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267, 1163–1166. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., and Chory, J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.G., Tanese, N., Pugh, B.F., and Tjian, R. (1990). Functional domains and upstream activation properties of cloned human TATA binding protein. Science 248, 1625–1630. Erratum. Science 249, 844. [DOI] [PubMed] [Google Scholar]
- Puzio, P.S., Lausen, J., Almeida-Engler, J., Cai, D., Gheysen, G., and Grundler, F.M. (1999). Isolation of a gene from Arabidopsis thaliana related to nematode feeding structures. Gene 239, 163–172. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.M., Xu, Y., and Wagner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Bastow, R.M., Solomon, K.S., Dowson-Day, M.J., Elumalai, R.P., and Millar, A.J. (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10, 859–868. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Smith, H. (1999). Phytochromes. Tripping the light fantastic. Nature 400, 710–713. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Fairchild, C.D., Kuhn, R.M., and Quail, P.H. (1996). Chromophore-bearing NH2-terminal domains of phytochromes A and B determine their photosensory specificity and differential light lability. Proc. Natl. Acad. Sci. USA 93, 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Hoecker, U., and Quail, P.H. (1997). RED1 is necessary for phytochrome B–mediated red light–specific signal transduction in Arabidopsis. Plant Cell 9, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., Cowl, J.S., and Harberd, N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta, M.T., Hicks, K.A., Jacobs, C.I., Young, J.C., Hangarter, R.P., and Meeks-Wagner, D.R. (1996). The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 10, 691–702. [DOI] [PubMed] [Google Scholar]