Abstract
The SOS3 (for SALT OVERLY SENSITIVE3) calcium binding protein and SOS2 protein kinase are required for sodium and potassium ion homeostasis and salt tolerance in Arabidopsis. We have shown previously that SOS3 interacts with and activates the SOS2 protein kinase. We report here the identification of a SOS3 binding motif in SOS2 that also serves as the kinase autoinhibitory domain. Yeast two-hybrid assays as well as in vitro binding assays revealed a 21–amino acid motif in the regulatory domain of SOS2 that is necessary and sufficient for interaction with SOS3. Database searches revealed a large family of SOS2-like protein kinases containing such a SOS3 binding motif. Using a yeast two-hybrid system, we show that these SOS2-like kinases interact with members of the SOS3 family of calcium binding proteins. Two-hybrid assays also revealed interaction between the N-terminal kinase domain and the C-terminal regulatory domain within SOS2, suggesting that the regulatory domain may inhibit kinase activity by blocking substrate access to the catalytic site. Removal of the regulatory domain of SOS2, including the SOS3 binding motif, resulted in constitutive activation of the protein kinase, indicating that the SOS3 binding motif can serve as a kinase autoinhibitory domain. Constitutively active SOS2 that is SOS3 independent also was produced by changing Thr168 to Asp in the activation loop of the SOS2 kinase domain. Combining the Thr168-to-Asp mutation with the autoinhibitory domain deletion created a superactive SOS2 kinase. These results provide insights into regulation of the kinase activities of SOS2 and the SOS2 family of protein kinases.
INTRODUCTION
Many extracellular stimuli, including hormones and such environmental signals as abscisic acid, gravity, light, salinity, drought, cold, oxidative stress, anoxia, and mechanical perturbation, cause changes in cytosolic free Ca2+ concentration (Poovaiah and Reddy, 1993; Bush, 1995; Trewavas and Malho, 1998; Knight, 2000). In addition, changes in cytosolic calcium also occur during the tip growth of root hairs and pollen tubes (Pierson et al., 1996). Cytosolic calcium changes could be brought about by release from internal stores and/or entry from outside the cell (Gilroy et al., 1993; Knight, 2000). The change can vary in magnitude, duration, frequency, and spatial arrangement within the cell, depending on the nature of the stimulus. These variables may contribute to the specificity of cytosolic calcium signals. In the cytosol, calcium functions as a second messenger, coupling the stimulus to cellular responses. The targets of second-messenger calcium in the cytosol are calcium-modulated proteins (Roberts and Harmon, 1992; Zielinski, 1998).
The superfamily of EF-hand helix-loop-helix calcium binding proteins represents the largest group of calcium-modulated proteins (Moncrief et al., 1990). More than 250 such proteins belonging to nearly 40 subfamilies have been found in various eukaryotic cells (Celio et al., 1996). Some of these proteins function in the buffering of cytosolic calcium concentration (Cox, 1990). Most, however, are presumed to function in the mediation of calcium signals in response to extracellular stimuli (Cohen and Klee, 1988; Cox, 1990). Upon binding to calcium, these proteins undergo conformational changes that subsequently regulate the activities of target proteins (Cohen and Klee, 1988; Cox, 1990; Roberts and Harmon, 1992; Celio et al., 1996; Zielinski, 1998). Various calcium binding proteins have been shown or suggested to be involved in nearly every aspect of cellular function, including cell division, expansion, and metabolism (Cohen and Klee, 1988; Cox, 1990; Celio et al., 1996). Despite the apparent importance of EF-hand calcium binding proteins, no specific function has been established for the majority of them (Celio et al., 1996). Elucidation of their function would require genetic studies of loss-of-function mutations and cell biological identification of their direct and indirect target proteins.
Calcium signaling often is coupled with protein phosphorylation. Phosphorylation by protein kinases is one of the most common and important regulatory mechanisms in signal transduction (Hardie, 1999). In plants, the calcium-dependent, calmodulin-independent protein kinases are modulated directly by calcium via their intrinsic calmodulin-like domain (Harper et al., 1991; Roberts and Harmon, 1992). The coupling between calcium and protein phosphorylation also can be indirect. For example, in calmodulin kinases, calcium binds to calmodulin, which in turn interacts with and activates the kinase catalytic subunit (Hardie, 1999). In the protein phosphatase type 2B (calcineurin), calcium controls the activity of the protein phosphatase catalytic subunit calcineurin A through two EF-hand calcium binding proteins, calmodulin and calcineurin B (Klee et al., 1988).
In Arabidopsis, the SOS2 (for SALT OVERLY SENSITIVE2) and SOS3 genes are required for potassium and sodium ion homeostasis and plant salt tolerance (Liu and Zhu, 1997; Zhu et al., 1998). SOS3 encodes a myristoylated EF-hand calcium binding protein (Liu and Zhu, 1998; Ishitani et al., 2000) that may sense the calcium signal elicited by salt stress (Lynch et al., 1989; Knight et al., 1997). SOS2 encodes a serine/threonine protein kinase with an N-terminal kinase domain similar to that of SNF1/AMPK (Hardie et al., 1998) and a novel C-terminal domain that is presumably regulatory (Liu et al., 2000). Recently, we found that SOS3 interacts physically with SOS2 in the yeast two-hybrid system as well as in vitro (Halfter et al., 2000). In the presence of calcium, SOS3 activates SOS2 protein kinase activity (Halfter et al., 2000). The interaction between SOS3 and SOS2 also is supported by sos2 sos3 double mutant analysis, which indicates that the two genes function in the same pathway (Halfter et al., 2000). Salt stress upregulation of the SOS1 gene encoding a Na+/H+ antiporter is partially under the control of the SOS3-SOS2 regulatory pathway (Shi et al., 2000).
In this article, we further characterize the interaction between SOS2 and SOS3 and identify SOS2 domains involved in kinase activation and autoinhibition. We show that a 21–amino acid motif in the regulatory domain of SOS2 is necessary and sufficient for the interaction with SOS3. We extend these protein interaction studies to a family of SOS2-like protein kinases and the SOS3 family of calcium binding proteins. We also found interaction between the N-terminal kinase domain and the C-terminal regulatory domain within the SOS2 protein. A constitutively active SOS2 kinase could be generated either by deletion of the C-terminal portion, including the SOS3 binding motif, or by a Thr168-to-Asp change in the activation loop of the SOS2 kinase domain. When the two constitutively active mutations were combined, a superactive SOS2 was created. Our results provide an important structural basis for understanding the regulation of the kinase activities of SOS2 and SOS2-like protein kinases.
RESULTS
Identification of an SOS3 Binding Motif in SOS2
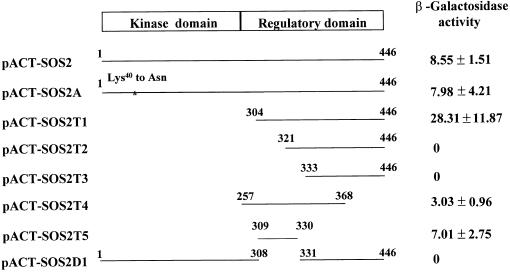
To identify amino acid residues in SOS2 that might be necessary for interaction with SOS3, we cloned various parts of SOS2 into the prey vector pACT2 and then tested for interaction with the bait pAS-SOS3 in the yeast two-hybrid system. Figure 1 shows that full-length SOS2 interacted strongly with SOS3 and that the Lys40-to-Asn mutation, which abolishes SOS2 kinase activity (Halfter et al., 2000), had little effect on this interaction. SOS2T1, which contains only the regulatory domain of SOS2, interacted more strongly with SOS3 (Figure 1). These results confirm our previous observation that the regulatory domain of SOS2 mediates its interaction with SOS3 (Halfter et al., 2000). The significantly stronger interaction exhibited by SOS2T1 is interesting and suggests that the kinase domain interferes with SOS3 binding, perhaps by blocking access to the regulatory domain.
Figure 1.
Deletion Analysis of SOS2 to Identify an SOS3 Binding Motif.
Depicted are yeast two-hybrid constructs of SOS2 kinase with various deletions that were tested in combination with bait pAS-SOS3 in yeast for LacZ activation and quantified as β-galactosidase activity (n = 3). The C-terminal regulatory domain has been shown previously to be sufficient for binding to SOS3, whereas the N-terminal kinase catalytic domain is not required (Halfter et al., 2000). N-terminal deletion of 320 or 332 amino acids results in the loss of β-galactosidase activity, whereas the N-terminal deletion of 303 amino acids leads to very strong interaction. Amino acid residues from Met309 to Arg330 are sufficient and necessary to activate the LacZ reporter and are at a level comparable to that of the intact kinase protein.
Serial deletions of the SOS2 regulatory domain were made and then tested for interaction with SOS3. The results show that residues between Asp304 and Tyr321 are required for SOS2 interaction with SOS3 (Figure 1). SOS2T4, which spans from Gly257 to Arg368, was found to be sufficient to interact with SOS3. We further delimited the region sufficient for SOS3 interaction to a stretch of 21 amino acids between Met309 and Arg330 of SOS2 (Figure 1). SOS2D1, in which this 21–amino acid motif was deleted, failed to interact with SOS3 at all (Figure 1). Therefore, we conclude that this 21–amino acid motif is necessary and sufficient for interaction with SOS3. This SOS3 binding sequence is designated the FISL motif for its conserved amino acid residues (see below) and for convenience.
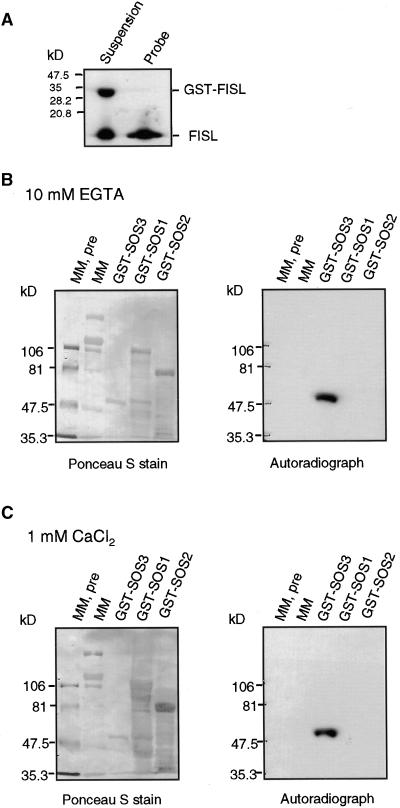
The FISL Motif Binds to SOS3 in Vitro
To determine whether the 21–amino acid FISL motif defined in the yeast two-hybrid system is sufficient to bind SOS3 in vitro, we expressed the peptide in Escherichia coli as a glutathione S-transferase (GST) fusion protein. The GST-FISL fusion protein was labeled with phosphorus-32, and then the GST portion was removed by protease cleavage (Figure 2A). The labeled FISL peptide was overlaid onto a protein blot of SOS3 and control proteins. Figures 2B and 2C show that FISL was able to bind to GST-SOS3. This binding is not dependent on Ca2+, because similar results were obtained when the reaction was depleted of Ca2+ (Figure 2B) or when Ca2+ was added (Figure 2C). In contrast, FISL did not bind to the control protein GST, GST-SOS2, or molecular mass markers. We also found that FISL was not capable of binding to the cytosolic tail of the sodium/proton antiporter SOS1 (Shi et al., 2000), which was expressed as a GST fusion protein. These results show that FISL binds specifically to SOS3.
Figure 2.
The FISL Motif Binds to SOS3 in Vitro in Gel Blot Overlay Assays.
(A) The 21–amino acid FISL motif identified in the yeast two-hybrid assay was produced as GST-FISL. GST-FISL was labeled with phosphorus-32 and, after cleavage with thrombin, which removed the GST portion, yielded 32P-FISL to be used as a probe.
(B) GST-SOS3 and control proteins. Molecular mass markers, both prestained (MM, pre) and unstained (MM), GST-SOS3, GST-SOS1, and GST-SOS2 were separated on a 7.5% SDS-PAGE gel, electroblotted onto a membrane, and stained with Ponceau S to reveal all proteins (left). The membrane was overlaid with 32P-FISL in the presence of 10 mM EGTA and subjected to autoradiography (right).
(C) As given in (B), except that the hybridization solution included 1 mM free Ca2+.
SOS2-like Protein Kinases
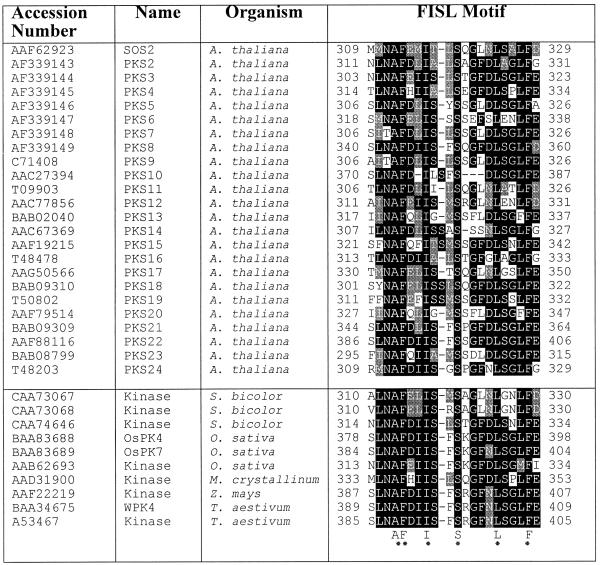
We searched the GenBank database and identified a large number of plant sequences that are highly similar to SOS2 in both the kinase catalytic domain and the regulatory domain. These SOS2-like proteins (protein kinase S [PKS] proteins) all contain a putative FISL motif. It seems that the FISL motif is found only in the SOS2 subfamily of protein kinases. Table 1 lists plant protein kinases that appear to contain an FISL motif. Sequence analyses of the putative FISL motifs indicate that residues A, F, I, S, L, and F are conserved absolutely (hence the name FISL motif). Twenty-three of the PKS proteins are from Arabidopsis and are named PKS2 to PKS24. PKS2 to PKS5 correspond to SIP1 (for SOS3-interacting proteins) to SIP4 (Halfter et al., 2000), respectively. PKS6 to PKS8 represent three other SOS3-interacting protein kinases that had been obtained from a two-hybrid library screening (Halfter et al., 2000) but were not included.
Table 1.
List of Plant Protein Kinases Containing the FISL Motif a
A 21–amino acid SOS3 binding sequence called the FISL motif was identified in SOS2. Other serine/threonine protein kinases appear to contain an FISL motif, with A, F, I, S, L, and F being conserved absolutely. The cDNAs for PKS2 to PKS8 were determined experimentally, and together with their predicted amino acid sequences, they have been submitted to GenBank.
Except for PKS3, which was cloned based on sequence homology with serine/threonine protein kinases (Mizoguchi et al., 1994), cDNAs were obtained for PKS2 to PKS8 by reverse transcription–polymerase chain reaction (PCR). Alignment of the amino acid sequences of SOS2 and seven PKS proteins shows that the kinase domains of these proteins are very conserved (Figure 3). In the superfamily of protein kinases, these proteins can be placed in the CaMKII/SNF1/AMPK family (Hanks and Hunter, 1995). Like SOS2, the PKS proteins contain the conserved FISL motif located near the kinase domain. When the seven PKS proteins were identified as SOS3-interacting proteins, all clones were truncated. Interestingly, these N-terminal truncations never extended into the conserved SOS3 binding motif (Table 2; Halfter et al., 2000). This is consistent with the FISL motif being necessary for the PKS proteins to interact with SOS3.
Figure 3.
Amino Acid Alignment of SOS2 with PKS2 to PKS8.
The N-terminal kinase catalytic domain is highly conserved. The C-terminal regulatory domain contains the conserved FISL motif (marked). Also marked is the activation loop between the conserved DFG and APE motifs (dots) and the Thr residue, which may be phosphorylated by an upstream protein kinase (asterisk). Dashed lines represent spaces that were introduced to maximize alignment.
Table 2.
Positions of N-Terminal Truncations of PKS6 to PKS8 Kinase Clones Obtained from Yeast Two-Hybrid Library Screening with SOS3 as Baita,b
| Clone No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| PKS6 | Δ12 | Δ29 | Δ56 | Δ243 | Δ244 | Δ253 | Δ268 | Δ290 | Δ302 |
| PKS7 | FLc | ||||||||
| PKS8 | Δ255 | Δ256 |
b The numbers of amino acids missing from the N-terminal part of the protein are shown.
c FL, full-length clone.
Expression of PKS Genes under Salt Stress
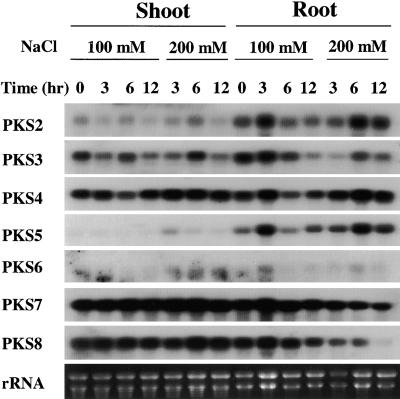
Steady state transcript levels of PKS genes in roots and shoots of young Arabidopsis seedlings were analyzed. Because of our interest in plant salt stress responses, potential PKS regulation by salt stress also was examined. Two-week-old seedlings were subjected to various durations of treatment with either 100 or 200 mM NaCl. The seedlings were separated into shoots and roots, and their mRNAs were analyzed using gene-specific probes for PKS2 to PKS8 (Figure 4). PKS2 was expressed at a higher level in the root than in the shoot. There was no substantial regulation of PKS2 by salt stress (Figure 4). Expression levels of PKS3 were comparable between the root and the shoot. After 12 hr of NaCl treatment, PKS3 transcript levels were reduced slightly (Figure 4).
Figure 4.
Expression of PKS2 to PKS8 in Arabidopsis Shoots and Roots in Response to Salt Stress.
Two-week-old Arabidopsis seedlings, grown on Murashige and Skoog (1962) nutrient agar plates, were treated with 100 or 200 mM NaCl and harvested at the specified times for RNA isolation. Twenty micrograms of poly(A)+ RNA was analyzed by RNA gel blotting. Ethidium bromide–stained rRNA is shown as a loading control. The blot was hybridized with gene-specific probes for PKS2 to PKS8.
The steady state level of PKS4 transcript was relatively high in both roots and shoots (Figure 4). There was slight upregulation of PKS4 expression by treatment with 200 mM NaCl. The PKS5 transcript level in the root was very low; its expression was higher in the shoot. In addition, its expression in the shoot was slightly upregulated by treatment with 200 mM NaCl (Figure 4).
PKS6 expression was extremely low in both roots and shoots (Figure 4). The salt stress treatment did not affect PKS6 expression significantly. In contrast, the levels of PKS7 and PKS8 expression in both the root and the shoot were very high (Figure 4). Interestingly, PKS7 and PKS8 expression in the root was repressed significantly by treatment with 200 mM NaCl. Both PKS7 and PKS8 expression in the shoot were maintained at high levels under NaCl treatment (Figure 4).
SOS3-like Calcium Binding Proteins
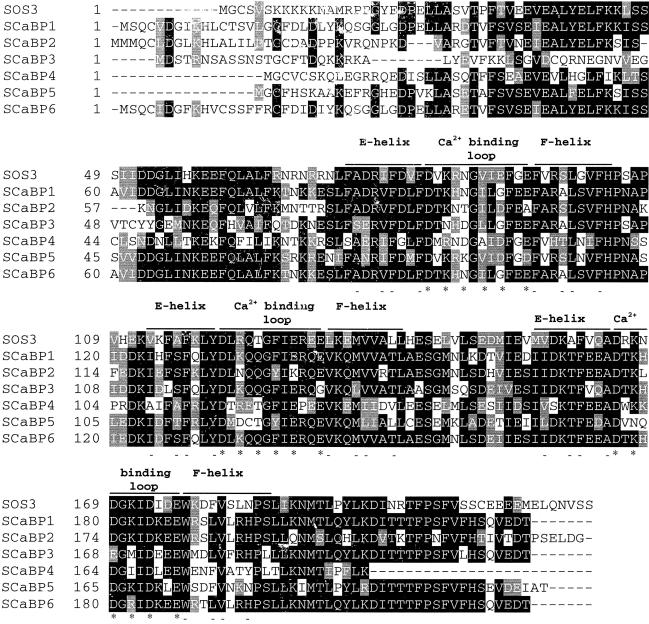
We also searched the GenBank database for SOS3-like calcium binding proteins (SCaBPs). Genomic DNA sequences are known for all of these proteins. cDNAs corresponding to the genomic sequences were obtained by reverse transcription–PCR. The translated amino acid sequences from the cDNAs are aligned with SOS3, as shown in Figure 5. SCaBP1, SCaBP5, and SCaBP6 were identified previously as calcineurin B–like proteins CBL2, CBL1, and CBL3, respectively, all with unknown functions (Kudla et al., 1999). As in SOS3, three EF-hand structures can be identified in the SCaBPs. However, some of the EF hands differ significantly from the canonical EF-hand domain (Moncrief et al., 1990). For example, the conserved acidic residue E or D in the -z position (i.e., the last asterisk in the calcium binding loops) is replaced with A in the first EF hand of SCaBP2 (Figure 5). In the second EF hand of SCaBP3, G is found in the -z position (Figure 5).
Figure 5.
Alignment of SOS3 with SCaBPs.
Database searches revealed sequences similar to SOS3, that is, SCaBP1 to SCaBP6. All contain three EF-hand calcium binding motifs. The calcium binding loops, flanked by E and F helices, are marked. Asterisks and dashes denote amino acid residues important for calcium binding and the EF-hand structure, respectively (Moncrief et al., 1990).
The LYD tripeptide at the junction of the E helix and the calcium binding loop in the second EF hand is deleted in the loss-of-function allele sos3-1 (Liu and Zhu, 1998). This sequence is conserved in all of the SCaBPs.
A consensus sequence for N-myristoylation (i.e., MGxxxS/T[K], where the letter x can be any amino acid) (Towler et al., 1988) can be found in SOS3, SCaBP4, and SCaBP5. We have shown recently that SOS3 is myristoylated and that the myristoylation is necessary for SOS3 function in plant salt tolerance (Ishitani et al., 2000). The potential significance of N-myristoylation for SCaBP4 or SCaBP5 is not known.
Interactions between PKS Proteins and SCaBPs
SOS3 interacts strongly with SOS2 in vitro as well as in the yeast two-hybrid system (Figure 1). To determine whether SCaBPs also interact with SOS2, we subjected the SCaBPs to yeast two-hybrid assays. These bait proteins by themselves did not cause any self-interaction (data not shown). When the SCaBP baits were introduced into yeast harboring the prey pACT-SOS2, β-galactosidase assays for LacZ reporter expression showed that SOS2 was able to interact very strongly with SOS3 and to a lesser extent with SCaBP1, SCaBP3, SCaBP5, and SCaBP6 as well (Figure 6).
Figure 6.
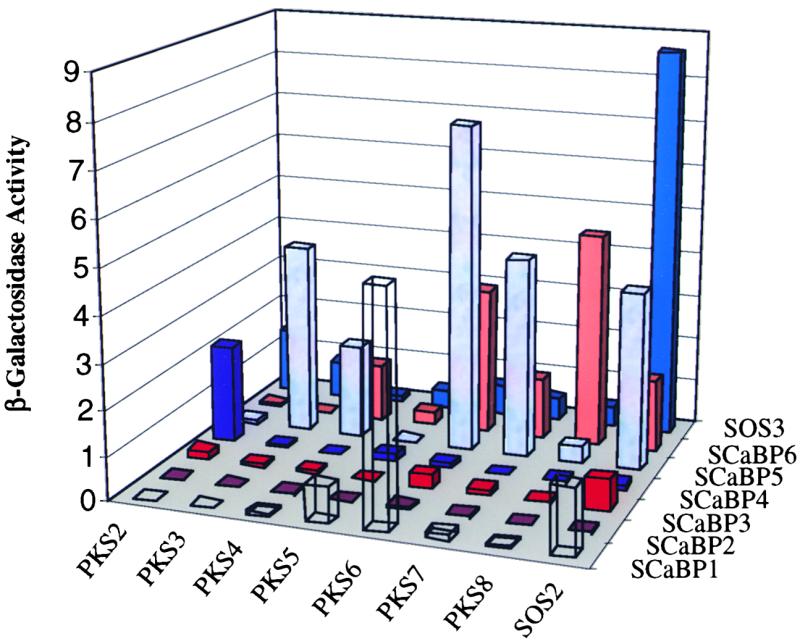
β-Galactosidase Activity of Various Combinations between PKS Proteins and SCaBPs in a Yeast Two-Hybrid Assay.
Yeast strain Y190 harboring SOS2 or PKS2 to PKS8 protein kinases in the pACT prey vectors were transformed with SOS3 or SCaBP1 to SCaBP6 in the bait vector pAS2. Their ability to activate the LacZ reporter gene, measured as β-galactosidase activity, was assayed as an indicator of the strength of their interaction. The activity values are the averages of three experiments. Standard deviations are not shown but are all within 5% of the respective average values. Numbers at left represent arbitrarily defined unit U.
To determine whether the PKS proteins interact with certain SCaBPs, we introduced the pAS-SOS3 and pAS-SCaBP baits into yeast strains harboring one of the pACT-PKS preys. Figure 6 shows that PKS2 interacted relatively strongly with SCaBP4 and SOS3. PKS3 interacted strongly with SCaBP5 and weakly with SOS3 (Figure 6). PKS3 did not show substantial interaction with any of the other SCaBPs. PKS4 showed interaction with SCaBP5 and SCaBP6 (Figure 6). PKS5 interacted weakly with SCaBP1 but showed no substantial interaction with SOS3 or any of the other SCaBPs (Figure 6). We suggest that PKS4 may interact strongly with SCaBPs that we have not yet identified.
PKS6 showed preferential interaction with SCaBP1, SCaBP5, and SCaBP6 (Figure 6). This protein kinase also interacted weakly with SOS3. Like PKS6, PKS7 also had the strongest interaction with SCaBP5 (Figure 6). However, PKS7 did not show interaction with SCaBP1. PKS7 showed some weak interaction with SCaBP6 as well as with SOS3. PKS8 exhibited a strong interaction with SCaBP6 (Figure 6). In fact, the interaction between SCaBP6 and PKS8 was stronger than that between SCaBP6 and any of the other protein kinases tested (Figure 6).
SCaBP2 did not show significant interaction with any of the protein kinases. Similarly, SCaBP3 had only weak interaction with SOS2 and little or no interaction with the other protein kinases. It is possible that strong interactions may be found between SCaBP2 or SCaBP3 and the other PKS proteins (e.g., PKS9 to PKS24) that were not tested. The strongest interaction for SCaBP1 was with PKS6. Interestingly, the strongest interaction for SCaBP5 was with PKS6 as well. However, unlike SCaBP5, which interacted substantially with several PKS proteins, SCaBP1 was not as promiscuous. SCaBP4 was very specific and interacted only with PKS2. SCaBP6 was similar to SCaBP5 in that both interacted with several of the protein kinase preys.
Interaction between the Kinase and the Regulatory Domains of SOS2
The observation that the SOS2 C-terminal regulatory domain interacted more strongly with SOS3 than did the full-length SOS2 prompted us to determine whether the SOS2 N-terminal kinase domain interacts with its C-terminal regulatory domain, thus blocking access to SOS3. The N-terminal 267–amino acid residues of SOS2 were cloned into the bait vector pAS2 and then introduced into yeast strains containing the various truncated SOS2 preys. Figure 7 shows that the SOS2 N-terminal bait interacted strongly with the SOS2 C-terminal prey, SOS2T1 (i.e., amino acid residues 304 to −446). The N-terminal bait also interacted strongly with a C-terminal region covering amino acid residues 257 to −368 but not with the full-length SOS2 or other SOS2 truncations (Figure 7). These results indicate that the N-terminal kinase domain–interacting sequence resides between amino acid residues 304 and −368. The FISL motif within this region appears to be required but not sufficient for interaction with the kinase domain (Figure 7). The interaction between the SOS2 kinase domain and its regulatory domain suggests that the kinase may be maintained in an inactive state by autoinhibition, that is, the regulatory domain may inhibit kinase activity by blocking substrate access to the active site.
Figure 7.
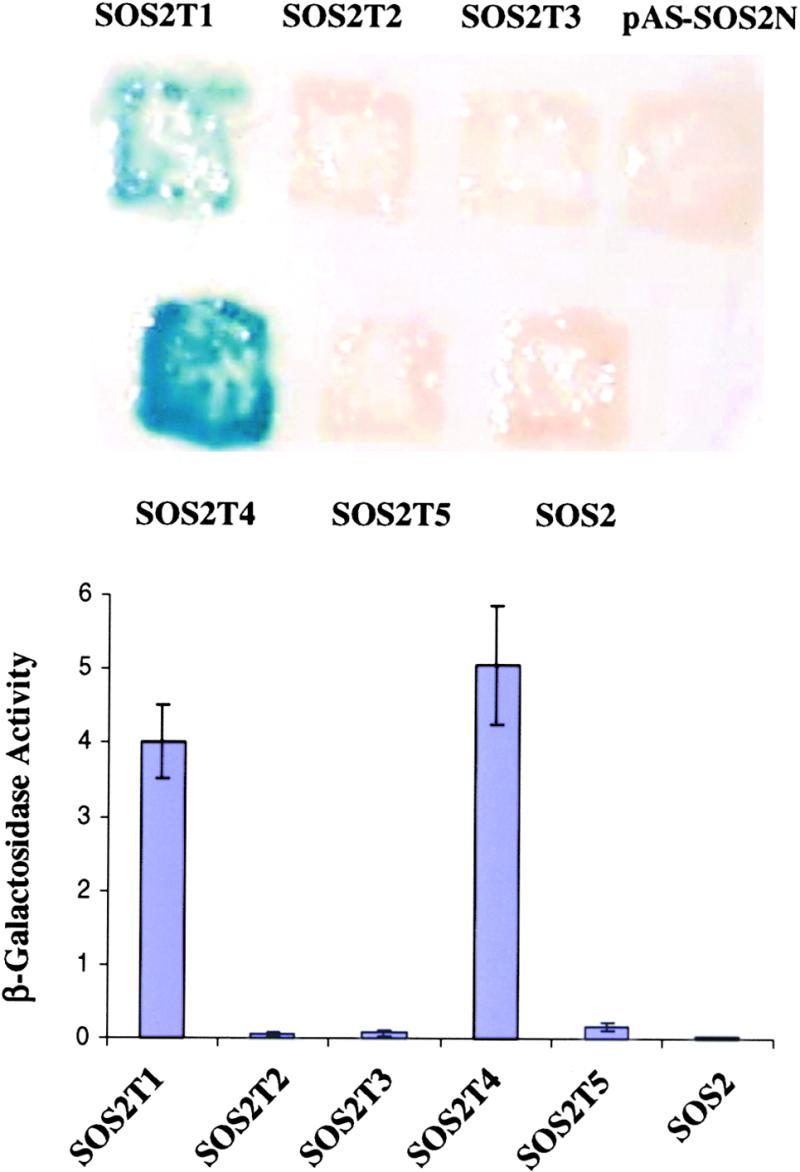
Interaction between the Kinase and Regulatory Domains within the SOS2 Protein.
Wild-type SOS2 and its deletion constructs depicted in Figure 1 were transformed into yeast strain Y190 harboring pAS-SOS2N as bait. Shown are β-galactosidase assays on a filter (top) or in liquid culture (bottom) for quantification of relative interaction strength. Numbers at left represent arbitrarily defined unit U. Error bars indicate ±sd.
The FISL Motif Is Inhibitory to SOS2 Kinase Activity
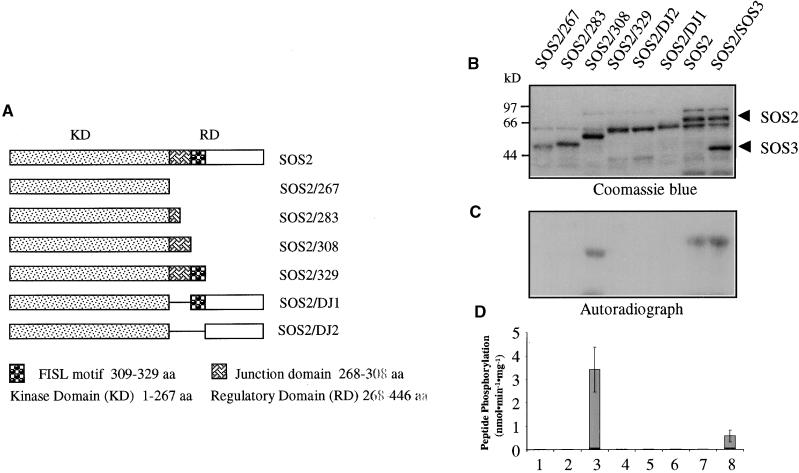
The SOS2 protein kinase is not active by itself but becomes active by binding to SOS3 via its FISL motif. Furthermore, the FISL motif is required for the regulatory domain to bind to the N-terminal kinase domain (Figure 7). Together, these results suggest that the FISL motif may inhibit SOS2 kinase activity. Alternatively, part of the junction region between the FISL motif and the kinase domain may be inhibitory, as is the case in calmodulin-dependent protein kinases (Meador et al., 1993). We tested these possibilities in an attempt to identify the autoinhibitory domain within SOS2. Mutant SOS2 proteins, in which the junction domain, the FISL motif, and/or the remaining C-terminal portion had been deleted (Figure 8A), were expressed in bacteria as GST fusion proteins and purified by glutathione column affinity chromatography (Figure 8B). The fusion proteins were then tested for autophosphorylation as well as phosphorylation of the peptide substrate p3 (Halfter et al., 2000).
Figure 8.
Identification of an Autoinhibitory Domain in SOS2 by Deletion Analysis.
C-terminal serial truncations or internal deletions of SOS2 were expressed in bacteria as GST fusion proteins and tested for autophosphorylation and phosphorylation of the peptide substrate p3. This analysis revealed that the SOS3-binding FISL motif can serve as the kinase autoinhibitory domain.
(A) Diagram of SOS2 mutant constructs. aa, amino acids.
(B) Coomassie blue–stained SDS-PAGE gel containing the SOS2 mutant proteins fused with GST.
(C) Autoradiograph of the gel shown in (B) to detect kinase autophosphorylation.
(D) Peptide phosphorylation activities of the proteins shown in (B). Error bars indicate ±sd (n = 3).
Removal of the entire regulatory domain (amino acids 268 to −446) or part or all of the junction domain abolished SOS2 autophosphorylation (Figure 8C) or phosphorylation of p3 (Figure 8D). SOS2 autophosphorylation was retained when the FISL motif and its downstream sequences were removed (Figure 8C, lane 3, SOS2/308). The level of autophosphorylation in the truncated protein SOS2/308 was similar to that in intact SOS2. However, the truncated SOS2 mutant was much more active than was the intact SOS2 in phosphorylating the p3 substrate (Figure 8D, lane 3). In agreement with previous findings (Halfter et al., 2000), SOS3 was necessary for SOS2 phosphorylation of p3 but not for SOS2 autophosphorylation (Figures 8C and 8D, lanes 7 and 8). The kinase activity in the truncated SOS2 mutant (SOS2/308) was not affected by SOS3 (data not shown), which is consistent with the lack of the FISL motif in the mutant kinase. When the FISL motif was added to the SOS2/308 mutant, the resulting mutant version, SOS2/329, became inactive in autophosphorylation and phosphorylation of p3 (Figures 8B to 8D, lanes 4). These results show that the junction domain is necessary for SOS2 kinase activity, whereas the FISL motif is sufficient to keep the kinase inactive.
A Mutation in the Activation Loop Activates SOS2 Kinase Activity
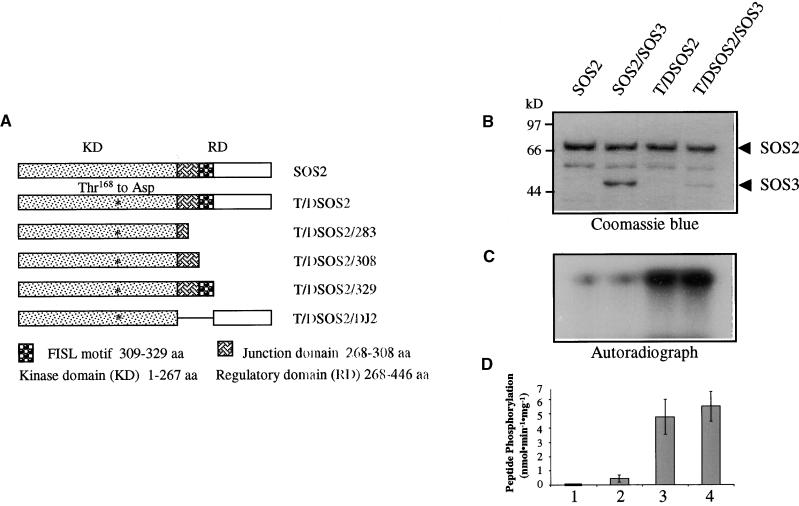
Many protein kinases contain an activation segment situated between the conserved Asp-Phe-Gly (DFG) and Ala-Pro-Glu (APE) motifs in the kinase domain (Johnson et al., 1996). Phosphorylation of this activation segment often is required for kinase activation by upstream protein kinases. We located a putative activation segment in SOS2 and the PKS proteins (Figure 3). This sequence was tested to determine whether it could function as an activation loop and whether a constitutively active SOS2 kinase could be engineered by a Thr-to-Asp change (to mimic phosphorylation by upstream kinases) in the putative activation loop. The Thr168-to-Asp change was made by site-directed mutagenesis of SOS2 cDNA. Figure 9 shows that the single amino acid substitution substantially increased SOS2 kinase activity. Both SOS2 autophosphorylation and phosphorylation of p3 substrate were enhanced by the Thr168-to-Asp change (Figures 9C and 9D). Furthermore, the mutation made SOS2 independent of SOS3. The addition of SOS3 did not alter the activity of the mutant kinase significantly.
Figure 9.
Activation of SOS2 Kinase Activity by the Thr168-to-Asp Mutation in the Activation Loop.
(A) Diagram of SOS2 mutants that were expressed in bacteria as GST fusion proteins. aa, amino acids.
(B) Coomassie blue–stained SDS-PAGE gel containing the relevant proteins.
(C) Kinase autophosphorylation.
(D) Phosphorylation of p3 by the kinases. Error bars indicate ±sd (n = 3).
Engineering a Superactive SOS2 Kinase by Combining the Deletion and Activation Loop Mutations
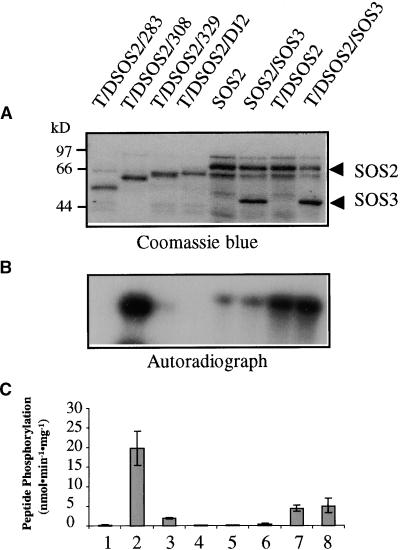
The data presented above show that a constitutively active, SOS3-independent SOS2 protein kinase can be created either by removing the FISL motif or by mutating the activation loop sequence (Figures 8 and 9). We also tested the effect of combining the activation loop mutation with various regulatory domain deletions. The results are presented in Figure 10. Combining the Thr168-to-Asp mutation with the SOS2/308 truncation resulted in a kinase that is more active than any of the previous single mutant versions (Figure 10). In the combined mutant form (T/DSOS2/308), the effect of the two single mutations appeared to be more than additive. The activation loop mutation also was able to overcome the inhibitory effect of the FISL motif in the SOS2/329 truncated protein (Figure 10, lane 3). The T/DSOS2/329 mutant was active in phosphorylating p3, although it was not very active in autophosphorylation. In contrast, the superactive T/DSOS2/308 had very high autophosphorylation activity in addition to being extremely active in p3 phosphorylation (Figure 10, lane 2).
Figure 10.
Creation of a Superactive SOS2 Kinase by Combining the Thr168-to-Asp Mutation with the Autoinhibitory Domain Deletion.
Construction of the SOS2 mutant proteins was as shown in Figure 9A.
(A) Coomassie blue–stained SDS-PAGE gel containing the relevant proteins.
(B) Kinase autophosphorylation.
(C) Phosphorylation of p3 by the kinases. Error bars indicate ±sd (n = 3).
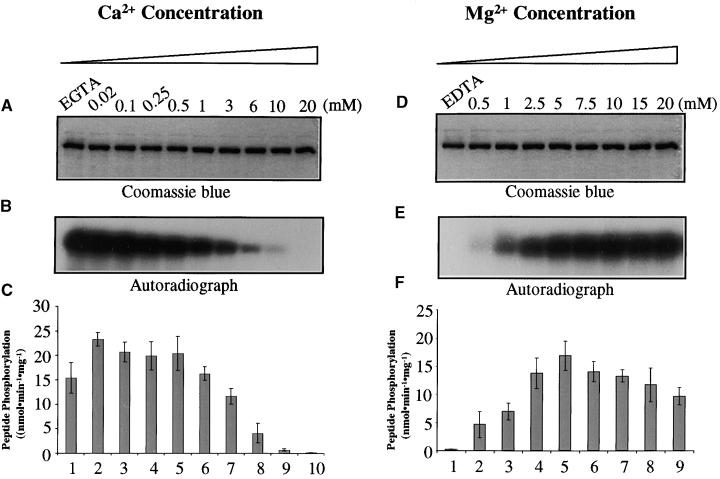
Effect of Divalent Cations on the Superactive SOS2 Mutant Kinase
Substrate phosphorylation by the intact SOS2 is dependent on both SOS3 and Ca2+ (Halfter et al., 2000; Ishitani et al., 2000). To determine whether the superactive SOS2 kinase mutant (T/DSOS2/308) is similarly dependent on Ca2+, we assayed autophosphorylation activities and p3 phos-phorylation activities of the mutant kinase at various Ca2+ concentrations. Figures 11A to 11C show that autophosphorylation of the mutant kinase was highest when there was no Ca2+ (i.e., with EGTA). The presence of Ca2+ inhibited the autophosphorylation activity. This inhibition appeared to be proportional to the Ca2+ concentration. This is in contrast to the effect in intact SOS2 kinase, in which the autophosphorylation as well as the p3 phosphorylation are not affected by Ca2+ (Halfter et al., 2000).
Figure 11.
Effect of Ca2+ and Mg2+ on the Superactive Mutant SOS2 Kinase.
(A) Coomassie blue–stained SDS-PAGE gel showing the mutant kinase protein (T/DSOS2/308). (B) Autophosphorylation activity of the kinase.
(C) p3 phosphorylation activity of the kinase.
(D) to (F) Same as (A) to (C), respectively, except for the indicated differences in Ca2+ and Mg2+ concentrations in the kinase reactions.
Error bars in (C) and (F) indicate ±sd (n = 3).
The mutant kinase was active in phosphorylating p3 when no Ca2+ was present (Figures 11A to 11C, lanes 1, EGTA). The addition of 0.02 μM Ca2+ resulted in a slight increase in p3 phosphorylation (Figures 11A to 11C, lanes 2). However, very high levels of Ca2+ were clearly inhibitory to the p3 phosphorylation activity of the mutant kinase (Figures 11A to 11C).
To determine whether the inhibitory effect of Ca2+ was specific, we assayed the mutant kinase at various Mg2+ concentrations. Figures 11D to 11F show that the mutant kinase was not active in either autophosphorylation or p3 phosphorylation when no Mg2+ was present (lanes 1, EDTA). The addition of Mg2+ to the assay solution restored kinase autophosphorylation and p3 phosphorylation activities. Very high levels of Mg2+ appeared to inhibit the p3 phosphorylation slightly but not the kinase autophosphorylation (Figures 11D to 11F).
DISCUSSION
SOS2 and SOS2-like proteins (i.e., PKS) represent a novel family of serine/threonine protein kinases. The PKS proteins all contain a highly conserved kinase domain in the N-terminal part, which is similar to that of SNF1/AMPK protein kinases (Figure 3). The C-terminal portions of the PKS proteins also are conserved but lack substantial sequence similarities to well-characterized proteins or protein domains. Through site-directed mutagenesis and deletion analysis, we discovered functional domains in the founding member of the PKS protein family, SOS2, that are critical in intramolecular and intermolecular protein interactions and kinase activation or suppression. These results provide important insights into the regulation of activities of the PKS family of protein kinases.
Protein–protein interactions are key to the understanding of protein function. Our finding of SOS2–SOS3 interaction led to the discovery that the SOS2 protein kinase functions in a salt stress–activated calcium-signaling pathway (Halfter et al., 2000). SOS2 kinase activity is dependent on SOS3 and calcium (Halfter et al., 2000). In this study, we found that SOS2 interaction with SOS3 is mediated by a stretch of 21 amino acid residues within the regulatory domain of SOS2. This short motif is both necessary and sufficient for interaction with SOS3. In a separate study, we also attempted to define a sequence motif within SOS3 that might mediate its interaction with SOS2 (Ishitani et al., 2000). However, no such motif was found to be sufficient for SOS2 binding. It appeared that various parts of SOS3 participate in its binding to SOS2.
The SOS3-binding FISL motif is located near the kinase domain of SOS2. The FISL motif is also found in all PKS proteins, located near the kinase domain (Figure 3). Interestingly, N-terminal truncations of PKS clones identified in the yeast two-hybrid screen with SOS3 never extended into the conserved SOS3 binding motif (Table 2; Halfter et al., 2000). This finding is consistent with the notion that the FISL motif is necessary for the PKS proteins to interact with SOS3 and most likely for interactions with the SCaBPs as well.
The sequence between the FISL motif and the kinase domain is not conserved in the PKS proteins. In calmodulin kinase I, the calmodulin binding domain also is near the kinase domain, and in between is the autoinhibitory domain (Goldberg et al., 1996). However, we found that the junction region between the SOS3 binding motif and the kinase domain does not function as the autoinhibitory sequence of SOS2. Rather, this junction region is necessary for SOS2 kinase activity because its deletion makes SOS2 inactive (Figure 8). We found that the FISL motif serves as the autoinhibitory domain: it is sufficient to keep SOS2 in an inactive state (Figure 8). The sequence after the SOS3 binding motif also is conserved between SOS2 and the PKS proteins. This region may mediate interaction with other regulators of the protein kinase.
The interaction between the kinase domain and the regulatory domain within SOS2 is interesting. Together with other findings in this study, it suggests the following model for the regulation of SOS2 kinase activity. In the inactive form, the kinase domain of SOS2 is inhibited by interaction with the regulatory domain on the same protein. In response to cytosolic Ca2+ increases elicited by salt stress, and together with SOS3 binding to the FISL motif in the regulatory domain, the kinase is liberated and relieved from inhibition. Because Ca2+ is not required for SOS3 binding to the FISL motif, its precise role in kinase activation remains to be defined.
Our results with the activation loop mutation suggest the possibility that SOS2 also may be activated via phosphorylation by an upstream protein kinase. Both animal AMPK and yeast SNF1 protein kinases can be activated by an upstream kinase, the molecular identity of which is still unknown (Hardie et al., 1998). Mutation of the conserved Thr168 in the activation loop to the acidic residue Asp to mimic Thr-P resulted in constitutive activation of SOS2 (Figures 9 and 10). It is interesting that the Thr-to-Asp change could overcome the inhibitory effect of the regulatory domain. Remarkably, the activation loop mutation and the autoinhibitory domain deletion had a synergistic effect on the SOS2 kinase activity. A novel superactive and constitutive SOS2 kinase was created when the two changes were combined. These constitutively active forms of the kinase provide valuable reagents for future explorations of the functions of SOS2 in phosphorylating its target proteins. Similar molecular manipulations probably can be applied to the other PKS proteins to obtain constitutively active forms to probe the functions of the protein kinases in plants.
The superactive form of SOS2 was inhibited by high concentrations of Ca2+ but not by Mg2+ (Figure 11). This is in sharp contrast to the intact SOS2 protein, which depends on Ca2+ for activation (Halfter et al., 2000; Ishitani et al., 2000). The underlying mechanism for such differential effects of Ca2+ is not known at present.
We have begun to apply some of the knowledge gained from the analysis of SOS2 domains to the PKS family of protein kinases. The SOS3 binding sequence is highly conserved in the PKS proteins. Thus, all of the PKS proteins examined interact with SOS3 to some extent. However, their interactions with SOS3 are weak compared with that between SOS2 and SOS3. We hypothesized that some of the PKS proteins showing weak interactions with SOS3 may interact strongly with SCaBPs. Indeed, we found that the PKS proteins interact differentially with the SCaBPs. We hope to determine which particular combinations between PKS family members and SCaBP family members give strong interactions, because strong interactions are likely to be meaningful for the in vivo function of these proteins. In the in vitro characterization of enzymes, several different molecules might be used as substrates, but only molecules with the highest affinities are considered to be true substrates. Among the protein kinases tested, SOS2 exhibited the strongest interaction with SOS3. The in planta significance of this strong interaction is consistent with genetic evidence showing that SOS3 and SOS2 function in the same pathway for plant salt tolerance (Halfter et al., 2000).
Our studies revealed very specific interactions. For example, PKS3 interacted strongly with SCaBP5 but weakly or not at all with other SCaBPs. In contrast, PKS8 showed specific interaction with SCaBP6 (Figure 6). These results suggest that PKS3 most likely functions together with SCaBP5 and that PKS8 probably acts in concert with SCaBP6. Similarly, preferential interaction between SCaBP4 and PKS2 (Figure 6) suggests the possibility that these two proteins function as a complex. The interaction studies can be complemented by future expression analysis to determine whether the interaction partners are co-expressed spatially and temporally. True interactions require at least some overlap in the spatial and temporal expression of the interaction partners.
In this regard, we have analyzed the expression of PKS2 to PKS8 in shoots and roots, and their regulation by salt stress. Interestingly, some were expressed preferentially in the root (e.g., PKS2 and PKS5), whereas some were expressed equally in roots and shoots (e.g., PKS3, PKS4, PKS7, and PKS8). PKS7 and PKS8 showed a substantial downregulation by NaCl stress in the root, whereas several others (e.g., PKS2, PKS4, and PKS5) were slightly upregulated by the stress (Figure 4). The very high levels of expression and downregulation of PKS7 and PKS8 by salt stress indicate that these two protein kinase genes may function in some processes associated with active growth or metabolism.
Some of the other interactions between the two families of proteins are not as specific. For example, PKS6 interacted strongly with SCaBP1, SCaBP5, and SCaBP6. Apart from its interaction with SOS3, SOS2 also had strong interaction with SCaBP5. These and other less specific interactions could be functionally relevant. Many if not most of the functions in plants are performed by redundant genes. The redundancies provide flexibility for evolutionary changes and are an important mechanism to protect against undesirable recessive mutations. In the case of PKS6, its interactors SCaBP1, SCaBP5, and SCaBP6 may have redundant functions. Alternatively, these SCaBPs may function in separate pathways and mediate different calcium signals. In the latter case, PKS6 would be a point of communication between the different pathways. Recently, Shi et al. (1999) reported that PKS13/CIPK1 is capable of interacting with SCaBP5/CBL1. The functional significance of this interaction is unknown because the functions of the PKS/CIPK and SCaBP/CBL proteins are not known.
Genome sequencing efforts are revealing an increasingly large number of protein kinases in plants (Arabidopsis Genome Initiative, 2000). An important aspect of the functional genomics of these kinases is to determine which proteins interact with them and which type of second messengers may regulate their activities. Because of their interaction with ScaBPs, as was found in this study, members of the PKS family are expected to function in mediating calcium signaling. Various hormonal, developmental, and environmental stimuli, such as salt stress, cold, drought, and wounding, have been shown to cause changes in cytosolic free calcium concentration (Poovaiah and Reddy, 1993; Bush, 1995; Pierson et al., 1996; Trewavas and Malho, 1998; Knight, 2000). SOS2 and SOS3 function specifically in regulating plant responses to high Na+ and Li+ stresses (Liu and Zhu, 1997; Zhu et al., 1998). We suggest that the other PKS proteins and SCaBPs may function in regulating plant responses to osmotic stress, cold, abscisic acid, and mechanical perturbations or in some growth and developmental processes in which calcium serves as a second messenger. The in planta function of these proteins awaits determination in genetic studies by using transgenic plants or knockout mutants.
METHODS
SOS2 Deletion and Site-Directed Mutant Constructs
Plasmids pAS-SOS3 and pACT-SOS2 were described by Halfter et al. (2000). For pACT-SOS2 deletion constructs, polymerase chain reaction (PCR) amplifications were performed with the following primer pairs. Forward primers were as follows: for pACT-SOS2T1, 5′- ATGGCCATGGATGAAGGGCCCCTGATGATGAATGC-3′; for pACT-SOS2T2, 5′-ATGGCCATGGGCTTAAATTTATCTGCACTATTTGA-3′; for pACT-SOS2T3, 5′-ATGGCCATGGATTTTGTTAAAAGGCAAACC-CGTTTTG-3′; for pACT-SOS2T4, 5′-ATGGCCATGGGAATCAAG-AAAGATCCTTGGTTCA-3′; and for pACT-SOS2T5, 5′-ATGGCC-ATGGAGATGATGAATGCCTTTGAGATGATTACC-3′ (the NcoI site is underlined in each primer). The reverse primer was 5′-CGGGAT-CCGGATCACCTGTCAAATAGTGCAGATAAA-3′ (the BamHI site is underlined) for all reactions. All PCR fragments were cloned into pACT2 between the NcoI and BamHI sites.
To introduce the Lys40-to-Asn mutation into pACT-SOS2 to make pACT-SOS2A, the following primer pairs were used for the first PCR: 5′-GCGGATCCGAATGACAAAGAAAATGAGAAGAGTGGGC-3′ (the BamHI site is underlined) and 5′-ATTGTACTCTTAGCCATAATG-TTGATGGCT-3′, and 5′-GTGATAATGTAGCCATCAACATTATGG-CTA-3′ and 5′-GCGAATTCTTAAGTTGGGATCAAAACGTGATTG-TTCTG-3′ (the EcoRI site is underlined). Using the PCR products as templates, we performed a second amplification with the primer pair 5′-GCGGATCCGAATGACAAAGAAAATGAGAAGAGTGGGC-3′ (the BamHI site is underlined) and 5′-GCGAATTCTTAAGTTGGGATC-AAAACGTGATTGTTCTG-3′ (the EcoRI site is underlined). The final product was cloned between the BamHI and EcoRI sites of pACT2.
For the deletion construct pACT-SOS2D1, primer pairs 5′-GCG-GATCCGAATGACAAAGAAAATGAGAAGAGTGGGC-3′ (the BamHI site is underlined) and 5′-CGGGTTTGCCTTTTAACAAAATCC-TGTCGCAGGGGCCCTTCATC-3′, and 5′-GAGAGAAATGATGAA-GGGCCCCTGCGACAGGATTTTGTTA-3′ and 5′-GCGAATTCTTAA-GTTGGGATCAAAACGTGATTGTTCTG-3′ (the EcoRI site is underlined), were used for the first PCR. The second PCR was performed as described above (pACT2-SOS2A). The final products were cloned between the BamHI and EcoRI sites of pACT2.
To express the SOS2 N-terminal kinase domain in the bait vector pAS2, we used primers 5′-ATGGCCATGGATGACAAAGAAAATGAGAAG-3′ (the NcoI site is underlined) and 5′-ACGCGTCGACGA-TTTAATCTGAACCAAGGATC-3′ (the SalI site is underlined) for PCR amplification of SOS2 cDNA. The product was cloned into the pAS2 vector between the NcoI and SalI sites.
To make deletion constructs for constitutively active SOS2 kinase, we used the following primer pairs for PCR amplification. The SOS2 forward primer was 5′-CGGGATCCATGACAAAGAAAATGAGAAG-3′ (the BamHI site is underlined). The reverse primers were as follows: 5′-GGAATTCTCAATTTAATCTGAACCAAGGATC-3′ for SOS2/267; 5′-GGAATTCTCAATCATCCAAATTCACTTCTTC-3′ for SOS2/283; 5′-GGAATTCTCACAGGGGCCCTTCATCATTTC-3′ for SOS2/308; and 5′-GGAATTCTCAGTCAAATAGTGCAGATAAATTTAAG-3′ for SOS2/329 (the EcoRI site is underlined in each primer). To make SOS2/DJ1 and SOS2/DJ2 constructs, we amplified first-round PCR products with the SOS2 forward primer and the reverse primers 5′-CATCTCAAAGGCATTCATCATATTTAATCTGAACCAAGGATC-3′ for SOS2/DJ1 and 5′-AACAAAATCCTGTCGCCTATTTAATCTGAACCAAGGATC-3′ for SOS2/DJ2 and the SOS2 reverse primer 5′-GGAATTCTCAAAACGTGATTGTTCTGAG-3′ (the EcoRI site is underlined) with forward primers 5′-GATCCTTGGTTCAGATTAAATATGATGAATGCC-TTTGAGATG-3′ for SOS2/DJ1 and 5′-GATCCTTGGTTCAGATTA-AATAGGCGACAGGATTTTGTT-3′ for SOS2/DJ2. The PCR products were then isolated from agarose gels. The two PCR fragments for SOS2/DJ1 were used together as a template to be amplified by SOS2 forward and reverse primers. Similarly, the two PCR fragments for SOS2/DJ2 were used together as a template to be amplified by SOS2 forward and reverse primers. All final PCR fragments were cloned into pGEX-2TK between the BamHI and EcoRI sites.
To introduce the Thr168-to-Asp mutation into SOS2 to make the T/DSOS2 construct, we used the SOS2 forward primer and 5′-GAG-TTCCACATGTGTCACGCAGAAGTTCTAC-3′ and the SOS2 reverse primer and 5′-GTAGAACTTCTGCGTGACACATGTGGAACTC-3′ for PCR amplification. The PCR products were isolated from agarose gels and then used as templates for amplification by SOS2 forward and reverse primers. The final fragment was cloned into pGEX-2TK vector between the BamHI and EcoRI sites.
For the T/DSOS2/283, T/DSOS2/308, T/DSOS2/329, and T/DSOS2/ DJ2 constructs, T/DSOS2 plasmid was used and amplified by SOS2/283, SOS2/308, SOS2/329, and SOS2/DJ2 primers. The final fragments were cloned into pGEX-2TK vector between the BamHI and EcoRI sites.
For all amplifications, Turbo-pfu DNA polymerase (Stratagene) was used on SOS2 cDNA as a template. All constructs were sequenced completely to verify that there were no PCR or cloning errors.
Yeast Two-Hybrid Interaction Assays
Saccharomyces cerevisiae strain 190 expressing the pAS-SOS3 bait was transformed with pACT-SOS2A, pACT-SOS2T1, pACT-SOS2T2, pACT-SOS2T3, pACT-SOS2T4, pACT-SOS2T5, and pACT-SOS2D1. Transformed yeast cells were selected on synthetic complete medium lacking tryptophan and leucine. Empty prey vector pACT2 was used as a negative control. Interaction was selected on synthetic complete medium lacking tryptophan, leucine, and histidine, and supplemented with 25 mM 3-amino-1,2,4-triazole.
For interaction analysis of SOS2 and protein kinase S (PKS) proteins with SOS3 and SOS3-like calcium binding proteins (SCaBP), yeast Y190 strains expressing the bait pAS-SOS3 or one of the pAS-SCaBPs were transformed with pACT-SOS2, pACT-PKS2, pACT-PKS3, pACT-PKS4, pACT-PKS5, pACT-PKS6, pACT-PKS7, or pACT-PKS8 in all combinations.
To analyze the interaction between SOS2 N-terminal and C-terminal domains, yeast Y190 cells expressing the bait pAS2-SOS2N were transformed with pACT-SOS2, pACT-SOS2T1, pACT-SOS2T2, pACT-SOS2T3, pACT-SOS2T4, or pACT-SOS2T5.
All bait proteins were tested for self-activation, and none of them was found to activate the two reporter genes His3 or LacZ.
β-Galactosidase Filter Assays and Quantification
Yeast cells were grown at 30°C and transferred to a nitrocellulose filter (BioTrace NT 0.45 μM; Gelman Sciences, Ann Arbor, MI) for β-galactosidase assay. The assay was performed as described by Halfter et al. (2000).
For quantitative assays, we used a procedure published at http://www.fhcrc.org/labs/gottschling/Bgal.sht. Yeast cells were grown in YEPD (for yeast extract, peptone, and dextrose) liquid medium until mid-log phase, pelleted, and resuspended in an equal volume of buffer Z (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol). The OD600 of the cell suspension was measured. To an aliquot of 0.8 mL of the suspension, we added 50 μL of 0.1% SDS and 100 μL of chloroform, mixed well, and incubated it for 15 min at 30°C. Then, 160 μL of 4 mg/mL o-nitrophenyl β-d-galactopyranoside was added, mixed well, and incubated at 30°C for 20 min. The reaction was stopped by adding 400 μL of 1 M Na2CO3 and pelleted, and the supernatant was measured at OD420 and OD550. β-Galactosidase activity (U) was calculated as U = 1000 × ([OD420] − [1.75 × OD550]) / ([time (min)] × [vol (mL)] × OD600).
cDNA Cloning of PKS and SCaBPs
cDNAs for PKS and SCaBP open reading frames were obtained by reverse transcription–PCR. Template mRNA was isolated from 2-week-old wild-type plants (ecotype Columbia). PKS cDNAs were cloned into pGEX-2TK between the BamHI and EcoRI sites. Final constructs were confirmed by sequencing. Primer pairs for PCR on first-strand cDNAs were as follows (the BamHI and EcoRI restriction sites are underlined): for PKS2, 5′-CGGGATCCATGGAAAATAAGCCAAGTG-3′ and 5′-GGAATTCAAAACTTCAATGGTTCTTC-3′; for PKS3, 5′-CGGGATCCATGGAGAAGAAAGGATCTG-3′ and 5′-GGA-ATTCAGTGCCAAGCTAATACAAAG-3′; for PKS4, 5′-CGGGATCCA-TGGTCGGAGCAAAACCGGTG-3′ and 5′-GGAATTCAAGCAGGT-GTAGAGGTCCAG-3′; for PKS5, 5′-CGGGATCCATGCCAGAGATCG-AGATTG-3′ and 5′-GGAATTCTTAAATAGCCGCGTTTGTTG-3′; for PKS6, 5′-CGGGATCCATGAGTGGAAGCAGAAGG-3′ and 5′-GGA-ATTCTTATTGCTTTTGTTCTTCAG-3′; for PKS7, 5′-CGGGATCCA-TGGAATCACTTCCCCAG-3′ and 5′-GGAATTCTTACATGATGTCATTGTGCCATG-3′; and for PKS8, 5′-CGGGATCCATGGCGGAGA-AAATCACGAG-3′ and 5′-GGAATTCTATTCAGTGTCAGACGGCA-3′.
For SCaBPs, primer pairs for PCR on first-strand cDNAs were as follows (the NcoI and SalI restriction sites are underlined): for SCaBP1, 5′-CATGCCATGGATGTCGCAGTGCGTTGAC-3′ and 5′-ACGCGTCGACAGGTATCTTCAACCTGAG-3′; for SCaBP2, 5′-CATGCCATGGATGATGATGCAATGTTTAG-3′ and 5′-ACGCGTCGACATCCATCCAGCTCACTAG-3′; for SCaBP3, 5′-CATGCCATGGAT-GGATTCAACAAGAAATTC-3′ and 5′-ACGCGTCGACAGGTATCTTCCACTTGCGAG-3′; for SCaBP4, 5′-CATGCCATGGATGGGATG-TGTTTGCAGCAAG-3′ and 5′-ACGCGTCGACTTACTTCAAGAAAG-GGATAGTC-3′; for SCaBP5, 5′-CATGCCATGGATGGGCTGCTTCCA-CTC-3′ and 5′-ACGCGTCGACTCATGTGGCAATCTCATC-3′; and for SCaBP6, 5′-CATGCCATGGATGTCGCAGTGCATAGAC-3′ and 5′-ACGCGTCGACTCAGGTATCTTCCACCTG-3′. SCaBP cDNAs were cloned into pAS2 between the NcoI and SalI sites for yeast two-hybrid assays.
Expression and Purification of GST Fusion Proteins in Escherichia coli
GST-SOS2 and GST-SOS3 were obtained as described by Halfter et al. (2000). All mutant SOS2-GST fusion constructs (glutathione S-transferase [GST] on the N-terminal side)—SOS2/267, SOS2/283, SOS2/308, SOS2/329, SOS2/DJ1, SOS2/DJ2, T/DSOS2, T/DSOS2/283, T/DSOS2/308, T/DSOS2/329, and T/DSOS2/DJ2—were transformed into Escherichia coli strain BL21(DE3) cells. Recombinant protein expression was induced at OD600 0.7 with 0.5 mM isopropylthio-β-galactoside. Cells were harvested and lysed with lysozyme and sonication. Recombinant proteins were affinity purified from bacterial lysates with glutathione-Sepharose (Amersham Pharmacia).
Protein Binding in Vitro
The SOS2 cDNA fragment corresponding to amino acids 309 to 330 (i.e., the FISL motif) was cloned from pACT-SOS2T5 into pGEX-2TK for recombinant protein expression in E. coli, yielding GST-FISL. The GST-FISL fusion protein, which contains a recognition sequence for protein kinase A, was labeled in a kinase reaction with protein kinase A and γ-32P-ATP, as described by Halfter et al. (2000). GST was cleaved off using Thrombin (Amersham Pharmacia). GST, GST-SOS3, GST-SOS1, and GST-SOS2 were fractionated on 7.5% SDS-PAGE gels and electroblotted onto a nitrocellulose membrane. The blot was incubated overnight with 32P-FISL in TBST (20 mM Tris-HCl, pH 7.8, 0.5 M NaCl, and 0.15% [v/v] Tween 20) at 4°C. After washing with TBST, 32P-FISL binding to the blot was visualized by autoradiography.
Oligopeptide Phosphorylation Assays
Phosphorylation assays using GST-SOS2 and mutant SOS2-GST fusion proteins as kinases and peptide p3 (ALARAASAAALARRR) as substrate were performed as described by Halfter et al. (2000). Control reactions were conducted either without peptide p3 or without kinase proteins. The kinase buffer included 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 1 mM CaCl2, 10 μM ATP, and 1 mM DTT. The kinase reaction was in a total volume of 20 μL and was started by adding 200 μM peptide p3 and 0.5 μL of γ-32P-ATP (5 μCi), and the mixture was transferred to 30°C for 30 min. All reactions contained 100 ng of SOS2 or SOS2 mutant proteins. For partially purified kinase proteins, their concentrations were adjusted on the basis of the intensity of Coomassie Brilliant Blue R-250–stained bands on SDS-PAGE gels compared with the intensity of BSA. The reaction was stopped by adding 0.5 μL of 0.5 M EDTA, and the GST fusion proteins bound to glutathione-Sepharose beads were pelleted. Ten microliters of the supernatant was spotted onto P81 cellulose phosphate paper (Whatman) for peptide phosphorylation analysis. The paper was washed with 1% phosphoric acid, and the phosphorylated peptide was quantified by Phosphorimaging (Molecular Dynamics, Sunnyvale, CA). To the remaining 10-μL reaction mixture, 3 μL of 3 × protein loading buffer (200 mM Tris-HCl, pH 6.8, 8% SDS, 30% glycerol, 1.5% β-mercaptoethanol, and 0.3% bromophenol blue) was added and denatured by boiling for 4 min; then the samples were run on a 10% SDS-PAGE gel. The gel was dried and exposed to x-ray film for kinase autophosphorylation analysis.
To determinate whether the T/DSOS2/308 mutant depends on Ca2+, phosphorylation assays were performed in kinase buffer (20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 10 μM ATP, and 1 mM DTT) with 0.02, 0.1, 0.25, 0.5, 1, 3, 6, 10, and 20 mM CaCl2 or 10 mM EGTA at 30°C for 30 min. Different Mg2+ concentrations (0.5, 1, 2.5, 5, 7.5, 10, 15, and 20 mM or 1 mM EDTA) also were tested in reactions with kinase buffer (20 mM Tris-HCl, pH 8.0, 1 mM CaCl2, 10 μM ATP, and 1 mM DTT).
RNA Gel Blot Analysis
Wild-type (ecotype Columbia) seedlings of Arabidopsis thaliana were grown on Murashige and Skoog (1962) nutrient agar plates that were positioned vertically. After 2 weeks, salt treatment was applied by immersing the roots of the seedlings on an agar surface with a Murashige and Skoog (1962) solution supplemented with 100 or 200 mM NaCl. At the given time points, shoot and root tissues were separated at the base of the hypocotyl and harvested for RNA isolation and RNA gel blot analysis (Liu et al., 2000). Twenty micrograms of poly(A)+ RNA was loaded in each lane, size-fractionated by electrophoresis, and blotted to a nylon membrane. 32P-labeled probes were from gene-specific fragments from the 3′ end of each gene, including 3′ untranslated sequences.
Acknowledgments
We thank Dr. Hans Bohnert for critical reading of the manuscript and Dr. D. Grahame Hardie for stimulating discussions. This work was supported by National Institutes of Health Grant R01GM59138 and United States Department of Agriculture National Research Initiative Grant 0001657 to J.-K.Z.
References
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bush, D.S. (1995). Calcium regulation in plant cells and its role in signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 95–122. [Google Scholar]
- Celio, M.R., Pauls, T., and Schwaller, B. (1996). Guidebook to the Calcium-Binding Proteins. (Oxford, UK: Oxford University Press).
- Cohen, P., and Klee, C.B. (1988). Calmodulin. (New York: Elsevier).
- Cox, J.A. (1990). The role of intracellular calcium binding proteins. In Stimulus Response Coupling, J.-R. Dedman and V.L. Smith, eds. (Boston: CRC Press), pp. 266–269.
- Gilroy, S., Bethke, P.C., and Jones, R.L. (1993). Calcium homeostasis in plants. J. Cell Sci. 106, 453–462. [DOI] [PubMed] [Google Scholar]
- Goldberg, J., Nairn, A.C., and Kuriyan, J. (1996). Structural basis for the autoinhibition of calcium/calmodulin–dependent protein kinase I. Cell 84, 875–887. [DOI] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks, S.K., and Hunter, T. (1995). The eukaryotic protein kinase superfamily. In The Protein Kinase Facts Book, Vol. 1, D. Hardie and S. Hanks, eds. (London: Academic Press), pp. 7–47.
- Hardie, D.G. (1999). Plant protein serine/threonine kinases: Classification and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 97–131. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., Carling, D., and Carlson, M. (1998). The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Harper, J.F., Sussman, M.R., Schaller, G.E., Putnam-Evans, C., Charbonneau, H., and Harmon, A.C. (1991). A calcium-dependent protein kinase with a regulatory domain similar to calmodulin. Science 252, 951–954. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.-S., Wei, M., and Zhu, J.-K. (2000). SOS3 function in plant salt tolerance requires myristoylation and calcium binding. Plant Cell 12, 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, L.N., Noble, M.E.M., and Owen, D.J. (1996). Active and inactive protein kinases: Structural basis for regulation. Cell 85, 149–158. [DOI] [PubMed] [Google Scholar]
- Klee, C.B., Draetta, G.F., and Hubbard, M.J. (1988). Calcineurin. Adv. Enzymol. 61, 149–200. [DOI] [PubMed] [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–324. [DOI] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B–like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 96, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1997). An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc. Natl. Acad. Sci. USA 94, 14960–14964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, J., Polito, V.S., and Lauchli, A. (1989). Salinity stress increases cytoplasmic calcium activity in maize root protoplasts. Plant Physiol. 90, 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador, W.E., Means, A.R., and Quiocho, F.A. (1993). Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262, 1718–1721. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Hayashida, N., Yamaguchi-Shinizaki, K., Kamada, H., and Shinozaki, K. (1994). Cloning and sequencing of a novel serine/threonine protein kinase in Arabidopsis thaliana. Plant Physiol. 106, 1229–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief, N.D., Kretsinger, R.H., and Goodman, M. (1990). Evolution of EF-hand calcium-modulated proteins. I. Relationships based on amino acid sequences. J. Mol. Evol. 30, 522–562. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Pierson, E.S., Miller, D.D., Callaham, D.A., van Aken, J., Hackett, G., and Hepler, P.K. (1996). Tip-localized calcium entry fluctuates during pollen tube growth. Dev. Biol. 174, 160–173. [DOI] [PubMed] [Google Scholar]
- Poovaiah, B.W., and Reddy, A.S.N. (1993). Calcium and signal transduction in plants. Crit. Rev. Plant Sci. 12, 185–211. [DOI] [PubMed] [Google Scholar]
- Roberts, D.M., and Harmon, A.C. (1992). Calcium-modulated protein targets of intracellular signals in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 375–414. [Google Scholar]
- Shi, H., Ishitani, M., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Kim, K.-N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., Luan, S., and Kudla, J. (1999). Novel protein kinases associated with calcineurin B–like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler, D.A., Adams, S.P., Eubanks, S.R., Towery, D.S., Jackson-Machelski, E., Glaser, L., and Gordon, J.I. (1988). Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263, 1784–1790. [PubMed] [Google Scholar]
- Trewavas, A.J., and Malho, R. (1998). Ca2+ signaling in plant cells: The big network! Curr. Opin. Plant Biol. 1, 428–433. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, R.E. (1998). Calmodulin and calmodulin binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725. [DOI] [PubMed] [Google Scholar]