Abstract
Salicylic acid (SA) has been shown to act as a signal molecule that is produced by many plants subsequent to the recognition of potentially pathogenic microbes. Increases in levels of SA often trigger the activation of plant defenses and can result in increased resistance to subsequent challenge by pathogens. We observed that the polyketide 6-methylsalicylic acid (6-MeSA), a compound that apparently is not endogenous to tobacco, can mimic SA. Tobacco leaves treated with 6-MeSA show enhanced accumulation of the pathogenesis-related (PR) proteins PR1, β-1,3-glucanase, and chitinase and also develop increased resistance to tobacco mosaic virus. We transformed tobacco with 6msas, the 6-methylsalicylic acid synthase (6MSAS) gene from Penicillium patulum, to generate plants that constitutively accumulate 6-MeSA. Analysis of primary transformants and the first generation progeny of 6MSAS tobacco revealed that plants can be engineered to accumulate significant amounts of 6-MeSA as a conjugate. Levels of total 6-MeSA increased with plant age. Increased 6-MeSA accumulation correlated with increased levels of PR1 and chitinase proteins and resulted in enhanced resistance of NN genotype 6MSAS tobacco to tobacco mosaic virus. Our results demonstrate that a multistep biosynthetic pathway can be engineered into plants using a single fungal polyketide synthase gene. The functional expression of 6msas can be used to activate disease resistance pathways that normally are induced by SA.
INTRODUCTION
The importance of salicylic acid (SA) as a signal molecule in plant disease resistance responses is well documented and has been the subject of a number of reviews (Raskin, 1992; Durner et al., 1997; Mauch-Mani and Métraux, 1998; Dempsey et al., 1999; Scott et al., 1999; Klessig et al., 2000). In many plants, tissue levels of SA increase upon invasion by certain incompatible pathogens and exposure to certain chemicals or environmental stresses (Yalpani et al., 1994; Leôn et al., 1995). This increase may occur not only around the site of inoculum penetration but in more distant tissues as well. Increases in SA frequently induce increased resistance to subsequent challenge by a spectrum of viral, bacterial, or fungal pathogens. SA can induce the expression of several classes of pathogenesis-related (PR) protein gene families (Ward et al., 1991; Van Loon and Van Strien, 1999). Although some, such as β-1,3-glucanases and chitinases, have antimicrobial hydrolytic activities, it remains unclear how most SA-induced PR proteins contribute to disease resistance. Other evidence indicates that SA may contribute to resistance by affecting the sensitivity of the triggers for defense activation (Shirasu et al., 1997). Although there is little doubt that SA has a key role in resistance, recent reports indicate that in certain plant–microbe interactions, jasmonic acid, ethylene, nitric oxide, and possibly other molecules also may be involved in the activation of local and systemic defenses (Pieterse and van Loon, 1999; Klessig et al., 2000). Additional work indicates that SA may play a regulatory role in other developmental processes. For example, it has been shown that SA may affect alternative oxidase activity, chilling resistance, stomatal opening, senescence, and cell growth (Raskin, 1992; Janda et al., 1999; Rate et al., 1999; Morris et al., 2000).
In plants, SA most likely is formed by 2-hydroxylation of benzoic acid or benzoyl conjugates. These are likely products of the β-oxidation of trans-cinnamic acid from the phenylpropanoid pathway (Leôn et al., 1993; Yalpani et al., 1993; Silverman et al., 1995; Chong et al., 2001). However, little is known about how SA biosynthesis is regulated. Exogenously applied SA is conjugated readily to form SA glucose ether and an SA glucose ester (Edwards, 1994; Lee and Raskin, 1998). These conjugates may serve as a storage pool from which SA may be liberated by β-glucosidases (Chen et al., 1995). An SA-inducible tobacco SA glucosyltransferase that forms both the ether and ester products when expressed in Escherichia coli has been characterized and described (Lee and Raskin, 1998, 1999). The broad substrate specificity of this enzyme included trans-cinnamic acid, benzoic acid, and 4-hydroxybenzoic acid, suggesting that this SA glucosyltransferase may be involved in regulating the levels of several intermediates in the SA biosynthetic pathway. In tobacco mosaic virus (TMV)-inoculated tobacco, SA is converted to methylsalicylate and its glucoside. Methylsalicylate vapor has been proposed to act as a potential airborne defense inducer (Shulaev et al., 1997). A gene for a SA carboxyl methyltransferase that generates this compound was isolated recently (Ross et al., 1999). Belles et al. (1999) also observed that SA can be further 5-hydroxylated to form gentisic acid, which may accumulate in a free or a conjugated form. They suggest that, in addition to SA, gentisic acid can act as a signal molecule and trigger defense protein accumulation in tomato.
Compelling evidence for the importance of SA in disease resistance was obtained from work with plants expressing the Pseudomonas putida gene for SA hydroxylase (nahG). Tobacco and Arabidopsis plants expressing this enzyme constitutively display reduced resistance against viral, bacterial, and fungal pathogens (Gaffney et al., 1993; Delaney et al., 1994). However, it is possible that SA hydroxylase affects the accumulation of other phenylpropanoids as well (Cameron, 2000). In microorganisms, SA can be produced from chorismic acid and isochorismic acid (Serino et al., 1995). Verberne et al. (2000) transformed tobacco with the chorismate:isochorismate isomerase gene (entC) from E. coli and the SA-forming isochorismate:pyruvate lyase gene (pmsB) from Pseudomonas fluorescence. Plants expressing both enzymes accumulated increased levels of SA, SA conjugates, and PR proteins. These plants also displayed increased resistance to TMV and the fungus Oidium lycopersicon. These data add further evidence for the important role of SA in plant disease resistance.
Here, we present a novel approach to the manipulation of SA response pathways using a fungal polyketide synthase (PKS) gene. Prokaryotic and eukaryotic PKSs form a wide variety of biomolecules, including long-chain fatty acids, pigments, immunosuppressants, and antibiotics (Monaghan and Tkacz, 1990; O'Hagan et al., 1992). Despite their diverse end products, PKSs are composed of very similar functional modules. The key reaction in the biosynthesis of polyketides involves repeated decarboxylative condensations between acylthioesters that build the product, two carbons at a time. After each condensation, structural variability is introduced into the carbon chain by one of three successive reactions involving keto reduction, dehydration, and enoyl reduction of the β-keto group formed after each condensation reaction.
Among the PKSs, aromatic PKSs consist of a group of iteratively used active sites contained either on a group of separate proteins encoded by a cluster of genes, as in bacteria, or on a large single protein, as in fungi. 6-Methylsalicylic acid synthase (6MSAS) is a well-characterized fungal aromatic PKS from Penicillium patulum Bainier (anamorph P. griseofulvum Dierckx). The 6msas gene, the first fungal PKS gene cloned (Beck et al., 1990), was identified as a 5322-bp open reading frame encoding a protein of 1774 amino acids and a molecular mass of 190,731 D. The native 6MSAS protein is made up of four identical subunits and has a molecular mass of ∼740 kD (Spencer and Jordan, 1992). 6MSAS catalyzes three successive condensation reactions to form 6-methylsalicylic acid (6-MeSA) from one molecule of acetyl-CoA and three molecules of malonyl-CoA and uses NADPH as a reducing cofactor (Dimroth et al., 1970).
All active sites for the 11 transformations required to produce 6-MeSA are carried on the single multifunctional protein. Catalysis involves the repeated use of some of these active sites. The expression of functional P. patulum 6msas in bacteria and yeast was demonstrated by Bedford et al. (1995) and Kealey et al. (1998). The latter suggested that heterologous expression of a phosphopantetheinyl transferase might be required to convert apo-6MSAS to holo-6MSAS. In P. patulum, 6-MeSA is a precursor of the antibiotic patulin. We were interested in 6MSAS because its product, 6-MeSA, bears structural resemblance to SA. We postulated that 6-MeSA might activate defense responses in plants similar to those activated by SA. Here we present evidence that 6MSAS can be expressed functionally in tobacco and that 6-MeSA mimics SA by enhancing the accumulation of PR proteins and resistance to TMV.
RESULTS
6-MeSA Treatment Activates Resistance Mechanisms in Tobacco
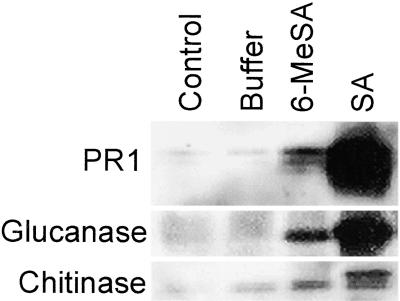
The polyketide product of the reactions catalyzed by 6MSAS from P. patulum shows structural resemblance to SA. To determine if 6-MeSA can mimic SA in planta and trigger similar defense responses, we infiltrated leaves of NN genotype, nontransgenic tobacco plants, with buffer in the absence or presence of either 2.5 mM SA or 2.5 mM 6-MeSA. As expected, by 7 days after infiltration, antiserum raised against acidic PR1, glucanase, and chitinase of tobacco allowed the detection of a significant accumulation of SA-induced proteins (Figure 1). 6-MeSA also induced these proteins, although not as strongly as SA. When an NN genotype tobacco cultivar is inoculated with TMV, it responds hypersensitively, and spread of the virus is restricted. Enhanced resistance in such plants is reflected by the reduced size of virus-induced necrotic lesions (Holmes, 1938).
Figure 1.
6-MeSA Induces PR Protein Accumulation in Tobacco.
The uppermost fully expanded leaves of 8-week-old NN genotype tobacco plants were left untreated (control) or infiltrated with buffer in the presence or absence of 2.5 mM 6-MeSA or SA. Seven days later, the treated and control leaves were sampled. Antiserum raised against tobacco PR1, glucanase, or chitinase was used for immunoblots prepared from the leaf samples.
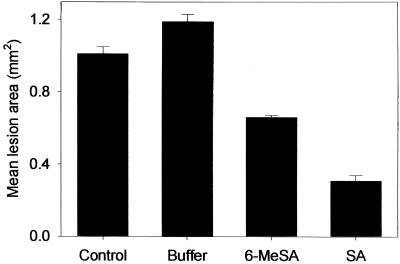
Consistent with its effect on PR proteins, 6-MeSA also induced enhanced resistance to TMV when leaves were inoculated 7 days after infiltration (Figure 2). The mean area of individual TMV lesions on leaves that had been pretreated with 6-MeSA was 0.66 ± 0.01 mm2, 55% of the area of lesions on buffer-treated control leaves. Again, SA appeared to be a more potent inducer than 6-MeSA. Although the lesions were smaller in the SA-treated leaves, the results indicate that 6-MeSA induces resistance to TMV. The difference in the efficacy of 6-MeSA compared with SA could be due to dissimilarities in bioactivity, the presence of impurities in the 6-MeSA preparation used, or the differential uptake, transport, or metabolism of the compounds. Using HPLC analysis, we confirmed that the 6-MeSA used was not contaminated by SA. We also determined that 6-MeSA treatment did not affect the levels of SA accumulation in tobacco significantly (data not shown). These results with exogenously applied 6-MeSA suggested that this compound could activate defense mechanisms similar to those activated by SA.
Figure 2.
6-MeSA Induces Resistance of Tobacco against TMV.
The youngest fully expanded leaves of 8-week-old NN genotype tobacco plants were left untreated (control) or infiltrated with buffer in the presence or absence of 2.5 mM 6-MeSA or SA. Seven days later, the control and infiltrated leaves were inoculated with TMV. The area of induced lesions (50 lesions per leaf for five plants per treatment) was determined 5 days later. Error bars represent ±se.
Transgenic 6MSAS Tobacco Accumulates 6-MeSA
Even though the results of the leaf infiltration experiments needed to be viewed with caution, we were encouraged sufficiently to try to generate transgenic tobacco expressing 6MSAS. To this end, the binary transformation vector pPHP12424 was constructed. The 6msas and nptII genes in this construct were designed for high level constitutive expression in plants using the SCP1 and UCP3 promoters, respectively, each with the Ω′ translation enhancement sequence. Because its substrates acetyl-CoA, malonyl-CoA, and NADPH were expected to be available in plastids, a ribulose bisphosphate carboxylase targeting sequence from petunia was used to direct the accumulation of 6MSAS protein to plastids. Primary leaf explants of NN and nn genotype tobacco were inoculated with Agrobacterium strain EHA105/PHP12424 and then cocultivated and selected on kanamycin-containing callus initiation medium. Fast-growing, kanamycin-resistant calli were transferred to selective regeneration medium for the production of transgenic tobacco shoots. Reverse transcription–mediated polymerase chain reaction analysis of callus lines was performed to verify the transcription of the 6msas gene (data not shown). Transgenic shoots initially were identified using NPTII ELISA of leaf samples. NPTII-positive shoots were established in the greenhouse, and developing transgenic plants were assayed for the maintenance of NPTII expression. Compared with nontransformed controls, the primary transgenic plants (T0 generation) had a normal phenotype.
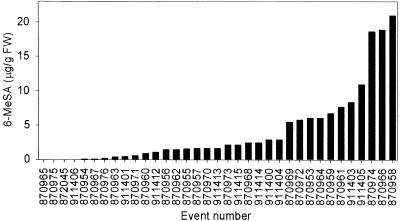
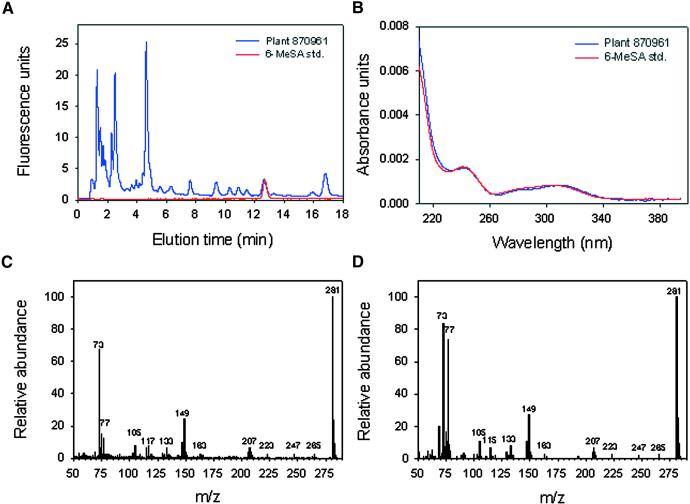
HPLC analysis of hydrolyzed extracts of leaf samples taken at flowering time identified a large number of T0 tobacco events that produced a metabolite that cochromatographed with authentic 6-MeSA (Figure 3). This substance was not detectable in untransformed tobacco and was identical to 6-MeSA, as confirmed by its fluorescence emission, its UV light absorption spectrum, and gas chromatography–mass spectral analysis (Figure 4). Four of these 6-MeSA–accumulating lines were in an NN genotype background. The levels of total 6-MeSA in leaves of some of the plants exceeded 20 μg/g fresh weight. 6-MeSA appears to accumulate primarily as a conjugate. For plant 917287, we measured 0.078 ± 0.002 μg of free 6-MeSA per gram fresh weight. This was near the fluorescence detection limit for 6-MeSA and insufficient to be confirmed by diode array analysis. Base hydrolysis followed by acid hydrolysis of tissue extracts (Enyedi et al., 1992) released 4.47 ± 0.06 μg of 6-MeSA per gram fresh weight, whereas treatment with almond β-glucosidase yielded 4.53 ± 0.08 μg of 6-MeSA per gram fresh weight. This finding suggests that 6-MeSA is accumulated primarily as a β-glucoside.
Figure 3.
Accumulation of 6-MeSA in T0 Generation 6MSAS Tobacco.
HPLC analysis was performed on hydrolyzed extracts of leaves from NN and nn genotype 6MSAS transgenic plants. Plants were sampled at flowering time. FW, fresh weight.
Figure 4.
Identification of 6-MeSA in 6MSAS Tobacco.
(A) HPLC fluorescence chromatogram of a leaf extract from plant 870,961 (blue trace) and the 6-MeSA standard (std.; red trace).
(B) UV light absorption spectrum of a leaf extract from plant 870,961 (blue trace) and the 6-MeSA standard (std.; red trace).
(C) Gas-liquid chromatography–mass spectroscopy analysis of a trimethylsilylated leaf extract from plant 870,961. m/z, mass-to-charge ratio.
(D) Gas-liquid chromatography–mass spectroscopy analysis of the trimethylsilylated 6-MeSA standard. m/z, mass-to-charge ratio.
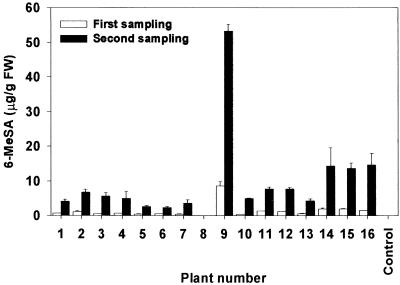
Accumulation of 6-MeSA is a heritable trait. The T0 plants were self-pollinated. Seed from the selfed plants were surface sterilized and sown on kanamycin-containing germination medium to select transgenic T1 individuals. Kanamycin-resistant, NPTII-positive T1 plants were tested for 6-MeSA, and five of seven selected events produced progeny that accumulated the compound. Of the 16 individual plants from event 911403 that were tested, 15 had detectable levels of 6-MeSA, whereas no 6-MeSA was detected in leaves of untransformed control NN genotype plants (Figure 5). Four weeks after transplanting to soil, 6-MeSA levels in fully expanded leaves of the transgenic plants ranged from less than 0.1 to nearly 9 μg/g fresh weight. By 8 weeks after transplanting, the levels had increased in all plants and reached as much as 53.3 ± 1.91 μg/g fresh weight. These results reveal that progeny from plants transformed with 6msas can inherit the trait of 6-MeSA accumulation and that levels of this compound can increase with plant age. It is unclear why the T1 selection 911403-9 accumulated exceptionally high levels of 6-MeSA.
Figure 5.
Leaf Levels of 6-MeSA Increase with Age in 6-MSAS Tobacco.
Accumulation of 6-MeSA in T1 progeny derived from selfed NN genotype plant 870956. Untransformed NN genotype tobacco was used as a control. FW, fresh weight. Error bars represent ±se.
Activation of Disease Resistance Mechanisms in 6MSAS Tobacco
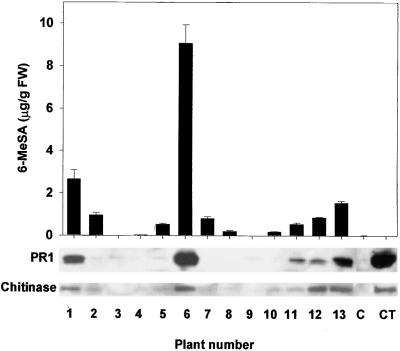
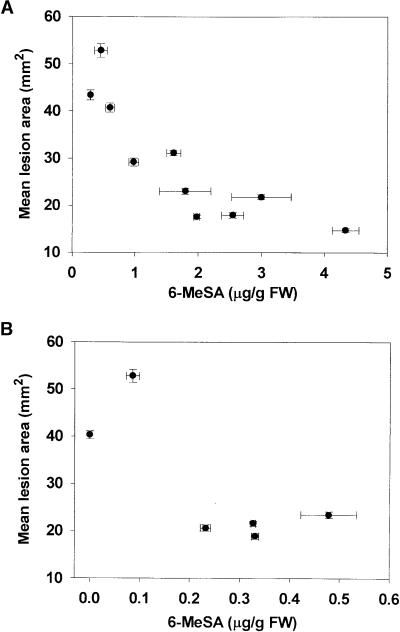
Extracts of leaf samples from NN genotype event 870856 were subjected to protein gel blot analysis to examine the effect of 6-MeSA accumulation on PR proteins. There appeared to be a rough correlation between total 6-MeSA levels and the accumulation of PR1 and chitinase proteins (Figure 6). To study the effect of 6-MeSA accumulation on disease resistance, we inoculated T1 plants derived from line 870955 and additional T1 selections from event 911403 with TMV. The mean area of individual lesions that developed subsequently was determined 5 days later and compared with 6-MeSA levels in a lower leaf of the same plant. The results shown in Figure 7A for derivatives of 911403 demonstrate an inverse relation between mean lesion area and levels of 6-MeSA. The mean area of individual TMV-induced lesions on the transgenic plant that accumulated 0.29 ± 0.01 μg of 6-MeSA per gram fresh weight was 43.4 ± 1.0 mm2, nearly three times larger than the 14.7 ± 0.4 mm2 lesions on the plant that accumulated 4.33 ± 0.21 μg of 6-MeSA per gram fresh weight. Six plants derived from line 870955 accumulated lower levels of 6-MeSA than plants from event 911403. Nevertheless, the plants that accumulated more than 0.23 ± 0.01 μg of 6-MeSA per gram fresh weight displayed increased TMV resistance (Figure 7B). Leaves of untransformed tobacco accumulated levels of total SA similar to those in 6MSAS-tobacco lines (0.10 ± 0.01 versus 0.08 ± 0.01 μg of total SA per gram fresh weight, respectively). Therefore, it is unlikely that the increased TMV resistance of the transgenic plants resulted from a breakdown of 6-MeSA to SA. The data obtained with the 6MSAS plants are consistent with the results of the leaf infiltration experiments and suggest that 6-MeSA acts as an inducer of defense proteins and a trigger of increased resistance to TMV infection.
Figure 6.
Effect of Tissue 6-MeSA Levels on PR Protein Accumulation in 6MSAS Tobacco.
Protein gel blot analysis of PR1 and chitinase proteins was performed on extracts from the youngest fully expanded leaves of 4-week-old T1 progeny from plant 870956. The same leaf was used as that sampled for 6-MeSA. Equivalent leaves from untransformed NN genotype tobacco (C) and plants that were sampled 4 days after inoculation with TMV (CT) were used as controls. FW, fresh weight. Error bars represent ±se.
Figure 7.
Effect of Tissue 6-MeSA Levels on Resistance against TMV in 6MSAS Tobacco.
(A) Virus resistance in T1 progeny from plant 911403.
(B) Virus resistance in T1 progeny from plant 870955.
The youngest fully expanded leaves of 4-week-old plants were inoculated with TMV, and the mean areas of individual TMV-induced lesions (50 lesions per leaf) were measured 5 days later and compared with 6-MeSA levels in a lower leaf. FW, fresh weight. Error bars represent ±se.
Effect of 6-MeSA on Plant Phenotype
T0 generation transgenic tobacco plants had a normal morphology and produced viable seed. All T1 generation plants looked normal until just before flowering. However, NN genotype T1 plants produced from three events (870955, 870956, and 911403) developed an odd leaf-curling phenotype on leaves that were forming as the floral bud appeared (Figure 8). As the inflorescence emerged, subsequent leaves looked normal. No abnormal morphology was observed with nn genotype T1 transgenic plants. Additional transgenic 6MSAS tobacco of the NN genotype will need to be generated to determine if the effect on leaf morphology is a transformation artifact or an effect of 6MSAS expression.
Figure 8.
Effect of 6MSAS Expression on Leaf Phenotype of NN Genotype Tobacco.
Leaves formed just before opening of the first inflorescence grow in a nonsymmetrical pattern. Foliage that formed subsequently appears normal.
DISCUSSION
In this work, we demonstrated successful plant transformation with a multifunctional fungal polyketide synthase gene. A complex biosynthetic pathway was introduced into tobacco using a single gene from P. patulum. Tobacco transformed with 6msas constitutively accumulated significant amounts of 6-MeSA. This trait is heritable. Results from treatment of leaves with 6-MeSA and work with 6MSAS transgenic plants indicated that 6-MeSA accumulation induces enhanced defense protein levels and virus resistance, probably by mimicking but not acting through SA, a natural defense hormone in tobacco. It is expected that the defense mechanisms that were activated by heterologous 6MSAS expression can affect resistance to other phytopathogenic taxa and can be exploited in other crops as well.
Although we were unable to detect 6MSAS protein by protein gel blot analysis (data not shown), transformed plants appeared to have sufficient functional enzyme activity to allow the production and accumulation of 6-MeSA in leaf tissues. Leaf 6-MeSA levels increased progressively with plant age, primarily in the form of conjugates (Figure 5). Further work is needed to determine the identity of the conjugates. At this time, we do not know if 6-MeSA conjugates have the ability to induce PR protein accumulation or if defenses are activated by free 6-MeSA only. Venkatasubbaiah and Chilton (1992) reported that 6-MeSA produced by a species of Phoma was phytotoxic and had in vitro inhibitory activity against various bacteria and fungi, including the producing organism. In our study, 6-MeSA–accumulating plants did not develop any necrotic symptoms, perhaps because a significant proportion of the 6-MeSA formed accumulated as conjugates. Levels of 6-MeSA exceeded 50 μg/g fresh weight in some leaves (Figure 5), which is of a similar order of magnitude as the amount of total SA that accumulates in TMV-inoculated tobacco leaves (Enyedi et al., 1992).
The substrates for 6MSAS (acetyl-CoA, malonyl-CoA) and NADPH are expected to be available in chloroplasts. Therefore, we targeted 6MSAS protein to plastids by linking the signal sequence from the petunia ribulose bisphosphate carboxylase small subunit gene to the 6msas gene. However, it is likely that for 6-MeSA to activate defense gene expression, it has to leave the plastids. By analogy to the fate of 4-hydroxybenzoic acid (Siebert et al., 1996), it is likely that SA and possibly 6-MeSA glycosides accumulate in the vacuole, with some forms also becoming incorporated into the cell wall. Although conjugates probably are not the defense-inducing molecules, release of the free SA from a pool of SA conjugates has been proposed and could be relevant in 6-MeSA–forming transgenic plants (Chen et al., 1995).
Work by Kealey et al. (1998) indicates that 6MSAS, like all polyketide synthases, fatty acid synthases, and nonribosomal peptide synthetases, must be converted to its active holo-form by post-translational transfer of the 4′-phosphopantetheinyl moiety of CoA to the side chain hydroxyl of a conserved serine residue in the acyl carrier protein domains. A number of bacterial phosphopantetheinyl transferases have been cloned and described (Lambalot et al., 1996; Reuter et al., 1999). Indeed, Kealey et al. (1998) demonstrated that 6-MeSA production in E. coli or yeast cells transformed with 6msas is increased dramatically if the cells also express the Bacillus subtilis sfp gene encoding a phosphopantetheinyl transferase. Our data suggest that tobacco produces a protein with an enzymatic activity that allows the formation of holo-6MSAS.
Phosphopantetheinylation of the acyl carrier domains of apo-6MSAS may be achieved by endogenous enzymes that are involved in catalyzing this reaction on the acyl carrier protein(s) of fatty acid biosynthesis. Interestingly, among the microbial phosphopantetheinyl transferases studied, the protein encoded by the B. subtilis sfp gene appears to be outstanding. Although most phosphopantetheinyl transferases seem to have a narrow substrate recognition spectrum, Sfp can efficiently phosphopantetheinylate a number of peptidyl carrier proteins, acyl carrier domains, or subunits of fatty acid synthases and polyketide synthases (Lambalot et al., 1996; Reuter et al., 1999). Our data suggest that certain plant phosphopantetheinyl transferases also have a broad substrate spectrum. It remains to be determined if 6-MeSA levels greater than those observed here can be achieved if 6MSAS plants also overproduce a phosphopantetheinyl transferase. Our results with 6MSAS tobacco also suggest that plants could be engineered to functionally express other wild-type or engineered polyketide synthases to produce molecules with pharmacological or other valuable properties.
METHODS
Plant Material and Treatments
Tobacco (Nicotiana tabacum) cv Xanthi-nc (NN genotype) and cv Xanthi (nn genotype) (seed kindly provided by Prof. Ilya Raskin, Rutgers University, New Brunswick, NJ) was raised in a greenhouse under conditions similar to those described by Simmons et al. (1998). Regenerated plants were self-pollinated. To study the effect of 6-methylsalicylic acid (6-MeSA) on pathogenesis-related (PR) proteins and tobacco mosaic virus (TMV) resistance, we syringe infiltrated the uppermost fully expanded leaves of 8-week-old plants with 5 mM bis-tris buffer, pH 6.5, in the absence or presence of 2.5 mM 6-MeSA or 2.5 mM salicylic acid (SA). Seven plants were used for each treatment. An additional set of plants were not subjected to infiltration and were used as controls. Seven days later, leaf tissue from two plants per treatment was sampled and pooled for analysis of PR protein accumulation, and control and treated leaves from the remaining plants were inoculated with a suspension of TMV (strain U1). The area of individual TMV-induced lesions was determined 5 days after inoculation. No visible phytotoxicity was associated with these treatments. All reagents used were of the highest purity available.
Construction of the Transformation Vector
Genomic clones of Penicillium patulum 6msas (6-methylsalicylic acid synthase) were obtained from Prof. Eckhardt Schweizer (University of Erlangen, Germany). He provided us with two clones, M41 and M1. Clone M41 was in pUC18, having the DNA insert EcoRI–SalI from the EcoRI restriction site at nucleotide 993 to the SalI restriction site at nucleotide 2544 of the 6msas gene. The M41 fragment was missing 87 bp of open reading frame at the 5′ end of the gene. This missing region contained exon 1. M1 had the DNA insert SalI–SstI from the restriction site at position 2544 to the SstI restriction site at the end of the 6msas gene (Beck et al., 1990). Because the M41 genomic fragment was missing exon 1 of the 6msas gene, the full-length 6msas open reading frame was assembled by ligating a synthetic DNA linker containing exon 1 of the 6msas gene to the 5′ end of the M41 fragment at the EcoRI site. The M1 (3′) fragment of the 6msas gene was ligated subsequently to the M41/synthetic linker fragment at the 3′ SalI site of the M41 fragment, generating the full-length 5325-bp open reading frame.
The resulting clone, pPHP11028, was used for subsequent cloning into transformation vectors. By means of subcloning and intermediate vectors, the 19.9-kb transformation vector pPHP12424 (SCP1::Ω′::CT::6msas::PINII/UCP3::Ω′::nptII::PINII) was generated. The 6msas DNA (with the intron removed) was fused to the chloroplast targeting sequence of the small subunit of ribulose bisphosphate carboxylase of petunia (Dean et al., 1987), and a polyadenylation signal sequence was obtained from the proteinase inhibitor II (PINII) gene of potato (Keil et al., 1986). Expression of 6msas and the kanamycin resistance marker NPTII was driven by the strong constitutive promoters SCP1 and UCP3 (Lu et al., 2000), respectively, each with the Ω′ translation enhancer element from TMV (Gallie et al., 1987).
Plant Transformation and Regeneration
Primary leaf explants of NN and nn genotype tobacco were transformed with Agrobacterium tumefaciens strain EHA105 containing pPHP12424. Transformation and regeneration were performed essentially as described by Bidney et al. (1992), except that leaves were simply bisected with a scalpel rather than subjected to microprojectile bombardment before inoculation with Agrobacterium.
Analysis of Salicylates and PR Proteins in Leaf Extracts
Free and conjugated forms of SA and 6-MeSA were extracted and quantified after chemical (base followed by acid) or β-glucosidase hydrolysis (Enyedi et al., 1992). Samples were analyzed with a liquid chromatography system (Waters, Milford, MA). Ten microliters of each sample was injected onto a Waters 5-μm Symmetry Shield RP8 4.6 × 250-mm column that was maintained at 50°C and equilibrated in 90% 35-mM ammonium acetate, pH 4.7, and 10% acetonitrile at a flow rate of 1 mL/min. Ten minutes after injection, a linear gradient was started and the acetonitrile concentration increased to 12% over 7 min. SA eluted at 7.1 min, and 6-MeSA eluted at 13 min. They were quantified with a scanning fluorescence detector (model 474; Waters) using excitation and emission wavelengths of 297 and 412 nm, respectively. The identity of 6-MeSA in tobacco extracts was confirmed by its coelution with the authentic standard and by analysis of the UV light absorption spectrum, as measured with a photodiode array detector (model 996; Waters).
For gas chromatography–mass spectrometry analysis, 100 μL of the hydrolyzed tobacco extracts used for HPLC analysis was dried in limited volume autosampler vials using a SpeedVac (Savant Instruments, Holbrook, NY). The dried residue was resuspended subsequently in 100 μL of bis(trimethylsilyl)trifluroacetamide (Supelco, Bellafonte, PA), and the vials were capped and incubated at 50°C for 15 min. Samples were analyzed using a Hewlett-Packard 5890 gas chromatograph interfaced to a model 5972 mass spectrometer. One-microliter samples were injected (splitless, 250°C inlet) into a 30-m Hewlett-Packard 5MS column with 0.25-mm i.d. and 0.25-μm film thickness. The column conditions were as follows: 170°C for 5 min, with the temperature increased at 5°C/min to 180°C. Finally, the temperature was increased at 20°C/min to 250°C and held for 2 min. The 50 to 550 mass-to-charge ratio range was scanned. The identity of 6-MeSA in tobacco extracts was confirmed by matching the mass spectrum with that of the authentic standard, which coeluted at 7.9 min.
Protein gel blot analysis of the accumulation of PR proteins was as described by Simmons et al. (1998). The analysis of resistance to TMV followed protocols reported previously (Yalpani et al., 1991).
Acknowledgments
The 6msas gene was kindly provided by Prof. Eckhardt Schweizer, and a sample of the 6-MeSA standard was obtained from Prof. Chaitan Khosla (Stanford University, Stanford, CA). Numerous people at Pioneer Hi-Bred International provided critical technical and other support. Among these we particularly thank Kelli Van Waus and Joni Heller for running protein gel blot analysis and Dr. Jan Hazebroek for operating the mass spectrometer. We also are indebted to Alejandra Pascal and her colleagues for expert support with nursing the plants in the greenhouse, and we thank Drs. Jon Duvick and Carl A. Maxwell for providing a critical review of the manuscript.
References
- Beck, J., Ripka, S., Siegner, A., Schiltz, E., and Schweizer, E. (1990). The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum: Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 192, 487–498. [DOI] [PubMed] [Google Scholar]
- Bedford, D.J., Schweizer, E., Hopwood, D.A., and Khosla, C. (1995). Expression of a functional fungal polyketide synthase in the bacterium Streptomyces coelicolor a3(2). J. Bacteriol. 177, 4544–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles, J.M., Garro, R., Fayos, J., Navarro, P., Primo, J., and Conejero, V. (1999). Gentisic acid as a pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Mol. Plant-Microbe Interact. 12, 227–235. [Google Scholar]
- Bidney, D., Scelonge, C., Martich, J., Burrus, M., Sims, L., and Huffman, G. (1992). Microprojectile bombardment of plant tissues increases transformation frequency by Agrobacterium tumefaciens. Plant Mol. Biol. 18, 301–314. [DOI] [PubMed] [Google Scholar]
- Cameron, R.K. (2000). Salicylic acid and its role in plant defense responses: What do we really know? Physiol. Mol. Plant Pathol. 56, 91–93. [Google Scholar]
- Chen, Z.X., Malamy, J., Henning, J., Conrath, U., Sanchezcasas, P., Silva, H., Ricigliano, J., and Klessig, D.F. (1995). Induction, modification, and transduction of the salicylic acid signal in plant defense responses. Proc. Natl. Acad. Sci. USA 92, 4134–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J., Pierrel, M.-A., Atanassova, R., Werck-Reichhart, D., Fritig, B., and Saindrenan, P. (2001). Free and conjugated benzoic acid in tobacco plants and cell cultures. Induced accumulation upon elicitation of defense responses and role as salicylic acid precursors. Plant Physiol. 125, 318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, C., van den Elzen, P., Tamaki, S., Black, M., Dunsmuir, P., and Bedbrook, J. (1987). Molecular characterization of the rbcS multi-gene family of Petunia (Mitchell). Mol. Gen. Genet. 206, 465–474. [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gutrella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Dimroth, P., Walter, H., and Lynen, F. (1970). Biosynthese von 6-Methylsalicylsäure. Eur. J. Biochem. 13, 98–110. [DOI] [PubMed] [Google Scholar]
- Durner, J., Shah, J., and Klessig, D.F. (1997). Salicylic acid and disease resistance in plants. Trends Plant Sci. 2, 266–274. [Google Scholar]
- Edwards, R. (1994). Conjugation and metabolism of salicylic acid in tobacco. J. Plant Physiol. 143, 609–614. [Google Scholar]
- Enyedi, A.J., Yalpani, N., Silverman, P., and Raskin, I. (1992). Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 89, 2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gallie, R., Sleat, D., Watts, J., Turner, P., and Wilson, T. (1987). The 5-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 15, 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, F.O. (1938). Inheritance of resistance to tobacco mosaic virus disease in tobacco. Phytopathology 28, 553–561. [Google Scholar]
- Janda, T., Szalai, G., Tari, I., and Paldi, E. (1999). Hydroponic treatment with salicylic acid decreases the effects of chilling injury in maize (Zea mays L.) plants. Planta 208, 175–180. [Google Scholar]
- Kealey, J.T., Liu, L., Santi, D.V., Betlach, M.C., and Barr, P.J. (1998). Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA 95, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, M., Sanchez-Serrano, J., Schell, J., and Willmitzer, L. (1986). Primary structure of a proteinase inhibitor II gene from potato (Solanum tuberosum). Nucleic Acids Res. 14, 5641–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig, D.F., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot, R.H., Gehring, A.M., Flugel, R.S., Zuber, P., Lacelle, M., Marahiel, M.A., Reid, R., Khosla, C., and Walsh, C.T. (1996). A new enzyme superfamily: The phosphopantetheinyl transferases. Chem. Biol. 3, 923–936. [DOI] [PubMed] [Google Scholar]
- Lee, H.I., and Raskin, I. (1998). Glucosylation of salicylic acid in Nicotiana tabacum cv. Xanthi-nc. Phytopathology 88, 692–697. [DOI] [PubMed] [Google Scholar]
- Lee, H., and Raskin, I. (1999). Purification, cloning, and expression of a pathogen inducible UDP-glucose: Salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 274, 36637–36642. [DOI] [PubMed] [Google Scholar]
- Leôn, J., Yalpani, N., Raskin, I., and Lawton, M.A. (1993). Induction of benzoic acid 2-hydroxylase in virus-inoculated tobacco. Plant Physiol. 103, 323–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leôn, J., Lawton, M.A., and Raskin, I. (1995). Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 108, 1673–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G., Bidney, D., Bao, Z., Hu, X., Wang, J., Vortherms, T., Scelonge, C., Wang, L., Shao, A., Bruce, W., and Duvick, J. (2000). Constitutive promoters and Sclerotinia disease resistance in sunflower. In Proceedings of the 15th International Sunflower Conference, Tolouse, France, K72–77.
- Mauch-Mani, B., and Métraux, J.P. (1998). Salicylic acid and systemic acquired resistance to pathogen attack. Ann. Bot. 82, 535–540. [Google Scholar]
- Monaghan, R.L., and Tkacz, J.S. (1990). Bioactive microbial products: Focus upon mechanism of action. Annu. Rev. Microbiol. 44, 271–301. [DOI] [PubMed] [Google Scholar]
- Morris, K., Mackerness, S.A., Page, T., John, C.F., Murphy, A.M., Carr, J.P., and Buchanan-Wollaston, V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23, 677–685. [DOI] [PubMed] [Google Scholar]
- O'Hagan, D., Rogers, S.V., Duffin, G.R., and Edwards, R.L. (1992). Biosynthesis of the fungal polyketide cubensic acid from Xylaria cubensis. Tetrahedron Lett. 33, 5585–5588. [Google Scholar]
- Pieterse, C.M.J., and van Loon, L.C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Raskin, I. (1992). Role of salicylic acid in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 439–463. [Google Scholar]
- Rate, D.N., Cuenca, J.V., Bowman, G.R., Guttman, D.S., and Greenberg, J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, K., Mofid, M.R., Marahiel, M.A., and Ficner, R. (1999). Crystal structure of the surfactin synthetase-activating enzyme Sfp: A prototype of the 4-phosphopantetheinyl transferase superfamily. EMBO J. 18, 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.R., Nam, K.H., D'Auria, J.C., and Pichersky, E. (1999). S-adenosyl-l-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch. Biochem. Biophys. 367, 9–16. [DOI] [PubMed] [Google Scholar]
- Scott, I.M., Dat, J.F., Lopez-Delgado, H., and Foyer, C.H. (1999). Salicylic acid and hydrogen peroxide in abiotic stress signaling in plants. Phyton Ann. Rei Bot. 39, 13–17. [Google Scholar]
- Serino, L., Reimmann, C., Baur, H., Beyeler, M., Visca, P., and Haas, D. (1995). Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249, 217–228. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Nakajima, H., Rajasekhar, V.K., Dixon, R.A., and Lamb, C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. [Google Scholar]
- Siebert, M., Sommer, S., Li, S.M., Wang, Z.X., Severin, K., and Heide, L. (1996). Genetic engineering of plant secondary metabolism: Accumulation of 4-hydroxybenzoate glucosides as a result of the expression of the bacterial ubiC gene in tobacco. Plant Physiol. 112, 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Métraux, J.P., and Raskin, I. (1995). Salicylic acid in rice: Biosynthesis, conjugation, and possible role. Plant Physiol. 108, 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, C., Hantke, S., Grant, S., Johal, G.S., and Briggs, S.P. (1998). The maize lethal leaf spot 1 mutant has elevated resistance to fungal infection at the leaf epidermis. Mol. Plant-Microbe Interact. 11, 1110–1118. [Google Scholar]
- Spencer, J.B., and Jordan, P.M. (1992). Purification and properties of 6-methylsalicylic acid synthase from Penicillium patulum. Biochem. J. 288, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C., and Van Strien, E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Venkatasubbaiah, P., and Chilton, W.S. (1992). An epoxydon-derived ester from a Phoma sp. pathogenic to rhubarb. J. Nat. Prod. 55, 639–643. [Google Scholar]
- Verberne, M.C., Verpoorte, R., Bol, J.F., Mercado-Blanco, J., and Linthorst, H.J.M. (2000). Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783. [DOI] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Métraux, J.P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N., Silverman, P., Wilson, T.M.A., Kleier, D.A., and Raskin, I. (1991). Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N., Leôn, J., Lawton, M.A., and Raskin, I. (1993). Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani, N., Enyedi, A.J., Leôn, J., and Raskin, I. (1994). Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 193, 372–376. [Google Scholar]