Abstract
We have identified and characterized a novel tobacco gene, called ZGT (from the Chinese phrase zhong guang tiaokong, or clock and light controlled), that is regulated by the circadian clock and light. ZGT transcripts have alternate forms that are differentially expressed in different tissues. ZGT is expressed rhythmically in light/dark cycles and in constant light. Constitutive expression of ZGT sustains the expression of the clock-controlled LHCB1*1 gene in constant darkness, when it would normally dampen, but does not affect LHCB1*1 expression in constant light. ZGT expression is induced rapidly by light, and overexpression of ZGT increases the sensitivity of the circadian oscillator to brief light pulses. The ZGT promoter includes a G-box motif that is found in many light-regulated promoters in plants and is the same as the E box described for rhythmically regulated promoters of animal circadian clock genes. The ZGT promoter also includes “evening element” motifs that are correlated with circadian control of plant genes. We postulate that light- and clock-regulated expression of ZGT acts as a coupling agent between the central circadian oscillator and rhythmic LHCB1*1 expression and that it may function as a component in plant phototransduction pathways.
INTRODUCTION
Circadian (daily) rhythms are a crucial adaptation of organisms to consistent changes in their environment, in particular to the daily changes of illumination, temperature, humidity, and so on. Circadian rhythms are governed by an internal, autonomous oscillator that can be entrained by environmental cues. The daily light/dark (LD) cycle is the dominant time cue that provides temporal information to circadian oscillators. In most animals, circadian rhythms are expressed in both constant darkness (DD) and constant light (LL; at least in dim constant light). In contrast, many observable rhythms in plants dampen rapidly in DD. Therefore, for plant rhythms, light is both a temporal cue that indicates the time of day and a conditional requirement for the expression of the output rhythms (Johnson et al., 1998; Johnson, 2001).
Among the plant rhythms that dampen rapidly in DD are rhythms of gene expression. For example, circadian rhythms of mRNA abundance that are expressed robustly in LL but dampen rapidly in DD are known for many genes, including those encoding the light-harvesting protein–pigment complex (LHC) gene family, catalase, the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, nitrate reductase, and many others (Deng et al., 1990; Pilgrim and McClung, 1993; Pilgrim et al., 1993; Watillon et al., 1993; Zhong et al., 1997; Fejes and Nagy, 1998). The case of catalase mRNA rhythms is particularly interesting. There are at least six different catalase isoenzymes in Arabidopsis, of which at least two, CAT2 and CAT3, are controlled by a circadian oscillator. In LD, both CAT2 and CAT3 mRNA abundances are robustly rhythmic, but upon transfer to DD, both mRNA rhythms dampen quickly. The intriguing observation is that CAT2 mRNA dampens rapidly to a low steady state abundance (as is true for most known circadian clock–regulated plant genes) but CAT3 mRNA dampens to a high steady state abundance (Zhong et al., 1997). Mutations in both the PHYA and CRY1 genes affect the damping characteristics of catalase mRNA rhythms (Zhong et al., 1997).
The plant gene family for which mRNA abundance rhythms were first reported and that remains the best characterized is the one encoding the proteins of the LHC. Daily variations in the mRNA level of LHC transcripts with a peak in the early daytime were first reported by Kloppstech (1985), and subsequent studies demonstrated that this rhythm persists in LL (Otto et al., 1988). It is now known that the expression of many members of the LHC family exhibits circadian rhythmicity that rapidly dampens in DD (Kellmann et al., 1993; Fejes and Nagy, 1998). Light stimulation of phytochrome and possibly other photoreceptors is necessary to maintain the robust expression of these LHC genes (Fejes and Nagy, 1998). Most of the members of the LHC family are regulated by transcriptional control, and the promoter regions of a few members of the family have been analyzed and found to bind a variety of DNA binding proteins (Anderson and Kay, 1995; Carré and Kay, 1995), including the critical clock protein CCA1 (Wang and Tobin, 1998).
Numerous lines of evidence, including the expression of certain genes such as CCR2/AtGRP7 (Heintzen et al., 1997; Strayer et al., 2000), indicate that central circadian oscillators continue to operate in plants in DD (Johnson et al., 1998; Johnson, 2001), yet most of the known outputs of plant circadian oscillators—including LHC gene expression—dampen in DD. We have discovered a new plant gene, which we call ZGT, from tobacco, whose overexpression maintains the expression of the LHCB1*1 promoter in DD and that affects the sensitivity of the circadian oscillator to phase-resetting light signals. The expression of ZGT is induced rapidly by light pulses and is controlled by the circadian clock. We postulate that ZGT acts as a light-dependent coupling factor between a central circadian oscillator and rhythmic LHCB1*1 expression and that it also may act as an amplifier of photoreceptive input to circadian oscillators.
RESULTS
Tobacco ZGT Is a New Gene That Is Expressed Rhythmically
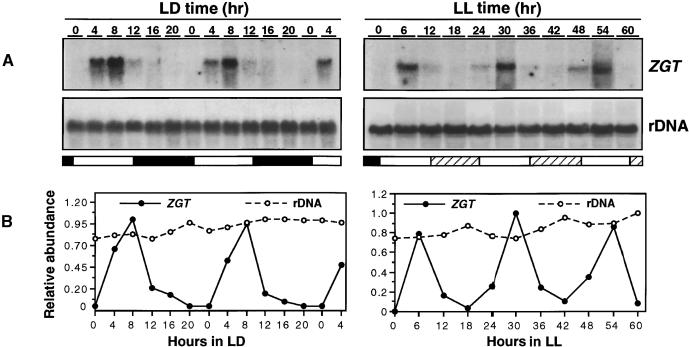
We identified a new gene from a tobacco genomic library that we have named ZGT, from the Chinese phrase zhong guang tiaokong (clock and light controlled). When its temporal expression pattern was analyzed by RNA gel blotting, we found a robust daily rhythm of ZGT mRNA abundance in plants grown in LD cycles and in LL. Figure 1 shows that in LD 12:12 (lights on from LD time 0 [LDT 0] to LDT 12, and lights off from LDT 12 to LDT 24/0), the peak levels of the ZGT transcript occurred at approximately LDT 8 (8 hr after dawn) and the mRNA abundance was at its lowest level at approximately LDT 0 (dawn) in a time-course experiment in which RNA was collected at 4-hr intervals. To determine if the expression of this transcript is circadian, we performed time-course collection of RNA from tobacco plants exposed to LL (after entrainment by an LD cycle). The RNA gel blot hybridized with a ZGT probe revealed a robust circadian rhythm of ZGT mRNA abundance for at least 2.5 days in LL (Figure 1). In the LL time course, the peak ZGT mRNA abundance was in the middle of subjective day (RNA was sampled every 6 hr in the LL experiment). The same blots probed for rRNA (as a loading control) showed a constant level. In a similar time-course experiment performed in DD, the level of ZGT mRNA expression was very low and appeared to be constitutive (data not shown). These data indicate that ZGT expression is controlled by a circadian biological clock and is light dependent.
Figure 1.
Daily and Circadian Rhythms of ZGT Transcript Abundance.
The daily rhythms are shown at left and the circadian rhythms at right.
(A) Tobacco plants were first grown in a greenhouse under natural LD conditions for 6 weeks and then entrained to LD 12:12 for three cycles before harvesting RNA in LD or released to LL at 50 μE m−2 sec−1 for harvesting RNA in LL. Total RNA was collected from leaves at the indicated LD or LL time points. RNA gel blots (20 μg/lane) were first probed with a ZGT fragment and then stripped and rehybridized with an rDNA probe to normalize the loading. The open and closed bars beneath the blots represent the light and dark periods, respectively, and the hatched bars indicate subjective nights in LL.
(B) Quantification of the data shown in (A). The maximum transcript level was taken as 1.0.
Because ZGT is expressed robustly with a rhythmic pattern, we were interested in further characterizing this gene and its possible role in plant circadian systems. On the basis of the original genomic clone, we first determined the nucleotide sequence of the ZGT DNA fragment and its 5′ and 3′ flanking regions. The full-length cDNA sequence of ZGT was determined by rapid amplification of cDNA ends (RACE). Overlapping 5′ and 3′ RACE cDNA sequences indicated that ZGT has a single open reading frame encoding a putative ZGT protein with 291 amino acids, whose structural features are depicted in Figure 2A. Database searches demonstrated that the deduced ZGT protein does not share significant sequence similarity to any known proteins, except a putative homolog in Arabidopsis that was released recently to the sequence database (GenBank accession number CAB87751; see Figure 2B). We will refer to this Arabidopsis sequence as pZGT (for putative ZGT); it shows 61% identity and 74% similarity in amino acid sequence to that of tobacco ZGT based on Gapped BLAST and PSI-BLAST analyses (Altschul et al., 1997). Near the C terminus of tobacco ZGT, we found an aspartate- and glutamate-rich (DE-rich or DE-repeat) region. These DE-rich sequences also are found in a number of other proteins, such as nucleolar transcription factors (GenBank accession numbers P25976, P25977, P25979, P25980, and P17480), nucleolins (GenBank accession numbers P13383, P15771, P19338, JH0148, and BAA86200), GTPase activating proteins (GenBank accession numbers P46061 and NP_010454), and some kinases (GenBank accession numbers T28145, CAA65449, O15066, and Q61771).
Figure 2.
Structure of the Tobacco ZGT Gene.
(A) Scheme of the ZGT gene is shown at the top. The coding region of the ZGT gene is denoted by an open box, and its translational initiation and termination codons are marked as ATG (starting at +74) and TAA (starting at +946), respectively. The transcriptional start site is numbered as +1 and indicated by an arrow. The hatched box represents the aspartate- and glutamate-rich region (DE Rich) near the C terminus. The location of the putative G/E box in the 5′ flanking region is depicted by a closed square, and the two closed circles denote the positions of two putative evening elements (EE1 and EE2). The two HindIII restriction sites are labeled. The lengths of the two transcripts, ZGT-l and ZGT-s, are shown immediately below. AGTTT indicates the conserved nucleotide motif in the 3′ end of these two cDNAs, which occurs just before the poly(A) sequence.
(B) Sequence alignment of the entire coding region of the tobacco ZGT gene (GenBank accession number AF368237) and a putative homolog found in the Arabidopsis database (GenBank accession number CAB87751) by using the FASTA algorithm (Pearson and Lipman, 1998) in the GCG package (Genetics Computer Group). Identical amino acids are indicated by vertical lines, and similar amino acids are indicated by dots. Spacers and gaps are indicated by dashes.
(C) Alignment of the flanking sequences of putative G boxes (or E boxes) in the promoters of tobacco ZGT (from −151 to −138; this study, GenBank accession number AF368237), a putative Arabidopsis ZGT homolog (pZGT; GenBank accession number CAB87751), Arabidopsis CCA1 (U28422), Arabidopsis LHY (AJ006404), Drosophila vri (Y11837), Drosophila per (D00009), Drosophila tim (U37018), and the mouse vasopressin gene (M88354). The central CACGTG cores are outlined in black. To clarify the alignments, the sequences labeled with asterisks are inverted relative to the orientation in their promoters. Identical nucleotides in at least four of the eight flanking sequences are shaded in gray.
(D) Comparison of putative evening elements (EE) in the tobacco ZGT and Arabidopsis pZGT promoter regions. The consensus sequence for the nine-nucleotide AAAATATCT EE that was found to be a conserved motif in the promoters of various cycling genes in Arabidopsis (Harmer et al., 2000) is shown at the bottom. Single asterisks denote the sequences that are on the antisense strand relative to their promoters. Nucleotide locations relative to the transcription start site are shown to the right (the coordinate with double asterisks indicates the distance from the translation start site, because the transcriptional start site for Arabidopsis pZGT is not yet determined). The EE cores are outlined in black. Identical nucleotides in the flanking regions of at least three of the seven sequences are shaded in gray.
The ZGT Promoter Contains Conserved Light- and Clock-Regulated Motifs
Between positions −147 and −142 of the ZGT 5′ flanking region, we found a palindromic hexanucleotide DNA sequence, CACGTG, that is one version of the light-regulated motif known as a G-box core (Chattopadhyay et al., 1998; Martínez-García et al., 2000). Within the 680-bp 5′ flanking region of the Arabidopsis pZGT gene, we also found four putative G-box motifs located at −84 to −79, −263 to −258, −334 to −321, and −675 to −670 relative to the first codon (numbered as +1) of its putative translational start site. The presence of this core target element for light signaling in the ZGT promoter is consistent with its playing a role in the light regulation of ZGT, which we demonstrate below (see Figure 5). Intriguingly, the same consensus CACGTG sequence functions as a circadian enhancer motif in Drosophila and mammalian circadian clock gene promoters—in which case it has been called an E box. In Figure 2C, we compare the flanking sequences of the G box or E box (G/E box) in the ZGT promoter with those from various clock- and/or light-regulated genes. In the case of the Arabidopsis pZGT gene, we chose the motif closest to its putative “evening element” motif (see below and Figure 2D) for comparison. The tobacco ZGT promoter has flanking sequences downstream of the G/E box that are highly conserved relative to those downstream of the G/E box in the promoter of the Arabidopsis circadian clock–associated gene (CCA1) encoding an myb-related transcription factor (Wang et al., 1997; Wang and Tobin, 1998).
Figure 5.
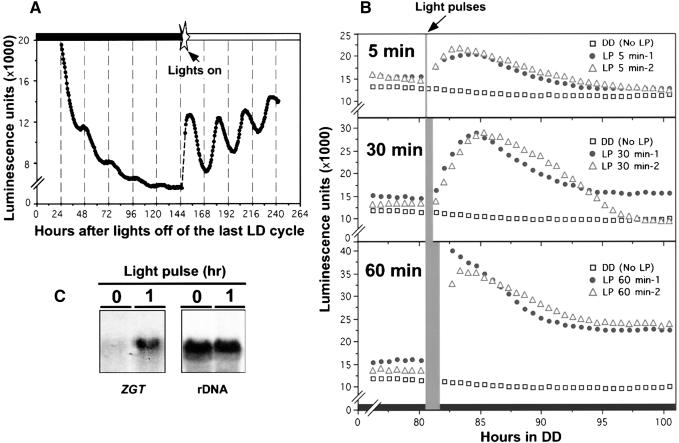
ZGT Gene Expression Is Light Inducible.
(A) The rhythmicity of ZGT promoter activity is light dependent. Reporter expression of the fusion gene PZGT::luc in transgenic seedlings that had been grown under LD 16:8 for 1 week showed significant damping of the circadian rhythm after the seedlings were placed in DD. The expression of PZGT::luc luminescence rapidly recovered stable rhythmicity upon transfer to white LL after 6 days in DD (at time 144 hr).
(B) Acute induction of PZGT::luc reporter expression by brief light pulses. One-week-old transgenic PZGT::luc seedlings grown under LD 16:8 cycles were transferred to DD, and the luminescence was measured by an automated photomultiplier apparatus (one seedling per channel). After the seedlings had been maintained in DD for 80 hr, they were irradiated with white light pulses (50 μE m−2 sec−1; LP) for 5, 30, or 60 min (triplicates for each). Controls were seedlings that did not receive a light pulse (No LP). After the light pulses, seedlings were returned to DD, and luminescence was measured. The basal levels were normalized to correspond with those of the control. Data for two representative seedlings are shown for each treatment.
(C) Light-induced accumulation of the ZGT transcripts. Wild-type seedlings were grown under LD 12:12 cycles for 3 weeks and then transferred to DD. After 56 hr in DD, the seedlings (five seedlings in each group) were irradiated or not irradiated with a 1-hr white light pulse (50 μE m−2 sec−1). One hour after the light pulse had ended, seedlings were harvested for isolation of total RNA. Analysis of the RNA gel blot (20 μg/lane) was as described in Figure 1.
In addition, putative evening elements AAAATATCT (EE1, −178 to −186) and AATATCT (EE2, −231 to −237) were found in an antisense orientation in the ZGT promoter (Figures 2A and 2D). This evening element, a conserved nine- or seven-nucleotide (AA)AATATCT motif in promoters for circadian control of plant gene expression, was recently identified by computational analysis of a number of cycling genes in Arabidopsis that have peak expression near dusk (Harmer et al., 2000). There are also four other putative evening elements in the ZGT promoter in a sense orientation (within −870 bp of the transcriptional start site) that differ by only one nucleotide from the core seven-nucleotide consensus sequence (Figure 2D). Arabidopsis pZGT has only the one putative evening element in its promoter, as shown in Figure 2D.
ZGT mRNA Has Alternate Termination Sites and Tissue-Specific Expression Patterns
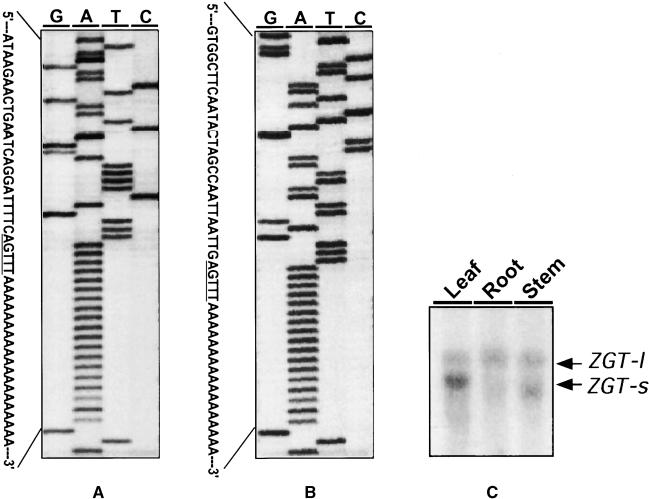
Our 3′ RACE analysis revealed two different termination sites for ZGT transcripts, each having a conserved AGTTT motif just before the poly(A) tail (Figures 2A, 3A, and 3B). These alternate terminations give rise to two isoforms of ZGT transcripts, ZGT-s and ZGT-l, that were detected in a long-running RNA gel blot assay (Figure 3C). ZGT-l is 147 bp longer than ZGT-s in the 3′ untranslated region. These alternate forms of the ZGT mRNA exhibited a tissue-specific pattern. At LDT 8, ZGT-l transcripts were expressed approximately equally in all tissues examined, whereas ZGT-s was transcribed differentially in different tissues (Figure 3C). In leaf tissues, levels of the ZGT-s transcript were much higher than those of the ZGT-l transcript. Both transcripts showed similar expression levels in stem tissues, whereas levels of the ZGT-s transcript were very low in root tissues. These results suggest that the 3′ untranslated region of the ZGT gene may play a regulatory role in development.
Figure 3.
Mapping and Expression of Two Transcripts of the ZGT Gene.
(A) and (B) The 3′ ends of cDNA sequences of the two ZGT transcript isoforms, ZGT-s and ZGT-l, respectively, determined by 3′ RACE from total RNA in leaf tissues collected at LDT 8. Lanes G, A, T, and C indicate the reactions for sequencing nucleotides. The conserved 3′ end motif of these two transcripts is underlined.
(C) Tissue-specific expression patterns of ZGT-s and ZGT-l were revealed by RNA gel blot analysis, using a ZGT hybridization probe. Total RNA was isolated at LDT 8 from root, leaf, or stem tissues. Equal amounts of total RNA (20 μg/lane) were loaded in each lane. The two ZGT transcripts are indicated by arrows.
ZGT Promoter Activity Is Rhythmic
Because the abundance of ZGT mRNA clearly cycles under both light/dark and free-running conditions (Figure 1), we were interested to know if its promoter was regulated by the circadian clock. To continuously monitor the circadian activity of the ZGT promoter, we created transgenic tobacco plants expressing the fusion gene PZGT::luc, in which the expression of a cDNA encoding the firefly luciferase structural gene (luc) is driven by a ZGT 5′ flanking region (from −950 to +73) as PZGT. ZGT promoter activity patterns were compared with that of LHCB promoter activity in transgenic tobacco expressing a circadian luminescence reporter, in which the expression of luc is driven by the promoter of the Arabidopsis LHCB1*1 gene (PLHCB::luc). LHCB1*1 encodes light-harvesting chlorophyll a/b binding proteins of photosystem II, and its control by a circadian clock has been studied extensively (Anderson and Kay, 1995; Fejes and Nagy, 1998).
Under free-running LL conditions, reporter expression of PZGT::luc confirmed a persisting circadian oscillation of luminescence in the transgenic seedlings (Figure 4). Although the luminescence rhythms of the PZGT::luc reporter exhibited a similar period (∼24.5 hr) to those of the PLHCB::luc reporter, cyclic expression of the ZGT promoter activity rhythm lagged ∼6 to 7 hr behind that of the LHCB promoter (Table 1). Also, the luminescence rhythms of the PZGT::luc reporter had an amplitude that was ∼2.8-fold lower than that of the PLHCB::luc reporter (Figure 4C and Table 1). The fact that the luminescence rhythms of the PZGT and the PLHCB reporters have the same period is consistent with but does not prove the hypothesis that the same central oscillator controls the cyclic expression of both genes. In comparing the phase of the ZGT mRNA rhythm shown in Figure 1 with the promoter activity rhythm shown in Figure 4C, note that the mRNA rhythm was measured after release from LD 12:12, whereas the promoter activity rhythm was measured after release from LD 16:8; this difference in pretreatment may account for the slightly different phase relationships of the ZGT promoter activity and the mRNA abundance rhythms.
Figure 4.
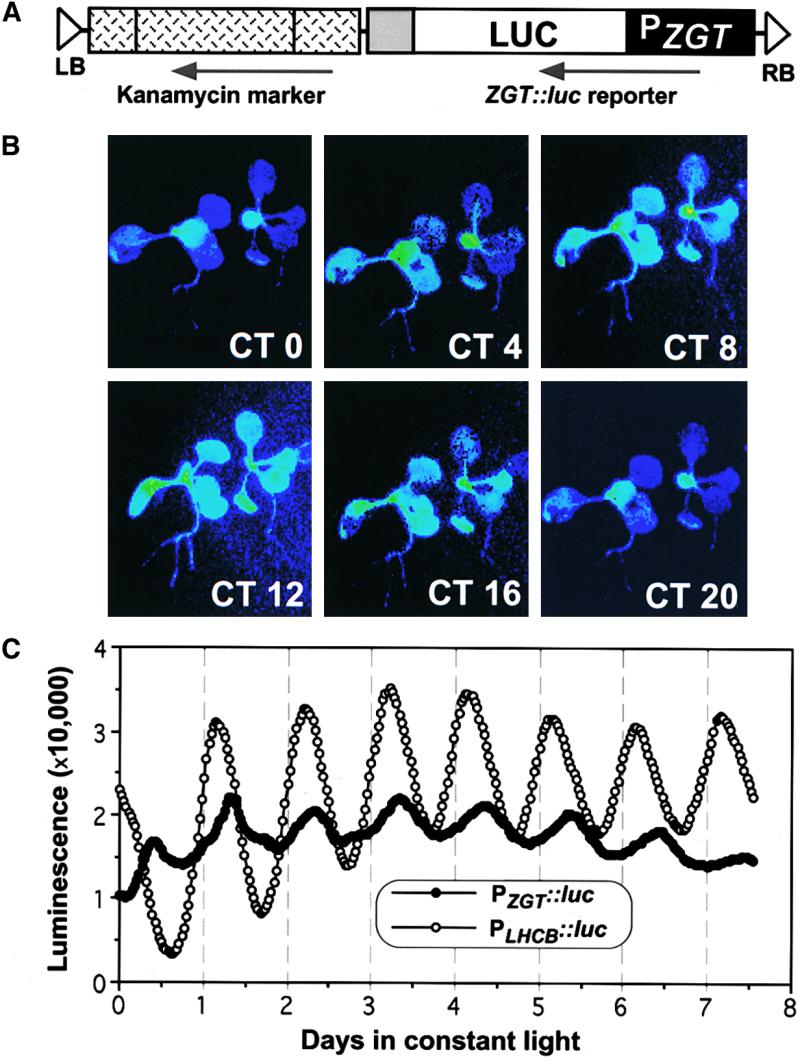
Circadian Rhythmicity of ZGT Promoter Activity.
(A) ZGT promoter activity was monitored with the luminescence reporter construct PZGT::luc. The diagram shows the T-region in the construct pZGT/Luc for PZGT::luc expression. The black box represents the 5′ flanking sequence of the ZGT gene from −950 to +73 (PZGT). The open box denotes the coding region for firefly luciferase (LUC). The gray box indicates the nopaline synthase polyadenylation region. LB and RB represent T-DNA left and right repeats, respectively. The stippled box to the left of the PZGT::luc cassette is an expression cassette of the neomycin phosphotransferase gene. Arrows indicate the direction of transcription.
(B) Images of transgenic tobacco seedlings (3 weeks old) expressing the fusion gene PZGT::luc captured every 4 hr in LL (labeled in circadian time [CT]). Seedlings had been entrained to LD 16:8 and then transferred to LL, and the images shown are for 24 to 44 hr in LL. Intensity is encoded in pseudocolor: blue is dim luminescence, and green/yellow is bright luminescence.
(C) Rhythmic luminescence of transgenic tobacco seedlings harboring PZGT::luc or PLHCB::luc reporter constructs in LL (seedlings had been entrained to LD 16:8 before release into white LL). At least six channels (one seedling per channel) were monitored continuously for luminescence for each transformant, and representative data are shown. Time 0 is the beginning of LL (subjective dawn; i.e., the end of the last dark interval in LD 16:8).
Table 1.
Comparison of the Luminescence Period, Amplitude, and Phase of Reporter Expression between PZGT::luc and PLHCB::luc Transgenic Seedlings in LL
| Period (hr)a
|
Phase Difference (hr)a
|
||||
|---|---|---|---|---|---|
| Promoter | Peak | Trough | Amplitudeb | Peak | Trough |
| ZGT | 24.9 ± 0.4 | 24.4 ± 0.5 | 1.4 ± 0.1 | ||
| 6.2 ± 0.6 | 7.0 ± 1.1 | ||||
| LHCB | 24.5 ± 0.3 | 24.6 ± 0.3 | 3.9 ± 0.3 | ||
Periods and phases of the luminescence rhythms were determined based on either peak or trough values as indicated. Phase differences were calculated as the difference between the PZGT::luc and the PLHCB::luc rhythms in hours. Values shown are averages ±sd (n 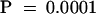 ).
).
Amplitude was evaluated as the ratio of the second peak value to the second trough value.
Expression of the ZGT Gene Is Light Regulated and Light Dependent
The rhythmic expression of many plant genes is dependent on light, and our RNA gel blot analyses showed that ZGT mRNA expression was light dependent, because ZGT expression in DD was very low compared with that in LD or LL. Therefore, we set out to determine if the activity of the ZGT promoter is affected by light. The rhythmicity of ZGT promoter activity as reported by a PZGT::luc construct in transgenic seedlings damped in constant darkness (Figure 5A). After LL was restored, however, the rhythm of PZGT::luc luminescence recovered immediately, indicating that a crucial light signal is involved in the expression of the ZGT gene. Because the luminescence of the PZGT::luc reporter responded so rapidly to the DD-to-LL transition (Figure 5A), we examined the response and sensitivity of ZGT gene expression to light pulses. As illustrated in Figure 5B, a single brief light pulse was able to rapidly activate the ZGT promoter activity of transgenic seedlings that had been in darkness for an extended period (80 hr). Even a 5-min light pulse elicited a significant increase in ZGT promoter activity that continued for at least 8 hr, and a 1-hr light pulse provoked a greater than twofold increase of luminescence that required many hours in darkness to dissipate (Figure 5B). Light-induced accumulation of ZGT transcripts was confirmed by RNA gel blot analysis (Figure 5C). These results show that the expression of the ZGT gene is light inducible and further suggest that the regulation of its photoresponsiveness occurs at least partially via changes in transcriptional rates. These results are consistent with the hypothesis that the G/E box in the ZGT promoter is recognized by light-regulated transcription factors such as PIF3 (Martínez-García et al., 2000) and HY5 (Chattopadhyay et al., 1998).
Expression of ZGT Sustains Circadian Oscillations of LHCB Expression in DD but Has Little Effect in LL
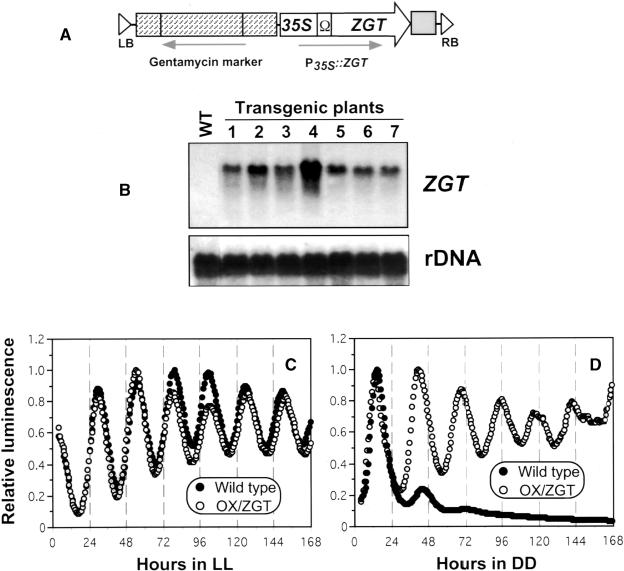
To determine if the ZGT gene plays a role in the plant circadian system, the ZGT structural gene was expressed under the control of the cauliflower mosaic virus 35S promoter fused to a tobacco mosaic virus translational enhancer (Ω) sequence (Figures 6A and 6B). Expression of the 35S promoter is constitutive in plants during the circadian cycle (Millar et al., 1992). The ZGT gene was expressed in transgenic tobacco harboring the PLHCB::luc luminescence reporter (Anderson and Kay, 1995) to determine whether the expression of ZGT would affect LHCB expression. We compared the rhythmic luminescence expression of the PLHCB::luc reporter in seedlings between single transformants with PLHCB::luc (wild type) and double transformants with PLHCB::luc and P35S::ZGT (OX/ZGT). PLHCB::luc reporter luminescence was monitored in either LL or DD after entrainment by LD 16:8 cycles. Under DD conditions, the circadian luminescence rhythms damped rapidly in wild type, whereas circadian oscillation was sustained stably in the ZGT gene–expressing seedlings (Figure 6D, OX/ZGT). However, constitutive expression of the ZGT gene did not significantly affect the circadian rhythms of LHCB promoter activity in LL conditions (Figure 6C).
Figure 6.
Effect of Constitutive Expression of the ZGT Gene on Circadian Rhythms of the Clock-Controlled LHCB Gene in Transgenic Seedlings.
(A) Scheme of the ZGT expression cassette in the construct pOX/ZGT. Between the left and right border sequences (LB and RB) of the T-DNA, the ZGT coding region and its 3′ flanking sequence (from +69 to +1236 relative to the transcription start site) were fused to the cauliflower mosaic virus 35S promoter (35S) with a tobacco mosaic virus 5′ untranslated leader sequence (Ω) followed by the poly(A) addition sequence from the nopaline synthase gene (gray box). A plant selection marker (a gentamycin resistance cassette) was included. Arrows indicate the direction of transcription.
(B) Overproduction of ZGT transcripts in the OX/ZGT transgenic plants. Total RNA (20 μg/lane) was prepared at CT 0 from the leaves of 6-week-old wild-type plants (WT) or six different randomly chosen OX/ZGT transgenic plants (lanes 1 to 7). The blot was probed sequentially with the ZGT probe, stripped, and reprobed with an rDNA probe.
(C) and (D) Overexpression of the ZGT gene sustains the rhythmic expression of the clock- and light-regulated LHCB1*1 promoter in DD but has no significant effect on its rhythms in LL. Rhythmic PLHCB::luc luminescence in wild-type or OX/ZGT transgenic seedlings was measured continuously in white LL (C) or DD (D) after release from LD 16:8 cycles. Representative data are shown as normalized luminescence from at least three independent experiments; in each experiment, 10 channels (three seedlings per channel) were used for both wild-type and OX/ZGT lines.
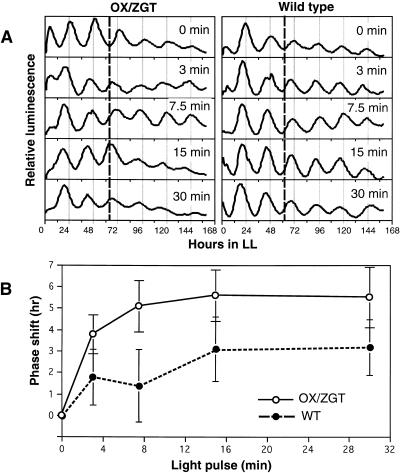
The fact that constitutive expression of ZGT sustains the rhythmic expression of the clock-controlled and light-responsive LHCB gene in DD encouraged us to determine whether the ZGT protein may be involved in the circadian system via either an input pathway and/or an output pathway. To determine if the expression of ZGT can affect the sensitivity of the circadian oscillator to light signals via the input pathway, we compared the magnitude of light-induced phase resetting in the wild-type versus the OX/ZGT background. First, we measured a phase response curve to 30-min white light pulses and found that both wild type and OX/ZGT exhibited very similar phase response curves (data not shown). However, when we chose a specific phase (circadian time 19[CT 19]) that gave reproducible phase shifts for both lines, we found that they showed different responses to light pulses whose duration ranged from 3 to 30 min. As shown in Figure 7, the magnitude of the light-induced phase shift increased with pulse duration. The phase shifts of OX/ZGT were statistically greater than those of wild type at the 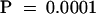 level, as analyzed by analysis of variance (randomized block design). Therefore, OX/ZGT seedlings are significantly more sensitive to phase resetting by brief light pulses than are wild-type seedlings.
level, as analyzed by analysis of variance (randomized block design). Therefore, OX/ZGT seedlings are significantly more sensitive to phase resetting by brief light pulses than are wild-type seedlings.
Figure 7.
Phase Resetting by Light Pulses in Wild-Type and OX/ZGT Transgenic Seedlings.
(A) Phase shifting of the PLHCB::luc luminescence rhythm by light pulses in the wild-type and OX/ZGT lines. As in Figure 6, the wild type is the PLHCB::luc strain and OX/ZGT is the PLHCB::luc strain that has been transformed with the P35S::ZGT construct. After LD, seedlings that had been in DD for 27 hr (i.e., at CT 19) were exposed to white light pulses (50 μE m−2 sec−1) of 3- to 30-min duration. After the light pulse ended, seedlings were maintained in darkness until CT 24 (i.e., 5.5 to 6 hr longer) and then transferred to constant red light, and their luminescence rhythms were measured. Vertical lines show the phase of the control trough on day 3 for comparison.
(B) Analysis of phase-shifting data from two independent experiments as described in (A). Periods and phases of the luminescence rhythms were determined based on the trough values extrapolated over four to six cycles, and there were triplicate samples for each treatment in both experiments. Phase shifts as a function of light pulse duration for the two strains are shown. Results from the two separate experiments were not statistically different at the 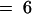 level (as assessed by analysis of variance), so the two experiments were treated as blocks in a randomized complete block design (
level (as assessed by analysis of variance), so the two experiments were treated as blocks in a randomized complete block design (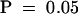 for each treatment). By this analysis, the phase shifts of OX/ZGT were significantly greater than those of the wild type (WT) at the
for each treatment). By this analysis, the phase shifts of OX/ZGT were significantly greater than those of the wild type (WT) at the 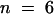 level (as assessed by an analysis of variance). Error bars indicate ±sd.
level (as assessed by an analysis of variance). Error bars indicate ±sd.
DISCUSSION
Plant Circadian Oscillators Can Run in DD
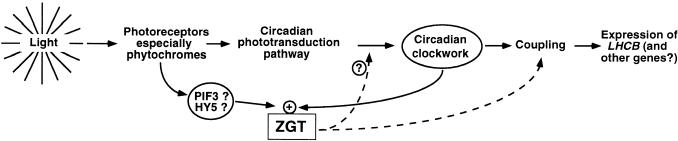
Many plant circadian rhythms exhibit sustained oscillations in LL but dampen in DD. Our data clearly show that under the appropriate conditions, these rhythms can be sustained, as by overexpression of ZGT. This result strongly suggests that the dramatic damping of rhythms in DD is not due to a concomitant damping of the oscillator itself but to reduction of the coupling between oscillator and rhythm. There is other evidence for the continued oscillation of central plant clockworks in DD. For example, another gene (CCR2) is expressed rhythmically for many cycles in DD (Strayer et al., 2000), and several other lines of evidence support the same conclusion (summarized in Johnson et al., 1998 and Johnson, 2001). Overexpression of ZGT can restore this coupling between clockwork and rhythm and allow sustained rhythms of PLHCB::luc luminescence. It has been shown that overexpression of phytochrome and/or stimulation of phytochrome by brief red light pulses can help to sustain LHCB expression rhythms (Kay and Millar, 1992; Johnson, 2001). Mutations in genes involved in phototransduction pathways, especially det1 and cop1, also can allow sustained rhythmicity of PLHCB::luc luminescence (Millar et al., 1995). In addition, mutations in PHYA and CRY1 sustain CAT mRNA rhythms (Zhong et al., 1997). However, one difference between the results with OX/ZGT and the det1/cop1 mutants is that ZGT overexpression does not alter the circadian period as do the det1 and cop1 mutations. In addition to the presence of a G/E box in PZGT that might be regulated by PIF3 and/or HY5, these correlations suggest that ZGT could interact with phototransduction pathways in plants. Aside from sustaining rhythms in DD, the other phenotype of ZGT overexpression that we have observed—increased sensitivity to light-induced phase shifting (Figure 7)— suggests that ZGT is involved directly in phototransduction (Figure 8).
Figure 8.
Model for the Action(s) of ZGT in the Circadian System That Drives LHCB Gene Expression.
ZGT expression is stimulated by light, possibly via PIF3/HY5/PHY modulation of ZGT's G/E box, and it is gated by the circadian oscillator (solid lines). In turn, ZGT couples the clock to outputs, such as the circadian gating of LHCB expression (dashed line). ZGT also may play a role in amplifying the sensitivity of the photoreceptive input to the clock (dashed line with question mark). The central circadian oscillator is labeled “Circadian clockwork.”
Why might plant output rhythms dampen in DD even though their central oscillator is running? Plant metabolism is absolutely dependent on light energy. Consequently, changes in light intensity profoundly alter the metabolic status of plant cells. However, for a timekeeping mechanism to be useful, it must be impervious to these changes in metabolism. Therefore, plants appear to have evolved a system in which the linkage between central oscillators and outputs is adjustable depending on metabolic status. The central clock oscillates under all conditions, as befits its role as an accurate timekeeper, but the degree of its coupling to outputs varies as a function of metabolism. For plants, light (and likely temperature as well) is probably an excellent predictor of metabolic capacity, so having the coupling between clock and outputs be dependent on light energy is logical. Thus, the amplitude of output rhythms is a function of light energy even while the underlying oscillator is relatively unaffected by light intensity (in wild-type plants). This hypothesis would be particularly applicable to plant output genes whose functions relate to the use of light energy, such as the LHC gene family. DD is an extreme case of low metabolic capacity that is rarely or never seen in nature (at least not after plants have passed the seedling stage); when light energy is absent, output gene expression becomes uncoupled from clock control. These speculations further imply that photoreceptive pathways might have evolved in such close conjunction with plant circadian oscillators that they became intrinsically linked in terms of expression of the output rhythms (Johnson, 1994; Johnson, 2001).
Light- and Clock-Regulated Motifs in the ZGT Promoter
As shown in Figure 5, we found that ZGT is robustly activated by light. This light regulation is likely to be mediated at least in part by the G/E-box motifs in the ZGT promoter. Martínez-García et al. (2000) demonstrated that PIF3, a nucleus-localized basic helix-loop-helix transcription factor that interacts with phytochrome, binds specifically to G-box–containing promoter sequences from various light-regulated plant genes, such as Arabidopsis CCA1, LHY, SPA1, and RBCS-1A. Phytochrome B binds reversibly to G-box–bound PIF3 upon light-triggered conversion of phytochrome B to its biologically active conformer. A similar story has been reported for the bZIP protein factor, HY5 (Chattopadhyay et al., 1998). The presence in the ZGT promoter of this core G-box element for light signaling is consistent with a role in the light regulation of ZGT by PIF3 and/or HY5. It is fascinating that this same motif functions in animal circadian feedback loops (there called an E-box element), where it is thought to be crucially important for rhythmic transcriptional regulation (Hao et al., 1997; Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Dunlap, 1999).
We also found evening element (EE) motifs in the ZGT promoter. This EE was recently identified in Arabidopsis to be a motif that regulates circadianly controlled promoters for peak expression near dusk (Harmer et al., 2000). Although the EE motives were found in an antisense orientation in both tobacco ZGT and Arabidopsis pZGT gene promoters, it is possible that they function as circadian enhancers partially mediating rhythmic expression of ZGT. The G/E and EE sequence information provide clues that can direct future research in analyzing the regulation of ZGT expression by light and the circadian clock.
The Role of ZGT in Plant Circadian Systems
Our evidence for a role of ZGT in plant circadian systems comes from experiments in which ZGT was overexpressed (Figures 6 and 7). Our results clearly show that when ZGT is expressed constitutively, the circadian oscillator that controls LHCB expression is “unmasked” and shown to be running in DD. We believe that ZGT plays a role in the endogenous circadian system as well. Although it is possible that the results of this overexpression provoke ZGT 's gene products into performing unusual functions, we postulate that its light and circadian regulation (Figures 1 and 5) argue for a physiological role. Ultimately, this question can be answered by knocking out ZGT and determining whether expression of light-dependent cycling genes is drastically altered. Our data indicate that in nature, the LD cycle and the circadian clock cooperate to cause a robust rhythm of ZGT mRNA during the daily cycle. Assuming that the ZGT protein is the relevant active gene product and that it has a turnover rate that allows significant accumulation, we postulate that the rhythmic abundance of ZGT plays a role in coupling central circadian oscillators to output rhythms and possibly amplifies light input signals into the clockwork. Although we have tested only the LHCB output rhythm, the pattern of many circadian outputs is similar to that of LHCB; therefore, it is reasonable to postulate that ZGT expression may affect other outputs.
What might the role of ZGT be in the maintenance of robust LHCB rhythmicity? Let us start by mentioning what ZGT is not. First, it is not maintaining the LHCB rhythm in DD by feeding back to the central oscillator (i.e., output feedback, as in other systems [Mrosovsky, 1996]) and thereby maintaining the oscillation of a central clockwork that normally damps in DD. This possibility can be eliminated because of the evidence that the central oscillator does not dampen rapidly in DD (Johnson et al., 1998; Strayer et al., 2000; Johnson, 2001). Second, rhythmic expression of ZGT is not the clock-regulated gate of LHCB expression postulated by Nagy, Kay, and co-workers (Kay and Millar, 1992; Fejes and Nagy, 1998), because constitutive expression of ZGT does not lead to constitutive expression of LHCB promoter activity.
ZGT promoter activity appears to phase-lag that of LHCB promoter activity (Figure 4C). On the basis of relatively infrequent sampling, ZGT mRNA abundance (Figure 1B) appears to follow ZGT promoter activity relatively closely in LL (Figure 4C). We have no information about the abundance of ZGT protein, but assuming that its half-life is a few hours, then it is unlikely that a rhythm of ZGT expression directly turns on and off the rhythm of LHCB promoter activity. Our working model postulates that a rhythm of ZGT abundance in natural LD cycles establishes a state during the light interval such that the coupling of the LHCB gene (and other genes) is maintained through the next cycle (Figure 8). This role of ZGT might be likened to the clutch of a vehicle: when engaged (by light-dependent expression of ZGT in LD and LL), clock and outputs are linked; when disengaged (by low expression of ZGT in DD), clock and outputs are unlinked. Thus, in this model, ZGT becomes the sensor of illumination that determines whether the clock gates the rhythmic expression of LHCB and possibly other genes. It is possible that ZGT is a link between PIF3/HY5/phytochrome (and other photoreceptors) and LHCB expression; possibly phytochrome overexpression and/or stimulation by brief red light pulses (Kay and Millar, 1992) leads to a concomitant increase in ZGT expression that maintains LHCB rhythms. In addition to its direct control by light, ZGT also is controlled by the central oscillator, as demonstrated by its expression patterns in LL (Figures 1 and 4).
On the basis of the results depicted in Figure 7, it is possible as well that ZGT acts as an amplifier of the circadian phototransduction pathway (Figure 8). The data in Figure 7 show that ZGT expression can increase significantly the sensitivity of the clock to light pulses, but the times at which light pulses are most effective in shifting phase are when ZGT mRNA levels are very low, namely, around dawn and dusk. Therefore, whether ZGT can act in the entrainment path of the circadian system in natural conditions depends largely on the turnover rate of the ZGT protein, which is unknown at present. If the turnover of ZGT is slow enough that significant levels are present at dawn and/or dusk, then it could play a significant role in the input signal to the clock. Until more is known about ZGT protein expression, however, we favor the interpretation that the primary effect of ZGT is in the coupling of clock to outputs. Nevertheless, the data shown in Figures 5 and 7 suggest that ZGT could function as a component in plant phototransduction pathways, possibly in pathways other than those leading to the circadian clock.
METHODS
Cloning of the ZGT Gene
The ZGT gene was originally isolated from a tobacco (Nicotiana tabacum) genomic DNA/λCharon 35 library using a probe containing the cyanobacterial kaiABC gene cluster (Ishiura et al., 1998) under conditions of low stringency. To clone the ZGT gene, we inserted ∼8.5 kb of SacI-digested genomic DNA harboring the ZGT fragment into pBluescript II KS+ (Stratagene, La Jolla, CA) to produce pSac8.5. On the basis of restriction analyses and DNA gel blot assays, a HindIII fragment (1.2 kb), a BamHI fragment (2.5 kb), and a BamHI-SacI fragment (6.0 kb) containing ZGT DNA sequences from pSac8.5 were then subcloned into pBluescript II KS+ or pGEM-7 (Promega, Madison, WI), yielding pHind1.2, pBam2.5, and pBS6.0, respectively.
Nucleotide sequencing of both DNA strands in the subclones was determined by the dideoxy chain termination method (Del Sal et al., 1989) or using an ABI automated sequencer (PE Applied Biosystems, Foster City, CA). The initial sequence information from pHind1.2 was used to design primers to determine the complete genomic or cDNA sequence of the ZGT gene along with other subclones. The DNA and protein sequences were analyzed using GCG software (Genetics Computer Group, Madison, WI). The GenBank database was searched with the putative amino acid sequence of ZGT, using the BLAST (Altschul et al., 1997) and FASTA (Pearson and Lipman, 1998) algorithms.
3′ and 5′ RACE Assay
The 3′- and 5′-ends of cDNA sequences of the ZGT gene were determined by 3′- or 5′-rapid amplification of cDNA ends (RACE), according to the manufacturer's directions (Life Technologies, Rockville, MD). Total RNA for RACE analysis was isolated from leaf tissues of 45-day-old tobacco plants 8 hr into the light/dark cycle. The 5′-RACE of ZGT cDNA was performed using the first and nested gene-specific primers 5′-TCATCGCTGCTATCTTCACCTTC-3′, 5′-TCTTCAAGCTCCACCTGC-3′, and 5′-TCTACCGCTAGGCGAACCG-3′. The 3′-RACE was conducted with the nested gene-specific primers 5′-TGATAACGGCGTTAACAGAC-3′, 5′-TGATCCGTG-TGAGCATC-3′, and 5′-ACGGAGCTTGTTGGCTG-3′. All of the 3′- and 5′-RACE fragments obtained were cloned into pBluescript II KS+ and then sequenced as described above.
Recombinant Plasmid Construction
A 1168-bp DNA fragment at positions +69 to +1236 containing the entire ZGT coding region and part of its 3′ flanking sequence was cloned into the SmaI site of pBluescript II KS+, and the insert was confirmed by sequence analysis. The SalI-SacI fragment containing a structural gene for firefly luciferase (luc) in pJD301 (Luehrsen et al., 1992), which was kindly provided by Dr. Virginia Walbot (Stanford University, Stanford, CA), was replaced with the ZGT coding sequence to produce pUC/35S::ZGT. The SphI (end-blunted)-BglII fragment from pUC/35S::ZGT was inserted into the SmaI-BamHI site of the binary vector pPZP122 (Hajdukiewicz et al., 1994), which was kindly provided by Dr. Pat Maliga (Rutgers, State University of New Jersey, Piscataway, NJ), to make pOX/ZGT. The BamHI-NcoI fragment in pJD301 was removed and then replaced with the 5′-flanking polymerase chain reaction fragment (−950 to +73) of the ZGT gene to create the PZGT::luc fusion gene. The BamHI-BagII fragment containing PZGT::luc from the resulting plasmid pUC/ZGT::luc was cloned into the BamHI site of the binary vector pPZP112 (Hajdukiewicz et al., 1994) to produce the plasmid pZGT/luc. Sequence-confirmed recombinant plasmids pOX/ZGT and pZGT/luc were introduced independently into competent cells of the host Agrobacterium tumefaciens LBA4404 (Life Technologies).
Generation of Transgenic Plants
Transgenic tobacco plants were obtained by Agrobacterium-mediated transformation of leaf discs with the binary constructs pOX/ZGT and pZGT/luc (Horsch et al., 1985). Tobacco (var Xanthi) expressing the fusion of the promoter for the LHCB1*1 gene from Arabidopsis thaliana and the firefly luciferase gene (PLHCB::luc), which was kindly provided by Drs. Shawn Anderson and Steve Kay (Anderson and Kay, 1995; Dr. Kay is presently at Scripps Research Institute, La Jolla, CA), was transformed by Agrobacterium harboring pOX/ZGT with a gentamycin-resistant selection marker. Tobacco (var NC TG22), which was kindly provided by Dr. Judith Thomas (North Carolina State University, Raleigh, NC), was transformed by Agrobacterium harboring pZGT/luc with a kanamycin-resistant selection marker. Antibiotic resistance of transgenic seedlings was tested by germinating them on half-strength Murashige and Skoog (1962) (MS) medium containing 200 μg/mL gentamycin sulfate (for OX/ZGT transformants) or kanamycin sulfate (for ZGT/luc transformants). Homozygous transformants were used in all experiments; these were determined by assaying transgene expression and by confirming nonsegregation of corresponding antibiotic resistance in the T2 or T3 seedling populations.
Seedling Growth Conditions
Seed were sterilized in 20% bleach (Clorox) containing 0.1% SDS for 15 min and washed three times with sterilized water. After the sterilized seed were soaked in 10 μM gibberellic acid overnight, they were germinated on 0.8% agar-solidified half-strength MS medium and grown in an incubator at 26°C under light/dark (LD) cycles of cool-white fluorescent light (50 μE m−2 sec−1) and darkness (either LD 12:12 or LD 16:8) for 1 week in the case of bioluminescence assays or for 3 to 4 weeks in the case of RNA isolation. In some experiments, the seedlings were then transferred to constant light (LL) or constant darkness (DD) as specified in the figure legends.
RNA Gel Blot Analysis
Total RNA was prepared from transgenic or wild-type tobacco (var Xanthi) plants and analyzed in RNA gel blots as described by Xu et al. (1996). An α-32P-dCTP–labeled 1.2-kb HindIII fragment from the ZGT gene was used as a probe for RNA gel blot assays. RNA gel blots were then stripped and rehybridized with a 2.5-kb EcoRI fragment probe containing an Arabidopsis rRNA gene for normalization of the RNA loading in each lane (Xu et al., 1996). Quantification of RNA gel blots was performed with an IS-1000 digital imaging system using Spot Density Tool software (Alpha Innotech, San Leandro, CA).
Analysis of Luminescence Rhythms in Transgenic Plants
Seedlings expressing firefly luciferase were germinated on half-strength MS agar medium and grown at 26°C in LD 16:8. Before lights off in the last LD cycle, individual 7-day-old seedlings were transplanted into a small scintillation vial filled with 0.4 mL of liquid MS medium containing 200 μM luciferin (Promega). For luminescence measurement in DD, the assay medium was supplemented with 10 μM ATP. This vial was then placed inside a 20-mL glass scintillation vial. After the dark interval was over, the vials were transferred to an automated multichannel photomultiplier tube apparatus (Johnson et al., 1995) for recording luminescence in either LL or DD. Periods and phases of the luminescence rhythms were determined based on the trough and/or peak values using LVA software supplied by Dr. Takao Kondo (Nagoya University, Nagoya, Japan) and the CHRONO program (Roenneberg and Taylor, 1999).
Imaging of Luminescent Seedlings
Three-week-old transgenic seedlings expressing the fusion gene PZGT::luc that had been grown in LD 12:12 cycles were transferred into a 100-mm-diameter Petri dish filled with 15 mL of liquid MS medium supplemented with 200 μM luciferin and incubated in continuous light. After 24 hr in LL, luminescent seedling images were captured with 80-sec exposures every 4 hr for 20 hr. Images were captured with a cooled charge-coupled device camera (TE/CCD512BKS; Princeton Instruments, Trenton, NJ) under the control of and analyzed by custom software designed by Dr. Takao Kondo (Kondo et al., 1994).
Assay of Phase Shifting
Seedlings of the PLHCB::luc reporter in either the wild-type or the OX/ZGT background were germinated and grown as described above. At dusk of the final LD cycle, seedlings were placed into darkness for 32 hr. For the phase response experiment (data not shown), seedlings were exposed to 30 min of white light pulses (50 μE m−2 sec−1) at different phases during the dark treatment (three samples for each phase and/or treatment). At the end of the 32 hr of darkness, seedlings were placed in constant red light (20 μE m−2 sec−1), and their luminescence rhythms were monitored as described above. For the experiments shown in Figure 7, seedlings were exposed to white light pulses (50 μE m−2 sec−1) of the indicated durations starting at hr 27 in DD (circadian time 19[CT 19]). After completion of the light pulse, seedlings were returned to DD until hr 32 and then transferred to constant red light, and their luminescence rhythms were measured. Periods and phases of the luminescence rhythms were determined based on the phases of the peaks and/or troughs.
Acknowledgments
We are grateful to many colleagues for their assistance in this study: to Drs. Takao Kondo and Masahiro Ishiura for sharing the kai DNA before publication of its sequence for use as a hybridization probe; to Drs. Matt Olson and Mark Stokes for assistance with statistical analyses; to Dr. Jiqing Sai for sharing his phase-shifting protocol (Figure 7) before publication; to Drs. Steve Kay and Shawn Anderson for generously providing PLHCB::luc transgenic tobacco seed; to Dr. Robert Hayes for the tobacco genomic library; to Dr. Virginia Walbot for the firefly luciferase (luc) construct; to Dr. Pat Maliga for binary vectors; to Dr. Judith Thomas for NC TG22 (Maryland Mammoth) tobacco seed; and to Dr. Takao Kondo for software for the acquisition and analysis of luminescence rhythms. This research was supported by grants from the National Institute of Mental Health (Nos. MH43836 and MH01179).
References
- Allada, R., White, N.E., So, W.V., Hall, J.C., and Rosbash, M. (1998). A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, S.L., and Kay, S.A. (1995). Functional dissection of circadian clock– and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc. Natl. Acad. Sci. USA 92, 1500–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré, I.A., and Kay, S.A. (1995). Multiple DNA–protein complexes at a circadian-regulated promoter element. Plant Cell 7, 2039–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.-H., Puente, P., Deng, X.-W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington, T.K., Wager-Smith, K., Ceriani, M.J., Staknis, D., Gekakis, N., Steeves, T.D.L., Weitz, C.J., Takahashi, J.S., and Kay, S.A. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Del Sal, G., Manfioletti, G., and Schneider, C. (1989). A common miniscale preparation of template DNA from phagemids, phages or plasmids suitable for sequencing. Biotechniques 7, 514–520. [PubMed] [Google Scholar]
- Deng, M.-D., Moureaux, T., Leydecker, M.-T., and Caboche, M. (1990). Nitrate-reductase expression is under the control of a circadian rhythm and is light-inducible in Nicotiana tabacum leaves. Planta 180, 257–261. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Fejes, E., and Nagy, F. (1998). Molecular analysis of circadian clock–regulated gene expression in plants: Features of the “output” pathways. In Biological Rhythms and Photoperiodism in Plants, P. Lumsden and A. Millar, eds (Oxford, UK: BIOS Scientific Publishers), pp. 99–118.
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hao, H., Allen, D.L., and Hardin, P.E. (1997). A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 17, 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.-S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., Nater, M., Apel, K., and Staiger, D. (1997). AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch, R.B., Fry, J.E., Hoffman, N.L., Eicholtz, D., Rogers, S.D., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C.R., Tanabe, A., Golden, S.S., Johnson, C.H., and Kondo, T. (1998). Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H. (1994). Illuminating the clock: Circadian photobiology. Semin. Cell Biol. 5, 355–362. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H. (2001). Endogenous timekeepers in photosynthetic organisms. Annu. Rev. Physiol. 63, 695–728. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H., Knight, M.R., Kondo, T., Masson, P., Sedbrook, J., Haley, A., and Trewavas, A.J. (1995). Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science 269, 1863–1865. [DOI] [PubMed] [Google Scholar]
- Johnson, C.H., Knight, M., Trewavas, A., and Kondo, T. (1998). A clockwork green: Circadian programs in photosynthetic organisms. In Biological Rhythms and Photoperiodism in Plants, P. Lumsden and A. Millar, eds (Oxford, UK: BIOS Scientific Publishers), pp. 1–34.
- Kay, S.A., and Millar, A.J. (1992). Circadian-regulated Cab gene transcription in higher plants. In Molecular Genetics of Biological Rhythms, M.W. Young, ed (New York: Marcel Dekker), pp. 73–89.
- Kellmann, J.-W., Merforth, N., Wiese, M., Pichersky, E., and Piechulla, B. (1993). Concerted circadian oscillations in transcript levels of nineteen Lhca/b (cab) genes in Lycopersicon esculentum (tomato). Mol. Gen. Genet. 237, 439–448. [DOI] [PubMed] [Google Scholar]
- Kloppstech, K. (1985). Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta 165, 502–506. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Tsinoremas, N.F., Golden, S.S., Johnson, C.H., Kutsuna, S., and Ishiura, M. (1994). Circadian clock mutants of cyanobacteria. Science 266, 1233–1236. [DOI] [PubMed] [Google Scholar]
- Luehrsen, K.R., DeWet, J.R., and Walbot, V. (1992). Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 216, 397–414. [DOI] [PubMed] [Google Scholar]
- Martínez-García, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element–bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Short, S.R., Chua, N.-H., and Kay, S.A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4, 1075–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.J., Straume, M., Chory, J., Chua, N.-H., and Kay, S.A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267, 1163–1166. [DOI] [PubMed] [Google Scholar]
- Mrosovsky, N. (1996). Locomotor activity and non-photic influences on circadian clocks. Biol. Rev. 71, 343–372. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Otto, B., Grimm, B., Otthersbach, P., and Kloppstech, K. (1988). Circadian control of the accumulation of mRNAs for light- and heat-inducible chloroplast proteins in pea (Pisum sativum). Plant Physiol. 88, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, W.R., and Lipman, D.J. (1998). Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, M.L., and McClung, C.R. (1993). Differential involvement of the circadian clock in the expression of genes required for ribulose-1,5-bisphosphate carboxylase/oxygenase synthesis, assembly, and activation in Arabidopsis thaliana. Plant Physiol. 103, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, M.L., Caspar, T., Quail, P.H., and McClung, C.R. (1993). Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol. Biol. 23, 349–364. [DOI] [PubMed] [Google Scholar]
- Roenneberg, T., and Taylor, W. (1999). Automated recordings of bioluminescence with special reference to the analysis of circadian rhythms. Methods Enzymol. 305, 104–119. [DOI] [PubMed] [Google Scholar]
- Rutila, J.E., Suri, V., Le, M., So, M.V., Rosbash, M., and Hall, J.C. (1998). CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93, 805–814. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Más, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watillon, B., Kettmann, R., Boxus, P., and Burny, A. (1993). Developmental and circadian pattern of rubisco activase mRNA accumulation in apple plants. Plant Mol. Biol. 23, 501–509. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Zhu, Q., Panbangred, W., Shirasu, K., and Lamb, C. (1996). Regulation, expression and function of a new basic chitinase gene in rice (Oryza sativa L.). Plant Mol. Biol. 30, 387–401. [DOI] [PubMed] [Google Scholar]
- Zhong, H.H., Resnik, A.S., Straume, M., and McClung, C.R. (1997). Effects of synergistic signalling by phytochrome A and cryptochrome 1 on circadian clock–regulated catalase expression. Plant Cell 9, 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]