Abstract
FPA is a gene that regulates flowering time in Arabidopsis via a pathway that is independent of daylength (the autonomous pathway). Mutations in FPA result in extremely delayed flowering. FPA was identified by means of positional cloning. The predicted FPA protein contains three RNA recognition motifs in the N-terminal region. FPA is expressed most strongly in developing tissues, similar to the expression of FCA and LUMINIDEPENDENS, two components of the autonomous pathway previously identified. Overexpression of FPA in Arabidopsis causes early flowering in noninductive short days and creates plants that exhibit a more day-neutral flowering behavior.
INTRODUCTION
To accurately gauge when to flower, plants use environmental cues such as daylength and temperature as well as developmental (often called autonomous) controls to coordinate synchronous flower production (Vince-Prue, 1975). As a result of physiological and genetic studies in Arabidopsis, many loci that specifically regulate flowering time have been identified (reviewed in Simpson et al., 1999).
Mutations in genes that promote flowering cause a late-flowering phenotype. Such genes have been grouped into two classes based on details of their mutant phenotypes. Mutations that result in late flowering that is independent of daylength define one group of genes that promotes flowering through inductive long-day photoperiods (Koornneef et al., 1991). In effect, these mutations are “blind” to inductive photoperiods; therefore, late-flowering, photoperiod pathway mutants flower as if they were grown in noninductive, short-day conditions. A second class of mutations results in delayed flowering that is still responsive to daylength (Koornneef et al., 1991). These mutants flower late in long days but even later in short days; therefore, these mutants define genes that promote flowering independently of daylength. Accordingly, these mutations define genes in a pathway often designated as autonomous to indicate that it is distinct from the photoperiod pathway.
The late-flowering phenotype of the autonomous-pathway mutants can be abolished by cold treatment of sufficient duration to promote flowering (i.e., vernalization), whereas mutants in the photoperiod pathway are relatively unaffected by vernalization (Koornneef et al., 1991). A number of genes that promote flowering in both the photoperiod and autonomous pathways have been cloned, including CONSTANS (CO) (Putterill et al., 1995), GIGANTEA (GI) (Fowler et al., 1999), LUMINIDEPENDENS (LD) (Lee et al., 1994a), FCA (Macknight et al., 1997), FT (Kardailsky et al., 1999), FHA/CRY2 (Guo et al., 1998), FLAVIN BINDING, KELCH REPEAT, F BOX (FKF1) (Nelson et al., 2000), SOC1 (Lee et al., 2000; Samach et al., 2000), and FLOWERING LOCUS C (FLC) (Michaels and Amasino, 1999; Sheldon et al., 1999).
The autonomous regulation of flowering time has been the focus of much research due to naturally occurring variation in this pathway. The majority of Arabidopsis accessions may be considered as either spring or winter annuals. Spring annual varieties do not require vernalization to flower rapidly. Without previous vernalization, winter annual varieties flower months later than spring varieties. However, winter varieties flower as rapidly as spring varieties when given a vernalization treatment (Napp-Zinn, 1969).
The late-flowering behavior of winter varieties is due to the presence of dominant alleles of two genes, FRIGIDA (FRI) and FLC (Napp-Zinn, 1979; Burn et al., 1993; Lee et al., 1993, 1994b; Clarke and Dean, 1994; Koornneef et al., 1994). Overexpression of FLC is sufficient to delay flowering in the absence of FRI (Michaels and Amasino, 1999; Sheldon et al., 1999) and FRI-containing plants have no discernible phenotype in the absence of FLC (Michaels and Amasino, 2001). Therefore, it appears that FRI functions solely to upregulate FLC, which in turn delays flowering.
FLC/FRI–containing plants exhibit delayed flowering that is reversed completely by vernalization (Lee and Amasino, 1995), similar to that of the autonomous-pathway mutants fca, fy, fve, ld, fld, and fpa. The phenotypic similarity of FRI-containing plants and autonomous-pathway mutants is a reflection of the antagonistic roles of FRI versus the wild-type function of autonomous-pathway genes in FLC regulation. Whereas FRI acts to increase FLC RNA levels, the autonomous-pathway genes act to decrease FLC. Thus, FLC message levels are increased in autonomous-pathway mutants similar to FRI-containing lines (Michaels and Amasino, 1999; Sheldon et al., 1999). Furthermore, the late-flowering phenotype of mutations in autonomous-pathway genes is abolished by the loss of FLC function, indicating that autonomous-pathway genes, like FRI, act solely to regulate FLC (Michaels and Amasino, 2001).
It is interesting that many genes that both promote and inhibit flowering appear to have evolved to regulate one gene, FLC. To understand the regulation of FLC expression, it is necessary to determine the components of the autonomous pathway that regulate FLC. FPA is one such component, and it was chosen for further investigation.
In this article, we describe the identification of FPA. FPA encodes an RNA recognition domain–containing protein. We show that the expression pattern for FPA is similar to that of other genes identified previously in the autonomous pathway. Additionally, we demonstrate that overexpression of this autonomous-pathway gene causes precocious flowering in short days.
RESULTS
Molecular Identification of FPA
A screen for embryo-defective auxotrophic mutants that were deficient in the synthesis of vitamins and amino acids was performed in the Columbia accession of Arabidopsis (Patton et al., 1998). The arrested embryos from embryo-defective mutants were grown on enriched medium and screened for compounds that would rescue the embryo-defective phenotype; one mutant that could be rescued by treatment with supplemental biotin defined a new biotin biosynthetic locus, BIO2 (Patton et al., 1998). To recover viable homozygous bio2 mutant seed, heterozygotes were treated daily with an external application of supplemental biotin (see Methods), and these biotin-treated heterozygotes yielded progeny that segregated for extremely late flowering plants (Patton et al., 1998). The bio2 lesion was found to be a deletion that removed the BIO2 gene, and the lack of BIO2 provided a polymerase chain reaction (PCR)–based assay to determine the bio2 genotype. In a segregating population of >300 plants from a biotin-rescued heterozygote, all plants heterozygous or wild type at the BIO2 locus flowered similarly to wild-type plants, whereas the bio2 homozygous mutants were late flowering. Thus, the lesion that removed BIO2 is tightly linked genetically to the mutation that caused the late-flowering phenotype.
BIO2 is located near the bottom of chromosome II (Patton et al., 1998). The late-flowering phenotype of bio2 homozygotes rescued with biotin was at first thought to result from loss of several linked genes within the deletion because none of the known flowering loci had been mapped to that chromosomal region (Patton et al., 1998). When subsequent map updates indicated that FPA might be closer to BIO2 than previously thought, complementation tests were performed with fpa to check for allelism. Because bio2 homozygotes are not viable without supplemental biotin, bio2 heterozygotes were crossed to fpa-2 and the progeny were scored for late flowering. Approximately 50% of the F1 progeny displayed wild-type flowering (after producing ∼7 to 10 leaves), whereas the other half of the population flowered after producing 45 to 55 leaves, similar to fpa. This result is consistent with noncomplementation of a heterozygous recessive mutation crossed to a homozygous recessive mutation in the same locus. Thus, the bio2 deletion also appeared to remove the known flowering locus FPA. The fpa allele linked to the bio2 deletion is referred to as fpa-5 in this article. Other mutant alleles (fpa-2 and fpa-3) are described by Meier et al. (2001).
To determine the degree of linkage of FPA to BIO2 and to evaluate the possibility that fpa-5 and bio2 were caused by the same deletion, we developed a screen for recombination between fpa-5 and bio2 based on the embryo-lethal phenotype of bio2 homozygotes without the addition of supplemental biotin. Plants heterozygous for the bio2 lesion were self-pollinated in the absence of biotin, and the subsequent generation was assayed for late flowering. The presence of late-flowering progeny that would not require supplemental biotin would indicate a recombination event between bio2 and fpa. No late-flowering recombinants were found among ∼10,000 plants, indicating that the linkage was extremely close and that FPA probably resided in the same deletion that removed BIO2.
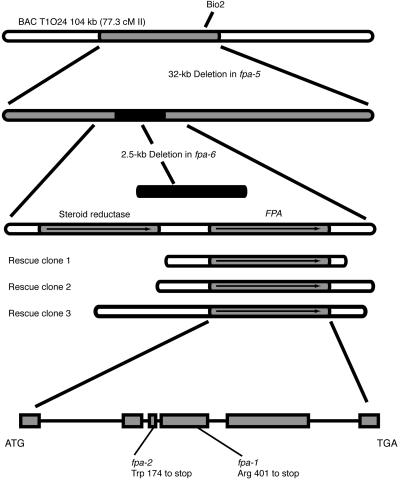
DNA gel blot analyses (see Methods) revealed that the bio2 and fpa-5 lesions were part of a 32-kb deletion of a region predicted to encode nine genes in addition to BIO2 (summarized in Figure 1). An additional allele of fpa (fpa-6) was isolated from a T-DNA–based mutagenesis, but the fpa-6 allele did not cosegregate with the T-DNA; therefore, the lesion in the FPA gene in fpa-6 did not contain a T-DNA insertion. Characterization of the fpa-6 allele revealed that there was a 2.5-kb deletion within the 32-kb region deleted in fpa-5; thus, fpa-6 narrowed the interval containing FPA to two candidate genes. The deletion in fpa-6 removed the 3′ end of a putative steroid reductase and the promoter and 5′ coding region of an RNA recognition motif (RRM)–containing protein (Figure 1). To determine which transcript encoded FPA, the steroid reductase and the RRM-containing protein were sequenced in two ethyl methanesulfonate–induced fpa alleles, fpa-1 and fpa-2. Point mutations were detected in both alleles in the gene that encodes the RRM-containing protein. The point mutation in fpa-1 converts arginine 401 to a stop codon, and the mutation in fpa-2 converts tryptophan 174 to a stop codon (Figure 1). Thus, FPA appears to be encoded by a putative RRM-containing protein.
Figure 1.
Cloning and Characterization of FPA.
Summary of the isolation of FPA based on deletion alleles (top). The deletion found in fpa-5 approximately encompasses nucleotides 24,500 to 57,000 of bacterial artificial chromosome (BAC) T1O24. BAC T1O24 is located 77.3 cM (centimorgan) from the top of chromosome II. Schematic representations of genomic clones that rescued fpa-6 mutants are shown in the middle. The genomic structure of FPA is shown at bottom; boxes designate exons and lines designate introns. Point mutations in fpa-1 and fpa-2 are shown.
Rescue of fpa-6 with Clones of the RRM-Containing Protein
To verify that the RNA recognition motif–containing protein was FPA, the smallest region that rescued the late-flowering phenotype was determined by transforming small clones in the region into fpa-6. A library with an insert size of 9 to 20 kb was created from three overlapping bacterial artificial chromosomes (T1O24, T4I14, and T6P16) that contained the FPA region. Transformation of this library into fpa-6 resulted in several wild-type-flowering T1 plants (data not shown). The inserts derived from the wild-type-flowering T1 transformants were amplified by PCR from genomic DNA using primers that flanked the insert site. The three smallest clones were chosen for sequencing. Nested primers were used to sequence the insert ends to define the boundaries of the clones. Diagnostic digests were performed on the clones to verify that the insert contained the entire region encompassed by the defined end sequence. The only complete gene product represented on the rescuing clones was the protein containing the putative RRM-containing protein (Figure 1). The smallest clone that was found to rescue the mutant contained ∼2.5 kb of 5′ DNA and 1 kb of 3′ DNA of the putative RRM-containing protein (Figure 1).
FPA Contains RRMs
Isolation of an FPA cDNA revealed that the FPA gene is composed of six exons (Figure 1) that encode a 901–amino acid residue protein. The predicted FPA protein contains three RRMs (Birney et al., 1993), which indicates that FPA may function as an RNA binding protein. Alignment of the individual FPA RRMs to the closest matches of RRMs in other proteins shows that the majority of amino acid residues in each RRM of FPA are represented in the same positions of one or more of the RRMs of other proteins (Figure 2). Two small regions known as RNP-2 and RNP-1 consensus sequences define RRMs (Burd and Dreyfuss, 1994) and are shown boxed in Figure 2. The closest relative in the Arabidopsis genome is a predicted protein on chromosome IV that is 38% similar to FPA (CAB78307). The similarity between FPA and CAB78307 is distributed equally throughout the protein, and 15 introduced gaps are required to achieve this level of similarity (data not shown). There is a citrus expressed sequence tag (EST) C24205 that exhibits greater similarity to FPA than does CAB78307, although the EST spans only a portion of the gene.
Figure 2.
Amino Acid Sequence Alignment of FPA RRMs.
Three RRMs of FPA are shown aligned to the most similar RRMs in the databases. Boxed regions show RNP-2 and RNP-1 subdomains of RRM regions. Consensus regions for RNP domains are defined by Burd and Dreyfuss (1994). Black boxes denote amino acid residues identical to those in FPA, and gray boxes denote similar amino acid residues. FPA RRM1 is shown aligned to Saccharomyces cerevisiae Negative Growth Regulatory Protein (S.c. Ngr1p), Rattus norvegicus C-terminal domain binding protein (R.n. rA4), Homo sapiens PTB-associated Splicing Factor (H.s. PSF), Mus musculus octamer binding protein (M.m. NonO), and Drosophila melanogaster No-on-transient A (D.m. NONA). FPA RRM2 is shown aligned to H. sapiens alternative splicing factor (H.s. ASF-2), H. sapiens cold inducible RNA binding protein (H.s. CIRP), H. sapiens cleavage stimulation factor (H.s. CSTF2), M. musculus RNA binding protein (M.m. TIA1), and Zea mays EAR1 protein (Z.m. EAR1). FPA RRM3 is shown aligned to S. cerevisiae U6 snRNP protein (S.c. PRP24), Caenorhabditis elegans hypothetical protein T07F10.3 (C.e. Hyp), D. melanogaster splicing protein snf (D.m. snf), M. musculus snRNP (M.m U2B), and Arabidopsis thaliana snRNP-specific protein (A.t. U1A).
Characterization of fpa Alleles
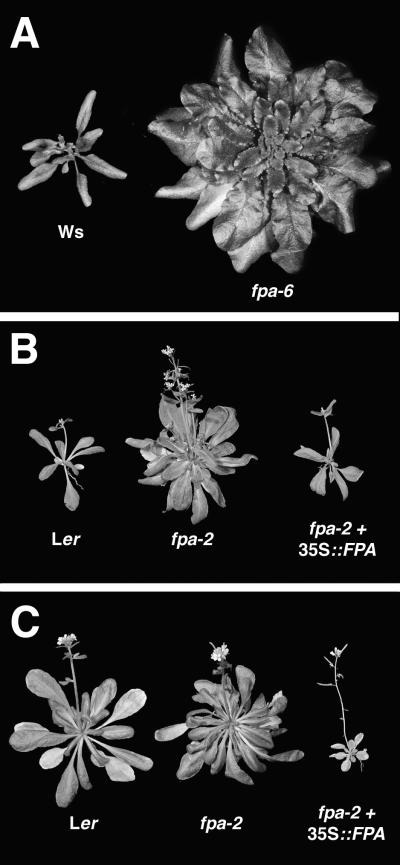
Alleles of fpa display different strengths of inhibition of floral induction. The fpa-6 mutant in Wassilewskija (Ws) displays the strongest delay of flowering (Figure 3). The fpa-6 deletion allele removes the entire 5′ regulatory region of FPA and the first exon of the coding region and is likely to be in a null allele. The two alleles in the Landsberg erecta (Ler) accession (fpa-1 and fpa-2) are not as late flowering as fpa-6 in Ws (flowering behavior is presented in Tables 1 and 2), because the Ler FLC allele is a partial suppressor of fpa mutants (Sanda and Amasino, 1996). Thus, a direct comparison of the fpa alleles cannot be made. However, fpa-2 is clearly later flowering than fpa-1, indicating that fpa-1 may not represent a null allele.
Figure 3.
Flowering Phenotypes of Lines Used in this Study.
(A) Ws and fpa-6 in Ws grown under long-day photoperiods.
(B) Ler, fpa-2 in Ler, and fpa-2 + 35S::FPA in Ler grown under long-day photoperiods.
(C) Ler, fpa-2 in Ler, and fpa-2 + 35S::FPA in Ler grown under short-day photoperiods.
Table 1.
Rosette Leaf Number at Flowering of fpa-6 and FPA Cosuppression Lines
| Genetic Background | Leaf Number at Flowering in Long Daysa |
|---|---|
| Ws | 7.5 ± 0.5b |
| Ws fpa-6 | 46.1 ± 2.0 |
| Ws FPA cosuppression line 1 | 43.3 ± 3.6 |
| Ws FPA cosuppression line 2 | 42.4 ± 4.1 |
a Leaf number was determined using only rosette leaves; cauline leaves were not counted.
Ten plants were measured to determine flowering time data. Error is displayed as 1 sd.
Table 2.
Rosette Leaf Number at Flowering of fpa Alleles and Overexpression Lines of FPA
| Genetic Background | Leaf Number at Flowering in Long Daysa | Leaf Number at Flowering in Short Daysa |
|---|---|---|
| Ler | 7.2 ± 0.6b | 23.9 ± 1.3 |
| Ler fpa-1 | 14.7 ± 1.0 | Not determined |
| Ler fpa-2 | 17.8 ± 1.4 | 49.2 ± 2.2 |
| Ler fpa-2 + 35S::FPA line 1 | 8.0 ± 0.7 | 10.3 ± 1.5 |
| Ler fpa-2 + 35S::FPA line 2 | 8.1 ± 0.6 | 10.5 ± 1.4 |
| Ler fpa-2 + 35S::FPA line 3 | 8.3 ± 1.1 | 10.2 ± 1.1 |
a Leaf number was determined using only rosette leaves; cauline leaves were not counted.
Ten plants were measured to determine flowering time data. Error is displayed as 1 sd.
Overexpression of FPA Results in Early-Flowering in Short Days
To determine the effect of increased FPA activity, we designed a construct for overexpression in which the genomic coding region (start to stop codon) was joined with the constitutive cauliflower mosaic virus 35S promoter and the nopaline synthase terminator region. Plants transformed with this construct, however, displayed one of two phenotypes, either later or earlier flowering than wild-type plants.
The frequency with which the late-flowering class arose depended on the accession that was transformed. In the first transformed generation (T1), late-flowering phenotypes were seen at the highest frequencies in accession Columbia. For example, in one experiment, 82% of the plants (59 of 72) in the T1 generation in accession Columbia were >10 leaves later flowering than the wild type. In Ws, <10% of the T1 population was more than five leaves later flowering than wild-type plants. However, the severity of the late-flowering phenotype in Ws transformants increased in the next generation; the progeny of many of these slightly late flowering T1 plants became severely late flowering and produced at least 30 more leaves than wild-type Ws (data from representative lines are shown in Table 1). Furthermore, this more severe late-flowering phenotype segregated in a fully dominant manner (3:1 late:early flowering), and late flowering cosegregated with the transgene (data not shown).
Thus, many lines transformed with a construct designed for overexpression yielded late-flowering plants that were phenotypically similar to fpa mutants, and the late-flowering phenotype was dominant and cosegregated with the transgene. Furthermore, the late-flowering phenotype displayed from sense expression constructs was suppressed by vernalization, similar to that of fpa mutants (data not shown). The existence of dominant mutants that contain a construct designed to increase mRNA levels but that exhibit a phenotype similar to loss-of-function mutants is likely to be a result of gene silencing (Fire, 1999).
To identify transgenic lines in which the expression of this construct resulted in increased FPA activity, we introduced the overexpression construct into fpa-2 mutants, which are in the Ler background. Transgenic lines in which the late-flowering phenotype of fpa-2 was rescued were identified (Table 2 and Figure 3). From 144 primary transformants, 55 of the plants displayed wild-type flowering when grown under long-day photoperiod conditions. Three representative lines were chosen for further characterization. When these 35S::FPA plants were grown under short-day conditions, they flowered much more rapidly than the wild type (Table 2 and Figure 3). Thus, FPA overexpression results in plants that flower after producing a similar number of leaves in either inductive or noninductive photoperiods.
FPA Expression Pattern Is Similar to That of FCA and LD and Is Expressed in Nonflowering Plants
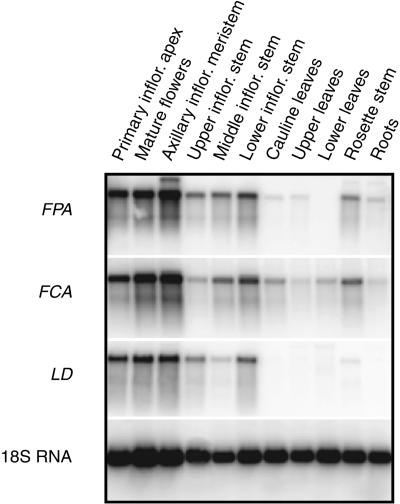
Because the FPA RNA could not be detected by RNA gel blotting, the spatial localization of FPA RNA was determined by reverse transcription–PCR of RNA samples isolated from various plant tissues (Figure 4). The expression of FPA was compared with that of FCA and LD, two other genes in the autonomous pathway. The expression patterns for the three genes are remarkably similar. FPA expression can be detected in most tissues, but it is found at the highest levels in the flower stem and meristematic regions of the plant.
Figure 4.
Reverse Transcription–PCR Analysis of FPA, FCA, and LD Expression Patterns.
FPA, FCA, and LD expression patterns were determined by reverse transcription–PCR. Twenty-five cycles of PCR amplification were used for each sample except the control 18S RNA product, which was detected at 17 cycles. Tissue for these experiments was obtained from the Ws accession at 22 days after germination. The inflorescence height at the time of tissue harvest was between 16 and 19 cm. The primary inflorescence (inflor.) apex consisted of the uppermost 3 mm of the inflorescence and contained immature floral buds. Flowers were taken on the day of anthesis. Axillary inflorescence meristem tissue was isolated with 2 mm of stem on either side of the meristem. Upper, middle, and lower inflorescence stem are defined as internode stem sections of the primary inflorescence. The lower stem refers to the first internode from the rosette followed by middle stem and upper stem, respectively. Stem sections did not contain shoot meristems. Cauline leaves are the oldest two leaves of the inflorescence. Upper leaves are defined as the youngest two leaves of the rosette, whereas the lower leaves are the oldest two rosette leaves. Rosette stem was obtained by removing roots, leaves, and the inflorescence from the rosette. Root tissue was isolated from plants grown on sterile medium (see Methods).
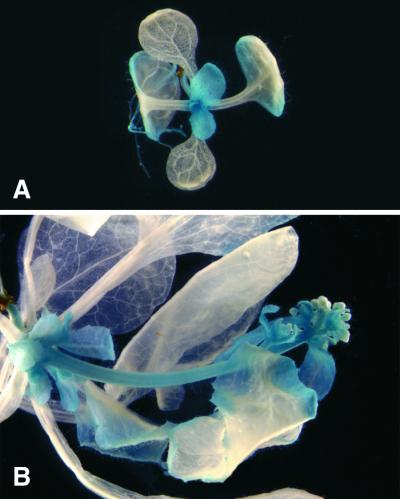
To further explore the spatial and temporal expression pattern of FPA, we fused a 1.8-kb fragment of genomic sequence 5′ to the start codon of FPA to the β-glucuronidase (GUS) reporter gene and determined the staining pattern of plants containing this construct (Figure 5). FPA expression based on GUS activity is similar to the reverse transcription–based PCR data except for floral buds that appear to possess weak GUS activity. The high FPA expression seen in flowers in the reverse transcription–based PCR DNA gel blot may be due to the expression of FPA in the flower peduncle, a tissue that was included in the samples for reverse transcription–PCR. Overall, FPA is expressed throughout the plant life cycle, indicating that activation of FPA expression is not a binary switch that controls floral induction; rather, FPA may be available to constantly promote flowering during development. In addition, FPA is most highly expressed in young and actively growing tissues, and as the tissue matures the expression decreases. For example, the young third and fourth leaves in the 13-day postgermination plant possess GUS activity, whereas the third and fourth leaves in the more mature 19-day postgermination plant do not possess any detectable GUS activity (Figure 5).
Figure 5.
FPA::GUS Fusion Analysis of FPA Expression
(A) FPA::GUS expression in Ws before flowering, 13 days after germination.
(B) FPA::GUS expression in Ws during flowering (20-mm inflorescence), 19 days after germination.
DISCUSSION
Genetic and physiological analysis of late-flowering mutants in Arabidopsis has revealed two phenotypic classes. One class does not respond to daylength differences and vernalization treatments (photoperiod-pathway mutants), whereas the second group (autonomous-pathway mutants) responds to these environmental cues. Therefore, the autonomous pathway is thought to represent a developmental flowering program. In this study, we report the molecular identification and characterization of the autonomous-pathway component FPA.
FPA is predicted to be a 901–amino acid residue protein that contains three N-terminal RRMs. RRMs contain a degenerate consensus sequence in which no single position is conserved completely; rather, RRMs are composed of a similar framework of uncharged and hydrophobic amino acid residues. In addition to amino acid sequence variability, RRMs also are variable in length (Birney et al., 1993). The crystal structure of the RNA binding protein Sex-Lethal from Drosophila demonstrates that RRMs interact directly with single-stranded RNA and therefore are likely to mediate RNA binding specificity (Handa et al., 1999). Thus, this sequence and length variability may confer unique RNA sequence binding specificities.
FPA may function similarly to other proteins that contain RRMs. RNA binding proteins are involved in various aspects of RNA metabolism such as alternate splicing, stabilizing and destabilizing mRNAs, and regulation of translation initiation. However, the primary sequence of FPA does not provide any clues as to the specific aspect of RNA metabolism that FPA might affect. Outside the RRM regions, FPA does not exhibit significant similarity with any proteins of known function. FCA is another RNA binding protein in the autonomous pathway (Macknight et al., 1997). However, there is only weak similarity between FPA and FCA; in fact, the RRM regions of several RNA binding proteins from animals are more similar to FPA than are the RRMs of FCA. Also, outside of the RRMs, there is no obvious similarity between FPA and FCA.
Attempts to ascribe a function to FPA may be aided by previous work that has shown that FLC is the predominant negative regulator of the autonomous-flowering pathway. FLC appears to be the convergence point of the inhibition of flowering that is mediated by dominant alleles of the FRI locus and the promotion of flowering by the autonomous-pathway genes FVE, LD, FCA, and FPA (Michaels and Amasino, 1999; Sheldon et al., 1999). The FRI-mediated inhibition of flowering is due to increased levels of FLC mRNA. The converse is true of autonomous-pathway genes; these genes promote flowering by acting to decrease the levels of FLC mRNA. Therefore, FLC message levels act as an internal rheostat for flowering time that is positively regulated by FRI and negatively regulated by autonomous-pathway genes, such as FPA (Michaels and Amasino, 2001).
There are many mechanisms by which an RNA binding protein such as FPA might decrease FLC mRNA levels. For example, FPA may interact directly with FLC mRNA to affect splicing or stability. Alternatively, FPA may regulate other autonomous-pathway genes that in turn regulate FLC gene expression. FPA is the second example of an RRM-containing RNA binding protein in the autonomous pathway. FCA is the other RRM-containing RNA binding protein (Macknight et al., 1997), and LD encodes a homeobox-containing protein that might function as an RNA binding protein (Aukerman et al., 1999). Like FPA, both FCA and LD negatively regulate FLC message levels. Because of the preponderance of RNA binding proteins in the autonomous pathway, it is tempting to speculate that the autonomous pathway regulates FLC message levels post-transcriptionally.
It is possible that FRI may interact directly with the autonomous pathway. The late-flowering phenotype of plants containing dominant alleles of FRI or autonomous-pathway mutations are identical; in both cases, FLC mRNA levels are high and vernalization causes early flowering and a decrease of FLC mRNA levels (Michaels and Amasino, 2000). Because FRI is epistatic to the autonomous pathway (i.e., FRI causes late flowering in a background that is wild type for autonomous-pathway genes such as FPA), FRI could function by blocking the ability of FPA or other autonomous-pathway genes to downregulate FLC mRNA levels.
Although FPA clearly is a negative regulator of FLC, the fact that overexpression of FPA in short days results in early flowering indicates that FPA may have other functions. Null mutants of FLC are only slightly earlier flowering than wild-type plants in short days (Michaels and Amasino, 2001). Overexpression of FPA results in much earlier flowering than does loss of FLC gene function. Thus, one possibility is that FPA is a negative regulator of FLC and other flowering repressors and that FPA overexpression negatively regulates a collection of flowering repressors. It is also possible that 35S::FPA may interact positively with the photoperiod pathway, or with genes downstream of the photoperiod pathway, to produce early-flowering plants in short days. Overexpression of either FT or CO, components of the photoperiod pathway, produce essentially day-neutral plants that flower as early in short days as they do in long days (Kardailsky et al., 1999; Samach et al., 2000). Additionally, overexpression of SOC1, a MADS-box transcription factor that acts downstream of CO, also results in day-neutral early flowering (Lee et al., 2000; Samach et al., 2000). Because FPA overexpression lines flower with a similar number of leaves regardless of photoperiod conditions, it is possible that FPA may interact with similar downstream targets of photoperiod pathway genes. It must be noted that ectopic overexpression of FPA may affect genes that are not normally affected by the wild-type FPA locus.
Recent genetic analyses also indicate that FPA may have a broad role in plant development and flowering time regulation. Analysis of all possible double mutant combinations of the late-flowering mutants revealed differences between fpa and other autonomous-pathway mutants (Koornneef et al., 1998). Most notably, mutants in fpa displayed the most extreme late-flowering phenotype of the autonomous-pathway mutants when combined with the photoperiod-pathway mutants fe and ft, indicating that FPA may promote flowering through multiple pathways. Additionally, the double mutant between autonomous-pathway mutants fpa-1 and fy was not attainable, presumably because of early lethality, indicating that FPA and FY are vital, but redundant, for a function in addition to flowering (Koornneef et al., 1998). Further investigation will be required to reveal the molecular basis of the role of FPA in flowering time regulation and possibly in other aspects of plant development.
METHODS
Biotin-Mediated Rescue of the Embryo-Defective Phenotype of bio2 Homozygotes
Arabidopsis thaliana plants heterozygous for the bio2 lesion were sprayed twice daily with a 1-mM aqueous biotin (Sigma, St. Louis, MO) solution until seed had matured. With daily biotin supplementation, homozygous bio2 mutants formed ∼65 leaves and then initiated flowering; however, even with continued biotin supplementation, bio2 mutants did not produce viable seed.
Determination of Deletion Size of Lesion in bio2
Bacterial artificial chromosome (BAC) T1O24 was used a probe for hybridization to genomic DNA isolated from the bio2 homozygous mutant to determine the size of the deletion. Briefly, bio2 and wild-type genomic DNA were digested with EcoRI and run on a 1% agarose gel, transferred to a nylon membrane, and probed with labeled T1O24 DNA. The radioactivity was detected using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA), and the bio2 lane was compared with the wild-type lane for the absence of bands (data not shown).
Generation of Rescuing Clone Library
BACs T1O24, T4I14, and T6P16 were partially digested with Sau3AI and ligated into the BamHI site of 3300s. A total of 3000 clones containing inserts ranging in size from 6 to 20 kb were isolated and transformed into the Agrobacterium tumefaciens strain ABI. fpa-6 mutant plants were then transformed with the entire library. Early-flowering plants were identified among the first-generation transformants. The inserts were amplified from the transformants with Takara LA Taq polymerase (Panvera, Madison, WI). Polymerase chain reaction (PCR) conditions were an initial denaturation of 3 min at 95°C, followed by 35 cycles of 20 sec at 98°C and 15 min at 68°C and then 10 min at 72°C (final extension). The primers used for the amplification step were 5′-ttgcatgcctgcaggtcgac-3′ and 5′-tacgaa-ttcgagctcggtac-3′. Sequencing primers were 5′-tgcttccggctcgtatgttg-3′ and 5′-tatattactaattaattggggacc-3′.
Molecular Markers for Genotypic Analysis of fpa-1 and fpa-2 Alleles
Cleaved amplified polymorphic sequences were designed for the alleles of fpa. fpa-1 can be identified using primers 5′-cacaaggtacga-ggcgccctatga-3′ and 5′-ccactgatccatctcttcctggaa-3′. These primers result in a 100-bp fragment that is cleaved with AclI (New England Biolabs, Beverly, MA) to yield 76- and 24-nucleotide fragments in the wild type but is not cleaved in fpa-1. fpa-2 can be identified using 5′-ttgtgttatcttcaggaacac-3′ and 5′-ctagtaacaagagacatactt-3′. These primers give a 100-bp PCR product that in fpa-2 cleaves at 22 bp with BfaI (New England Biolabs) but that is not cleaved in the wild type.
Generation of Overexpression Construct of FPA
The overexpression construct of FPA was created by PCR amplification of FPA from the start codon to the stop codon from genomic DNA. The primers used to generate overexpression of FPA were 5′-aaaggatccatggcgttatctatgaagccattcagagcc-3′ and 5′-aaagagctctca-aggcccctgtccagccggagtacc-3′ (restriction sites shown in boldface, sequence corresponding to FPA is underlined), which generated a 4593-bp FPA fragment that was digested with BamHI and SacI and ligated into the BamHI and SacI sites of pRAM1. pRAM1 was produced by ligating the HindIII-EcoRI fragment of pBI121 containing the 35S promoter and nopaline synthase terminator sequences into the HindIII and EcoRI sites of pPZP211 (Hajdukiewicz et al., 1994). The construct was transferred into the Agrobacterium strain ABI and transformed into Arabidopsis through floral dipping (Clough and Bent, 1998).
mRNA Detection by Reverse Transcription–Mediated PCR
RNA was prepared from tissue frozen in liquid nitrogen. RNA was isolated with TRI reagent (Sigma) according to instructions for subsequent reverse transcription reactions. Five micrograms of total RNA was annealed to 500 ng of random decamer oligonucleotides. Superscript II reverse transcriptase (Gibco Life Technologies, Gaithersburg, MD) was used to generate cDNA. Takara ExTaq (Panvera) was used for PCR amplification of cDNA for desired products. PCR cycle conditions were as follows: 26 cycles of 15 sec at 95°C, 30 sec at 55°C, and 2 min at 72°C. Primers were used to detect full-length message of LD, FPA, and FCA-γ. Primers used for PCR amplification of FPA were 5′-atggcgttatctatgaagccattcagagcc-3′ and 5′-tcaagg-cccctgtccagccggagtacc-3′; primers used for LD were 5′-ctcatgtac-tggctattcccttgg-3′ and 5′-tcgatcagctccaagatgtcgtcg-3′; primers used for FCA were 5′-atgaatggtcccccagatagagtag-3′ and 5′-tcatcaagcttt-attcttccacatgagttc-3′; primers used for 18S rRNA were obtained from Ambion (Austin, TX; catalog number 1718). Amplified DNA was transferred to nylon membranes (Gelman, Ann Arbor, MI) using standard protocols (Sambrook et al., 1989). DNA probes for hybridization were labeled with 32P using the Prime-a-Gene labeling system, as described by the manufacturer (Promega, Madison, WI).
Histochemical β-Glucuronidase Assays
Whole plants, grown under long-day photoperiods, were fixed in 90% acetone for 1 hr at 4°C and were washed three times for 30 min in 50 mM NaPO4, pH 7.0. Staining for β-glucuronidase (GUS) activity was performed by incubating plants in 50 mM NaPO4 with 0.5% Triton X-100, 0.5 mM X-gluc (Research Organics Inc., Cleveland, OH), 0.5 mM K3(Fe[CN]6), and 0.5 mM K4(Fe[CN]6) for 12 hr. The plants were cleared with three 12-hr washes of 70% ethanol. The construct to determine the GUS expression pattern was generated by PCR amplification of the FPA 5′ regulatory region using the primers 5′-aaactgcagtgagaagtctgatgacacaatcattcaatc-3′ and 5′-aaaggatccccc-atcgggattgtttcaattgacgatcctatgg-3′; the boldface type indicates restriction sites used for cloning. The resulting amplification product was digested with PstI and BamHI and ligated to a PstI-BamHI–digested pPZPGUS vector (HindIII to EcoRI fragment of pBI101.2 in pPZP211).
Plant Growth Conditions
Plants were grown under fluorescent light (100 μE m−2 sec−1; cool-white Sylvania [Danvers, MA]) at 24 ± 1°C. Plants were fertilized with 2 g/L Dyna-Grow 7-9-5 fertilizer (Dyna-Grow Corp., San Pablo, CA) at 2-week intervals. Daylengths were 8 hr of light and 16 hr of dark for short-day and 16 hr of light and 8 hr of dark for long-day conditions. Plants grown in sterile media for the isolation of root tissue were grown on 5.5% Agar plates containing 2 g/L Dyna-Grow 7-9-5 fertilizer and 0.5 g MES (Sigma) and pH adjusted to 5.7. Light conditions for plants grown on sterile media were the same as those used for soil-grown plants.
Acknowledgments
We thank Ann Hu (Syngenta) for sequencing of the fpa-1 allele. This research was supported by the College of Agricultural and Life Sciences of the University of Wisconsin and by grants to R.M.A. from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the National Science Foundation.
References
- Aukerman, M.J., Lee, I., Weigel, D., and Amasino, R.M. (1999). The Arabidopsis flowering-time gene LUMINIDEPENDENS is expressed primarily in regions of cell proliferation and encodes a nuclear protein that regulates LEAFY expression. Plant J. 18, 195–203. [DOI] [PubMed] [Google Scholar]
- Birney, E., Kumar, S., and Krainer, A.R. (1993). Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., and Dreyfuss, G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Burn, J.E., Smyth, D.R., Peacock, W.J., and Dennis, E.S. (1993). Genes conferring late flowering in Arabidopsis thaliana. Genetica 90, 147–155. [Google Scholar]
- Clarke, J.H., and Dean, C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Sam, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Handa, N., Nureki, O., Kurimoto, K., Kim, I., Sakamoto, H., Shimura, Y., Muto, Y., and Yokoyama, S. (1999). Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Blankestijn-de Vries, H., Hanhart, C., Soppe, W., and Peeters, T. (1994). The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 6, 911–919. [Google Scholar]
- Koornneef, M., Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C.J., and Peeters, A.J. (1998). Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., and Amasino, R.M. (1995). Effect of vernalization, photoperiod and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Bleecker, A., and Amasino, R. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Lee, I., Aukerman, M.J., Gore, S.L., Lohman, K.N., Michaels, S.D., Weaver, L.M., John, M.C., Feldmann, K.A., and Amasino, R.M. (1994. a). Isolation of LUMINIDEPENDENS—A gene involved in the control of flowering time in Arabidopsis. Plant Cell 6, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Michaels, S.D., Masshardt, A.S., and Amasino, R.M. (1994. b). The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6, 903–909. [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, L., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89, 737–745. [DOI] [PubMed] [Google Scholar]
- Meier, C., Bouquin, T., Eggert, M.E., Nielsen, M.E., Raventos, D., Mattsson, O., Rocher, A., Schomburg, F., Amasino, R.A., and Mundy, J. (2001). Gibberellin response mutants identified by luciferase imaging. Plant J. 25, 509–519. [DOI] [PubMed] [Google Scholar]
- Michaels, S., and Amasino, R. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S., and Amasino, R. (2000). Memories of winter: Vernalization and the competence to flower. Plant Cell Environ. 23, 1145–1154. [Google Scholar]
- Michaels, S., and Amasino, R. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous-pathway mutations, but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn, K. (1969). Arabidopsis thaliana (L.) Heynh. In The Induction of Flowering: Some Case Histories, L.T. Evans, ed (Melbourne, Australia: MacMillan), pp. 291–304.
- Napp-Zinn, K. (1979). On the genetical basis of vernalization requirement in Arabidopsis thaliana (L.) Heynh. In La Physiologie de la Floraison, P. Champagnat and R. Jaques, eds (Paris: Colloques Internationaux du Centre National de la Recherche Scientifique), pp. 217–220.
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Patton, D.A., Schetter, A.L., Franzmann, L.H., Nelson, K., Ward, E.R., and Meinke, D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanda, S.L., and Amasino, R.M. (1996). Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet. 251, 69–74. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene. A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., Gendall, T., and Dean, C. (1999). When to switch to flowering. Annu. Rev. Cell Dev. Biol. 15, 519–550. [DOI] [PubMed] [Google Scholar]
- Vince-Prue, D. (1975). Photoperiodism in Plants (London: McGraw-Hill).